Abstract
Coeliac disease, a prevalent immune-mediated enteropathy driven by dietary gluten, provides an exceptional human model to dissect the genetic, environmental and immunologic factors operating in autoimmunity. Despite the causative antigen being an exogenous food protein, coeliac disease has many features in common with autoimmune disease including a strong HLA class II association and the presence of pathogenic CD4+ T cells and autoantibodies. CD8+ intraepithelial lymphocytes specifically target and destroy intestinal epithelium in response to stress signals and not a specific antigen. A unique feature of coeliac disease is the ability to remove gluten to induce disease remission and reintroduce it to trigger a memory response. This provides an unparalleled opportunity to study disease-relevant CD4+ T cells that have been expanded in vivo. As a result, the causative peptides have been characterised at a level unprecedented for any autoimmune disease. Despite the complexity of the gluten proteome, resistance to gastrointestinal proteolysis and susceptibility to post-translational modification by transglutaminase help shape a restricted repertoire of immunogenic gluten peptides that have high affinity for disease-associated HLA. The critical steps in coeliac disease pathogenesis have been broadly elucidated and provide the basis for experimental therapies in pre-clinical or clinical development. However, little is known about how and why tolerance to gluten sometimes breaks or fails to develop. Understanding the interactions between genes, the environment, gluten immunity and the microbiome may provide novel approaches for the prevention and treatment of disease.
Introduction
Coeliac disease (CD) is a chronic immune-mediated enteropathy precipitated by exposure to dietary gluten in genetically predisposed individuals.1 The seminal work of Dutch paediatrician Willem Dicke in the 1940s established a component of wheat, subsequently shown to be gluten, was the environmental driver of CD, and that removal of wheat from the diet led to prompt clinical recovery. The dietary trigger and prominent clinical phenotype of malabsorption influenced the view that CD is primarily a gastrointestinal illness. However, advances in the understanding of its genetic and immunologic basis now firmly position CD as an immune illness with systemic manifestations and features more in common with autoimmune disease (AID), where a pathogenic adaptive immune response targets self antigens. In common with many AID, genetic and environmental factors are important in CD development, inheritance is polygenic, a strong association with specific histocompatibility leucocyte antigen (HLA) genes exists, and both pathogenic CD4+ T cells and autoantibodies are present.2 Circulating autoantibodies directed against the endogenous enzyme tissue transglutaminase 2 (TG2) are a feature of active CD, and notably, their formation is dependent on and driven by the exogenous antigen gluten. Anti-TG2 antibodies can be detected in the intestine before overt tissue damage occurs, and have several pathogenic effects. Furthermore, recent insights into a key effector role for CD8+ intraepithelial lymphocytes (IELs) in the targeted killing of intestinal enterocytes that express IL-15 and stress-induced molecules has prompted some experts to consider this cell auto-reactive.2
Despite many similarities with AID, CD is unique in that the driving antigen, gluten, is exogenous. Several other features set it apart from other more ‘classical' AID, including the ability to easily access and sample the main target organ (intestine) by endoscopy, and that disease-specific CD4+ T cells can be readily isolated from the intestine and blood following gluten ingestion. Furthermore, the HLA association in CD, one of the strongest of all human HLA-linked diseases, shapes a restricted repertoire of immunogenic gluten peptides. These features mean that gluten has been better characterised than any other antigen implicated as causative in AID, and also make CD an ideal model to dissect the genetic and immune pathways potentially relevant in AID pathogenesis. Here, we review the genetic, environmental and immunologic factors that contribute to broken tolerance to gluten and why CD is of significance to the AID field.
A global clinical problem on the rise
CD affects 1–2% of the Western population and, like many chronic inflammatory diseases and AID, is substantially increasing in prevalence.3 There is a modest gender bias favouring females. The clinical effects of CD are broad and include gastrointestinal upset, chronic fatigue, nutrient deficiencies, other AID, osteoporosis, liver disease, infertility, sepsis and lymphoproliferative malignancy.1 Diagnosis rests on demonstrating the characteristic intestinal damage of villous atrophy, crypt hyperplasia and intraepithelial lymphocytosis.1 Circulating antibodies to TG2, endomysium (which contains the target antigen TG2) and deamidated gliadin peptides (DGP) are highly sensitive for CD and are useful screening tests in the clinic, but the broad presentation of CD means detection rates remain suboptimal.4
Treatment of CD is strict and lifelong removal of the offending antigen, a gluten-free diet (GFD). Gluten describes the storage proteins (prolamins) from wheat, barley and rye. In some countries such as Australia and New Zealand, oats are also excluded from a GFD as they contain gluten-like proteins that are immunogenic in some CD patients.5 A strict GFD resolves symptoms and normalises CD-specific antibodies and enteropathy. However, maintaining satisfactory GFD compliance is challenging and many patients fail to achieve full disease remission. Therefore, the development of more effective treatment and diagnostic approaches are major research goals.
Genetic insights highlight the central role of the immune system
CD is a highly heritable polygenic disease with the strongest genetic risk linked to the HLA region.6, 7 The specific HLA susceptibility genes encode for HLA-DQ2.5 (DQA1*05 and DQB1*02), HLA-DQ8 (DQA1*03 and DQB1*03:02) and HLA-DQ2.2 (DQA1*02:01 and DQB1*02:02).8 The DQA1 and DQB1 alleles encode the DQ αβ-heterodimer protein that resides on antigen-presenting cells (APCs) and facilitates gluten peptide and CD4+ T-cell interaction. In a large European study, 99.6% of 1008 patients with CD expressed HLA-DQ2.5, HLA-DQ8, HLA-DQ2.2 or HLA-DQ7 (DQA1*05+, DQB1*02−).8 This contrasts with a prevalence of these haplotypes of ~30–50% in the general Western population.4
The high concordance rate for monozygotic twins (~80%) compared with the HLA-identical siblings (~30%) and dizygotic twins (~10%) underscores the importance of non-HLA genes in CD risk.9 More than 70 candidate genes in 42 non-HLA loci have been implicated in CD heritability based on genome-wide association studies6, 10 and follow-up studies using Immunochip fine mapping or targeted analysis of disease-associated loci11, 12, 13 (Table 1). The individual contribution from each of these regions is small (odds ratio <1.5) and collectively they account for ~15% of the additional disease risk.14 Despite the small contribution, these loci highlight the importance of several immune pathways in CD pathogenesis, including roles for T- and B-cell activation, chemokine receptor activity and cell migration, cytokine binding, thymic differentiation of CD4+ and CD8+ T cells, stress pathways and innate immunity (Table 1). Underscoring the systemic nature of immune dysregulation in CD, only one gene has been shown to be relatively gut specific (RGS1).11 RGS1 expression is specific to the intestinal IEL compartment, and is believed to regulate chemokine receptor signalling and B-cell activation, and have a pivotal role in cell trafficking and tissue immunopathology (see below for the role of IELs in the intestinal lesion of CD). Most of the CD risk loci are shared in common with AID, such as rheumatoid arthritis (RA), multiple sclerosis (MS) and type 1 diabetes (Table 1).10, 12, 15 However, natural killer (NK)-cell activation and interferon (IFN)-γ-production gene pathways appear to be selectively enriched in CD.16 Abadie et al.16 showed that the network of interactions between CD susceptibility genes can generate a range of functional outcomes, such as high IL-15 and/or IFN-α levels, with the specific combination of susceptibility genes potentially contributing to the heterogeneity of CD phenotypes.
Table 1. Gene candidates and loci associated with coeliac disease and their overlap with autoimmune diseases.
Interestingly, ~90% of the identified risk loci map to non-coding regions (promoter regions, enhancers or non-coding RNA genes). This suggests that it is regulation of gene expression rather than changes at the protein-coding level that is more important for CD susceptibility and development.14 Recently, lnc13 was identified as a long non-coding RNA harbouring six CD-associated single-nucleotide polymorphisms.17 Notably, lnc13 regulates the expression of CD-associated inflammatory genes and its dysregulation may contribute to intestinal inflammation and autoimmunity.
Testing for HLA-DQ2.5/8/2.2 in the clinic is now common because it is an effective tool to exclude CD when these genotypes are absent.18 However, the test has limited positive predictive value for CD given the high prevalence of these genes in the general population. Tools that incorporate non-HLA single-nucleotide polymorphisms associated with CD may improve risk stratification and could also provide prognostic information.7, 19, 20 In collaboration with Inouye and colleagues, we showed that a genomic tool that generates a weighted score based on the presence of ~200 CD-associated single-nucleotide polymorphisms identified in previous studies6, 10, 12 provides a more fine-scaled estimation of CD risk than HLA typing,7 and better predicts CD development in first-degree relatives.20 More studies are needed to establish the clinical utility and role for genomic testing.
HLA and the gluten-specific CD4+ T-cell response
The strong HLA class II association in CD underscores the critical role of CD4+ T cells. Gluten-specific CD4+ T cells can be readily isolated and cloned from the intestinal lesion in CD and react to gluten peptides in an HLA-restricted manner. The gluten proteome is large and complex but the repertoire of gluten peptides that drive disease is highly restricted because of the specific binding requirements dictated by HLA-DQ2.5, DQ8 and DQ2.2. Indeed, there are more than 16 000 12mer gluten peptides from wheat, barley and rye that could potentially activate T cells21 but only just over 30 gluten-specific T-cell epitopes have been identified.22
The gluten peptides harbouring T-cell epitopes are high in proline, rendering them resistant to gastrointestinal proteolysis as humans lack endogenous prolyl endopeptidases. A 33mer that contains six overlapping copies of immunodominant T-cell epitopes from wheat α-gliadin (the alcohol soluble fraction of wheat gluten) is particularly rich in proline, resistant to intestinal degradation and highly immunogenic in HLA-DQ2.5+ CD.23 Although proposed as a major driver of wheat gluten toxicity, other important epitopes from protease resistant regions of ω-gliadin also elicit strong responses in most CD patients.21
Gluten-specific T cells preferentially respond to post-translationally modified gluten.24 This modification, deamidation, is mediated by the ubiquitous enzyme TG2 (Figure 1). TG2 has a role in wound repair, is abundant in the small intestine and its activity is increased by tissue stress and inflammation.25 TG2 is considered the autoantigen of CD as it is the target of IgA autoantibodies26 and TG2 antibodies may have direct pathogenic effects (see below). TG2 has a critical role in CD pathogenesis by catalysing the conversion of glutamine (Q) to the more negatively charged glutamate (E). The specificity of TG2 is determined by the amino acids that flank glutamine residues, with preferential deamidation of glutamine in peptides conforming to the sequence QXP or QXXP (X denotes any amino acid).27 This site selectivity is crucial as the negative charge introduced by glutamate serves as an anchor in the pocket of the peptide-binding groove of the HLA molecule. The greater stability of the major histocompatibility complex (MHC):peptide complex means that deamidated gluten peptides are more immunogenic. Post-translational modification is an important feature of AID such as RA, where both B- and T-cell responses are directed to specific citrullinated proteins introduced by the action of peptidylarginine deiminase enzymes. Although genetic variants in loci encoding these enzymes predispose to RA, a genetic link between TG2 and CD risk has not been reported.
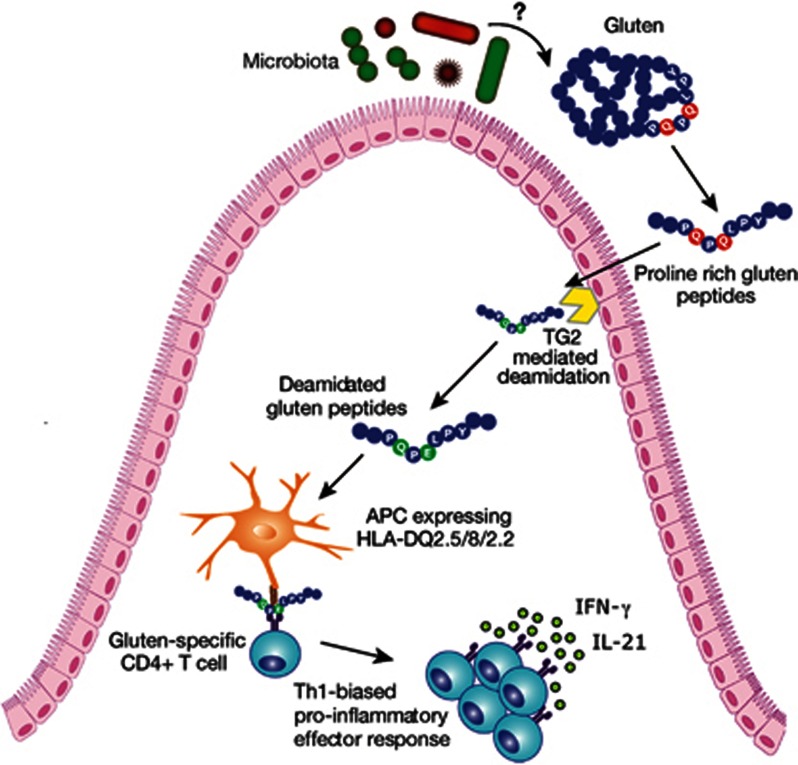
Figure 1.
Key features of the CD4+ T-cell response to gluten. Gluten peptides containing T-cell epitopes resist gastrointestinal degradation due to their high proline content. Tissue TG2 catalyses the deamidation of gluten peptides, which can then bind more efficiently to the disease-relevant HLA-DQ molecules on APCs. Activated gluten-specific T cells secrete a variety of pro-inflammatory cytokines such as IFN-γ and IL-21 that contribute to the intestinal lesion (Figure 3). The microbiota may have several effects, including modifying gluten proteolysis and the net production of immunogenic peptides.
Fine mapping of HLA-DQ2.5-restricted gluten T-cell epitopes has identified a preference for glutamate at anchor positions P4 or P6, and occasionally P7, corresponding to glutamine residues susceptible to deamidation by TG2.27, 28, 29, 30 Most T-cell reactivity in HLA-DQ2.5-associated CD is directed toward the immunodominant wheat gluten T-cell epitopes DQ2.5-glia-α1a (PFPQPELPY) and DQ2.5-glia-α2 (PQPELPYPQ) contained within an α-gliadin peptide.28, 31 These epitopes are dependent on the deamidation of a single glutamine to glutamate (represented by E) at P4 or P6 for optimal T-cell reactivity. Most CD patients mount responses to α-gliadin and ω-gliadin epitopes but responses to γ-gliadins are less frequent.21 Thus, the effect of TG2 is to generate more immunogenic T-cell epitopes, and through its site-selective targeting of gluten peptides, shape the immunogenic T-cell repertoire. Structural studies have provided additional insight into the gluten-HLA-T-cell receptor (TCR) complex and their interactions, explaining how deamidation enhances binding, and providing a structural basis for biased TCR gene usage.32, 33 In HLA-DQ8 CD binding favours glutamate at P1 and P9 to generate a more stable peptide:MHC complex.34
The importance of generating an activation threshold in the peptide:MHC complex is exemplified by the gene–dose effect in HLA-DQ2.5 CD. Gluten presented by HLA-DQ2.5 homozygous APCs can induce at least a fourfold higher T-cell response than HLA-DQ2.5 heterozygous APCs due to the higher number of DQ2.5 αβ-heterodimers available to the present gluten.35 This appears to have clinical consequences, as HLA-DQ2.5 homozygous patients have the highest risk for CD development and a more severe phenotype demonstrated by earlier disease onset, greater villous atrophy and clinical features at presentation, and a slower rate of villous healing on a GFD.36 Thus, genetic factors clearly have a role in the clinical expression of disease.
Access to gluten-specific T cells in vivo has advanced T-cell epitope discovery
A major aim of AID research is to characterise the causative antigen driving the auto-reactive T-cell response but realising this goal has remained elusive. In CD, the relative ease in accessing small intestinal tissue by endoscopy has facilitated the isolation and cloning of gluten-specific T cells from intestinal biopsies. T-cell clones and lines are a well established and useful tool to identify T-cell epitopes, however the use of long-term culture with potent mitogens could change the composition of the T-cell population of interest and potentially favour the expansion of subpopulations that are uninformative.37 Furthermore, this approach requires a costly and invasive procedure and has limited throughput. A complementary method to comprehensively define disease-specific T-cell epitopes relevant in vivo was needed.
This was realised when Anderson et al.31 showed that a 3-day oral gluten challenge could induce gluten-specific CD4+ T cells in the peripheral blood of treated CD patients, but not healthy controls, six days after commencing the challenge (Figure 2). These T cells are disease HLA-restricted, express the gut-homing β7 integrin indicating an intestinal origin, and mostly express the CD45RO+ memory phenotype.37, 38 By greatly expanding a memory-recall population in vivo, it became possible to directly sample antigen-specific CD4+ T cells from peripheral blood using sensitive functional assays such as IFN-γ ELISpot,21, 31, 37, 38, 39 IFN-γ/IP-10 ELISA40 or with tetramers.37, 41 This means long-term in vitro culture can be avoided and endoscopy is not required. The ability to remove dietary gluten and reintroduce it (that is, oral gluten challenge) to provide a source of polyclonal gluten-specific T cells in vivo is unique to CD and has enabled unprecedented characterisation of the antigen-specific T-cell response.
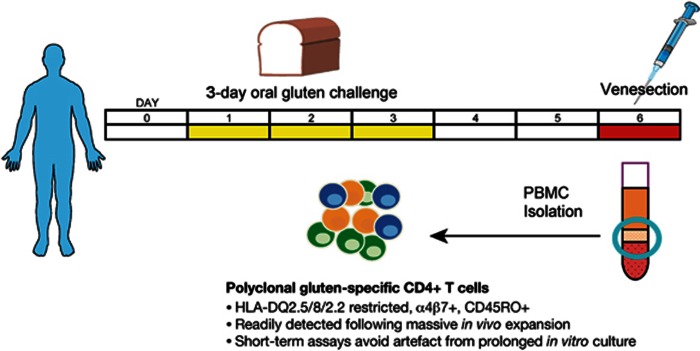
Figure 2.
Exploiting oral gluten challenge to elicit disease-specific T cells in vivo. Short-term oral gluten challenge in treated CD mobilises a polyclonal gluten-specific CD4+ T-cell population in vivo. Patients with CD consume a gluten-containing cereal for 3 days, and blood is collected on day 6 to isolate peripheral blood mononuclear cells. By greatly expanding the gluten-specific T-cell population in vivo, a range of functional and immunophenotyping approaches can be readily employed without the need for prolonged in vitro expansion. Overnight functional assays such as IFN-γ ELISpot can be used for high throughput T-cell epitope mapping. Single-cell sorting by flow cytometry can be applied to the antigen-specific CD4+ T cells identified by proliferation or IFN-γ secretion, and these cells can be analysed for TCR repertoire or cultured to generate T-cell clones.
Use of tetramers has comparable sensitivity to IFN-γ ELISpot and confirms the massive expansion of gluten-specific CD4+ T cells induced by gluten challenge, with T cells reacting to the DQ2.5-glia-α1a and DQ2.5-glia-α2 epitopes ranging from 1:1000 to 1:5000 among CD4+ T cells, equivalent to 1:5000 to 1:25 000 among peripheral blood mononuclear cells.37 Using magnetic bead enrichment with tetramers, it is possible to detect gluten-specific T cells in the blood of treated and untreated CD patients without the need to expand this population by gluten challenge.42 Interestingly, 3-day gluten challenge concomitantly mobilises CD8+ and γδ T cells into peripheral blood.43 These T cells express intestinal homing markers and have a memory or effector phenotype, but their precise role and specificity for gluten is not defined.
The ability to readily isolate gluten-specific T cells has afforded an opportunity to assess the TCR repertoire expressed by T cells specific for the immunodominant α- and ω-gliadin epitopes. These studies highlight a biased and public TCR repertoire, with preferential TCR alpha and beta variable (TRAV and TRBV, respectively) gene use shaped by the specific epitope, specifically TRAV26/TRBV7-2 (DQ2.5-glia-α2),33, 44, 45, 46 TRAV26/TRBV9 (DQ8-glia-α1)47, 48 and TRAV4/TRBV4 (DQ2.5-glia-ω2).49 There is a high level of binding affinity of TCRs for their cognate ligand, greater than that reported in more traditional autoimmune reactions. A non-germline encoded arginine residue in the CDR3 region is found in most TCRs specific for DQ2.5-glia-α2 and DQ8-glia-α1.45, 48 This conserved residue appears to be a critical element in TCR recognition by interacting with the deamidated epitope. This suggests that TCR selection is shaped in vivo by a post-translationally modified antigen based on the structural requirements for binding.
We exploited the oral gluten challenge approach to undertake comprehensive T-cell epitope mapping studies in HLA-DQ2.5+ CD using gluten peptide libraries spanning the wheat, barley, rye and oat gluten proteome.5, 21 The hierarchy of immunogenic peptides was highly consistent between CD patients with the same HLA type and was dependent on the cereal consumed. Three distinct peptides, encompassing the DQ2.5-glia-α1a/α2, DQ2.5-glia-ω1/ω2 and DQ2.5-hor-3 epitopes, accounted for most of the T-cell response to whole deamidated gluten. Although α-gliadin epitopes have been regarded as the most important driver of CD, this work showed these are only relevant after wheat is consumed and not after barley or rye. In fact, it is the ω-gliadin epitopes that are immunodominant after consumption of any of these cereals.21 Despite sequence similarity of the α- and ω-gliadin epitopes, their TCR repertoires are unique and show biased use of different TRAV and TRBV genes.49 Appreciating the uniqueness of these distinct immunodominant epitopes has been crucial for the development of peptide-specific applications, including a novel CD diagnostic based on detection of whole blood IFN-γ release after gluten challenge40 and a tolerogenic therapeutic vaccine (Nexvax2).50
Employing this unbiased approach to T-cell epitope characterisation allowed us to confirm that the specificity and hierarchy of the T-cell response to dominant T-cell epitopes after wheat ingestion in children is comparable to that in adults with CD.46 This suggests that by the time CD is diagnosed, irrespective of the age of the patient, the T-cell epitopes that drive disease are well established, subsequently remain stable over time and significant epitope spreading does not occur.
CD4+ T cells and CD8+ IELs are required for villous atrophy
The intestinal lesion in untreated (active) CD is characterised by infiltration with gluten-specific CD4+ T cells exhibiting a Th1-biased phenotype, with release of pro-inflammatory cytokines dominated by IFN-γ (Figure 3).51 IL-21 and IL-15 are also increased and drive development of the intestinal lesion in part by promoting activation of cytotoxic CD8+ IELs. IL-21 regulates both adaptive and innate responses, drives production of IFN-γ, stimulates B-cell responses, upregulates the cytotoxic activity of IELs and renders effector T cells resistant to regulatory T-cell (Treg) suppression.52 IL-21 also supports development of Th17 cells, which are involved in the pathogenesis of a number of AID, but their relevance in CD is unclear. High IL-17 production from gluten-specific CD4+ T cells in active CD mucosa has been demonstrated53, 54 but others have not been able to replicate this.51 IL-15 drives IEL cytotoxicity and loss of tolerance (discussed below) and can also promote the transformation of IELs into a T-cell lymphoma, a rare but highly fatal malignant complication of CD.55
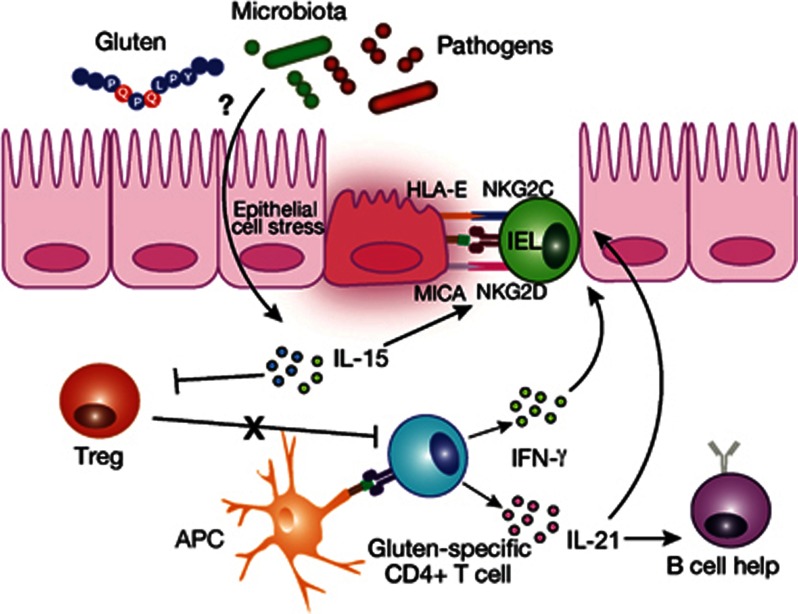
Figure 3.
Cytotoxic IELs mediate killing of stressed intestinal enterocytes. The induction of an NK cell-like phenotype in CD8+ IELs mediates cytotoxic killing of stressed intestinal enterocytes. Stress-inducible molecules such as MIC and HLA-E are the ligands for the activating NKG2D and CD94/NKG2C receptors that, under the influence of IL-15, are highly expressed on IELs in active disease. Only intestinal enterocytes expressing IL-15 and the ligands for activating NK receptors will be killed. The drivers of epithelial stress may include gluten or inflammatory stimuli such as gastrointestinal infections. Gluten-specific CD4+ T cells produce IFN-γ and IL-21 that promote activation of IELs and stimulate B-cell responses. IL-15 renders effector T cells resistant to the suppressive effects of Tregs and, in the lamina propria, endows mucosal DCs with inflammatory properties promoting pro-inflammatory responses and preventing Treg differentiation.
Protease resistant gluten peptides transit across the epithelium via transcellular and paracellular pathways.56 Increased epithelial permeability may be mediated by the tight-junction disruptor zonulin.57 Gluten peptides are deamidated and activate gluten-specific T cells, most likely within the lamina propria (Figure 1). Dendritic cells (DCs) are presumed to have a major role in presentation of these peptides to CD4+ T cells, however less is known about their identity and the location of this activation. After a 3-day gluten challenge there is an infiltrate of CD14+CD11c+ DCs in the intestinal mucosa and a reduction of CD11c+CD103+ DCs possibly due to migration to draining lymph nodes.58 It is possible that both of these DC populations are involved in gluten presentation, to intestinal resident T cells, such as effector memory cells, or central memory T cells in the secondary lymphoid organs.
Activated gluten-specific CD4+ T cells are implicated in mediating intestinal damage but the relevant effector pathways for a long time remained unclear. Mediators upregulated by IFN-γ such as the matrix metalloproteinases,59 which influence tissue remodelling, appear important but alone do not seem to fully account for the extent of enteropathy. Elevated numbers of CD8+ TCRαβ+ IELs are observed in the active CD lesion but until recently their significance was unclear. We now appreciate they play a key effector role in tissue destruction after they expand and adopt an NK cell-like phenotype (Figure 3). Distinct to the CD4+ T-cell response to gluten, this process is triggered by recognition of altered ‘self' tissue (epithelial stress) rather than a specific antigen.
In the healthy state, IELs express the inhibitory receptor CD94/NKG2A, but in active CD they express high levels of the activating NKG2D and CD94/NKG2C receptors.60, 61 At the same time, ‘stressed' intestinal epithelial cells in active CD patients express high levels of the stress-inducible MHC class I chain-related protein (MIC) molecules and non-classical MHC class I molecule HLA-E, which are the main ligands for NKG2D and CD94/NKG2C, respectively (Figure 3).61, 62 IL-15 plays a key role by upregulating the activating NKG2D receptor and acting as a co-stimulatory molecule, the effect being to license cytotoxic IELs with the ability to kill intestinal epithelial cells expressing the stress-induced MIC molecules.
The causes of epithelial stress are not well defined. Gluten may trigger an innate response, however, the specific peptides and pattern recognition receptors have not been defined. An innate immune stimulatory effect of a gliadin sequence (A-gliadin p31-43) has been reported but this work has not been replicated.63 Natural pest resistance proteins in wheat, the amylase trypsin inhibitors, have the capacity to bind TLR4 and stimulate an innate immune response in the intestine but their role in human disease is unclear.64 Other drivers could include microbes, which might explain the epidemiological observation that gastrointestinal infection, such as rotavirus, is associated with risk for CD development and can precede disease onset.65
Jabri and colleagues66 offer an elegant model to explain how adaptive gluten immunity acts in synergy with epithelial stress to drive villous damage. They showed that some family members of CD patients with negative CD serology and normal small intestinal histology have stressed epithelium as demonstrated by ultrastructural alterations and elevated expression of stress markers and IL-15. However, the expression of high levels of inhibitory NK-cell receptors on cytotoxic IELs appears to prevent extensive epithelial cell killing. In contrast, patients with latent (potential) CD who have positive CD serology, that is, evidence of adaptive immunity to gluten, but normal small intestine, exhibit no epithelial stress response and activating NK-cell receptors are not upregulated. Consistent with a ‘two-hit' model, the development of coeliac disease appears to depend on an adaptive immune response to gluten acting in synergy with epithelial stress to drive cytotoxic IELs and villous atrophy.
Emerging insights into the B-cell response to transglutaminase and gluten
The contribution of B cells and TG2- and DGP-specific antibodies to CD pathogenesis has received less scrutiny than the role of CD4+ gluten-specific T cells. Anti-TG2 antibodies have a variety of in vitro effects including inhibiting intestinal epithelial cell proliferation, activating monocytes and inhibiting angiogenesis.67 Interestingly, anti-TG2 IgA deposits are detectable in intestinal tissue before the development of overt CD, suggesting antibody production occurs early in disease alongside the gluten-specific T-cell response.68 Deposits are also found in extra-intestinal sites such as muscle, liver, kidney and brain showing that TG2 is widely accessible to gut-derived circulating autoantibodies and may be responsible for some clinical manifestations in CD.69 For example, known pregnancy complications in CD such as infertility, early pregnancy loss and intra-uterine growth retardation may be caused by impaired endometrial angiogenesis and placental damage mediated by anti-TG2 antibodies.70 More research is needed to define the precise role of anti-TG2 antibodies in contributing to the enteropathy and extra-intestinal manifestations of CD.
The recent work of Sollid and colleagues has shed substantial light on TG2 and gluten-specific antibody production in CD. They showed a massive expansion of plasma cells targeting TG2 and gluten in the intestine of patients with active CD, with 5–25% of all IgA plasma cells specific for TG2.71, 72 A GFD induces a substantial reduction in intestinal plasma cells and normalises plasma TG2 and DGP levels often within months, suggesting that long-lived plasma cells are not formed. Interestingly, they showed that antibody responses to gluten73 and TG274 show restricted use of variable heavy (VH) and variable light (VL) chain gene segments with few somatic hypermutations despite high binding affinity. The low level of mutations could suggest that B-cell activation occurs extrafollicularly and is non T-cell dependent. The lack of an exceptionally long-lived plasma cell response would support this notion as these cells are generally formed in germinal centres in a T-cell dependent manner. Further work is required to understand the significance of these findings.
The co-occurrence of autoantibodies and HLA-restricted self-reactive T cells is common in AID and these components often localise together in affected tissue, for example, type 1 diabetes and thyroiditis. Close interactions between CD4+ T cells and B cells may be important for amplifying the inflammatory response, as B cells can present antigen to T cells and they in turn can provide help for autoantibody production (Figure 4). In CD, T-cell dependent antibody production is supported by the observation that TG2 and DGP antibody formation is strictly dependent on the presence of CD-associated HLA types as well as gluten.
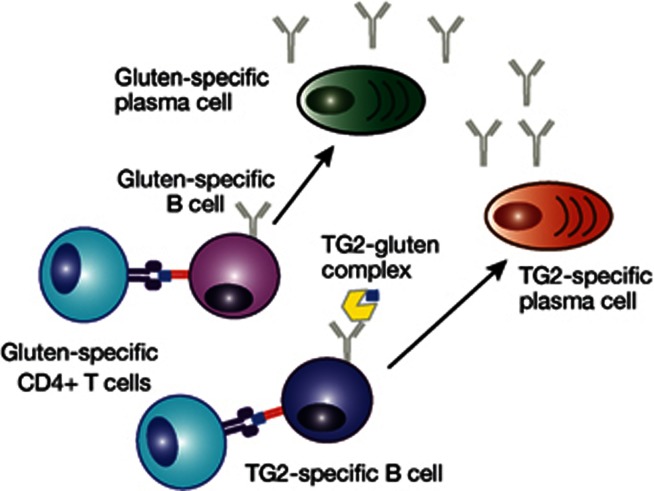
Figure 4.
CD4+ T-cell and B-cell interaction may amplify the immune response to gluten and generate circulating antibodies. Circulating TG2- and DGP-specific antibodies are a feature of active CD and disappear when dietary gluten is excluded. Their occurrence requires the presence of CD-associated HLA types implicating a key role for T cells in their formation. The proposed model is that TG2-specific B cells internalise TG2-gluten complexes and then present them to gluten-specific CD4+ T cells, which in turn, provide help for antibody production. Gluten-specific B cells may receive help from gluten-specific CD4+ T cells if the B-cell epitope is linked to a T-cell epitope. B cells may enhance presentation of gluten peptides to CD4+ T cells to amplify the pro-inflammatory response to gluten.
As TG2-reactive CD4+ T cells have not been identified, it is proposed that TG2-specific B cells internalise TG2-gluten complexes and then present them to gluten-specific CD4+ T cells, which then provide help for antibody production75 (Figure 4). T cell help may be important for loss of tolerance to TG2. As B-cell engagement will promote internalisation and efficient presentation at very low antigen concentrations, the result is a powerful amplification loop where B cells act as highly efficient APCs enhancing the gluten-specific CD4+ T-cell response. Antigen-presenting B cells could potentially modify the T-cell epitope repertoire displayed to T cells and influence functional responses. Further insight is needed into how TG2- and gluten-specific B cells are activated, where this occurs and how gluten-specific T and B cells interact in vivo.
In contrast to the detailed knowledge of T-cell epitopes in gluten, mapping of gluten-specific B-cell epitopes is restricted to wheat gliadin. Several linear epitopes have been defined in α- and γ-gliadin by screening phage-display and synthetic peptides in adult CD patients,76 and a peptide microarray approach has defined several discontinuous gliadin epitopes.77 Notably, several B-cell epitopes lie within or are in close proximity to immunodominant T-cell epitopes, for example, the B-cell epitope QPQQPF in γ-gliadin is encompassed within DQ2.5-glia-ω1/2.78 This raises the possibility that antibodies could influence T-cell recognition of the related epitope and modify gluten-specific responses in vivo.
Why and how does gluten tolerance fail?
The Western diet contains a large amount of gluten (~15–20 g day−1) and HLA genetic susceptibility to CD is seen in almost half the population. Why only 1–2% of the population develop CD and the remainder do not is of substantial interest. Importantly, in HLA-DQ2.5+ volunteers without CD, T-cells targeting immunodominant gluten epitopes (DQ2.5-glia-α1a and -α2, DQ2.5-glia-ω1 and -ω2 and DQ2.5-glia-γ2) are not present in intestinal biopsies and few are present in the peripheral blood.79 The tetramer-binding T cells show lower gluten-specific proliferation, do not express gut-homing markers, do not display a Treg phenotype and do not have a biased TCR repertoire typical of responses to certain gluten epitopes in CD patients. Thus, healthy individuals with HLA susceptibility to CD generally do not mount pro-inflammatory or regulatory T-cell responses to the dominant gluten epitopes that cause CD.
The situation in CD is quite different. Perhaps counter-intuitively, high levels of induced Foxp3- IL-10-producing type 1 (Tr1) Tregs and naturally occurring Foxp3+ Tregs are present in the CD intestinal lesion and peripheral blood, with increased levels associated with active disease.80, 81, 82 The antigen specificity of these reported Tregs is unknown. Accumulation of Foxp3+ Tregs in target organ tissue is noted in AID such as RA and inflammatory bowel disease and may be due to attraction of Tregs to the inflamed tissue in a non-antigen-specific manner. Gianfrani et al.83 showed that IL-10-producing gliadin-specific Tr1 clones expanded from treated CD intestine are able to inhibit the proliferation of gliadin-specific CD4+ effector T-cell clones. Furthermore, IL-10 and TGF-β can be detected in high amounts in the CD intestinal lesion.84, 85
Failure of the suppressive capacity of Tregs in CD could be one explanation for broken tolerance, and although some studies have supported this concept,86, 87 others have shown function is retained, at least in vitro.82, 88 These conflicting results highlight the difficulty in identifying Treg populations that have true suppressive capacity in vivo. As commonly employed Treg markers such as CD25 and Foxp3 are not solely expressed on Tregs and can be upregulated in activated T cells, identifying more specific markers is of interest. Reduced IL-7 receptor (CD127) expression can identify CD4+ T cells that have a suppressive Foxp3+ phenotype even when CD25 is low or absent,89 but this is yet to be examined in detail in CD. CD134 (OX40) has been used as a marker for antigen-specific T cells and CD39 can be used to identify antigen-specific Tregs.90, 91 This approach has been used to show CD39+ Tregs compose a substantial proportion of T cells induced by 3-day oral wheat ingestion in CD, and they remain functionally suppressive (Cook et al., in submission). The ability to effectively monitor antigen-specific Tregs will provide a powerful research tool and may be useful in assessing tolerogenic strategies for CD in clinical trials.
IL-15 promotes loss of tolerance to gluten
As the defect in gluten tolerance in CD is not readily explained by a defect in the number or recruitment of Tregs, attention has turned to IL-15. This pleiotropic cytokine can block the ability of TGF-β to suppress activation of mucosal T cells84 and impair the ability of CD25+ Foxp3+ Tregs from the blood and intestinal biopsies of CD patients to suppress effector CD4+ T cells in vitro.88 In an HLA-DQ8 mouse model overexpressing IL-15 in the lamina propria, IL-15 in combination with retinoic acid altered the tolerogenic phenotype of intestinal DCs and promoted a pro-inflammatory response characterised by the release of IL-12 and IL-23.92 Along with retinoic acid, these cytokines then promote Th1 and Th17 polarisation, and the failure of induction of Foxp3+ Tregs to dietary gliadin. The authors note that although retinoic acid is considered important in the generation of Tregs and oral tolerance, its adjuvant effect is consistent with its usage as a pro-inflammatory adjuvant in anti-tumour immunity. Notably, anti-TG2 IgG and IgA antibodies are generated in the transgenic mice but no enteropathy develops, supporting the concept that in the absence of the epithelial stress and increased IL-15, adaptive gluten immunity alone is insufficient to induce tissue damage.
The emerging link between environment, the microbiome and gluten immunity
Environmental factors contribute to the development of CD in genetically susceptible individuals but what they are, and how they do this, is poorly understood. Population studies associate several factors with increased CD risk such as perinatal and childhood infections, use of antibiotics and proton-pump inhibitors, higher socioeconomic status and better hygiene, and elective Caesarean section.3 Delayed introduction of dietary gluten may reduce the time of onset of development CD but it remains unclear if there is an optimal time for gluten introduction.93, 94 These controlled studies failed to demonstrate a protective role for breast feeding in reducing CD risk. Many of the implicated environmental factors may act by altering the composition of the microbiome.95
Dysbiosis or perturbations in the composition of resident commensal microbial communities away from those found in healthy individuals, are reported in a range of autoimmune and inflammatory diseases, including CD. The dysbiosis is characterised by an abundance of Proteobacteria and a decrease in Lactobacillus.95 Epidemiological data support an association between dysbiosis and increased risk of CD but there is little understanding of how it might influence gluten-specific immunity in vivo. In vitro data supports a broad range of influences of microbes on immune responses to gluten.95 This includes roles in modifying Treg induction, epithelial cell stress and IEL activation, phenotypic and functional maturation of DCs and pro-inflammatory cytokine production, intestinal permeability, and the induction of CD4+ T-cell responses.95 There is no convincing evidence of a role for molecular mimicry in CD, and no microbe-specific T cells cross-reactive with gluten have been identified. Verdu and colleagues96 recently showed that gluten metabolism in the small intestine of gnotobiotic mice is differentially affected by opportunistic pathogens such as Pseudomonas aeruginosa (Psa) and commensal bacteria such as Lactobacillus spp. We showed that the peptides generated from the proteolytic action of these bacteria show strikingly different levels of immunogenicity to gluten-specific T cells induced in CD patients after 3-day wheat challenge in vivo, with Psa increasing and Lactobacillus spp. reducing the load of immunogenic peptides.96 An imbalance of Psa over Lactobacillus spp. in vivo could favour the net generation of immunogenic gluten peptides and promote disease (Figure 1). Microbe–dietary–host interactions may be an important modulator of autoimmune risk in genetically susceptible people. Targeting this interaction may provide novel approaches to reduce disease risk or modify the activity of established disease.
Translational significance of understanding coeliac disease pathogenesis
A range of experimental therapies for patients with CD are in development.50 Some aim to reduce the load of immunogenic gluten peptides contacting the mucosal immune system by enzymatically degrading gluten, sequestering it or reducing its passage across the mucosa. These quantitative approaches may reduce the adverse effects of small amounts of dietary gluten but would not replace the need for this treatment. Blockade of IL-15 may be useful in refractory CD, where IL-15 drives inflammation despite adequate gluten removal. Pre-clinical approaches to selectively inhibit TG2 or block HLA are also being explored.50
Restoring tolerance to the causative antigen in AID is highly attractive but complicated by the need to sufficiently characterise the driving antigen and monitor auto-reactive T cells.97 In contrast to the more ‘classical' AID, development of epitope-specific approaches in CD are feasible because the causative T-cell epitopes in gluten are well characterised.21 The therapeutic vaccine Nexvax2 that targets the T-cell epitopes relevant in HLA DQ2.5+ CD is entering Phase 2 clinical trials to assess efficacy.50 Induction of tolerance may be achieved through several mechanisms, such as deletion of naive T cells specific for the dominant peptides composing the vaccine or the induction of Treg differentiation.97 Tregs may inhibit pro-inflammatory T cells and exert linked suppression by blocking the differentiation of naive T cells that recognise self antigens released as a result of tissue damage (epitope spreading). They may also act directly at the level of memory T cells to promote anergy and cause deletion.97 Pharmacogenetics will be important in the selection of peptides relevant to the CD HLA-DQ type. Modifying gluten immunity using the hookworm Necator americanus has shown some promise and larger controlled studies are underway.98
Conclusion
Our view of CD has evolved from a gastrointestinal malabsorptive illness to an immune disease characterised by HLA-dependence, gluten-specific CD4+ T cells, autoantibodies to TG2 and systemic clinical manifestations. CD exemplifies how an illness can have autoimmune-like features without having to be driven by an endogenous antigen. Indeed, CD challenges many traditional notions of what might be reasonably considered an ‘autoimmune disease'. For example, formation of autoantibodies occurs to a self-antigen despite the driver being an exogenous dietary protein. The tissue destruction of CD is mediated by cytotoxic IELs directed against ‘altered-self' in a highly targeted manner, as only the enterocytes expressing IL-15 and the stress-induced ligands for NK receptors will be killed. On this basis, Jabri and Sollid2 argue that IELs in CD could be regarded as self-reactive. If one is to accept this designation, CD could be reasonably considered as a model of organ-specific autoimmunity.
So how well does CD fulfil the definition of an AID? The modified Witebsky's postulates employ three-tiers of evidence to establish defining criteria for AID.99 This includes evidence that is direct, for example, transfer of disease by pathogenic autoantibody; indirect, for example, identifying autoantibodies or T cells within the target lesion or reproduction of disease in experimental animals; or circumstantial, for example, clinical clues, such as clustering of AID within the same patient. In practice, most AID, including those regarded as ‘bona fide' AID, do not fulfil all of the criteria. It is notable that CD fulfils several, the seeming distinction being the requirement for an exogenous driving antigen. However, Jabri and Sollid2 raise an interesting point—could exogenous drivers be important in the pathogenesis of more traditional AID? Exploring the role of non-self antigens as drivers in other AID may be informative. As in the case for CD, this may be via a mechanism distinct from molecular mimicry.
Although diagnosis of CD still rests on demonstrating intestinal villous atrophy,1 this may underestimate the true burden of disease. Some patients with positive CD serology and a CD-susceptible HLA type but minimal enteropathy suffer gluten-induced symptoms and morbidity, and clinically benefit from a GFD.100 This supports the notion that CD is a systemic illness, the hallmark of which is aberrant immunity to gluten, and that gastrointestinal damage is an important but not necessarily exclusive manifestation. In the future it will be important to determine if other approaches, such as determining the presence of a pathogenic gluten-specific T-cell response, might have greater clinical utility and accuracy in diagnosing CD.
The rise in CD prevalence makes approaches to improve diagnosis and enhance treatment outcomes a pressing clinical need. Several novel therapies for CD are in development, and their success will test the accuracy of our knowledge of CD pathogenesis. Uncovering how environmental factors and the microbiome shape the balance between gluten tolerance and immunity in genetically susceptible individuals may reveal entirely new approaches for disease prevention and management.
Footnotes
MYH and JAT-D are co-inventors of patents pertaining to the use gluten peptides in therapeutics, diagnostics and non-toxic gluten in coeliac disease. JAT-D is a shareholder of Nexpep Pty. Ltd and a consultant to ImmusanT Inc. Nexpep Pty. Ltd and ImmusanT Inc. were formed to develop novel diagnostics and treatments for coeliac disease.
References
- Ludvigsson JF, Bai JC, Biagi F, Card TR, Ciacci C, Ciclitira PJ et al. Diagnosis and management of adult coeliac disease: guidelines from the British Society of Gastroenterology. Gut 2014; 63: 1210–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol 2013; 13: 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebwohl B, Ludvigsson JF, Green PH. The unfolding story of celiac disease risk factors. Clin Gastroenterol Hepatol 2014; 12: 632–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RP, Henry MJ, Taylor R, Duncan EL, Danoy P, Costa MJ et al. A novel serogenetic approach determines the community prevalence of celiac disease and informs improved diagnostic pathways. BMC Med 2013; 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy MY, Tye-Din JA, Stewart JA, Schmitz F, Dudek NL, Hanchapola I et al. Ingestion of oats and barley in patients with celiac disease mobilizes cross-reactive T cells activated by avenin peptides and immuno-dominant hordein peptides. J Autoimmun 2015; 56: 56–65. [DOI] [PubMed] [Google Scholar]
- van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet 2007; 39: 827–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham G, Tye-Din JA, Bhalala OG, Kowalczyk A, Zobel J, Inouye M. Accurate and robust genomic prediction of celiac disease using statistical learning. PLoS Genet 2014; 10: e1004137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karell K, Louka AS, Moodie SJ, Ascher H, Clot F, Greco L et al. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European genetics cluster on celiac disease. Hum Immunol 2003; 64: 469–477. [DOI] [PubMed] [Google Scholar]
- Greco L, Romino R, Coto I, Di Cosmo N, Percopo S, Maglio M et al. The first large population based twin study of coeliac disease. Gut 2002; 50: 624–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 2010; 42: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 2008; 40: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 2011; 43: 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Achury J, Zhernakova A, Pulit SL, Trynka G, Hunt KA, Romanos J et al. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat Genet 2015; 47: 577–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withoff S, Li Y, Jonkers I, Wijmenga C. Understanding celiac disease by genomics. Trends Genet 2016; 32: 295–308. [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 2008; 359: 2767–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadie V, Sollid LM, Barreiro LB, Jabri B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol 2011; 29: 493–525. [DOI] [PubMed] [Google Scholar]
- Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH et al. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016; 352: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye-Din JA, Cameron DJ, Daveson AJ, Day AS, Dellsperger P, Hogan C et al. Appropriate clinical use of human leukocyte antigen typing for coeliac disease: an Australasian perspective. Intern Med J 2015; 45: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanos J, van Diemen CC, Nolte IM, Trynka G, Zhernakova A, Fu J et al. Analysis of HLA and non-HLA alleles can identify individuals at high risk for celiac disease. Gastroenterology 2009; 137: 834–840. [DOI] [PubMed] [Google Scholar]
- Abraham G, Rohmer A, Tye-Din JA, Inouye M. Genomic prediction of celiac disease targeting HLA-positive individuals. Genome Med 2015; 7: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye-Din JA, Stewart JA, Dromey JA, Beissbarth T, van Heel DA, Tatham A et al. Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med 2010; 2: 1–14. [DOI] [PubMed] [Google Scholar]
- Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012; 64: 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM et al. Structural basis for gluten intolerance in celiac sprue. Science 2002; 297: 2275–2279. [DOI] [PubMed] [Google Scholar]
- Molberg O, McAdam SN, Korner R, Quarsten H, Kristiansen C, Madsen L et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med 1998; 4: 713–717. [DOI] [PubMed] [Google Scholar]
- Wang Z, Griffin M. TG2, a novel extracellular protein with multiple functions. Amino Acids 2012; 42: 939–949. [DOI] [PubMed] [Google Scholar]
- Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med 1997; 3: 797–801. [DOI] [PubMed] [Google Scholar]
- Vader LW, de Ru A, van der Wal Y, Kooy YM, Benckhuijsen W, Mearin ML et al. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J Exp Med 2002; 195: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentz-Hansen H, Korner R, Molberg O, Quarsten H, Vader W, Kooy YM et al. The intestinal T cell response to alpha-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med 2000; 191: 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentz-Hansen H, McAdam SN, Molberg O, Fleckenstein B, Lundin KE, Jorgensen TJ et al. Celiac lesion T cells recognize epitopes that cluster in regions of gliadins rich in proline residues. Gastroenterology 2002; 123: 803–809. [DOI] [PubMed] [Google Scholar]
- Qiao SW, Bergseng E, Molberg O, Jung G, Fleckenstein B, Sollid LM. Refining the rules of gliadin T cell epitope binding to the disease-associated DQ2 molecule in celiac disease: importance of proline spacing and glutamine deamidation. J Immunol 2005; 175: 254–261. [DOI] [PubMed] [Google Scholar]
- Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV. In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med 2000; 6: 337–342. [DOI] [PubMed] [Google Scholar]
- Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci USA 2004; 101: 4175–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Montserrat V, Mujico JR, Loh KL, Beringer DX, van Lummel M et al. T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat Struct Mol Biol 2014; 21: 480–488. [DOI] [PubMed] [Google Scholar]
- Henderson KN, Tye-Din JA, Reid HH, Chen Z, Borg NA, Beissbarth T et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 2007; 27: 23–34. [DOI] [PubMed] [Google Scholar]
- Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci USA 2003; 100: 12390–12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karinen H, Karkkainen P, Pihlajamaki J, Janatuinen E, Heikkinen M, Julkunen R et al. Gene dose effect of the DQB1*0201 allele contributes to severity of coeliac disease. Scand J Gastroenterol 2006; 41: 191–199. [DOI] [PubMed] [Google Scholar]
- Raki M, Fallang LE, Brottveit M, Bergseng E, Quarsten H, Lundin KE et al. Tetramer visualization of gut-homing gluten-specific T cells in the periph eral blood of celiac disease patients. Proc Natl Acad Sci USA 2007; 104: 2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RP, van Heel DA, Tye-Din JA, Barnardo M, Salio M, Jewell DP et al. T cells in peripheral blood after gluten challenge in coeliac disease. Gut 2005; 54: 1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarca A, Radano G, Di Mase R, Terrone G, Maurano F, Auricchio S et al. Short wheat challenge is a reproducible in vivo assay to detect immune response to gluten. Clin Exp Immunol 2012; 169: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros N, Tye-Din JA, Hardy MY, Anderson RP. Ex vivo whole blood secretion of interferon (IFN)-gamma and IFN-gamma-inducible protein-10 measured by enzyme-linked immunosorbent assay are as sensitive as IFN-gamma enzyme-linked immunospot for the detection of gluten-reactive T cells in human leucocyte antigen (HLA)-DQ2.5(+ -associated coeliac disease. Clin Exp Immunol 2014; 175: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brottveit M, Raki M, Bergseng E, Fallang LE, Simonsen B, Lovik A et al. Assessing possible celiac disease by an HLA-DQ2-gliadin tetramer test. Am J Gastroenterol 2011; 106: 1318–1324. [DOI] [PubMed] [Google Scholar]
- Christophersen A, Raki M, Bergseng E, Lundin KE, Jahnsen J, Sollid LM et al. Tetramer-visualized gluten-specific CD4+ T cells in blood as a potential diagnostic marker for coeliac disease without oral gluten challenge. United European Gastroenterol J 2014; 2: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Newell EW, Glanville J, Fernandez-Becker N, Khosla C, Chien YH et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ alphabeta T cells and gammadelta T cells in celiac disease. Proc Natl Acad Sci USA 2013; 110: 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao SW, Christophersen A, Lundin KE, Sollid LM. Biased usage and preferred pairing of alpha- and beta-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int Immunol 2014; 26: 13–19. [DOI] [PubMed] [Google Scholar]
- Broughton SE, Petersen J, Theodossis A, Scally SW, Loh KL, Thompson A et al. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 2012; 37: 611–621. [DOI] [PubMed] [Google Scholar]
- Hardy MY, Girardin A, Pizzey C, Cameron DJ, Watson KA, Picascia S et al. Consistency in polyclonal T-cell responses to gluten between children and adults with celiac disease. Gastroenterology 2015; 149: 1541–1552 e1542. [DOI] [PubMed] [Google Scholar]
- Petersen J, van Bergen J, Loh KL, Kooy-Winkelaar Y, Beringer DX, Thompson A et al. Determinants of gliadin-specific T cell selection in celiac disease. J Immunol 2015; 194: 6112–6122. [DOI] [PubMed] [Google Scholar]
- Qiao SW, Raki M, Gunnarsen KS, Loset GA, Lundin KE, Sandlie I et al. Posttranslational modification of gluten shapes TCR usage in celiac disease. J Immunol 2011; 187: 3064–3071. [DOI] [PubMed] [Google Scholar]
- Dahal-Koirala S, Risnes LF, Christophersen A, Sarna VK, Lundin KE, Sollid LM et al. TCR sequencing of single cells reactive to DQ2.5-glia-alpha2 and DQ2.5-glia-omega2 reveals clonal expansion and epitope-specific V-gene usage. Mucosal Immunol 2016; 9: 587–596. [DOI] [PubMed] [Google Scholar]
- Wungjiranirun M, Kelly CP, Leffler DA. Current status of celiac disease drug development. Am J Gastroenterol 2016; 111: 779–786. [DOI] [PubMed] [Google Scholar]
- Bodd M, Raki M, Tollefsen S, Fallang LE, Bergseng E, Lundin KE et al. HLA-DQ2-restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunol 2010; 3: 594–601. [DOI] [PubMed] [Google Scholar]
- Peluso I, Fantini MC, Fina D, Caruso R, Boirivant M, MacDonald TT et al. IL-21 counteracts the regulatory T cell-mediated suppression of human CD4+ T lymphocytes. J Immunol 2007; 178: 732–739. [DOI] [PubMed] [Google Scholar]
- Monteleone I, Sarra M, Del Vecchio Blanco G, Paoluzi OA, Franze E, Fina D et al. Characterization of IL-17A-producing cells in celiac disease mucosa. J Immunol 2010; 184: 2211–2218. [DOI] [PubMed] [Google Scholar]
- Fernandez S, Molina IJ, Romero P, Gonzalez R, Pena J, Sanchez F et al. Characterization of gliadin-specific Th17 cells from the mucosa of celiac disease patients. Am J Gastroenterol 2011; 106: 528–538. [DOI] [PubMed] [Google Scholar]
- Ettersperger J, Montcuquet N, Malamut G, Guegan N, Lopez-Lastra S, Gayraud S et al. Interleukin 15-dependent T cell-like Innate intraepithelial lymphocytes develop in the intestine and transform into lymphomas in celiac disease. Immunity 2016; 45: 610–625. [DOI] [PubMed] [Google Scholar]
- Menard S, Lebreton C, Schumann M, Matysiak-Budnik T, Dugave C, Bouhnik Y et al. Paracellular versus transcellular intestinal permeability to gliadin peptides in active celiac disease. Am J Pathol 2012; 180: 608–615. [DOI] [PubMed] [Google Scholar]
- Fasano A, Not T, Wang W, Uzzau S, Berti I, Tommasini A et al. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 2000; 355: 1518–1519. [DOI] [PubMed] [Google Scholar]
- Beitnes AC, Raki M, Brottveit M, Lundin KE, Jahnsen FL, Sollid LM. Rapid accumulation of CD14+CD11c+ dendritic cells in gut mucosa of celiac disease after in vivo gluten challenge. PLoS ONE 2012; 7: e33556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Di Sabatino A, Bauer M, Della Riccia DN, Bizzini F, Biagi F et al. Matrix metalloproteinase pattern in celiac duodenal mucosa. Lab Invest 2005; 85: 397–407. [DOI] [PubMed] [Google Scholar]
- Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 2004; 21: 357–366. [DOI] [PubMed] [Google Scholar]
- Meresse B, Curran SA, Ciszewski C, Orbelyan G, Setty M, Bhagat G et al. Reprogramming of CTLs into natural killer-like cells in celiac disease. J Exp Med 2006; 203: 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue S, Mention JJ, Monteiro RC, Zhang S, Cellier C, Schmitz J et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 2004; 21: 367–377. [DOI] [PubMed] [Google Scholar]
- Maiuri L, Ciacci C, Ricciardelli I, Vacca L, Raia V, Auricchio S et al. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003; 362: 30–37. [DOI] [PubMed] [Google Scholar]
- Junker Y, Zeissig S, Kim SJ, Barisani D, Wieser H, Leffler DA et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J Exp Med 2012; 209: 2395–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stene LC, Honeyman MC, Hoffenberg EJ, Haas JE, Sokol RJ, Emery L et al. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am J Gastroenterol 2006; 101: 2333–2340. [DOI] [PubMed] [Google Scholar]
- Setty M, Discepolo V, Abadie V, Kamhawi S, Mayassi T, Kent A et al. Distinct and synergistic contributions of epithelial stress and adaptive immunity to functions of intraepithelial killer cells and active celiac disease. Gastroenterology 2015; 149: 681–691 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo I, Barone MV, Martucciello S, Lepretti M, Esposito C. Tissue transglutaminase in celiac disease: role of autoantibodies. Amino Acids 2009; 36: 693–699. [DOI] [PubMed] [Google Scholar]
- Salmi TT, Collin P, Jarvinen O, Haimila K, Partanen J, Laurila K et al. Immunoglobulin A autoantibodies against transglutaminase 2 in the small intestinal mucosa predict forthcoming coeliac disease. Aliment Pharmacol Ther 2006; 24: 541–552. [DOI] [PubMed] [Google Scholar]
- Korponay-Szabo IR, Halttunen T, Szalai Z, Laurila K, Kiraly R, Kovacs JB et al. In vivo targeting of intestinal and extraintestinal transglutaminase 2 by coeliac autoantibodies. Gut 2004; 53: 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tersigni C, Castellani R, de Waure C, Fattorossi A, De Spirito M, Gasbarrini A et al. Celiac disease and reproductive disorders: meta-analysis of epidemiologic associations and potential pathogenic mechanisms. Hum Reprod Update 2014; 20: 582–593. [DOI] [PubMed] [Google Scholar]
- Di Niro R, Snir O, Kaukinen K, Yaari G, Lundin KE, Gupta NT et al. Responsive population dynamics and wide seeding into the duodenal lamina propria of transglutaminase-2-specific plasma cells in celiac disease. Mucosal Immunol 2016; 9: 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snir O, Mesin L, Gidoni M, Lundin KE, Yaari G, Sollid LM. Analysis of celiac disease autoreactive gut plasma cells and their corresponding memory compartment in peripheral blood using high-throughput sequencing. J Immunol 2015; 194: 5703–5712. [DOI] [PubMed] [Google Scholar]
- Steinsbo O, Henry Dunand CJ, Huang M, Mesin L, Salgado-Ferrer M, Lundin KE et al. Restricted VH/VL usage and limited mutations in gluten-specific IgA of coeliac disease lesion plasma cells. Nat Commun 2014; 5: 4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Niro R, Mesin L, Zheng NY, Stamnaes J, Morrissey M, Lee JH et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat Med 2012; 18: 441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid LM, Molberg O, McAdam S, Lundin KE. Autoantibodies in coeliac disease: tissue transglutaminase—guilt by association? Gut 1997; 41: 851–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman AA, Gunnel T, Dietl A, Uhlig HH, Amin M, Fleckenstein B et al. B cell epitopes of gliadin. Clin Exp Immunol 2000; 121: 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choung RS, Marietta EV, Van Dyke CT, Brantner TL, Rajasekaran J, Pasricha PJ et al. Determination of B-cell epitopes in patients with celiac disease: peptide microarrays. PLoS ONE 2016; 11: e0147777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorum S, Steinsbo O, Bergseng E, Arntzen MO, de Souza GA, Sollid LM. Gluten-specific antibodies of celiac disease gut plasma cells recognize long proteolytic fragments that typically harbor T-cell epitopes. Sci Rep 2016; 6: 25565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophersen A, Risnes LF, Bergseng E, Lundin KE, Sollid LM, Qiao SW. Healthy HLA-DQ2.5+ subjects lack regulatory and memory T cells specific for immunodominant gluten epitopes of celiac disease. J Immunol 2016; 196: 2819–2826. [DOI] [PubMed] [Google Scholar]
- Tiittanen M, Westerholm-Ormio M, Verkasalo M, Savilahti E, Vaarala O. Infiltration of forkhead box P3-expressing cells in small intestinal mucosa in coeliac disease but not in type 1 diabetes. Clin Exp Immunol 2008; 152: 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjova T, Uibo O, Heilman K, Rago T, Honkanen J, Vaarala O et al. Increased FOXP3 expression in small-bowel mucosa of children with coeliac disease and type I diabetes mellitus. Scand J Gastroenterol 2009; 44: 422–430. [DOI] [PubMed] [Google Scholar]
- Frisullo G, Nociti V, Iorio R, Patanella AK, Marti A, Assunta B et al. Increased CD4+CD25+Foxp3+ T cells in peripheral blood of celiac disease patients: correlation with dietary treatment. Hum Immunol 2009; 70: 430–435. [DOI] [PubMed] [Google Scholar]
- Gianfrani C, Levings MK, Sartirana C, Mazzarella G, Barba G, Zanzi D et al. Gliadin-specific type 1 regulatory T cells from the intestinal mucosa of treated celiac patients inhibit pathogenic T cells. J Immunol 2006; 177: 4178–4186. [DOI] [PubMed] [Google Scholar]
- Benahmed M, Meresse B, Arnulf B, Barbe U, Mention JJ, Verkarre V et al. Inhibition of TGF-beta signaling by IL-15: a new role for IL-15 in the loss of immune homeostasis in celiac disease. Gastroenterology 2007; 132: 994–1008. [DOI] [PubMed] [Google Scholar]
- Forsberg G, Hernell O, Melgar S, Israelsson A, Hammarstrom S, Hammarstrom ML. Paradoxical coexpression of proinflammatory and down-regulatory cytokines in intestinal T cells in childhood celiac disease. Gastroenterology 2002; 123: 667–678. [DOI] [PubMed] [Google Scholar]
- Hmida NB, Ben Ahmed M, Moussa A, Rejeb MB, Said Y, Kourda N et al. Impaired control of effector T cells by regulatory T cells: a clue to loss of oral tolerance and autoimmunity in celiac disease? Am J Gastroenterol 2012; 107: 604–611. [DOI] [PubMed] [Google Scholar]
- Granzotto M, dal Bo S, Quaglia S, Tommasini A, Piscianz E, Valencic E et al. Regulatory T-cell function is impaired in celiac disease. Dig Dis Sci 2009; 54: 1513–1519. [DOI] [PubMed] [Google Scholar]
- Zanzi D, Stefanile R, Santagata S, Iaffaldano L, Iaquinto G, Giardullo N et al. IL-15 interferes with suppressive activity of intestinal regulatory T cells expanded in Celiac disease. Am J Gastroenterol 2011; 106: 1308–1317. [DOI] [PubMed] [Google Scholar]
- Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med 2006; 203: 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddiki N, Cook L, Hsu DC, Phetsouphanh C, Brown K, Xu Y et al. Human antigen-specific CD4(+ CD25(+ CD134(+ CD39(+ T cells are enriched for regulatory T cells and comprise a substantial proportion of recall responses. Eur J Immunol 2014; 44: 1644–1661. [DOI] [PubMed] [Google Scholar]
- Zaunders JJ, Munier ML, Seddiki N, Pett S, Ip S, Bailey M et al. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40). J Immunol 2009; 183: 2827–2836. [DOI] [PubMed] [Google Scholar]
- DePaolo RW, Abadie V, Tang F, Fehlner-Peach H, Hall JA, Wang W et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature 2011; 471: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezinga SL, Auricchio R, Bravi E, Castillejo G, Chmielewska A, Crespo Escobar P et al. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med 2014; 371: 1304–1315. [DOI] [PubMed] [Google Scholar]
- Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med 2014; 371: 1295–1303. [DOI] [PubMed] [Google Scholar]
- Verdu EF, Galipeau HJ, Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol 2015; 12: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminero A, Galipeau HJ, McCarville JL, Johnston CW, Bernier SP, Russell AK et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology 2016; 151: 670–683. [DOI] [PubMed] [Google Scholar]
- Anderson RP, Jabri B. Vaccine against autoimmune disease: antigen-specific immunotherapy. Curr Opin Immunol 2013; 25: 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croese J, Giacomin P, Navarro S, Clouston A, McCann L, Dougall A et al. Experimental hookworm infection and gluten microchallenge promote tolerance in celiac disease. J Allergy Clin Immunol 2015; 135: 508–516. [DOI] [PubMed] [Google Scholar]
- Rose NR, Bona C. Defining criteria for autoimmune diseases (Witebsky's postulates revisited). Immunol Today 1993; 14: 426–430. [DOI] [PubMed] [Google Scholar]
- Kurppa K, Collin P, Viljamaa M, Haimila K, Saavalainen P, Partanen J et al. Diagnosing mild enteropathy celiac disease: a randomized, controlled clinical study. Gastroenterology 2009; 136: 816–823. [DOI] [PubMed] [Google Scholar]
- Coleman C, Quinn EM, Ryan AW, Conroy J, Trimble V, Mahmud N et al. Common polygenic variation in coeliac disease and confirmation of ZNF335 and NIFA as disease susceptibility loci. Eur J Hum Genet 2016; 24: 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KA, Mistry V, Bockett NA, Ahmad T, Ban M, Barker JN et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature 2013; 498: 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostensson M, Monten C, Bacelis J, Gudjonsdottir AH, Adamovic S, Ek J et al. A possible mechanism behind autoimmune disorders discovered by genome-wide linkage and association analysis in celiac disease. PLoS ONE 2013; 8: e70174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CP, Murray JA, Ding YC, Tien Z, van Heel DA, Neuhausen SL. Replication of celiac disease UK genome-wide association study results in a US population. Hum Mol Genet 2009; 18: 4219–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut 2009; 58: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn's disease and celiac disease. PLoS Genet 2011; 7: e1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]