Abstract
Brown adipose tissue (BAT) thermogenesis relies on a high abundance of mitochondria and the unique expression of the mitochondrial Uncoupling Protein 1 (UCP1), which uncouples substrate oxidation from ATP synthesis. Adrenergic stimulation of brown adipocytes activates UCP1-mediated thermogenesis; it also induces the expression of Ucp1 and other genes important for thermogenesis, thereby endowing adipocytes with higher oxidative and uncoupling capacities. Adipocyte mitochondrial biogenesis and oxidative capacity are controlled by multiple transcription factors, including the estrogen-related receptor (ERR)α. Whole-body ERRα knockout mice show decreased BAT mitochondrial content and oxidative function but normal induction of Ucp1 in response to cold. In addition to ERRα, brown adipocytes express ERRβ and ERRγ, 2 nuclear receptors that are highly similar to ERRα and whose function in adipocytes is largely unknown. To gain insights into the roles of all 3 ERRs, we assessed mitochondrial function and adrenergic responses in primary brown adipocytes lacking combinations of ERRs. We show that adipocytes lacking just ERRα, the most abundant ERR, show only mild mitochondrial defects. Adipocytes lacking ERRβ and ERRγ also show just mild defects. In contrast, adipocytes lacking all 3 ERRs have severe reductions in mitochondrial content and oxidative capacity. Moreover, adipocytes lacking all 3 ERRs have defects in the transcriptional and metabolic response to adrenergic stimulation, suggesting a wider role of ERRs in BAT function than previously appreciated. Our study shows that ERRs have a great capacity to compensate for each other in protecting mitochondrial function and the metabolic response to adrenergic signaling, processes vital to BAT function.
Brown adipose tissue (BAT) generates heat in response to changes in environmental temperature or diet, in a process called adaptive thermogenesis. The thermogenic capacity of brown adipocytes relies on their high mitochondrial content and oxidative capacity and the presence of the mitochondrial inner membrane protein Uncoupling Protein 1 (UCP1). When activated, UCP1 dissipates the mitochondrial proton gradient, resulting in rapid substrate oxidation that is uncoupled from ATP synthesis. BAT thermogenesis is activated in response to cold exposure by the local release of norepinephrine (NE) from sympathetic neurons. NE, via cAMP signaling, stimulates lipolysis. The released free fatty acids carry dual roles: they activate the UCP1 protein and fuel substrate oxidation (1). NE and cAMP also drive the acute induction of transcription of genes important for thermogenesis, including Ucp1, Dio2 (a deiodinase that generates local triiodothyronine), Pgc-1α (a transcriptional coactivator that enhances mitochondrial biogenesis and Ucp1 expression), and Gadd45γ (an activator of the p38 MAPK and Ucp1 expression) (2–7). Repeated exposure to cold or chronic stimulation of β-adrenergic signaling leads to sustained increases in mitochondrial biogenesis, oxidative capacity and UCP1 levels, and thereby enhanced brown adipocyte thermogenic capacity (1). Conversely, thermoneutral environments, obesity, and ageing lead to the attenuation of thermogenic capacity (8, 9). Thermogenically active BAT protects rodents, and possibly humans, from obesity and/or obesity-associated metabolic dysfunction (10–14). Understanding the pathways that generate and maintain brown adipocyte thermogenic capacity may enable approaches that counteract obesity and obesity-related diseases.
The nuclear receptor estrogen-related receptor (ERR)α (NR3B1) regulates the expression of a broad range of genes driving mitochondrial biogenesis, the tricarboxylic acid (TCA) cycle and substrate oxidation and is important for mitochondrial oxidative function in BAT (15–19). ERRα whole-body knockout (KO) mice show a decrease in BAT mitochondrial content and oxidative capacity and become hypothermic when exposed to cold (17). The ERRα KO mice show no defects in Ucp1 expression or cold-induced transcriptional responses in BAT (17), suggesting that ERRα is important for driving the high mitochondrial content rather than the adrenergic-induced gene expression program. The lack of a role for ERRα in BAT adrenergic responses is surprising considering that the transcriptional activity of ERRα is potently stimulated by the coactivator PGC-1α, which is induced and activated by adrenergic signaling, and that ERRα itself can bind and regulate Ucp1 in cultured adipocytes (4, 6, 20–23). The use of a complete ERRα KO model for determining the role of ERRα in BAT raises also a few so far unanswered questions. First, it is not clear whether the observed phenotype is entirely BAT autonomous and not impacted by the loss of ERRα in other cell types, such as sympathetic neurons or endothelial cells that interact with brown adipocytes. Second, it is possible that functions of ERRα in the adipocytes are masked by other transcription factors or signaling regulators that are expressed in BAT and compensate for the early loss of ERRα.
Besides ERRα, brown adipocytes express lower but significant levels of 2 other members of the ERR subfamily, ERRβ and ERRγ (7, 24). ERRβ has been implicated in the maintenance of embryonic stem cell pluripotency and the control of ion homeostasis and cell type-specific functions in the inner ear and retina (25–27). ERRγ has functions that overlap those of ERRα and ERRβ. Similar to ERRα, ERRγ promotes oxidative capacity in skeletal and cardiac muscle (28–30). Similar to ERRβ, ERRγ regulates genes important for ion homeostasis and cell type-specific differentiated function in heart, kidney, stomach, skeletal muscle, and trophoblasts (31–33). All 3 ERRs share nearly identical DNA binding domains, suggesting they have similar genomic occupancies when coexpressed; indeed, ERRα and ERRγ bind the same genomic sites in heart (18). Notably, ERRβ and ERRγ are closer to each other than to ERRα in terms of amino acid sequence similarity (particularly in the N-terminal region and ligand binding domains), their ability to be activated by agonist ligands, their transcriptional capacity (eg, ERRβ and ERRγ can activate genes or programs that ERRα does not, such as CDH1 and pluripotency), and their regulation by kinases, such as p38 (7, 34–36). In summary, all 3 ERRs are very close to each other in sequence and function, suggesting they are likely to share functions in cells where they are coexpressed, with ERRβ and ERRγ sharing some features that distinguish them from ERRα.
To clarify the roles of the 3 ERRs in brown adipocytes, we have generated and characterized primary brown adipocytes lacking different combinations of ERRs for their mitochondrial content, respiratory capacity, and adrenergic responses. We show here that the 3 ERRs are not required for cell survival or the differentiation of preadipocytes to brown adipocytes. The ERRs act in a complementary and compensatory fashion to control mitochondrial biogenesis and oxidative function. Moreover, ERRs are collectively important for the transcriptional response to adrenergic stimulation, suggesting a wider role of ERRs in brown adipocyte function than previously appreciated based on the ERRα KO mouse model. Our findings show that the ERRs have overlapping and redundant roles in protecting core mitochondrial function, which has interesting implications for the potential use of ERR ligands.
Materials and Methods
Isolation and differentiation of primary brown adipocytes from ERR floxed mice
Mice with floxed ERR alleles were generated at the Mouse Clinical Institute in Strasbourg, kindly provided by Johan Auwerx, and described in previous studies (32, 37). CRE-mediated recombination in each case deletes the second exon of the gene, which encodes the first zinc finger of the DNA binding domain, and causes a frameshift, leading to the loss of detectable protein (32, 37). The ERR floxed mice have been backcrossed to the C57BL/6JBom background (Taconic Biosciences) more than 10 times, then crossed to each other to produce mice with double and triple floxed genes. Mice were raised at 22°C on a 12-hour light, 12-hour dark cycle with free access to food (LM-485; Harland Tekland) and water. The Institutional Animal Care and Use Committee of The Scripps Research Institute approved animal procedures.
Primary brown adipocyte isolation and differentiation has been described (7). Briefly, BAT depots were isolated from the interscapular area of neonate mice, minced and digested by shaking for 40 minutes at 37°C in buffer containing 61.5mM NaCl, 2.5mM KCl, 0.65mM CaCl2, 2.5mM glucose, 50mM HEPES, 50-U/mL penicillin, 50-μg/mL streptomycin, 2% wt/vol BSA, and 1.5-mg/mL collagenase type I (Worthington). Digestion was assessed to be complete when the suspension was cloudy and remaining tissue fragments were small and limited. Cells were then filtered through a 70-μm cell strainer, resuspended, and plated in high glucose DMEM (Gibco; Invitrogen Life Technologies) with 20mM HEPES, 20% fetal bovine serum (FBS) (Gemini Bio-Products), and 50-U/mL penicillin and 50-μg/mL streptomycin. When cells reached confluency, they were switched first to induction media (DMEM, 10% FBS, 20nM insulin, 1nM triiodothyronine, 0.5mM 3-isobutyl-1-methylxanthine, 2-μg/mL dexamethasone, and 0.25mM indomethacin) for 2 days and then to differentiation media (DMEM, 10% FBS, 20nM insulin, and 1nM triiodothyronine) for 5–6 days.
Generation of ERR KO primary brown adipocytes
To induce recombination of floxed gene loci, primary brown preadipocytes at 70% confluency were incubated for 16 hours with green fluorescent protein (GFP)-expressing (control) or CRE-expressing lentiviruses in media containing 4-μg/mL polybrene (American Bioanalytical). The lentiviruses expressing GFP or CRE were generated in large batches by transfecting HEK 293T cells with psPAX2 (Addgene 12260), pMD2.G (Addgene 12259), and either pLVTHM (GFP control, Addgene 12247) or LV CRE pLKO.1 (Addgene 25997), centrifuging the supernatants at 25 000 rpm at 4°C for 2 hours, resuspending in 1-mL PBS per 15-cm plate, aliquoting, and storing at −80°C. Lentiviral doses were titrated to ones that induced optimal recombination (typically ∼95%) in cells with floxed ERR alleles without affecting brown adipocyte differentiation in wild type (WT) cells. Recombination efficiency of floxed alleles by CRE was determined by isolating total DNA and quantifying WT alleles by quantitative PCR, using primers specific to the nonrecombined genomic region (Table 1). The percent of nonrecombined alleles was determined by normalizing to the copy number of a control gene, Nrip1, in the same samples.
Table 1.
Oligonucleotide Primer Sequences
| PCR primers for quantitation of DNA | ||
|---|---|---|
| mt-CoxII | TCTCCCCTCTCTACGCATTCTA | ACGGATTGGAAGTTCTATTGGC |
| Esrra | CAGCTTCACCCCACCCTTTT | CCTTTGGGCCTGGCACTAAT |
| Esrrb | GCAAGCTGGATTATCTCAGAGCTAAG | GGCGGTCCCATCTAAAGTATGATTCC |
| Esrrg | GTTTTAAAGGCCCTTGGTGATCTCGC | CTGCAACCCTTGGACTGCCAGAAC |
| Nrip1 | TCCCCGACACGAAAAAGAAAG | ACATCCATTCAAAAGCCCAGG |
| PCR primers for quantitative RT-PCR | ||
| Aco2 | TCTCTAACAACCTGCTCATCGG | TCATCTCCAATCACCACCCACC |
| Adrb3 | CAGCCAGCCCTGTTGAAG | CCTTCATAGCCATCAAACCTG |
| Fabp4/aP2 | TGTGTGATGCCTTTGTGGGAACC | CTTCACCTTCCTGTCGTCTGCGG |
| Chchd10 | TGCCTTCAGTGGGGGAAAT | CAGGGCAGGGAGCTCAGAC |
| Cidea | AAACCATGACCGAAGTAGCC | AGGCCAGTTGTGATGACTAAGAC |
| mt-CoxII | TCTCCCCTCTCTACGCATTCTA | ACGGATTGGAAGTTCTATTGGC |
| Cpt1b | CTCCGAAAAGCACCAAAACAT | AGGCTCCAGGGTTCAGAAAGT |
| Cycs | ATTTCAACCCTTACTTTCCCG | CCACTTATGCCGCTTCATGGC |
| Dio2 | CTGCGCTGTGTCTGGAAC | GGAGCATCTTCACCCAGTTT |
| Endog | CCACCAATGCGGACTACC | AGGCATTCTGGTTGAGGTGT |
| Esrrα | GGAGGACGGCAGAAGTACAAA | GCGACACCAGAGCGTTCAC |
| Esrrβ | CCGGCCACCAATGAATGT | ATCCAGCCGTCGCTTGTACT |
| Esrrγ | TCCCCGACAGTGACATCAAA | GTGTGGAGAAGCCTGGAATA |
| Gabpα | CCGCTACACCGACTACGATT | ACCTTCATCACCAACCCAAG |
| Gadd45g | TTCGTGGATCGCACAATGACT | GGACTTTGGCGGACTCGTAGA |
| Idh3A | AGGACTGATTGGAGGTCTTGG | ATCACAGCACTAAGCAGGAGG |
| Ldhb | CCAGCAGGAGGGGGAGAGT | TGCTTAGGTAGCCCGCTCAG |
| Lpl | AACTTGTGGCCGCCCTGTA | TGGACGTTGTCTAGGGGGTAGT |
| Lxh8 | GAGCTCGGACCAGCTTCA | TTGTTGTCCTGAGCGAACTG |
| Nrf1 | CCACGTTGGATGAGTACACG | CTGAGCCTGGGTCATTTTGT |
| Pck1 | ATCTTTGGTGGCCGTAGACCT | GCCAGTGGGCCAGGTATTT |
| Pdk4 | GTTCCTTCACACCTTCACCAC | CCTCCTCGGTCAGAAATCTTG |
| Pgc-1α | GGAGCCGTGACCACTGACA | TGGTTTGCTGCATGGTTCTG |
| Pgc-1β | AGTGGGTGCGGAGACACAGAT | AAAGCTCCACCGTCAGGGACT |
| Ppara | AAGGCTATCCCAGGCTTTGC | TTTAGAAGGCCAGGCCGATCTC |
| Pparγ | AGGCCGAGAAGGAGAAGCTGTT | TGGCCACCTCTTTGCTCTGCTC |
| Ppia | CAAGACTGAATGGCTGGATG | ATGGGGTAGGGACGCTCTCC |
| Prdm16 | CAGCACGGTGAAGCCATTC | GCGTGCATCCGCTTGTG |
| Rplp0 | CTGTGCCAGCTCAGAACACTG | TGATCAGCCCGAAGGAGAAG |
| Sdhb | TACCGATGGGACCCAGACA | CGTGTGCACGCCAGAGTAT |
| Sirt3 | TTTCTTTCACAACCCCAAGC | ACAGACCGTGCATGTAGCTG |
| Tfam | CAAAGGATGATTCGGCTCAG | AAGCTGAATATATGCCTGCTTTTC |
| Tfb1m | ACCGAGGGCTTGGAATGTTA | TGGATCAATGTCTGCCAACTGT |
| Tfb2m | TTTGGCAAGTGGCCTGTGA | CCCCGTGCTTTGACTTTTCTA |
| Ucp1 | TGGAGGTGTGGCAGTGTTCAT | TGACAGTAAATGGCAGGGGAC |
| Zic1 | CTGTTGTGGGAGACACGATG | CCTCTTCTCAGGGCTCACAG |
Mitochondrial DNA (mtDNA) content
Total DNA was isolated using TRIzol (Invitrogen) according to the manufacturer's protocol. mtDNA was determined by quantifying the copy number of the mitochondrial gene mt-Co2 (CoxII), relative to the copy number of the nuclear gene Nrip1 (primer sequences in Table 1).
Gene expression
RNA was extracted from cells using TRIzol (Invitrogen) according to manufacturer's protocol and as reported (35). Quantitative RT-PCR was performed using gene-specific primers. Relative mRNA expression was normalized using Rplp0 (36B4) or Ppia (cyclophilin A) as reference genes. Primer sequences are in Table 1.
Protein analysis
Whole-cell lysates were prepared using radioimmunoprecipitation assay buffer (150mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1mM EDTA, and 50mM Tris; pH 7.5) with 1μM phenylmethylsulfonyl fluoride and 5-μL/mL protease inhibitor cocktail P8340 (Sigma). Proteins were resolved using SDS-PAGE and detected by Western blotting, using the following antibodies: anti-ERRα (ab76228; Abcam); anti-ERRγ (polyclonal serum from rabbits immunized with peptides SNKDRHIDSSC and CSSTIVEDPQTK [ERRγ amino acids 25–35 and 104–115, respectively] and purified for binding to SNKDRHIDSSC); anti-Ucp1 (Abcam); anti-Rt/Ms Total oxidative phosphorylation (OxPhos) complex mix (Invitrogen Life Technologies); anti-Ras-related Nuclear protein (RAN) (BD Biosciences); anti-ERRα (ab76228; Abcam). Protein abundance was quantified using ImageJ and normalized to RAN protein or Ponceau levels.
O2 consumption assay
O2 consumption rates (OCRs) of primary brown adipocytes were determined using a Seahorse XFe96 Flux analyzer as previously described (7). Briefly, adipocytes were trypsinized on day 4 of differentiation (5 d after infection with CRE or GFP lentivirus) and plated at 4000 cells/well in a Seahorse 96-well plate. OCRs were measured 48 hours later. Cells were exchanged to assay media (DMEM [10569; Invitrogen Life Technologies] supplemented with 143mM NaCl, 10mM glucose, 10mM sodium pyruvate, 2mM GlutaMAX, and 5mM HEPES buffer; pH 7.4) 1 hour before measuring OCRs. Maximal respiration was determined after injecting 0.5μM carbonyl cyanide-4-(trifluoromethoxy)phenyl-hydrazone (FCCP), and nonmitochondrial respiration was determined after injection of 2μM rotenone and 2μM antimycin A (RAA). For experiments where differences in numbers between WT and KO cells plated were seen, OCRs were normalized by the average number of adipocytes (lipid droplet containing cells) in representative wells of each genotype. Basal respiration was calculated by subtracting the OCR in the presence of RAA from that of the initial OCR, before addition of drugs. Maximal respiratory capacity was also corrected by subtracting OCR in the presence of RAA from the OCR in the presence of FCCP. Space respiratory capacity was determined by subtracting the basal OCR from the maximal OCR.
Statistical analysis
Data are expressed as the mean ± SD or SEM, as indicated, and were analyzed with a 2-tailed Student's t test for single variables. For comparisons between multiple variables, P values were determined using a two-way ANOVA test, followed by a Bonferroni analysis.
Results
ERRs are not required for brown adipocyte differentiation or survival
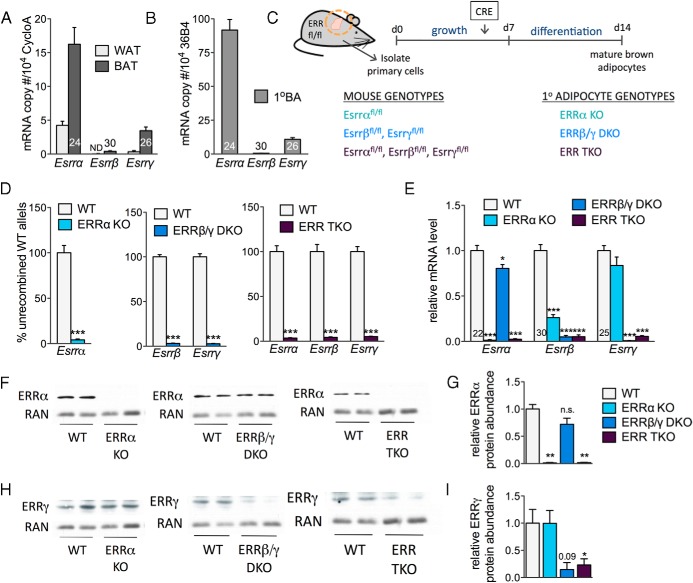
All 3 ERRs are expressed in BAT and enriched in BAT vs white adipose tissue (WAT) (Figure 1A). ERRβ and ERRγ show the highest BAT enrichment (>10-fold), whereas ERRα is the most abundant isoform in both BAT and WAT. The 3 ERRs are also expressed in differentiated primary brown adipocytes, at levels comparable with those seen in BAT (Figure 1B), suggesting that these cells provide a valid model system to define the roles of endogenous ERRs for brown adipocyte function. Thus, we generated brown adipocytes, lacking 1, 2, or all 3 endogenous ERRs, by isolating primary brown preadipocytes from neonate mice carrying ERR loci flanked by loxP sites, inducing recombination with lentiviruses that express the CRE recombinase (or GFP, as control), and then differentiating the cells to mature adipocytes (see scheme in Figure 1C). Expression of CRE led to the efficient and specific deletion of the floxed ERR exons (Figure 1D). We chose to focus our study on adipocytes of 3 genotypes: 1) the deletion of just ERRα alone (ERRα KO), the ERR that is most abundant in BAT, WAT, and primary brown adipocytes, and most dependent on PGC-1 coactivators for function (7); 2) deletion of ERRβ and ERRγ [ERRβ/γ double KO (DKO)], the 2 ERRs that have higher sequence similarity to each other than to ERRα, show highest enrichment in BAT vs WAT, and are transcriptionally active in the absence of PGC-1 coactivators (7, 38); and 3) the simultaneous deletion of all 3 ERRs [ERR triple KO (TKO)], to gain insights into the collective role of ERRs in brown adipocytes. In each case, CRE-mediated recombination led to a greater than 95% reduction in mRNA levels of the targeted isoform(s) (Figure 1E). In addition, ERRβ mRNA levels were significantly reduced (∼4-fold) in ERRα KO cells, and ERRα mRNA expression was modestly decreased (by 20%) in the ERRβ/γ DKO cells (Figure 1E). At the protein level, we confirmed the reduced expression of ERRα and ERRγ in cells having deleted ERRα and/or ERRγ alleles, respectively (Figure 1, F–I). The residual signal in the ERRγ Western blottings of DKO and TKO cells is likely a nonspecific band of similar molecular weight, given the more than 95% recombination of ERRγ alleles and the more than 20-fold decrease in ERRγ mRNA levels in these cells (Figure 1, D and E). We were unable to reliably detect ERRβ protein with the currently available antibodies. Despite the modest decrease of ERRα mRNA, ERRα protein levels were not significantly decreased in cells lacking ERRβ and ERRγ (ERRβ/γ DKO) (Figure 1, F and G). ERRγ protein levels were also not affected by the deletion of ERRα (Figure 1, H and I). In summary, we were able to generate 3 types of adipocytes: ones lacking ERRα and expressing decreased ERRβ mRNA and normal ERRγ levels, ones lacking just ERRβ and ERRγ, and ones lacking all 3 ERRs.
Figure 1.
Generation of primary brown adipocytes lacking ERRs. A, ERR mRNA levels in mouse gonadal WAT and interscapular BAT depots, expressed as copy number relative to control gene (cyclophilin A [Ppia]). B, ERR mRNA levels in differentiated primary brown adipocytes (1°BA), expressed as copy number relative to control gene (36B4 [Rplp0]). A and B, Data are expressed as mean ± SD (n = 3). Numbers within or above bars indicate threshold cycle (Ct) values for each gene. C, Scheme depicting generation of primary brown adipocytes with deleted ERRs. D, The percent of WT (unrecombined) ERR alleles in differentiated adipocytes derived from mice of the indicated genotypes and infected with GFP- or CRE-expressing lentivirus. Data are expressed as mean ± SEM (n = 6–12). E, Expression levels of ERRs in adipocytes generated from the 3 genotypes after infection with GFP- or CRE-expressing lentivirus. Data are expressed as mean ± SEM (n = 6–12). F and H, ERRα and ERRγ protein levels in adipocytes of the 3 genotypes compared with levels in WT adipocytes on day 7 of differentiation. G and I, Quantification of Western blottings, showing ERRα and ERRγ protein abundance (expressed relative to RAN levels); *, P < .05; ***, P < .001.
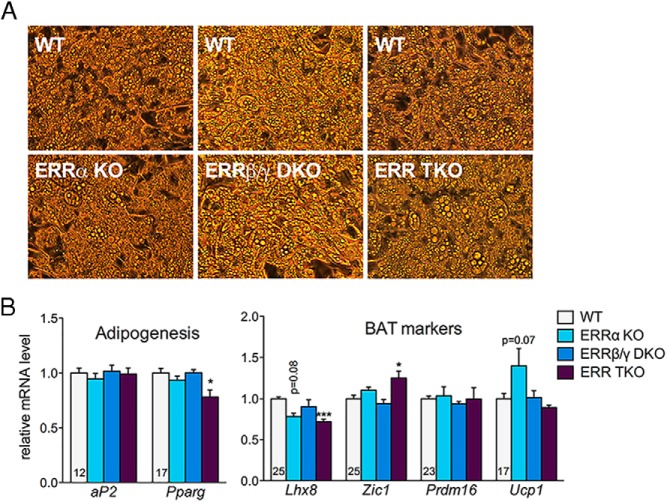
ERRα and ERRγ have been implicated in adipocyte differentiation (39, 40). We thus first assessed ERR KO cells for their ability to differentiate. Morphologically, cells of all 3 genotypes differentiated efficiently into mature brown adipocytes with multilocular lipid droplets (Figure 2A). Cells of all genotypes also expressed comparable levels of adipocyte differentiation markers (aP2 [Fabp4] and peroxisome proliferator-activated receptor γ [Pparg]), brown adipocyte enriched markers (Lhx8, Pdrm16, and Ucp1), and brown adipocyte-specific markers (Zic1) (Figure 2B) (41). Some differences were seen in specific genes (eg, a small decrease in Lhx8 in the TKO), but these likely reflect specific requirements of ERRs for the expression of such genes, rather than a general decrease in brown adipocyte differentiation, as other markers were similar or even higher in TKO cells (eg, Prdm16 and Zic1, respectively). In conclusion, ERRs are not required for the differentiation of brown preadipocytes into mature adipocytes. Moreover, we did not observe any decreases in cell numbers, suggesting that ERRs are not essential for the survival of brown adipocytes.
Figure 2.
ERRs are not required for the differentiation of preadipocytes into mature brown adipocytes. A, Representative bright-field images of WT and ERR KO primary brown adipocytes on day 7 of differentiation. B, Relative mRNA levels in WT and ERR KO primary brown adipocytes on day 7 of differentiation. Data are the mean ± SEM (n = 5–12) and expressed relative to the levels of each gene in WT cells; *, P < .05; ***, P < .001. Numbers within bars indicate threshold cycle (Ct) values for each gene in the WT control brown adipocytes. The Ct values of the brown adipocyte markers Lhx8, Zic1, Prdm16, and Ucp1 were more than 35 in differentiated 3T3-L1 cells, consistent with very low to undetectable expression of these genes in white adipocytes.
ERRs act in a complementary manner to regulate brown adipocyte gene expression
ERRα and ERRγ have been shown to affect the expression of genes important for mitochondrial biogenesis and function; for many of these genes, ERRα and ERRγ bind directly at ERR response elements near the genes, consistent with them being direct targets (38). Thus, we next assessed the expression levels of known ERR target genes whose function is important for mitochondrial oxidative function. We also assessed the expression levels of genes that are not known or predicted to be ERR targets (eg, CoxII, Adrb3) but can give insights into the altered state of adipocytes lacking ERRs.
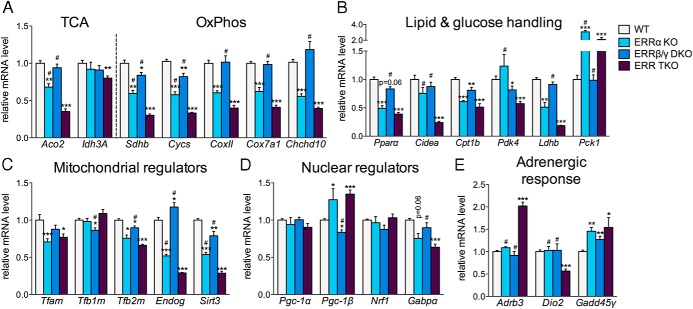
Deletion of just ERRα (ERRα KO cells) led to significant decreases in the expression of several known ERR targets (eg, Aco2, Sdhb, Cycs, Gabpα, Cpt1b, Pparα, and Ldhb) (Figure 3) (15–18, 42, 43). Given the very high levels of ERRα in the adipocytes (Figure 1B), the observed decreases seem relatively modest for some of these genes. In addition, the levels of some well-characterized ERRα targets, such as Idh3a, Cidea, and Pdk4, were not affected by the loss of ERRα (17, 44–46). Similarly, deletion of ERRβ and ERRγ (ERRβ/γ DKO) led to modest decreases in some ERR targets (eg, Sdhb, Cycs, Cpt1b, Sirt3, and Pdk4) but had no effect on many others (Aco2, Idh3a, Gabpa, Cidea, and Ldhb) (Figure 3). In contrast, deletion of all 3 ERRs led to significant and more pronounced decreases of a broader range of ERR target genes, suggesting that at many of these genes, ERRα, ERRβ, and ERRγ act together and can partially compensate for each others' loss (eg, Aco2, Sdhb, Cycs, Cidea, Ldhb, and Pdk4) (Figure 3).
Figure 3.
ERRs act in a complementary manner to regulate brown adipocyte gene expression. A–E, Relative mRNA levels in WT and ERR KO primary brown adipocytes on day 7 of differentiation. Data are the mean ± SEM (n = 5–12) and expressed relative to the levels of each gene in WT cells. Mitochondrial (C) and nuclear (D) regulators refer to genes important for mitochondrial biogenesis and acting in the mitochondria or nucleus, respectively; *, P < .05; **, P < .01; ***, P < .001, compared with WT cells; #, P < .05, compared with ERR TKO cells.
Deletion of ERRs led to pronounced decreases in genes involved in several metabolic oxidative pathways, including the TCA cycle, OxPhos, and lipid and glucose handling (Figure 3, A and B). Furthermore, many mitochondrial regulators important for mtDNA replication and expression were significantly decreased (Tfam, Tfb2m, Endog, and Sirt3) (Figure 3C). We also determined the levels of other nuclear transcriptional regulators that influence oxidative metabolism and mitochondrial biogenesis and found that although ERR TKO cells had modest decreases in Gabpa, there were no decreases in Nrf1, Pgc-1α, or Pgc-1β (Figure 3D). For all pathways, ERRα had a more prominent role than ERRβ and ERRγ; however, cells lacking all ERRs had the most pronounced gene expression defects.
Besides the expected decreased expression of known ERR targets, we observed that the expression of specific genes harboring ERREs, such as Pck1 and Pgc-1β, was enhanced in cells lacking ERRs, and in particular in cells lacking ERRα, consistent with ERRα acting as a repressor at such genes, as has been reported for Pck1 in hepatocytes (Figure 3, B and D) (47, 48). Moreover, we found that the expression of other genes that have been described as ERR targets, such as Pgc-1α, was not affected by the deletion of ERRs, suggesting their regulation in brown adipocytes is driven by factors other than ERRs (Figure 3D) (47, 49).
Finally, we assessed the expression of other genes whose function is important for brown adipocyte function, and in particular for the ability of adipocytes to respond to adrenergic stimulation, such as Dio2, Adrb3, and Gadd45γ. The levels of Dio2, which encodes the type II iodothyronine deiodinase and drives the local generation of bioactive T3, were decreased in the triple ERR KO adipocytes, suggesting that Dio2 is an ERR target (Figure 3E). The expression levels of the β3-adrenergic receptor Adrb3 and the signaling regulator Gadd45γ were modestly up-regulated in the same cells, consistent with the ERR KO adipocytes having intact adrenergic signaling (Figure 3E).
In summary, the gene expression studies showed a complex pattern of dependency of ERR targets on different ERR isoforms; some ERR target genes were dependent on ERRα, the most abundant ERR isoform in these cells, whereas many genes showed significant and pronounced decreases only when all 3 ERRs were deleted. ERRs were clearly not required for brown adipocyte specification, because the expression of Adrb3, Pgc-1α, and Pgc-1β (typically high in these cells) was either not affected or in some cases increased.
ERRs drive collectively brown adipocyte mitochondrial biogenesis and oxidative capacity
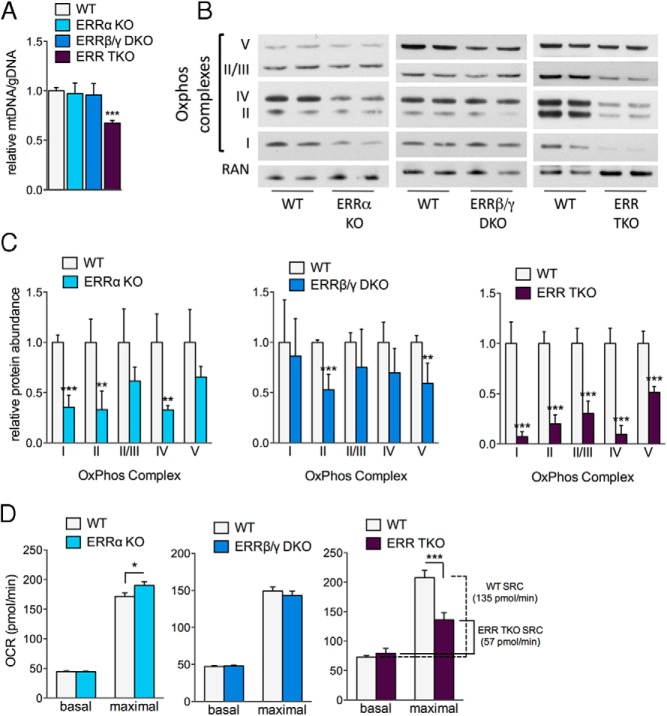
The decreased expression of TCA cycle, OxPhos, and mitochondrial biogenesis genes (Figure 3) suggests that adipocytes lacking ERRs have decreased mitochondrial content and oxidative function. Indeed, the ERR TKO cells had a significant reduction in the mitochondrial genome copy number, when compared with WT control cells (Figure 4A). Interestingly, neither the ERRα KO nor the ERRβ/γ DKO had reduced mitochondrial genome content, suggesting the ERRs functionally compensate for each other in maintaining mtDNA. Similarly, the protein content of all 5 OxPhos complexes was dramatically reduced in the ERR TKO adipocytes, with close to 10-fold losses for some of the complexes (eg, complexes I and IV) (Figure 4, B and C). Cells lacking just ERRα or ERRβ/ERRγ showed more modest, although significant, reductions in some of the OxPhos complexes (Figure 4, B and C).
Figure 4.
ERRs collectively control mitochondrial abundance and oxidative capacity of brown adipocytes. A, Relative mitochondrial genome content in WT and ERR KO primary brown adipocytes. Data are the mean ± SEM (n = 10–12). B, OxPhos complex protein levels in WT and ERR KO primary brown adipocytes on day 7 of differentiation. C, Quantification of Western blottings, showing indicated OxPhos complex abundance, after normalization to RAN levels. Data are from 2 separate experiments and are the mean ± SD (n = 4). D, OCRs of WT and ERR KO primary brown adipocytes measured by Seahorse flux analysis on day 6 of differentiation. Data are the mean ± SEM (n = 12–24 from 2 or more experiments); *, P < .05; **, P < .01; ***, P < .001.
To determine the functional consequences of changes in mitochondrial content or composition, we next measured the oxidative capacities of mature ERR KO adipocytes. Despite the reduction in mitochondrial gene expression and protein content, adipocytes lacking either ERRα alone or ERRβ/ERRγ together, showed no decreases in basal or maximal (ie, in presence of the uncoupler FCCP) OCRs (Figure 4D and Supplemental Figure 1, A and B). In contrary, ERRα KO cells had an unexpected, modest but reproducible and significant increase in their maximal OCR (Figure 4D and Supplemental Figure 1A). Adipocytes lacking all 3 ERRs were the only ones that showed a significant and dramatic defect in oxidative capacity (Figure 4D and Supplemental Figure 1C); although their basal respiration rate was unchanged, their maximal respiration rate was significantly reduced. Consequently, the spare respiratory capacity (SRC) of ERR TKO cells was just 42% of that seen in WT control cells (Figure 4D). Altogether, these data show, first, that the 3 ERRs are collectively important for brown adipocyte mitochondrial oxidative capacity, and second, that the different ERR isoforms can act largely redundantly with each other to preserve oxidative capacity.
Loss of ERRs impairs the transcriptional response of brown adipocytes to adrenergic signaling
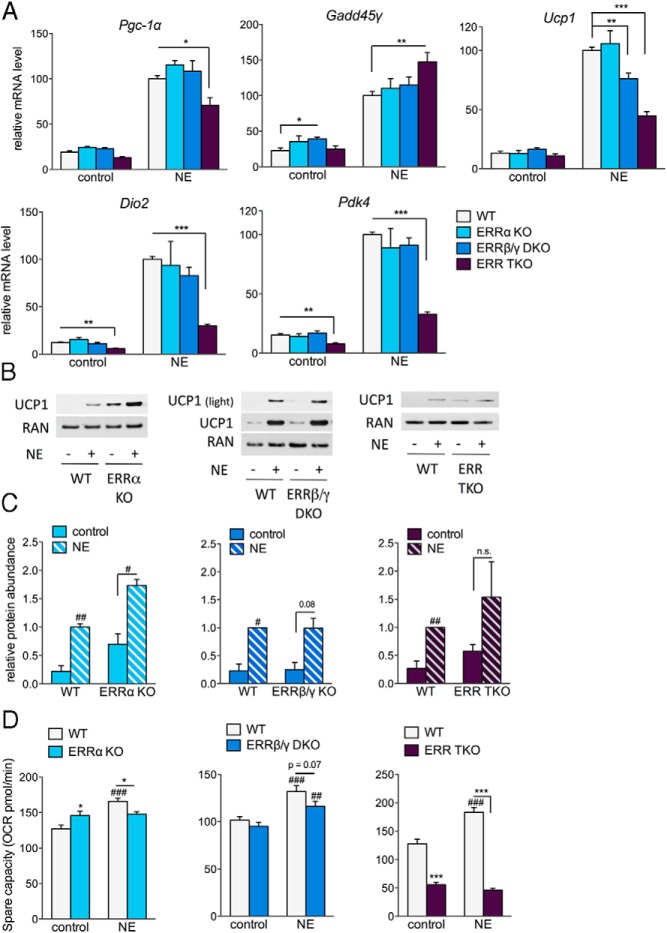
Adrenergic stimulation of brown adipocytes induces the expression of genes important for thermogenesis and enhances cellular oxidative capacity. To assess the possible role of ERRs in the acute transcriptional response to adrenergic stimulation, primary brown adipocytes were treated with NE for 4 hours. Although the expression levels of the ERRs themselves did not change in response to acute NE treatment (Supplemental Figure 1D), activators of the ERRs, Pgc-1α and Gadd45γ, were strongly induced in WT cells (Figure 5A), as previously reported (4, 7). NE also induced the expression of Ucp1, Dio2, and Pdk4 in WT cells (Figure 5A). In the TKO cells, the induction by NE was severely blunted for Ucp1, Dio2, and Pdk4 and moderately reduced for Pgc-1α. The induction of Gadd45γ was not decreased, suggesting that the ERR TKO adipocytes retain NE signaling but lose the induction of specific NE-regulated targets. Looking at the contribution of the different ERR isoforms in the NE-induced acute responses, neither ERRα KO nor ERRβ/γ DKO adipocytes showed pronounced defects, except for a modest decrease in the induction of Ucp1 in ERRβ/γ DKO cells (Figure 5A).
Figure 5.
Loss of ERRs impairs the transcriptional and cellular response to adrenergic stimulation. A, Relative mRNA levels in WT and ERR KO adipocytes treated for 4 hours with or without 0.1μM NE on day 7 of differentiation. Data are normalized to the levels of each gene in WT cells treated with NE (set at 100) and are expressed as the mean ± SEM (n = 5–9). B, UCP1 protein levels in WT and ERR KO cells treated with 0.1μM NE for 18 hours, collected on day 7 of differentiation. Blots shown are representative of 2 experiments. C, Quantification of Western blottings, showing UCP1 protein levels normalized to RAN or protein abundance determined by Ponceau staining. Data are expressed relative to the levels of UCP1 in WT cells treated with NE (set at 1) and are the mean ± SD (n = 2–3). D, SRC of WT and ERR KO cells treated with 0.1μM NE for 16 hours, measured on day 6 of differentiation. Data are the mean ± SEM (n = 13–15); *, P < .05; **, P < .01; ***, P < .001 compared with similarly treated WT; #, P < .05; ##, P < .01; ###, P < .001 compared with control (no NE) cells of the same genotype.
Next, we asked how the ERRs affected UCP1 protein levels. Despite having similar Ucp1 mRNA levels in basal conditions, both ERRα KO and ERR TKO cells showed unexpectedly higher UCP1 protein levels (Figure 5, A–C). UCP1 protein was further increased by NE in all KO cells, but the fold increase and the significance of the increase were compromised in ERR TKO cells. ERRβ/γ DKO cells showed a similar induction of UCP1 protein levels by NE as in WT cells (P = .08 in DKO and 0.02 in WT cells) (Figure 5, B and C), despite the reduced induction of Ucp1 at the transcriptional level (Figure 5A).
Finally, to determine the ability of the cells to remodel their mitochondrial capacity in response to an energetic demand we challenged the adipocytes with NE overnight. Overnight NE treatment of WT cells led to an approximately 25% increase in the SRC (maximal minus basal OCR) (Figure 5D), without having a consistent impact on the basal respiration rate (Supplemental Figure 1E), suggesting that WT cells remodel their mitochondrial function in response to adrenergic stimulation. The ERR TKO cells were defective in this response; their already low SRC did not increase after overnight exposure to NE (Figure 5D). Both the ERRα KO and the ERRβ/γ DKO cells showed reduced SRCs after NE treatment, compared with WT cells, but neither defect was as dramatic as the one seen in TKO cells (Figure 5D). Taken together, these data indicate that the ERRs are important for NE-induced cellular adaptations, both the acute transcriptional induction of genes and the increase in mitochondrial oxidative capacity.
Discussion
The capacity of ERRs (and in particular ERRα) to drive the expression of genes important for oxidative metabolism has been demonstrated in earlier studies (15, 16, 18, 38, 50). The current study shows that in cells coexpressing multiple ERR isoforms, such as primary brown adipocytes, the different ERRs act in a highly complementary fashion to control mitochondrial biogenesis and cellular oxidative capacity. Moreover, in addition to the expected effects of deletion of ERRs on mitochondrial content and function, we find that ERRs are important for the transcriptional and metabolic response of brown adipocytes to adrenergic stimulation. Our findings underscore the importance of a collective cell-autonomous role of ERRs for mitochondrial oxidative capacity in brown adipocytes and highlight that new roles of ERRs can become apparent by the deletion of multiple ERR isoforms.
Dufour et al first demonstrated that cardiac ERRα and ERRγ have similar genomic DNA occupancies, suggesting that the 2 ERRs target the same genes and have largely overlapping functions (18). According to this view, the relative levels of ERRs in different cell types are likely to determine their relative contribution to the expression of ERR targets. In BAT and cultured brown adipocytes, ERRα is the most abundant ERR isoform at the mRNA level, as well as the one ERR that is readily detected at the protein level by multiple antibodies. ERRα has also been shown to be important for BAT mitochondrial content and thermogenic function in vivo (17). Consequently, it was somewhat surprising that deletion of ERRα alone in primary brown adipocytes caused only mild decreases in expression of ERR targets, and no loss of mtDNA content or oxidative capacity, whereas deletion of all 3 ERRs caused dramatic defects at all levels (Figures 3 and 4). Notably, ERRα KO cells show significant decreases in ERRβ expression, suggesting that ERRγ is sufficient to maintain the mtDNA content and oxidative capacity in these cells. Similarly, ERRα alone is capable of supporting brown adipocyte oxidative function in ERRβ/γ DKO adipocytes, highlighting the redundancy of ERRα and ERRγ for mitochondrial function.
The lack of a detectable decrease in oxidative capacity in ERRα KO brown adipocytes is also surprising when considering the decreases in the mRNA levels of at least a few important TCA cycle and OxPhos genes and in the protein levels of OxPhos complexes I, II, and IV (Figures 3 and 4). This finding suggests that the levels of the genes and protein complexes affected by ERRα are not rate limiting for maximal oxidative capacity in cultured brown adipocytes and that the rate limiting factors (eg, possibly substrate supply to mitochondria) are not affected by the single deletion of ERRα. It also implies that even though we did not detect decreases in the oxidative capacity of ERRα KO cells, there may be situations, particularly in vivo, where OxPhos expression levels are rate limiting and loss of ERRα alone compromises cellular oxidative capacity.
The BAT depot of whole-body ERRα KO mice shows decreased mtDNA content and oxidative capacity (17), raising the question of why these defects are not seen in ERRα KO cells. Beyond the reasoning presented above, ie, that different factors may be rate limiting under different physiological states, there are other possible explanations. First, whole-body ERRα KO mice also have a decrease in BAT ERRβ and ERRγ levels (see Ref. 17 and our unpublished data), thus potentially behaving more similarly to triple ERR TKO adipocytes. Second, the loss of ERRα function in other cell types (eg, neurons or endothelial cells) may impact BAT activity and contribute to the decreased oxidative and thermogenic capacity in the whole-body ERRα KO mice. A similar phenomenon is seen in whole-body ERRγ KO mice, which have cardiac defects that are not seen in mice with just cardiac-specific loss of ERRγ (29–31). Interestingly, combining the cardiac-specific loss of ERRγ with a whole-body ERRα KO leads to severe defects in cardiac mitochondria, oxidative metabolism, and function, suggesting that ERRα and ERRγ carry complementary roles in the heart, like in brown adipocytes (29). Lastly, some of the differences observed between the current study and the whole-body ERRα KO model may be the consequence of the timing of ERR deletion. Here, ERRα is deleted just before adipocyte differentiation, whereas in the KO mouse ERRα is missing throughout development.
ERRα and ERRγ have also been implicated in the control of adipogenesis. ERRα KO animals have reduced white adipose depots (51) and suppression of ERRα or ERRγ in mesenchymal cells and 3T3-L1 preadipocytes impairs adipogenesis (39, 40, 52). However, we see no defects in brown adipocyte differentiation, even when all 3 ERRs are deleted. One possible explanation is that ERRs are needed for early commitment stages of adipogenesis, and that brown preadipocytes isolated from neonatal BAT no longer require ERRs for differentiation. Alternatively, because white and brown adipocytes are derived from different lineages, it is possible that ERRs are required for the differentiation of myf5− but not myf5+ derived adipocytes (53). Future work with mesenchymal cells isolated from ERR floxed mice can clarify the specific roles of ERRs in adipogenesis.
BAT thermogenic function relies on high mitochondrial oxidative capacity but also on the levels and activity of UCP1 (1). The roles of ERRs for regulating UCP1 expression appear complex. The induction of Ucp1 mRNA by NE is partially dependent on ERRs, consistent with previously described transcriptional roles of ERRs at this gene (7, 23, 24). However, the defect in transcriptional induction at the mRNA level is counterbalanced by an unexpected increase of UCP1 at the protein level. The increase in UCP1 protein is most prominent in ERRα KO cells but also seen in ERR TKO adipocytes. A possible explanation for these findings is that although ERRs activate Ucp1 transcription, they also activate the expression of a currently unknown gene that suppresses UCP1 at a posttranscriptional level (eg, at the level of translation or protein stability). Ultimately, what matters the most for thermogenesis is UCP1 activity and not just protein levels. We did not observe dramatic changes in the percentage of uncoupled basal respiration, except for a decrease in uncoupling in ERR TKO adipocytes treated overnight with NE (Supplemental Figure 1F). However, measurements of uncoupled respiration in intact cells are confounded by factors other than UCP1 (54). Future experiments with permeabilized cells, which allow control of substrate availability and the use of UCP1 activators and inhibitors, will better clarify the effects of ERRs on UCP1 activity.
Besides the defects in mitochondrial oxidative pathways and transcriptional responses to NE, ERR TKO brown adipocytes showed greatly altered expression of genes important for lipid and glucose handling, (eg, Ppara, Cidea, Cpt1b, Pck1, and Ldhb), suggesting additional cellular metabolism defects. Future studies will be needed to define the roles of ERRs for lipolysis, lipid oxidation and reesterification, and glycolysis. Furthermore, although the current study focuses on the collective, common role of ERR isoforms for mitochondrial oxidative function, it is possible that the 3 ERRs act in an isoform-specific manner to control other genes and cellular functions, as seen in other cell types (32, 35, 36). Even among the commonly regulated mitochondrial function genes, there are subtle differences on how they are affected by ERRs. Loss of ERRβ and ERRγ affected the basal levels of Pdk4, mitochondrial complexes II and V, and the induction of Ucp1 by NE, whereas loss of ERRα led to the increased expression of select genes (Pck1 and Pgc-1β) and a modest but reproducible increase in the maximal OCR (Figures 3 and 4). The ability of ERRα to repress target genes has been previously reported and may reflect the interaction of ERRα with corepressors (48, 55). Finally, the 3 ERR isoforms may still carry distinct physiological roles by being activated by distinct signals, and thus being active in different contexts. ERRβ and ERRγ are transcriptionally active in cells where ERRα shows no transcriptional activity, suggesting ERRβ and ERRγ can integrate different signals (7). It is becoming increasingly clear that BAT is activated by a multitude of signals besides NE (56); some of these signals may be relayed by select ERR isoforms.
The coexpression of all 3 ERRs in BAT may help protect vital oxidative functions. The ERRs are also coexpressed in other cell types that rely on high oxidative capacity, such as cardiomyocytes, type I skeletal myofibers, and kidney cells (57). The capacity of ERRs to functionally compensate for each other in such cell types may present interesting therapeutic windows for the application of ERR selective inhibitors, such as ERRα inverse agonists that restrict tumor growth (58, 59). Tumor-suppressive ERRα selective inhibitors may have no harmful effects in tissues with high energy demands that express and are protected by ERRβ and ERRγ. Furthermore, ERRβ/γ agonists used in combination with ERRα inverse agonists may boost the protective function of ERRβ and ERRγ and enhance therapeutic benefits of ERRα inhibition. The overlapping and isoform-specific functions of ERRs may therefore have important implications not only for BAT thermogenic capacity but also for the use of ERR ligands in cancer treatment.
Acknowledgments
We thank Johan Auwerx for providing ERR floxed mouse strains and Yoshitake Cho, Enrique Saez, Cristina Godio, Andrea Galmozzi, and Josep Villena for discussions and advice.
Present address for M.L.G.: The Lowy Medical Research Institute, La Jolla, CA 92037. Present address for E.L.B. and A.K.: Johns Hopkins University School of Medicine, Baltimore, MD 21205.
This work was supported by the National Institutes of Health Grant R01DK095686 (to A.K.), the Ruth L. Kirschstein National Research Service Award 5F31AG033487 (to M.L.G.), and the Shared Instrumentation Grant 1S10D16357.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BAT
- brown adipose tissue
- ERR
- estrogen-related receptor
- DKO
- double KO
- FBS
- fetal bovine serum
- FCCP
- carbonyl cyanide-4-(trifluoromethoxy)phenyl-hydrazone
- GFP
- green fluorescent protein
- KO
- knockout
- mtDNA
- mitochondrial DNA
- NE
- norepinephrine
- OCR
- O2 consumption rate
- OxPhos
- oxidative phosphorylation
- PPAR
- peroxisome proliferator-activated receptor
- RAA
- rotenone and antimycin A
- RAN
- Ras-related Nuclear protein
- SRC
- spare respiratory capacity
- TCA
- tricarboxylic acid
- TKO
- triple KO
- UCP1
- Uncoupling Protein 1
- WAT
- white adipose tissue
- WT
- wild type.
References
- 1. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. [DOI] [PubMed] [Google Scholar]
- 2. Ricquier D, Mory G, Bouillaud F, Thibault J, Weissenbach J. Rapid increase of mitochondrial uncoupling protein and its mRNA in stimulated brown adipose tissue. Use of a cDNA probe. FEBS Lett. 1984;178(2):240–244. [DOI] [PubMed] [Google Scholar]
- 3. Silva JE, Larsen PR. Potential of brown adipose tissue type II thyroxine 5′-deiodinase as a local and systemic source of triiodothyronine in rats. J Clin Invest. 1985;76(6):2296–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. [DOI] [PubMed] [Google Scholar]
- 5. Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. [DOI] [PubMed] [Google Scholar]
- 6. Cao W, Daniel KW, Robidoux J, et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24(7):3057–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gantner ML, Hazen BC, Conkright J, Kralli A. GADD45γ regulates the thermogenic capacity of brown adipose tissue. Proc Natl Acad Sci USA. 2014;111(32):11870–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9(2):203–209. [DOI] [PubMed] [Google Scholar]
- 9. Ouellet V, Routhier-Labadie A, Bellemare W, et al. Outdoor temperature, age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. J Clin Endocrinol Metab. 2011;96(1):192–199. [DOI] [PubMed] [Google Scholar]
- 10. Bartelt A, Bruns OT, Reimer R, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17(2):200–205. [DOI] [PubMed] [Google Scholar]
- 11. Lee P, Smith S, Linderman J, et al. Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes. 2014;63(11):3686–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu X, Zheng Z, Zhu X, et al. Brown adipose tissue transplantation improves whole-body energy metabolism. Cell Res. 2013;23(6):851–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stanford KI, Middelbeek RJ, Townsend KL, et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest. 2013;123(1):215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoneshiro T, Aita S, Matsushita M, et al. Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest. 2013;123(8):3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schreiber SN, Emter R, Hock MB, et al. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA. 2004;101(17):6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mootha VK, Handschin C, Arlow D, et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101(17):6570–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villena JA, Hock MB, Chang WY, Barcas JE, Giguère V, Kralli A. Orphan nuclear receptor estrogen-related receptor α is essential for adaptive thermogenesis. Proc Natl Acad Sci USA. 2007;104(4):1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dufour CR, Wilson BJ, Huss JM, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab. 2007;5(5):345–356. [DOI] [PubMed] [Google Scholar]
- 19. Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71(Can't get issue from record):177–203. [DOI] [PubMed] [Google Scholar]
- 20. Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1α (PGC-1α) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-α and -γ. Identification of novel leucine-rich interaction motif within PGC-1α. J Biol Chem. 2002;277(43):40265–40274. [DOI] [PubMed] [Google Scholar]
- 21. Kamei Y, Ohizumi H, Fujitani Y, et al. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci USA. 2003;100(21):12378–12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα). J Biol Chem. 2003;278(11):9013–9018. [DOI] [PubMed] [Google Scholar]
- 23. Debevec D, Christian M, Morganstein D, et al. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor α. Mol Endocrinol. 2007;21(7):1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dixen K, Basse AL, Murholm M, et al. ERRγ enhances UCP1 expression and fatty acid oxidation in brown adipocytes. Obesity (Silver Spring). 2013;21(3):516–524. [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Zhang J, Wang T, Esteban MA, Pei D. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. J Biol Chem. 2008;283(51):35825–35833. [DOI] [PubMed] [Google Scholar]
- 26. Chen J, Nathans J. Estrogen-related receptor β/NR3B2 controls epithelial cell fate and endolymph production by the stria vascularis. Dev Cell. 2007;13(3):325–337. [DOI] [PubMed] [Google Scholar]
- 27. Onishi A, Peng GH, Poth EM, et al. The orphan nuclear hormone receptor ERRβ controls rod photoreceptor survival. Proc Natl Acad Sci USA. 2010;107(25):11579–11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narkar VA, Fan W, Downes M, et al. Exercise and PGC-1α-independent synchronization of type I muscle metabolism and vasculature by ERRγ. Cell Metab. 2011;13(3):283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang T, McDonald C, Petrenko NB, et al. Estrogen-related receptor α (ERRα) and ERRγ are essential coordinators of cardiac metabolism and function. Mol Cell Biol. 2015;35(7):1281–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alaynick WA, Kondo RP, Xie W, et al. ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6(1):13–24. [DOI] [PubMed] [Google Scholar]
- 31. Alaynick WA, Way JM, Wilson SA, et al. ERRγ regulates cardiac, gastric, and renal potassium homeostasis. Mol Endocrinol. 2010;24(2):299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gan Z, Rumsey J, Hazen BC, et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J Clin Invest. 2013;123(6):2564–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo Y, Kumar P, Mendelson CR. Estrogen-related receptor γ (ERRγ) regulates oxygen-dependent expression of voltage-gated potassium (K+) channels and tissue kallikrein during human trophoblast differentiation. Mol Endocrinol. 2013;27(6):940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuercher WJ, Gaillard S, Orband-Miller LA, et al. Identification and structure-activity relationship of phenolic acyl hydrazones as selective agonists for the estrogen-related orphan nuclear receptors ERRβ and ERRγ. J Med Chem. 2005;48(9):3107–3109. [DOI] [PubMed] [Google Scholar]
- 35. Tiraby C, Hazen BC, Gantner ML, Kralli A. Estrogen-related receptor γ promotes mesenchymal-to-epithelial transition and suppresses breast tumor growth. Cancer Res. 2011;71(7):2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng B, Jiang J, Kraus P, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11(2):197–203. [DOI] [PubMed] [Google Scholar]
- 37. LaBarge S, McDonald M, Smith-Powell L, Auwerx J, Huss JM. Estrogen-related receptor-α (ERRα) deficiency in skeletal muscle impairs regeneration in response to injury. FASEB J. 2014;28(3):1082–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29(6):677–696. [DOI] [PubMed] [Google Scholar]
- 39. Ijichi N, Ikeda K, Horie-Inoue K, Yagi K, Okazaki Y, Inoue S. Estrogen-related receptor α modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem Biophys Res Commun. 2007;358(3):813–818. [DOI] [PubMed] [Google Scholar]
- 40. Kubo M, Ijichi N, Ikeda K, Horie-Inoue K, Takeda S, Inoue S. Modulation of adipogenesis-related gene expression by estrogen-related receptor γ during adipocytic differentiation. Biochim Biophys Acta. 2009;1789(2):71–77. [DOI] [PubMed] [Google Scholar]
- 41. de Jong JM, Larsson O, Cannon B, Nedergaard J. A stringent validation of mouse adipose tissue identity markers. Am J Physiol Endocrinol Metab. 2015;308(12):E1085–E1105. [DOI] [PubMed] [Google Scholar]
- 42. Mirebeau-Prunier D, Le Pennec S, Jacques C, et al. Estrogen-related receptor α modulates lactate dehydrogenase activity in thyroid tumors. PLoS One. 2013;8(3):e58683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Summermatter S, Santos G, Pérez-Schindler J, Handschin C. Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A. Proc Natl Acad Sci USA. 2013;110(21):8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hallberg M, Morganstein DL, Kiskinis E, et al. A functional interaction between RIP140 and PGC-1α regulates the expression of the lipid droplet protein CIDEA. Mol Cell Biol. 2008;28(22):6785–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1α coactivates PDK4 gene expression via the orphan nuclear receptor ERRα: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25(24):10684–10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Ma K, Sadana P, et al. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J Biol Chem. 2006;281(52):39897–39906. [DOI] [PubMed] [Google Scholar]
- 47. Arany Z, Lebrasseur N, Morris C, et al. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5(1):35–46. [DOI] [PubMed] [Google Scholar]
- 48. Herzog B, Cardenas J, Hall RK, et al. Estrogen-related receptor α is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem. 2006;281(1):99–106. [DOI] [PubMed] [Google Scholar]
- 49. Wang L, Liu J, Saha P, et al. The orphan nuclear receptor SHP regulates PGC-1α expression and energy production in brown adipocytes. Cell Metab. 2005;2(4):227–238. [DOI] [PubMed] [Google Scholar]
- 50. Villena JA, Kralli A. ERRα: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19(8):269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguère V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol Cell Biol. 2003;23(22):7947–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Delhon I, Gutzwiller S, Morvan F, et al. Absence of estrogen receptor-related-α increases osteoblastic differentiation and cancellous bone mineral density. Endocrinology. 2009;150(10):4463–4472. [DOI] [PubMed] [Google Scholar]
- 53. Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Keipert S, Jastroch M. Brite/beige fat and UCP1 - is it thermogenesis? Biochim Biophys Acta. 2014;1837(7):1075–1082. [DOI] [PubMed] [Google Scholar]
- 55. Yamamoto H, Williams EG, Mouchiroud L, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147(4):827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Villarroya F, Vidal-Puig A. Beyond the sympathetic tone: the new brown fat activators. Cell Metab. 2013;17(5):638–643. [DOI] [PubMed] [Google Scholar]
- 57. Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chang CY, McDonnell DP. Molecular pathways: the metabolic regulator estrogen-related receptor α as a therapeutic target in cancer. Clin Cancer Res. 2012;18(22):6089–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Deblois G, Giguère V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer. 2013;13(1):27–36. [DOI] [PubMed] [Google Scholar]