Abstract
The post-transcriptional modification of RNA by the addition of one or more chemical groups has been known for over 50 years. These chemical modifications, once thought to be static, are now being discovered to play key regulatory roles in gene expression. The advent of massive parallel sequencing of RNA (RNA-seq) now allows us to probe the complexity of cellular RNA and how chemically altering RNA structure expands the RNA vocabulary. Here we present an overview of the various strategies and technologies that are available to profile RNA chemical modifications at the cellular level. These strategies can be characterized as targeted and untargeted approaches: targeted strategies are developed for one single chemical modification while untargeted strategies are more broadly applicable to a range of such chemical changes. Key for all of these approaches is the ability to locate modifications within the RNA sequence. While most of these methods are built upon an RNA-Seq pipeline, alternative approaches based on mass spectrometry or conventional DNA sequencing retain value in the overall analysis process. We also look forward toward future opportunities and technologies that may expand the types of modifications that can be globally profiled. Given the ever increasing recognition that these RNA chemical modifications play important biological roles, a variety of methods, preferably orthogonal approaches, will be required to globally identify, validate and quantify RNA chemical modifications found in the transcriptome.
INTRODUCTION
While chemical modifications to RNA have been known since the 1950s,1 it is only over the past several years where RNA modifications have become more ‘mainstream’ and a prime focus of biological research.2 This interest (some would say renewed interest) was generated, in large part, by the discovery that particular RNA modifications were reversible and there are dedicated proteins that can generate (‘writers’), recognize (‘readers’), and remove (‘erasers’) the modification (Figure 1). The concept of reversible modifications suggested the possibility that some chemical changes to RNA structure would play a key role in gene expression in a manner reminiscent of DNA epigenetic changes. Not surprisingly, these dynamic changes in RNA modification status became referred to as the ‘epitranscriptome’.4
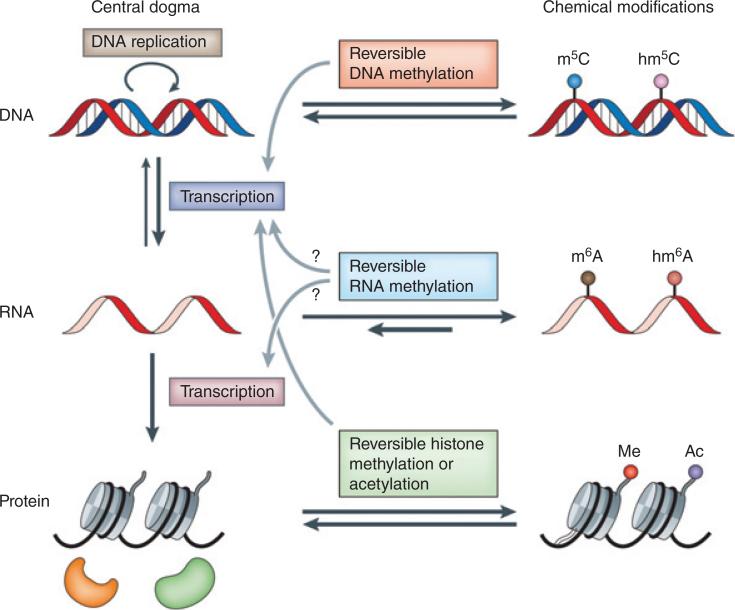
FIGURE 1.
Reversible chemical modifications that regulate the flow of genetic information. In the central dogma, genetic information is passed from DNA to RNA and then to protein. Epigenetic DNA modifications (e.g., the formation of 5-methylcytosine (m5C; also known as 5mC) and 5-hydroxymethylcytosine (hm5C; also known as 5hmC)) and histone modifications (e.g., methylation (me) and acetylation (ac)) are known to have important roles in regulating cell differentiation and development. Reversible RNA modifications (e.g., the formation of N6-methyladenosine (m6A) and N6-hydroxymethyladenosine (hm6A)) add an additional layer of dynamic regulation of biological processes. (Reprinted with permission from Ref 3. Copyright 2014 Nature Publishing Group)
Determining the biological function and significance of these modifications requires tools and technologies that can provide insights into the types and distributions of RNA modifications across the transcriptome.5 Leveraging advances made in DNA sequencing technologies, the use of high-throughput RNA-Seq6 has enabled the study of some RNA modifications at the organism level. Yet despite the excitement generated by these RNA-Seq approaches, this platform has limitations and for the field to grow, new and improved techniques will be required.
This review will examine the variety of technologies and approaches that are available for examining RNA modifications at a global level. After a brief overview of chemical modifications to RNA (themselves a subset of RNA post-transcriptional modifications), the article will examine the variety of strategies adapted for RNA-Seq that can generate high-throughput data on chemical modifications. Next, we discuss alternatives to RNA-Seq, with a focus on mass spectrometry (MS), which remains the only technology capable of directly identifying nearly all possible chemical modifications in RNA. The article concludes by examining new and improved technologies and strategies on the horizon that may become important contributors to the field of RNA modification.
CHEMICAL MODIFICATIONS TO RNA
After transcription, the cell is able to modify RNA in a number of ways. Post-transcriptional modifications that impact the sequence or stability of RNA include capping, splicing, trimming, G-cap, CCA tail addition (for tRNAs), polyadenylation, and polyuridylation.7 While these post-transcriptional modifications are biologically important, this article will focus on modifications that result in a change in the chemical structure of the RNA transcript, most commonly by the addition of small chemical groups (e.g., methyl) or the replacement of one chemical group for another (e.g., sulfur replacement of oxygen) to one or more individual nucleosides present within the already transcribed RNA. These chemical modifications are genome encoded processes, as one or more enzymes are utilized to effect such structural changes. Another class of chemical modifications to the transcribed RNA, those caused by oxidative damage,8–11 will not be discussed further although many of the techniques described below could be adapted to analyze RNA lesions.
The field of RNA chemical modifications is the subject of numerous reviews, to which the interested reader is directed.2,5,12–16 As these modifications have an impact on the techniques described below, a brief summary of the types and distribution is warranted. At present, ~150 enzymatically generated RNA chemical modifications have been identified.17 While the pace of ‘new’ nucleoside discovery has slowed, there is no evidence to suggest the current list is exhaustive as new organisms are studied and the technology for modification detection improves. Modifications of particular interest in this review are shown in Figure 2.
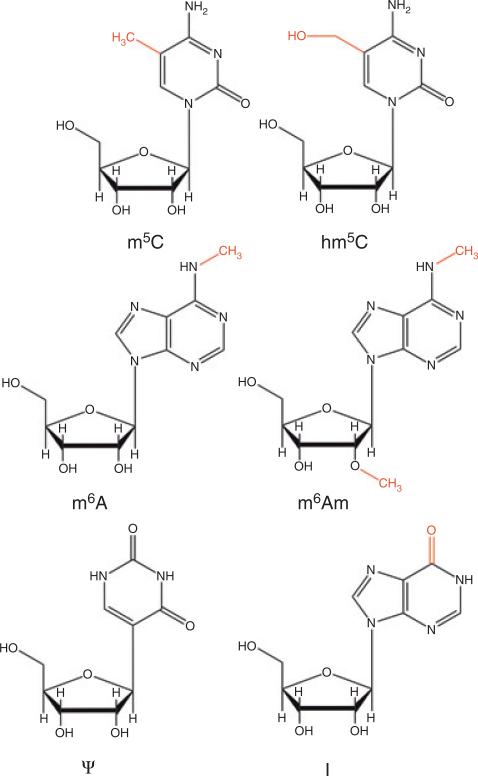
FIGURE 2.
Representative RNA chemical modifications. [m5C] 5-methylcytidine; [hm5C] 5-hydroxymethylcytidine; [m6A] N6-methyladenosine; [m6Am] N6,2′-O-dimethyladenosine; [Ψ] pseudouridine; [I] inosine.
The single most abundant modified nucleoside in RNA is pseudouridine, originally considered the ‘fifth’ nucleoside due to its relatively high (~5%) cellular abundance.18,19 Pseudouridine is an isomer of uridine, generated enzymatically by uracil base removal while on the RNA strand, rotation, and reattachment. This isomerization reaction leads to an additional hydrogen bond as compared to uridine as well as a carbon-carbon glycosidic bond (unlike carbon-nitrogen glycosidic bonds in all other nucleosides), features that have been exploited in developing technologies for pseudouridine detection. Pseudouridine is found across all types of RNA, being particularly abundant in eukaryotic rRNA.
The single most abundant class of modifications is methylation. Methylation of RNA takes many forms—the nucleobase can be methylated (sometimes multiply), the ribose sugar can be methylated at the 2′-hydroxyl, and both base and sugar methylations can be found on the same nucleoside. As with pseudouridine, methylations are found in all classes of RNA.
Inosine, a chemical modification that results from the enzymatic deamination of adenosine, is a special category of chemical modifications called RNA editing. Historically this term was coined to denote the insertions of uridine in mRNAs of two trypanosomatids protozoans and was later broadened to refer to a change in RNA sequence or base structure that changes the coding property from its gene sequence that included uridine insertions and deletions and cytosine to uridine and adenosine to inosine deamination.20 Given recent discoveries of other transcriptome modifications that change the coding property of a gene (vida supra), the utility of considering editing as a specialized class of RNA modifications may soon be a thing of the past. Inosine is the only editing event that results in a non-canonical nucleoside and is a prevalent RNA chemical modification, being concentrated in dsRNA in higher organisms. The translational machinery reads inosine as guanosine, thus base pairing it with cytidine. Such recoding events can lead to different protein sequences with profound impacts on protein function.
Beyond pseudouridine, methylations and inosine, the majority of other chemical modifications that have been identified arise from the multi-enzymatic processing of RNAs. These chemical changes can be simple (S for O in cytidine and uridine) or more complex, such as the addition of amino acids (e.g., N6-threonylcarbamoyladenosine [t6a]), sugars (e.g., galactosyl queuosine [galQ]) or fatty acids (e.g., 2-geranylthiouridine [ges2U]), or the generation of multi-ring structures such as the wybutosine family of modifications. At present, the vast majority of these more complex modification structures have only been found in tRNAs,17 although this limitation to that RNA class may be influenced by the relatively high amounts of tRNA per cell, which simplifies detection and identification of low abundance, complex chemical modifications.
CHARACTERIZATION OF RNA MODIFICATIONS—AN OVERVIEW
Techniques for identifying and characterizing chemical modifications to RNA have existed ever since the canonical nucleosides were discovered. Historically, modifications were first characterized by paper chromatography.19 In the late 1960s, the development of MS approaches for structural characterization were rapidly applied to unknown nucleoside structures, such that nearly all of the known RNA chemical modifications were determined by this approach.1 Once a structure has been determined, the generation of suitable standards—either synthetically or biosynthetically—enables the use of other strategies for identification of modified nucleosides. Most common in biochemistry labs is the use of thin layer chromatography (TLC)21 or high performance liquid chromatography (HPLC).22 Both approaches are based on the measurement of some relative (rather than absolute) characteristic of the modified nucleoside, and identification is made by comparison against an authentic standard or compiled library of relative migration distances (TLC) or retention times (HPLC).23 Additional specificity in nucleoside detection was enabled by coupling HPLC with MS, where the combination of HPLC retention time and molecular mass (plus structural data) from MS is now the most powerful platform for the identification and characterization of chemically modified nucleosides from an RNA sample.24,25
While nucleoside analysis by techniques such as liquid chromatography-mass spectrometry (LC-MS) is invaluable in determining what chemically modified nucleosides are present within an RNA sample, nucleoside analysis alone cannot reveal the sequence location within an RNA that is chemically modified. For that, approaches are required that retain sufficient sequence context to place the modification onto a particular nucleotide or limit the region that would contain the modification. The earliest approach for placing modifications within an RNA sequence involved a combination of ribonuclease (RNase) digestion and electrophoretic separation to fingerprint or map modifications on specific RNAs.26 This approach took advantage of the anomalous migration of RNase digestion products containing a modification, and often required subsequent sequencing of RNase digestion products to specifically locate the sequence position of the modification.
These cumbersome RNase fingerprinting approaches were replaced with improved techniques starting in the mid 1980s. The discovery of reverse transcriptase (RT), which when combined with the polymerase chain reaction (PCR), was used as an effective tool for locating the positions of certain chemical modifications, when those modifications inhibited the RT reaction (so-called ‘RT stops’). While an indirect method of detection, RT-PCR enabled the mapping of chemical modifications onto diverse RNAs with better sensitivity and specificity than historical RNase fingerprinting approaches.27 In addition, the RNase fingerprinting approach was adapted for use with LC-MS to eliminate the need for radioactive labeling and improve the speed and selectivity of analysis.28 Unlike an RT-PCR approach, RNA modification mapping by MS is a direct detection method, as the chemical modification is revealed by the anomalous mass of the modified nucleoside in the RNA sequence.
Without question, the methods and techniques described above—along with others not mentioned—have been key in improving our understanding of RNA modifications. However, these techniques nearly all are predicated on the analysis of one RNA sequence at a time. Thus, one could not envision determining the global RNA modification profile for a cell even when limiting to the modification profile for a particular class of RNA. As will be discussed below, a number of new technical advances have changed the paradigm from one RNA sequence at a time into a true ‘omics’ approach to RNA modification profiling. While these techniques are not without their own limitations, these modern approaches enable a greater picture of RNA modification profiles to be determined from less sample and far quicker than previously possible.
GLOBAL PROFILING OF MODIFICATIONS—COMMON THEMES
As the interest in understanding global profiles of chemical modifications to RNA has grown, new methods and technologies have been developed or adapted to simplify interpretation of the potential large datasets and to improve the accuracy of profiling results. As will become evident below, where these particular methods are discussed in more depth, a few common themes underlay method development (Figure 3). The vast majority of profiling approaches are built around the use of RT to create the cDNA for subsequent sequence analysis (e.g., RNA-Seq approaches). Many chemical modifications lead to RT stops or misincorporation errors. Thus, one theme that continues to emerge is to treat the sample, preferably in some selective fashion, to enhance RT stops. By selectively enhancing RT stops, the sites/locations of modified nucleosides are more precisely defined with an added benefit of higher modification detection sensitivity. Often used concurrently with sample treatment, another theme is the advantage that arises from differential sample analysis. By comparing untreated and treated samples, the researcher is looking for a difference in the datasets to reveal modification profiles. Such differential analyses also improve the precision and sensitivity of the experimental approach. Lastly, techniques that can incorporate an enrichment step before analysis benefit by removing unmodified RNAs from the background. Not only does enrichment allow the method to focus on the modified RNAs of interest, it can also improve the sensitivity and dynamic range of the measurement. Methods that incorporate one or more of these common themes have high utility and provide a template for future developments in the field.
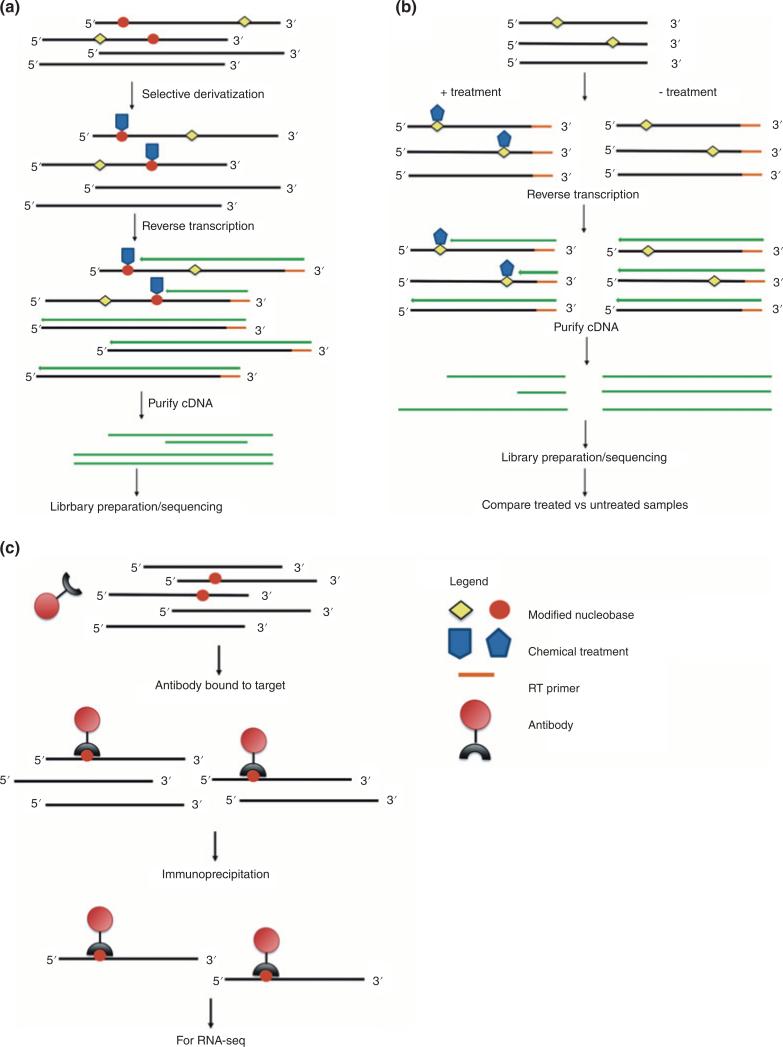
FIGURE 3.
Common themes in global profiling by RNA-Seq approaches. (a) Chemical modification derivatization strategies are used to enhance reverse transcriptase (RT) stops. (b) Differential analysis takes advantage of varying sensitivity of RNA-Seq to the presence of chemical modifications. (c) Affinity purification—usually through antibodies targeting specific chemically modified nucleosides—enriches the RNA pool prior to next-generation sequencing.
MODIFICATION PROFILING BY RNA-SEQ APPROACHES
It is hard to understate the impact of modern genomic sequencing technologies on science. Beyond their impacts in reducing the time and cost for whole genome sequencing analysis, the benefits of these technologies spilled over into numerous related areas. One such area—that of whole transcriptome sequencing, also referred to as RNA-Seq, served as the catalyst for transforming RNA modification mapping from a slow, cumbersome, one modification at one site approach into a field where the entire transcriptome—both coding and non-coding RNAs—can be examined for potential chemical modifications.
Bioinformatics Data from RNA Seq Only
A common theme of the RNA-Seq approaches—either specific for one modification or more general for a modification class—is the manipulation of the RNA sample into a form more conducive for modification analysis. These manipulations, whether chemical derivatization/treatment or selective enrichment via purification, improve the accuracy and specificity of analysis. However, there are well-established reports that the direct analysis of RNA-Seq data enables a computational analysis of possible RNA chemical modification sites. One such approach is RNA and DNA Differences (RDD). Inosine is the representative example here. Inosine arises from the deamination of adenosine and is typically recognized as guanosine during RT-PCR. In RDD, RNA-Seq data is aligned against the reference genome (DNA) to identify differences arising from A→G conversions seen with inosine.29 This RDD approach has improved greatly since it was first introduced through advances in computational tools to improve RDD identifications that can be attributed specifically to inosine.30,31
In addition, over a half dozen years ago, it was noted that ‘errors’ found in RNA-Seq datasets may not arise due to the technology/methodology, but rather are manifestations of chemically modified nucleosides in the RNA.32–34 Not surprisingly, these strictly computational analyses of RNA-Seq datasets focused on small RNAs, including microRNAs and tRNAs, as they are easier to characterize computationally and both RNA classes are characterized by rather robust chemical modifications (as well as 5′→3′-editing in the case of miRNAs). In these computational approaches, the key is to develop software with sufficient discriminatory power to identify possible sequencing ‘errors’ arising from the modification/polymerase interaction as opposed to standard experimental error or polymorphisms. While these computational approaches have been shown to reveal putative modification locations and therefore may have a role in allowing researchers to limit the scope of investigations, they do require validation by the other approaches discussed in this review.
Specific Modified Nucleosides
At present, the most successful RNA-Seq based approaches for profiling RNA modifications either utilize a specific derivatization reaction that changes the targeted modified nucleoside into a moiety that is amenable to detection during the RT step or take advantage of antibody recognition of the modified nucleoside for enrichment and mapping. Due to the limited number of specific derivatization chemistries or antibodies, these type of targeted approaches are only available for a small subset of modifications that include 5-methylcytidine (m5C), 5-hydroxymethylcytidine (hm5C), N6-methyladenosine (m6A), N6, 2′-O-dimethyl adenosine (m6Am), pseudouridine (Ψ), inosine (I), and 1-methyladenosine (m1A).
5-Methylcytidine [m5C] and 5-Hydroxymethylcytidine [hm5C]
One of the first approaches adapted to RNA-Seq technology was the use of bisulfite modification of RNA to identify methylation sites. Bisulfite sequencing has a rich history in both DNA and RNA methylcytidine analysis, and the strengths and weaknesses of this approach are now well appreciated.35 The essence of the chemistry is that bisulfite treatment of RNA leads to deamination of C, generating a U in its place. However, 5-methylcytidine undergoes much slower deamination relative to cytidine, thus it is read as a C during reverse transcription. By comparing control (untreated) samples against bisulfite-treated samples, one can—in principle—identify sites of methylation. As adapted with deep sequencing technologies, the sequencing depth can be used to identify RNA sites that are hypomodified (i.e., a non-stoichiometric amount of modified nucleoside is present within the sample). RNA bisulfite sequencing offers the advantage of directly detecting m5C during reverse transcription but the harsh reaction conditions (high pH) may lead to RNA degradation. A number of publications reporting on 5-methylcytidine in RNA are available that use this approach, and optimized protocols exist.36
An alternative, antibody-based strategy has been introduced more recently.37 In this strategy, Aza-IP, an RNA cytosine methyltransferase (RCMT) inhibitor, 5-azacytidine, is used to generate a covalent adduct of the RCMT and RNA at the methylation site. Immunoprecipitation (IP) of this adduct using an antibody against the particular RCMT under study is used to enrich the RNA sample prior to analysis. After treatment to break the enzyme-substrate adduct, the RNA can be sequenced as before. Khoddami and Cairns found that sequencing of these products also lead to a characteristic C > G transversion site that could be used to pinpoint 5-methylcytidine locations within the RNA.36 Unlike bisulfite treatment, this approach does not require additional sequencing of an untreated control, and the IP enrichment step improves sequencing depth. Another similar but chemically distinct immunoprecipitation approach, miCLIP (methylation individual-nucleotide resolution crosslinking immunoprecipitation) was developed by Hussain and co-workers.38 Similar to Aza-IP, miCLIP forms an irreversible covalent crosslink between the m5C and methyltransferase followed by immunoprecipitation, but differs in the way the bond is formed. In miCLIP, a highly conserved cysteine residue in the catalytic core of Nsun2 is mutated to alanine (C271A), forming a stable RNA–protein complex that can be detected by Western blots. Upon immunoprecipitation, the m5C enriched RNA pool is followed by standard NGS library preparation and sequencing. This is in contrast to Aza-IP where 5-azacytidine is intentionally incorporated in the nascent RNA by polymerases during the transcription process. Antibody-based technology are highly specific to the target and subsequent immunoprecipitation step facilitate detection of low abundance methylated RNAs. Other putative m5C methyltransferases in mammals (Nsun1-Nsun7) can also be explored to enhance detection as most of these proteins are known to bind to mRNAs as well.39,40
An antibody approach has also been developed for hm5C41,42 and recently used to examine RNA hydroxymethylation patterns in Drosophila.43 With this antibody-based strategy, only regions of RNA transcripts enriched in hm5C can be determined with the majority of hm5C peaks being detected in coding regions exhibiting UC-rich sequences containing a UCCUC repeating motif. Additional technique developments will be required to begin identifying the specific residues containing hm5C along with quantitative information on this modification.
N6-methyladenosine [m6A and N6, 2′-O-dimethyladenosine [m6Am]
Although m6A was known to be present in RNA transcripts over 40 years ago, the development of m6A-seq and related approaches has enabled organism-level studies on the abundance, distribution and biological function of this modification. The m6A-Seq (or MeRIP-Seq) approach is based on the use of highly specific antibodies to m6A, which are used to purify RNA fragments that contain this modification (Figure 4).45 The sample generated for sequencing is enriched in this specific modification, and identification of m6A consensus sites depends on sufficient enrichment. Site-specific identification is performed by comparing IP-enriched sample reads against non-enriched samples. Thus, specific detection of m6A is indirect and limited to methylation stoichiometries sufficient to meet data analysis parameters for identification. This approach revealed that m6A is particularly enriched in the 3′ UTR region of the transcript, near the stop codon and within long exons but cannot identify specific m6A residues.44 The sites of methylation can be also be predicted computationally by searching for R-A*-C sequence motif (where R is G/C/A; A* potential methylation site) and a recently discovered consensus motif DRACH (where D could be A/G or U; H could be A/C/U. Searching for such motifs in areas near the highest m6A signal can be used to limit the methylation site.3
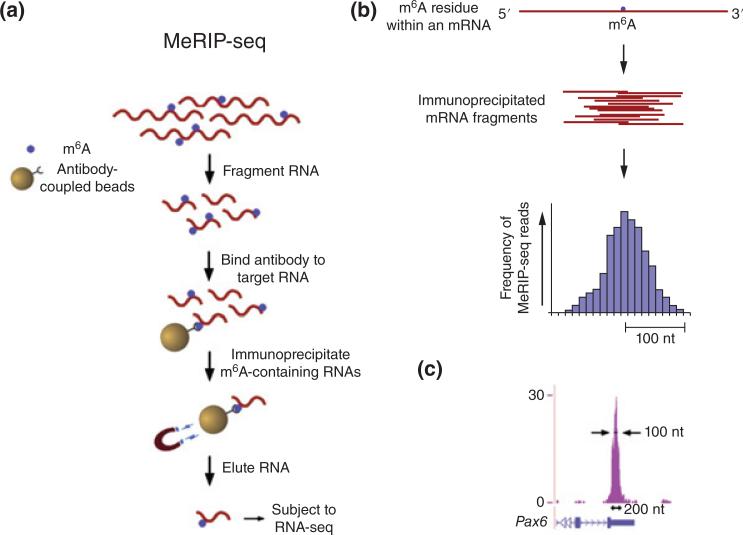
FIGURE 4.
Outline of MeRIP-Seq protocol and distribution of sequencing reads. (a) Schematic representation of MeRIP-Seq. Total RNA is subjected to RiboMinus treatment to remove rRNA species. RNAs containing m6A are then immunoprecipitated by mixing the RNA with m6A antibody-coupled Dynabeads. m6A-containing RNAs are then eluted from the antibody-coupled beads and subjected to a second round of m6A immunoprecipitation. The resulting RNA pool, which is highly enriched for m6A-containing RNAs, is then subjected to next-generation sequencing. (b) Schematic of sequencing reads and their alignment to locations in the genome surrounding an m6A site. (Top) An mRNA that contains a single m6A residue along its length. (Middle) Individual 100 nt wide mRNA fragments that are isolated following m6A immunoprecipitation, each of which contains the same m6A residue from the mRNA depicted above. (Bottom) Histogram showing predicted frequency of MeRIP-Seq reads obtained by sequencing individual immunoprecipitated fragments. Read frequency is predicted to increase with closer proximity to the m6A site, forming a ‘peak’ that is roughly 200 nt wide at its base and 100 nt wide at its midpoint. (c) Sequencing reads from MeRIP-Seq converge over m6A sites. Representative UCSC Genome Browser plot from MeRIP-Seq data, which demonstrates typical read frequency peak formation surrounding a site of m6A (shown here is the 30 UTR of Pax6). Peak height is displayed as reads per base per million mapped reads (BPM). (Reprinted with permission from Ref 44. Copyright 2012 Cell Press)
UV-CLIP (cross-linking immunoprecipitation) was adapted for use with the m6A-seq approach to enable direct identification of m6A sites within RNA fragments.46 The essence of this approach is that by direct UV-crosslinking of the antibody to m6A, subsequent reverse transcriptase treatment enables the precise location of the methylation to be identified. In addition, another related modification, N6-2′-O-dimethyl adenosine (m6Am) can also be mapped at the 5′ end of mRNAs. Unlike m6A which can cluster at different sites in the transcript, m6Am seem to be exclusively localized at the transcription start site (TSS).46 While this protocol does not address difficulties related to modification stoichiometry, it has significantly simplified locating m6A motifs to specific sites—essentially increasing the resolution (to ~200 nt) for detecting m6A at the transcriptome level.47 Chen and coworkers developed another photocrosslinking assay, photo-crosslinking-assisted m6A-sequencing (PA-m6A-seq), which further improved resolution of m6A in the transcript to ~20 nt.48
One can also couple these high throughput sequencing techniques with the writers, readers and erasers to validate m6A localization. As demonstrated by the work of Wang and co-workers, the two m6A reader proteins, YTHDF1 and YTHDF2, pulled down during PAR-CLIP experiments have significant overlap with m6A sites identified previously by m6A-seq.49,50 Interestingly, the two readers have opposing roles for the fate of a transcript. The former promotes translation by interacting with the protein machinery and the latter facilitates transcript decay. Other YTH domain family proteins in humans (YTHDF3 and YTHDC1) also have RNA binding properties, which can be explored in pull down experiments, and the m6A eraser FTO or Alkhb5 could be used for differential analysis.
Pseudouridine [Ψ]
Transcriptome-wide detection of Ψ by RNA-Seq was introduced nearly simultaneously by multiple groups.51–53 This approach is built off the long-standing selective chemical derivatization of Ψ by 1-cyclohexyl-3-(2-(4-morpholinyl)ethyl) carbodiimide tosylate (CMCT). As originally developed by Ofengand and co-workers, CMC can be selectively retained on Ψ generating an RT stop during RT-PCR.54 To adapt this to RNA-Seq, sequencing reads of small RNA fragments (typically generated during RT stops) are aligned against the reference genome to identify uridines that are likely derivatized. Schwartz and co-workers used spiked-in standards (in vitro transcribed RNAs) to quantify Ψ stoichiometries.53 Ψ-seq revealed a complex landscape of pseudouridylation in mRNA and ncRNA. Pseudouridine reveals no localization in distribution, being detected in coding regions of the transcript as well as 5′ and 3′ UTRs. However, it is noted that pseudouridylation is highly inducible both by dependent (requires H/ACA Ribonucleoprotein) or independent (Pus enzyme family) mechanisms.55
Inosine [I]
While inosine can be identified by RDD, as mentioned above, a number of technical concerns limit the utility of RDD at the transcriptome level. Cattenoz and co-workers developed an inosine specific sequencing protocol or iSeq that selectively enriched the samples with inosine prior to high throughput sequencing.56 This approach relies on the specificity of RNase T1, which is known to cleave at unmodified guanosine and inosine residue. In the presence of borate, guanosine forms a stable adduct with glyoxal. Glyoxalated guanosines are resistant to RNase T1 cleavage, generating 3′-inosine containing digestion products. One of the limitations of this method is the challenge in mapping small RNase T1 digestion products.
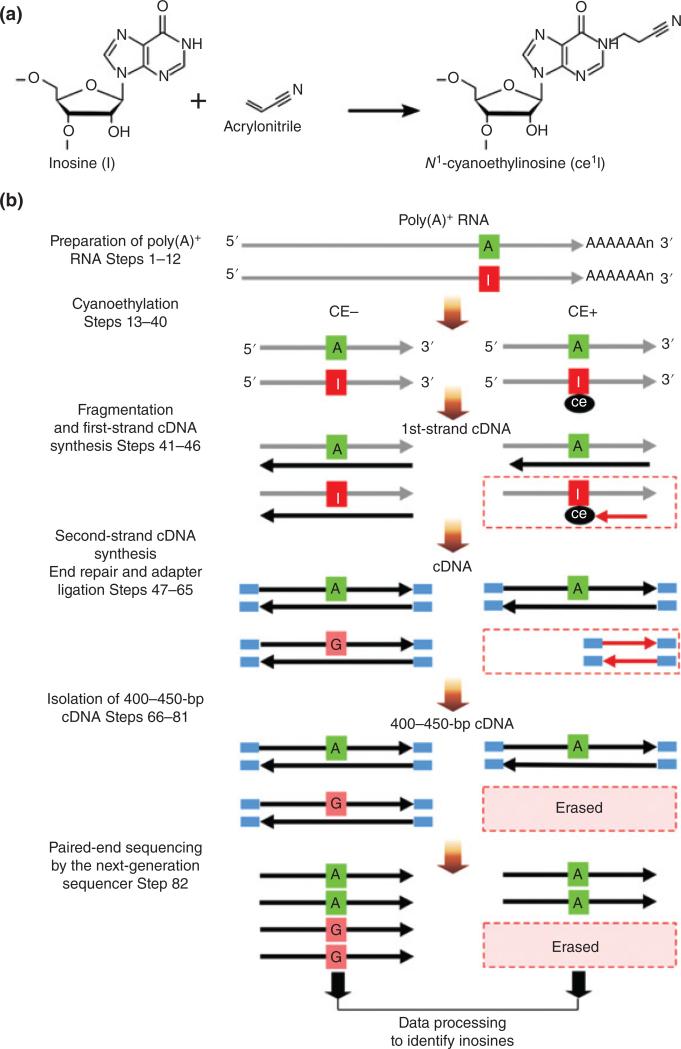
Suzuki and co-workers developed a differential approach called ICE-Seq (inosine chemical erasing).57 Inosine is selectively derivatized by cyanoethylation, which introduces an RT stop that leads to truncated RT-PCR products that are subsequently eliminated during sample processing (Figure 5). By comparing treated RNA fragments against untreated fragments, the combination of ‘erased’ reads with A→G conversions in the untreated sample enables more accurate determination of inosine at the transcriptome level. A low frequency of editing sites may give unreliable results as the ICE score is based on the ratio of G base reads between treated and untreated samples. Furthermore, differentiating an authentic editing site from systematic artifacts like SNPs, sequencing/errors and short sequencing reads is still a challenge.58
FIGURE 5.
Chemistry and outline of ICE-seq. (a) Chemistry of inosine a cyanoethylation. Inosine (I) on an RNA strand is cyanoethylated with acrylonitrile to form N1-cyanoethylinosine (ce1I). (b) ICE-seq procedure. O Schemes without (CE− condition) or with (CE+ condition) cyanoethylation of RNA are shown on the left and right, respectively. RNA and cDNA are O indicated by gray and black arrows, respectively. The I in the RNA strand Inosine (I) is specifically cyanoethylated to form ce1I (CE+). In both conditions, RNA bearing A at the editing site is converted to T in the cDNA during the first-strand synthesis. In the CE− condition, RNA bearing I is transcribed to C in the cDNA. In the CE+ condition, first-strand cDNA extension is arrested at the ce1I site (red arrow). Second strands of cDNA are synthesized to obtain double-stranded cDNA that is then subjected to the end-repair reaction and adapter ligation. The amplified cDNA with 400–450 bp is gel-purified. The cDNAs for the CE− and CE+ conditions are sequenced from both ends using next-generation sequencing. After data processing of sequence reads, A-to-I RNA editing sites can be identified by detecting erased G-containing reads upon cyanoethylation. (Reprinted with permission from Ref 57. Copyright 2015 Nature Publishing Group)
1-Methyladenosine [m1A]
The chemically modified nucleoside 1-methyl adenosine at position 58 of tRNAs is found in all three domains of life. It is also present in rRNA in a specific, highly conserved residue. While the role of m1A in ncRNAs is well established (i.e., structural stabilization),59 its presence and role in mRNA is still unexplored. Two research groups have recently published the global profiling of m1A in eukaryotic mRNA.60,61 Dominissini et al. developed m1A-seq, an antibody based approach to immunoprecipitate fragments containing the modified nucleoside. They also exploited the known m1A to m6A conversion (Dimroth rearrangement) as an orthogonal method to verify and validate the m1A sites resulting from immunoprecipitation. Li et al. used a differential approach by erasing the methyl group (using AlkB from E. coli) in m1A enriched immunoprecipitated fragments. By comparing the treated and untreated samples, the m1A-containing region was identified. Both groups reported that m1A is enriched in 5′-UTR and coding sequence regions.
The unique RT behavior of 1-methyladenosine has also been used for selective identification.62 Hauenschild et al. noted limited RT read-through with this modification, and that m1A residues have a distinct signature in reverse transcription which includes the identity of nucleotide 3′ to the modification (sequence dependent) and structure dependence. These signatures were used with a machine learning algorithm to predict the location of m1A in trypanosomal tRNAs. To date, this approach has been focused on tRNAs and rRNAs, as those RNA classes are known to contain m1A. However, the described protocol is compatible with any RNA class.
Profiling by Class of Modifications
Related to the modification-specific RNA-Seq approaches described above, there are several other recently developed high-throughput approaches that are more appropriately described as ‘modification class’ techniques. These approaches are typically directed against the specific chemical group(s) that are altered prior to reverse transcription. Thus, the nucleoside identity can vary in these cases providing a greater coverage of modification profiles present in the transcriptome.
Methyl Modifications
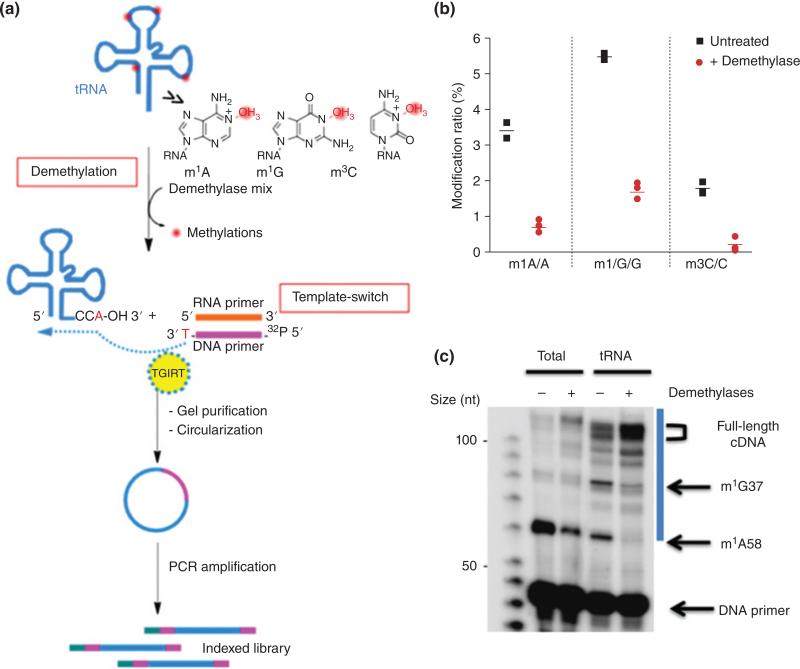
Arm-Seq (AlkB-facilitated RNA methylation sequencing)63 and demethylase-thermostable group II intron RT tRNA sequencing (DM-tRNA Seq)64 both are predicated on the removal of base methylations that are known to typically cause RT stops. ARM-seq uses a wild type E Coli AlkB demethylase while DM-tRNA Seq uses a genetically engineered AlkB (D151S), which has improved activity for m1G than the wild type. ARM-Seq ligates adapters at both ends prior to reverse transcription. DM-tRNA Seq uses a thermostable group II intron reverse transcriptase, which does not require ligation. It switches from an adapter to the 3′ end of the tRNA and synthesizes the cDNA (Figure 6).64 RNA fragments that are untreated will tend to create truncated products due to the interfering methylation. By comparing sequencing reads of untreated with treated samples, the locations of AlkB-sensitive base methylations (e.g., m1A, m3C and m1G) can be determined.
FIGURE 6.
Schematic representation of demethylase-thermostable group II intron RT tRNA sequencing (DM-tRNA-seq). (Reprinted with permission from Ref 64. Copyright 2015 Nature Publishing Group)
To date, these methods have been used to characterize methylations in tRNA and rRNA classes. Importantly, this concept demonstrates that differential enzymatic treatment using ‘erasers’ can be a general approach to large-scale examination of RNA chemical modifications. The utility of such a concept will depend, in part, on the further discovery of additional enzymes that are found to remove particular chemical modifications from RNA substrates.
2′-O-methyl Modifications
The RiboMeth-Seq approach has been developed for global detection of 2′-O ribose methylations.65 This approach takes advantage of the lability of 2′-hydroxyls to alkaline hydrolysis. Thus, unmodified RNAs treated under limited alkaline conditions should generate fragments that fully represent the entire RNA sequence. In contrast, a 2′-methyl hinders alkaline hydrolysis, leading to RNA fragments whose hydrolysis pattern is offset due to the 2′-methyl group. By aligning RNA-Seq data to a reference genome, the read ends for 2′-methyl modified nucleosides are revealed as underrepresented in the data set, enabling site-specific detection of this class of methylations.
Nicotinamide Adenine Dinucleotide (NAD) Caps
While technologies and approaches for characterizing 5′-capping modifications are not the focus of this review, the recent demonstration of NAD capture-Seq for NAD end-modified bacterial RNAs is worthy of discussion.66 This approach is the first example of chemical derivatization prior to library generation and deep sequencing that uses a chemical biology approach to modification detection. Cahova et al. used chemical derivatization to purify only RNA fragments that contain the desired modification class (here NAD 5’-capped RNAs). Moreover, derivatization was an enzymatically based modification to NAD that allowed subsequent biotinylation and purification.
RNA-SEQ ALTERNATIVES FOR MODIFICATION PROFILING AND MEASUREMENT
Mass Spectrometry
Clearly the last five years have seen the emergence of targeted approaches for the global profiling of RNA chemical modifications. Concurrent with those developments, there have also been advances in untargeted approaches that move these technologies beyond the one RNA at a time paradigm as well. The most significant advances have occurred in LC-MS based techniques. MS has always played an important role in the characterization of chemical modifications.67 Beyond nucleoside-level characterization, MS has proven a useful technique for characterizing modified nucleosides within a given RNA sequence. The general strategy for mapping modifications—RNA modification mapping—involves purification of a single RNA, digestion with an appropriate endonuclease (e.g., RNase T1, which cleaves specifically at unmodified guanosines), and separation and analysis using LC-MS. As all known chemical modifications except pseudouridine result in a mass increase to the canonical nucleoside, modifications are mapped to specific sites by monitoring the mass increase determined during collisionally induced dissociation tandem mass spectrometry (CID MS/MS).68–70 The advantages of LC-MS/MS strategies for RNA modification mapping include broad applicability (i.e., either by RNA class or modification type) and high accuracy (as mass can be measured with high precision by MS). The primary disadvantages include the general limitation to serial analysis of one RNA at a time, the relatively poorer sensitivity for RNA detection, and the technical skill required for LC-MS/MS instrumentation.
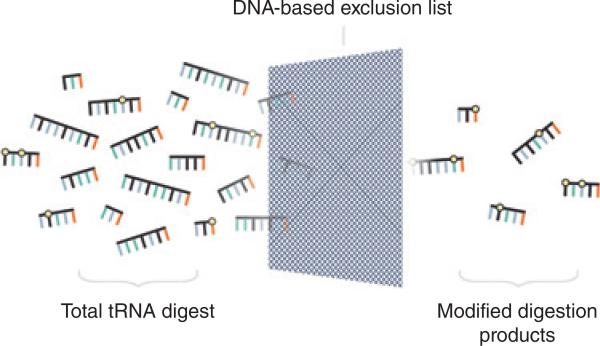
Recently, new developments in the field of RNA modification mapping and LC-MS/MS have addressed some of the historic limitations of MS in RNA modification profiling.71–75 Several approaches have built upon successes in the area of proteomics to move away from rigorous single RNA purification into the direct RNA modification mapping of multiple RNAs (e.g., the entire set of bacterial tRNAs from an organism, Figure 7).76 Here, LC-MS/MS technologies can provide robust quantitative mapping of RNA chemical modifications through better chromatography conditions and reproducible and defined fragmentation of endonuclease digestion products.77 As new computational methods are developed, data interpretation becomes more automated and amenable to increased throughputs in sample complexity and volume.78–80
FIGURE 7.
Schematic representation of DNA-based exclusion list for enhanced detection of modified RNAs by LC-MS/MS. (Reprinted with permission from Ref 76. Copyright 2015 American Chemical Society)
Despite these advances, LC-MS/MS approaches are still being applied to either more highly modified sRNAs, including tRNAs, or rRNAs. The application of LC-MS/MS to mRNAs or the entire transcriptome will require further improvements in the dynamic range of this technology. An unexplored but likely important role for MS in future global modification profiling studies will be as an independent and orthogonal technique to validate multiple modifications found in RNA-Seq approaches described earlier. When combined with relative or absolute quantification methods, this platform should provide a middle ground between RNA-Seq techniques and the targeted approaches described below that are still based on RT-PCR, TLC or other conventional biochemical approaches.
SCARLET (m6A)
Liu and coworkers developed SCARLET—Site-specific Cleavage And Radioactive labeling followed by Ligation-assisted Extraction and Thin-layer chromatography—to enable any particular m6A modification site to be examined quantitatively.81 The approach is built on several conventional historic biochemical approaches used for modified nucleoside identification/quantification including 33P post-labeling and TLC separation and detection of the targeted chemical modification.21,82 Site-specificity is enabled by RNase H-directed cleavage of the target RNA using specifically constructed chimeric oligonucleotides.83 A key point to emphasize is that SCARLET is based on the direct measurement of m6A in the RNA—no derivatization, enrichment, or amplification of the RNA is involved. However, the challenge is that a direct detection strategy does require highly sensitive methods82 (e.g., radioactivity) to enable determination of low abundance m6A sites. While SCARLET is a low-throughput approach, it is adaptable to other chemical modifications as the RNase H-step can be tailored to any RNA location.
RTL-P
Another example of quantitative measurement of site-specific modifications is RTL-P [Reverse Transcription at Low deoxyribonucleoside triphosphate (dNTP) concentrations followed by PCR] for 2′-O-methyl modifications.84 Here, the use of primers upstream and downstream of a 2′-O-methylation site in RNA allows for a comparative analysis of PCR products to measure modification levels. This approach is low throughput and involves the indirect determination of modification levels. However, as with any method based on differential analyses, the sensitivity of this approach is higher than methods using only a single input for analysis.
CONCLUSION
High throughput RNA-seq methods have enabled researchers to have an unprecedented view of the transcriptome. While second-generation DNA sequencing technologies have proven invaluable in global profiling of specific RNA chemical modifications, these technologies still suffer from the inability to directly detect RNA or RNA modifications and methods, outside of LC-MS/MS, are limited to only a handful of the more prevalent modifications (primarily methylations) in RNA. The inherent systematic error in the analysis can lead to false positive or negative results, which impact the accuracy of assigning modification sites especially those of low abundance. Short reads are prone to mapping/alignment errors due to the inherent repeats/duplication in the genome. Additional ambiguity in data interpretation can be caused by splice junctions, polymorphism between individuals and highly similar paralogous genes. Although there is no panacea for all of these limitations, increasing the read length, sequence depth and direct detection of modifications (bypassing the RNA to DNA conversion) would be significant improvements. In addition, the large data sets generated from NGS warrant a user-friendly bioinformatics pipeline that includes steps to lower false positives, increase sensitivity and selectivity.
One of the risks involved in IP-based methods is antibody quality and specificity. For accurate global profiling using antibodies, the antibody should capture only its cognate target. Co-precipitation of fragments with modifications other than the target may lead to false positives. All investigators in the field are cautioned to conduct appropriate validation assays—especially when using vendors who do not provide such data—and conduct appropriate controls to rule out multiple targets or limited specificity prior to embarking on time-, sample- and resource-consuming NGS studies. Moreover, the field would benefit by greater inclusion of antibody validation data within global profiling publications.
Newer technologies are on the horizon that offer the potential for direct RNA sequencing with the possibility that such direct analysis can also differentiate canonical nucleosides from chemically modified nucleosides. Nanopore-based sequencing has the potential to globally characterize RNA chemical modifications directly, obviating the need to make cDNA constructs. The nanopore sequencing platform allows detection of bases by the disruption in the current that flows through the protein channel.85,86 It would not be surprising to discover that chemical modifications would each have a unique current signature that would allow direct detection and sequencing. Ayub and co-workers have recently demonstrated RNA nanopore exosequencing that can discriminate the four canonical bases as well as inosine, m6A and m5C.85 Though the method has not yet been applied to the global profiling of transcripts, one can already envision an array of chip-based nanopores with engineered proteins optimized for specific modifications allowing parallel sequencing and data collection of RNA modification profiles directly and with high selectivity.
Another possibility is to use native error-prone or genetically optimized reverse transcriptases that can recognize RNA modifications and insert an appropriate nucleotide in the DNA strand that would serve as a direct modification marker during DNA sequencing. Alternatively, rate-based measurements, which have already been examined,87 could be improved by using these next generation reverse transcriptases, providing greater selectivity and differentiation when measuring incorporation across a modified nucleoside. One can also exploit the differential chemical reactivity of modified nucleotides with a variety of reagents to be coupled with high throughput sequencing. A wealth of information is available in literature for the orthogonal reactivity of modified bases if one is willing to revisit the organic chemistry of nucleobases. In this fashion, one can foresee a combination of direct detection of RNA chemical modifications that is validated by site- or modification-specific derivatization analyzed using older RT-cDNA based NGS.
Finally, additional possibilities exist with alternative platforms such as MS. While LC-MS/MS has become a mature, powerful technology, the methods available, limits of detection and ability to handle complex mixtures of modified RNAs are only in their infancy. If one uses the growth of LC-MS/MS within the field of proteomics as a reasonable guide for the possibilities of this technology, one readily notes that applications moving beyond RNA class into whole transcriptome studies are feasible, if just impractical at present.
ACKNOWLEDGMENTS
The authors thank Madeleine Lyons for assistance with the graphics. This work was supported in part by funding from the National Science Foundation (CHE1507357), the National Institutes of Health (GM058843) and the University of Cincinnati.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
FURTHER READING
Useful web sites
RNAcentral: http://rnacentral.org/
RNA-seq blog: http://www.rna-seqblog.com/
Modomics: http://modomics.genesilico.pl/
tRNAmodviz: http://genesilico.pl/trnamodviz
REFERENCES
- 1.Limbach P, Crain P, McCloskey J. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, Mason CE. The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet. 2014;15:127–150. doi: 10.1146/annurev-genom-090413-025405. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;14:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 4.Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellner S, Burhenne J, Helm M. Detection of RNA modifications. RNA Biol. 2010;7:237–247. doi: 10.4161/rna.7.2.11468. [DOI] [PubMed] [Google Scholar]
- 6.Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 7.Licatalosi D, Darnell R. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellacosa A, Moss EG. RNA repair: damage control. Curr Biol. 2003;13:R482–R484. doi: 10.1016/s0960-9822(03)00408-1. [DOI] [PubMed] [Google Scholar]
- 9.Falnes PO, Klungland A, Alseth I. Repair of methyl lesions in DNA and RNA by oxidative demethylation. Neuroscience. 2007;145:1222–1232. doi: 10.1016/j.neuroscience.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Fimognari C. Role of oxidative RNA damage in chronic-degenerative diseases. Oxid Med Cell Longev. 2015;2015:358713. doi: 10.1155/2015/358713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunomura A, Moreira PI, Castellani RJ, Lee HG, Zhu X, Smith MA, Perry G. Oxidative damage to RNA in aging and neurodegenerative disorders. Neurotox Res. 2012;22:231–248. doi: 10.1007/s12640-012-9331-x. [DOI] [PubMed] [Google Scholar]
- 12.Helm M, Alfonzo JD. Posttranscriptional RNA modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol. 2014;21:174–185. doi: 10.1016/j.chembiol.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. WIREs RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phizicky EM, Hopper AK. tRNA processing, modification, and subcellular dynamics: past, present, and future. RNA. 2015;21:483–485. doi: 10.1261/rna.049932.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machnicka M, Milanowska K, Osman O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother K, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2012;41:D262–267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C-T, Allen FW. Studies on an isomer of uridine isolated from ribonucleic acids. Biochim Biophys Acta. 1959;32:393–405. doi: 10.1016/0006-3002(59)90612-2. [DOI] [PubMed] [Google Scholar]
- 19.Cohn WE. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta. 1959;32:569–571. doi: 10.1016/0006-3002(59)90644-4. [DOI] [PubMed] [Google Scholar]
- 20.Maydanovchy O, Beal P. Breaking the central dogma by RNA editing. Chem Rev. 2006;106:3397–3411. doi: 10.1021/cr050314a. [DOI] [PubMed] [Google Scholar]
- 21.Grosjean H, Keith G, Droogmans L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol Biol. 2004;265:357–391. doi: 10.1385/1-59259-775-0:357. [DOI] [PubMed] [Google Scholar]
- 22.Pomerantz SC, McCloskey JA. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 23.Banoub J, Limbach P. Mass Spectrometry of Nucleosides and Nucleic Acids. CRC Press; Boca Raton, FL: 2010. p. 492. [Google Scholar]
- 24.Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YLJ, Prestwich EG, Lim KS, et al. A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol. 2015;560:29–71. doi: 10.1016/bs.mie.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su D, Chan CTY, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protocols. 2014;9:828–841. doi: 10.1038/nprot.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanger F, Brownlee GG, Barrell BG. A two-dimensional fractionation procedure for radioactive nucleotides. J Mol Biol. 1965;13:373–398. doi: 10.1016/s0022-2836(65)80104-8. [DOI] [PubMed] [Google Scholar]
- 27.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNA by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 28.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4584. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picardi E, Gallo A, Galeano F, Tomaselli S, Pesole G. A novel computational strategy to identify A-to-I RNA editing sites by RNA-Seq data: de novo detection in human spinal cord tissue. PLoS One. 2012;7:e44184. doi: 10.1371/journal.pone.0044184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picardi E, D'Erchia AM, Montalvo A, Pesole G. Using REDItools to detect RNA editing events in NGS datasets. Curr Protoc Bioinformatics. 2015;49:12.12.11–12.12.15. doi: 10.1002/0471250953.bi1212s49. [DOI] [PubMed] [Google Scholar]
- 31.Torres AG, Pineyro D, Rodriguez-Escriba M, Camacho N, Reina O, Saint-Leger A, Filonava L, Batlle E, Ribas de Pouplana L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015;43:5145–5157. doi: 10.1093/nar/gkv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res. 2009;37:2461–2470. doi: 10.1093/nar/gkp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Findeiss S, Langenberger D, Stadler PF, Hoffmann S. Traces of post-transcriptional RNA modifications in deep sequencing data. Biol Chem. 2011;392:305–313. doi: 10.1515/BC.2011.043. [DOI] [PubMed] [Google Scholar]
- 34.Iida K, Jin H, Zhu J-K. Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics. 2009;10:155. doi: 10.1186/1471-2164-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khoddami V, Yerra A, Cairns BR. Experimental approaches for target profiling of RNA cytosine methyltransferases. Methods Enzymol. 2015;560:273–296. doi: 10.1016/bs.mie.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:1–10. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussain S, Sajini A, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson J, Odom D, Ule J, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain S, Bashir Z. The epitranscriptome in modulating spatiotemporal RNA translation in neuronal post-synaptic function. Front Cell Neurosci. 2015;9:420. doi: 10.3389/fncel.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kallin EM, Rodriguez-Ubreva J, Christensen J, Cimmino L, Aifantis I, Helin K, Ballestar E, Graf T. Tet2 facilitates the derepression of myeloid target genes during CEBPα-induced transdifferentiation of pre-B cells. Mol Cell. 2012;48:266–276. doi: 10.1016/j.molcel.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 44.Meyer K, Saletore Y, Zumbo P, Elemento O, Mason C, Jaffrey S. Comprehensive analysis of mRNA methylation reveals enrichment in 30 UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominissini D, Moshitch-Moshkovitz S, Amariglio N, Rechavi G. Transcriptome-wide mapping of N6-methyladenosine by m6A-Seq. Methods Enzymol. 2015;560:131–147. doi: 10.1016/bs.mie.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. A majority of m 6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015;29:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen K, Lu Z, Wang X, Fu Y, Luo G, Liu N, Han D, Dominissini D, Dai Q, Pan T, et al. High-resolution N6-methyl adenosine (m6A) map using photo-cross-linking-assisted m6A sequencing. Angew Chem Int Ed Engl. 2015;54:1587–1590. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Zhike L, Gomez A, Hon G, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladeno-sine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–138. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zhao S, Roundtree I, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakin A, Kowalak JA, McCloskey JA, Ofengand J. The single pseudouridine residue in Escherichia coli 16S RNA is located at position 516. Nucleic Acids Res. 1994;22:3681–3684. doi: 10.1093/nar/22.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu G, Huang C, Yu Y-T. Pseudouridine in mRNA: incorporation, detection, and recoding. Methods Enzymol. 2015;560:187–217. doi: 10.1016/bs.mie.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cattenoz P, Taft R, Westhof E, Mattick J. Transcriptome-wide identification of A > I RNA editing sites by inosine specific cleavage. RNA. 2013;19:257–270. doi: 10.1261/rna.036202.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki T, Ueda H, Okada S, Sakurai M. Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nat Protoc. 2015;10:715–732. doi: 10.1038/nprot.2015.037. [DOI] [PubMed] [Google Scholar]
- 58.Kim M, Hur B, Kim S. RDDpred: a condition-specific RNA-editing prediction model from RNA-seq data. BioMedCentral Genomics. 2016;17(Suppl 1):5. doi: 10.1186/s12864-015-2301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roovers M, Wouters J, Bujnicki J, Tricot C, Stalon V, Grosjean H, Droogsman L. A primordial RNA modification enzyme: the case of tRNA m1A:methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N-methyladenosine methylome. Nat Chem Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 62.Hauenschild R, Tserovski L, Schmid K, Thüring K, Winz M-L, Sharma S, Entian K-D, Wacheul L, Lafontaine DLJ, Anderson J, et al. The reverse transcription signature of N-1-methyladenosine in RNASeq is sequence dependent. Nucleic Acids Res. 2015;43:9950–9964. doi: 10.1093/nar/gkv895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng G, Qin Y, Clark W, Dai Q, Yi C, He C, Lambowitz A, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12:835–839. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem Int Ed Engl. 2015;54:451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 66.Cahová H, Winz M-L, Höfer K, Nübel G, Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2014;519:374–377. doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 67.McCloskey JA, Nishimura S. Modified nucleosides in transfer RNA. Acc Chem Res. 1977;10:403–410. [Google Scholar]
- 68.Gaston KW, Limbach PA. The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry. RNA Biol. 2014;11:1568–1585. doi: 10.4161/15476286.2014.992280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Björkbom A, Lelyveld VS, Zhang S, Zhang W, Tam CP, Blain JC, Szostak JW. Bidirectional direct sequencing of noncanonical rNA by two-dimensional analysis of mass chromatograms. J Am Chem Soc. 2015;137:14430–14438. doi: 10.1021/jacs.5b09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wetzel C, Limbach PA. Mass spectrometry of modified RNAs: recent developments. Analyst. 2016;141:16–23. doi: 10.1039/c5an01797a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Limbach P. Method for comparative analysis of ribonucleic acids using isotope labeling and mass spectrometry. Anal Chem. 2012;84:8607–8613. doi: 10.1021/ac301638c. [DOI] [PubMed] [Google Scholar]
- 72.Li S, Limbach P. Mass spectrometry sequencing of transfer ribonucleic acids by the comparative analysis of RNA digests (CARD) approach. Analyst. 2013;138:1386–1394. doi: 10.1039/c2an36515d. [DOI] [PubMed] [Google Scholar]
- 73.Puri P, Wetzel C, Saffert P, Gaston KW, Russell SP, Cordero Varela JA, van der Vlies P, Zhang G, Limbach PA, Ignatova Z, et al. Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol Microbiol. 2014;93:944–956. doi: 10.1111/mmi.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wetzel C, Limbach P. The global identification of tRNA isoacceptors by targeted tandem mass spectrometry. Analyst. 2013;138:6063–6072. doi: 10.1039/c3an01224g. [DOI] [PubMed] [Google Scholar]
- 75.Taoka M, Nobe Y, Hori M, Takeuchi A, Masaki S, Yamauchi Y, Nakayama H, Takahashi N, Isobe T. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucleic Acids Res. 2015;43:e115. doi: 10.1093/nar/gkv560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao X, Limbach PA. Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LCMS/MS. Anal Chem. 2015;87:8433–8440. doi: 10.1021/acs.analchem.5b01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Popova AM, Williamson JR. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J Am Chem Soc. 2014;136:2058–2069. doi: 10.1021/ja412084b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sample PJ, Gaston KW, Alfonzo JD, Limbach PA. RoboOligo: software for mass spectrometry data to support manual and de novo sequencing of post-transcriptionally modified ribonucleic acids. Nucleic Acids Res. 2015;43:e64. doi: 10.1093/nar/gkv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakayama H, Takahashi N, Isobe T. Informatics for mass spectrometry-based RNA analysis. Mass Spectrom Rev. 2011;30:1000–1012. doi: 10.1002/mas.20325. [DOI] [PubMed] [Google Scholar]
- 80.Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T. Ariadne: a database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data. Nucleic Acids Res. 2009;37:e47. doi: 10.1093/nar/gkp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu N, Pan T. Probing RNA modification status at single-nucleotide resolution in total RNA. Methods Enzymol. 2015;560:149–159. doi: 10.1016/bs.mie.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 82.Zhao X, Yu Y. Detection and quantitation of RNA base modifications. RNA. 2004;10:996–1002. doi: 10.1261/rna.7110804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 84.Dong Z-W, Shao P, Diao L-T, Zhou H, Yu C-H, Qu L-H. RTL-P: a sensitive approach for detecting sites of 2’-O-methylation in RNA molecules. Nucleic Acids Res. 2012;40:e157. doi: 10.1093/nar/gks698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ayub M, Hardwick SW, Luisi BF, Bayley H. Nanopore-based identification of individual nucleotides for direct RNA sequencing. Nano Lett. 2013;13:6144–6150. doi: 10.1021/nl403469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith AM, Abu-Shumays R, Akeson M, Bernick DL. Capture, unfolding, and detection of individual tRNA molecules using a nanopore device. Front Bioeng Biotechnol. 2015;3:1–11. doi: 10.3389/fbioe.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vilfan ID, Tsai Y-C, Clark TA, Wegener J, Dai Q, Yi C, Pan T, Turner SW, Korlach J. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J Nanobiotechnol. 2013;11:1–1. doi: 10.1186/1477-3155-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]