Abstract
Evidence indicates that hypoxia and oxidative stress can control metabolic reprogramming of cancer cells and other cells in tumor microenvironments and that the reprogrammed metabolic pathways in cancer tissue can also alter the redox balance. Thus, important steps toward developing novel cancer therapy approaches would be to identify and modulate critical biochemical nodes that are deregulated in cancer metabolism and determine if the therapeutic efficiency can be influenced by changes in redox homeostasis in cancer tissues. In this review, we will explore the molecular mechanisms responsible for the metabolic reprogramming of tumor microenvironments, the functional modulation of which may disrupt the effects of or may be disrupted by redox homeostasis modulating cancer therapy.
Introduction
Otto Warburg and co-workers showed in the 1920s that cancer tissues can metabolize, even in aerobic conditions, ~10-fold more glucose to produce lactate than normal tissues, and this is known as aerobic glycolysis or the Warburg effect.1 For the biosynthetic demands of continuous proliferation and survival of cancer cells to be satisfied, many genetic and epigenetic changes in oncogenes and tumor suppressor genes, such as HIF-1α, AKT, Myc, PI3K/AKT, Ras, p53, PTEN and LKB1, responsible for tumorigenesis, can directly regulate the expression, regulation and activity of important components of metabolic pathways and exert a direct impact on metabolism, most remarkably, on glucose uptake and aerobic glycolysis.2, 3, 4, 5, 6 Although cancer cells display a diverse range of metabolic profiles, the metabolic phenotype resulting from the Warburg effect is considered a widespread cancer-associated trait. This increased aerobic glycolysis has been erroneously thought to be a consequence, not a cause or contributing factor, in cancer and has been suggested as evidence that mitochondrial function in cancer cells is compromised. Despite enhanced glycolysis, most cancer cells also maintain mitochondrial respiration to produce a significant fraction of ATP,7, 8, 9 and functional mitochondria are essential for the survival of cancer cells.2, 10
In recent years, therapeutic strategies targeting substantially altered cellular metabolism as a possible area for the development of novel anti-cancer agents have again been intensively investigated. The basis of metabolism correction strategies for cancer therapy, exploiting the heavy reliance of cancer cells on specific metabolic enzymes or processes, and finding a possible therapeutic window that we can use to eradicate cancer cells rather than normal non-transformed cells, involves the manipulation of redox homeostasis in cancer cells. The principles of current strategies exploiting metabolic perturbations for cancer therapy are nicely explained in a recent review.8
Studies have identified that cancer cells can become vulnerable to high reactive oxygen species (ROS) levels.11 Indeed, increased sensitivity and apoptosis of cancer cells from increased ROS in response to depletion of ATP from manipulation of glycolytic enzymes and chemotherapeutics or radiotherapy have been reported from preclinical studies,12, 13, 14 and the design of new therapeutic approaches combining chemotherapeutic reagents and glycolysis inhibitors is being studied. Therefore, the general application of combined therapies targeting cancer metabolism with inhibitors of metabolic enzymes or oncogenes and disabling key anti-oxidant systems with ROS inducers such as chemotherapeutic reagents appears to show promise and is speculated to be a promising strategy to overcome drug resistance.
However, we still lack solid evidence that such approaches are widely applicable in human patients, especially on a long-term basis. As far as we are aware, much of our current understanding of ROS is from studies with isolated mitochondria and cells in vitro, and the influence of high ROS levels induced by therapeutic interventions on cancer cellular metabolism with respect to cancer cell survival appears to have been studied surprisingly little.15, 16 Studies have indicated that metabolic changes and ROS production are intertwined in cancer cells.11 Notably, evidence indicates that oxidative stress and hypoxia can control metabolic reprogramming of tumor microenvironments and that perturbed metabolic pathways can also alter redox balance.10, 17, 18, 19, 20 Furthermore, we know that cancer cells can develop heightened anti-oxidant systems to survive in high oxidative stress environments (Figure 1).21, 22 For example, Ras- and Myc-driven cancer cells with their heightened anti-oxidant system are among the most difficult to treat.23 These findings indicate that decisions to use particular reagents, such as chemotherapeutics to increase intracellular ROS to reach a toxic level or anti-oxidants to disturb redox balance, may depend on the type and stage of cancer and the level of endogenous ROS, the activation of ROS-induced survival pathways and metabolic perturbations in cancer.
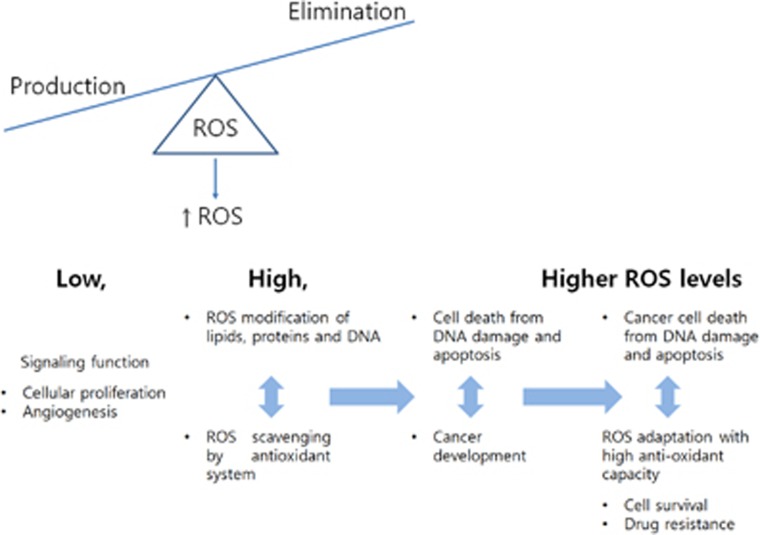
Figure 1.
REDOX adaptation in cancer. Low levels of ROS can activate various signaling pathways to stimulate cell proliferation and survival. Adaptation to persistent and high levels of ROS can promote cancer development, survival of cancer cells and resistance to chemotherapeutics.
Thus, it would be logical to assume that altered glycolytic metabolism owing to therapeutic interventions combining ROS inducers and specific inhibitors of metabolic targets can be both an oncogenesis and treatment resistance mechanism and also that successful cancer therapy would require a thorough understanding of the molecular mechanisms responsible for the metabolic reprogramming of the tumor microenvironment, which may be different between multiple neoplasms and even between individuals diagnosed with the same cancer type.
In this review, we provide a summary of published reports on the molecular mechanisms responsible for the metabolic reprogramming of tumor microenvironments, the functional modulation of which may disrupt the effects of or may be disrupted by redox homeostasis modulating cancer therapy.
Glucose metabolism in cancer cells
It is now clear that the spectrum of metabolic reprogramming in cancer cells goes beyond the Warburg effect and includes increased flux through the pentose phosphate pathway (PPP), high glutamine consumption, reduction/oxidation (redox) imbalance (either oxidative or reductive depending on cancer type,24, 25 and elevated rates of lipid biosynthesis including fatty acid synthesis and the mevalonate pathway (Figure 2).5, 8, 24, 25, 26 Although recent studies have revealed the importance of fatty acids and proteins as fuel sources for cancer cells to proliferate and survive,27, 28, 29, 30 extensive studies have been conducted on the use of glycolysis as a major fuel source for cancer cells. To address metabolic reprogramming, we will focus on glucose metabolism in this review. Notably, activation of oncogenes and suppression of tumor suppressors are mostly responsible for metabolic rewiring of cancer cells, and functional mutations found in metabolic genes are relatively scarce.14
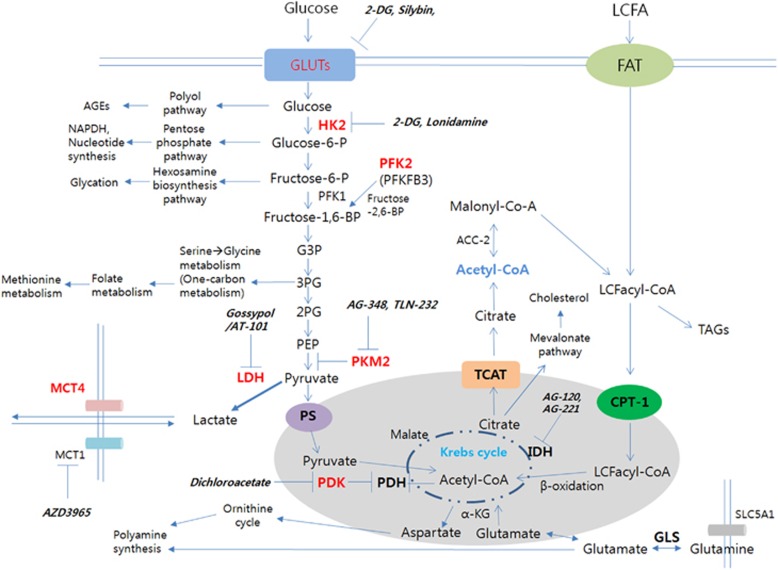
Figure 2.
Metabolic targets at a glance. GLUTs, HK2, PFK2, PKM2, LDH, PDK and MCT4 shown in red can be upregulated from stabilization of HIF-1α. Representative inhibitors of metabolic nodes entering clinical trials are shown in bold. ACC, acetyl-CoA carboxylase; AGE, advanced glycation end product; CPT-1, carnitine palmitoyltransferase 1; FAT, fatty acid translocase; G3P, glyceraldehyde 3-phosphate; GLS, glutaminase; GLUT, glucose transporter; HK2, hexokinase 2; LCFA, long chain fatty acid; LCFacyl-CoAs, long chain fatty acyl-CoAs; MCT, monocarboxylate transporter; PEP, phosphoenolpyruvate; PDH, pyruvate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PFK, phosphofructokinase; PS, pyruvate symporter; SLC5A1, sodium glucose co-transporter; TAG, triacylglyceride; TCAT, tricarboxylic acid transporter; 2-DG, 2-deoxyglucose; 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate.
The conversion of fructose-6-phosphate (F6P) to fructose 1,6-bisphosphate (F1,6BP) by phosphofructokinase-1 (PFK1), a rate-limiting and irreversible reaction in glycolysis, is a primary control point in glycolysis (Figure 2). PFK1, the prominent rate-limiting enzyme in glycolysis, is an allosteric enzyme. Fructose 2,6-bisphosphate (F2,6BP), the most potent allosteric activator of PFK1, is produced by PFK2 and critically regulates the glycolytic rate. The affinity of PFK1 for F6P is increased upon binding of F2,6BP to PFK1, and ATP-mediated inhibition of PFK1 is lost. Among all PFK2 enzymes, 6-phosphofructo-2-kinase (PFKFB3), with high kinase activity and almost no phosphatase activity, is generally found to be overexpressed in human cancers.31, 32 Thus, PFKFB3 preferentially drives the synthesis of F2,6BP. Fructose-1,6-biphosphatase 1 (FBP1), a rate-limiting enzyme in the gluconeogenesis pathway, catalyzes the hydrolysis of F1,6BP to F6P. The FBP1-mediated reaction is the reverse of the rate-limiting glycolytic reaction catalyzed by PFK1, and FBP1 can antagonize glycolysis. In cancer cells with overexpressed PFKFB3, negative feedback from the Krebs cycle is not effective on glycolysis, and cancer cells can keep high levels of PFK1 activity and glycolysis. PFKFB3 and F2,6BP promote cell cycle progression and suppress apoptosis via Cdk1-mediated phosphorylation and subsequent degradation of p27, a tumor suppressor gene, indicating that a certain glycolysis enzyme can also function as an oncogenic driver.31 Snail represses FBP1 expression by binding to the FBP1 promoter, thereby enhancing glycolysis.33 Thus, Snail not only induces the epithelial–mesenchymal transition but also suppresses mitochondrial oxidative metabolism.2, 34
Enhanced glycolysis in cancer cells does not necessarily increase glycolytic flux in downstream metabolic pathways. Cancer cells allow the buildup of glycolytic intermediates for biosynthesis by slowing down the last step of glycolysis that is catalyzed by pyruvate kinase M2 (PKM2). The increased generation of the PKM2 isoform from alternative splicing of the PK gene, an event shown to be under the regulation of the oncogenic transcription factor Myc commonly deregulated in cancer, indicates that PKM2 expression may be a potential oncogenic driver.3, 35 Unlike PKM1 that can efficiently promote glycolysis, PKM2, preferentially expressed in cancer cells in an inactive dimeric state, is inefficient at promoting glycolysis.6, 36 PKM2 exists as either an inactive dimer or a more active tetramer, and the transition between the two conformations is subject to post-translational modifications (PTMs).37 Oncogenic tyrosine kinases phosphorylate tyrosine 705 on PKM2 and promote the formation of the inactive dimer.38 High levels of glucose-induced acetylation of PKM239 and ROS-mediated direct oxidation of a cysteine residue on PKM240 decreases its activity. These data suggest that increased expression of inactive PKM2 in cancer increases glucose flux into PPP flux, NADPH and glutathione levels and cancer cell proliferation. Notably, the inhibitory function of PKM2 recently came under dispute; there are reports that inhibition or activation of PKM2 can increase or decrease cell proliferation and tumorigenesis, respectively.12, 37, 41
Isocitrate dehydrogenase (IDH) is an enzyme that catalyzes the reversible oxidative decarboxylation of isocitrate, producing α-ketoglutarate and carbon dioxide in the Krebs cycle, also known as the tricarboxylic acid cycle. In cancer cells, acetyl-coA enters the Krebs cycle and generates citrate, which is utilized by malic enzyme and IDH1 to produce NADPH.14 Interestingly, oncogenic IDH mutations result in complex genetic and epigenetic changes including DNA methylation and metabolism.24, 42 Gain-of-function mutations of IDH found in gliomas have the capacity to catalyze the NADPH-dependent reduction of α-ketoglutarate to 2-hydroxyglutarate and not to produce NADPH.43, 44 2-Hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases, including histone demethylases and the TET family of 5-methylcytosine hydroxylases, ultimately leading to genome-wide histone and DNA methylation alterations.45, 46
HIF-1 is a transcription factor comprised of HIF-1α and HIF-1β subunits. Under a hypoxic environment, HIF-1α is stabilized and activates its target genes. HIF-1 critically determines the metabolic shift from glucose oxidation to aerobic glycolysis in cancer. HIF-1 transcriptionally activates pyruvate dehydrogenase kinases (PDKs).47, 48, 49 Inactivation of pyruvate dehydrogenase (PDH) from phosphorylation by PDKs prevents pyruvate from entering the mitochondrial Krebs cycle, reduces mitochondrial respiration and prevents the excessive production of ROS.50 HIF-1 also enhances lactate dehydrogenase (LDH)-mediated pyruvate-to-lactate conversion.51 LDH-A is a direct transcriptional target of HIF and is highly inducible by hypoxia.52 HIF-1 also increases GLUT1, GLUT3, MCT4 and PKM2 to increase glucose uptake, to increase the export of lactate and to uncouple glycolysis and oxidative phosphorylation, respectively (Figure 2).53, 54 Thus, HIF1 can suppress mitochondrial respiration and also ROS production. HIF-1 induced expression of NADH dehydrogenase (ubiquinone) 1 subcomplex subunit 4-like 2 (NDUFA4L2), and suppression of electron transport chain complex I activity can inhibit oxygen consumption and mitochondrial ROS production.55 HIF-1-mediated metabolic reprogramming is responsible for the survival of metastatic cancers during their colonization in lungs by reducing cytotoxic ROS levels.50 It is also known that ROS can activate the HIF-1α promoter via a functional NF-κB site, indicating the negative feedback regulation of ROS by HIF-1.56, 57 Genetic ablation of adenosine monophosphate-activated protein kinase activates mTOR signaling with stabilization of HIF-1α and results in activation of aerobic glycolysis.58 This finding indicates that the cellular energy status is closely linked to aerobic glycolysis.
A recent report that tumor-associated mutant p53 drives the Warburg effect indicates that p53 mediates suppression of the Warburg effect.59 p53 regulates genes that can balance the utilization of respiratory and glycolytic pathways, and p53-deficient cells show higher rates of glycolysis and decreased mitochondrial respiration than those of wild-type cells.60 p53 downregulates expression of glucose transporter 1 (GLUT1) and GLUT4.61 In addition, p53 also downregulates PDK2 to increase the entry of pyruvate into mitochondria for oxidative metabolism,62 and p53-altered cellular metabolism is an important cause of increased ROS production in cancer cells.11 Physical interaction of p53 with glucose 6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme of the PPP, to inhibit the formation of the active G6PD dimer suppresses glucose consumption and NADPH and GSH production.63, 64 p53-induced glycolysis and apoptosis regulator (TIGAR) is a fructose-2,6-bisphosphatase. By lowering F2,6BP levels, p53 via p53-induced glycolysis and apoptosis regulator reduces the activity of PFK1. Thus, p53 can decrease the rate of glycolysis65 and counteracts the Warburg effect.62
Myc activates PDK1 and LDH-A66, 67 under normoxic conditions or in conjunction with HIF-1α under hypoxic conditions. Myc also drives glucose metabolism in triple-negative breast cancer cells by direct repression of thioredoxin-interacting protein (TXNIP).68 TXNIP is a potent negative regulator of glucose uptake, aerobic glycolysis and glycolytic gene expression. Thus, its repression by Myc provides an alternate route to Myc-driven glucose metabolism. The mutation of TP53, which is a defining molecular feature of triple-negative breast cancer, has been shown to enhance the correlation between Myc suppression of TXNIP and death from breast cancer. A Myc-driven metabolic shift to glycolytic, pentose-phosphate and glutaminolysis pathways has also been found in activated T lymphocytes in addition to cancer cells.69, 70
Targeting metabolism in cancer cells
Several cancer-associated alterations in catabolic and anabolic metabolic pathways, including glycolysis, the Krebs cycle, glutaminolysis and fatty acid oxidation, have been studied as potential drug targets, and identification of agents that selectively eradicate cancer cells based on metabolic alterations has gained immense interest.3, 71, 72 These approaches have led to the development of several molecules that are now entering clinical trials (Table 1),2, 3, 5, 6, 12, 73, 74 and readers are referred to excellent reviews with detailed summaries of metabolic targets for cancer therapy.5, 6 Proteins that are possible therapeutic targets include the glycolytic enzymes6, 75 (hexokinase-2,76 phosphoglycerate kinase-1,77, 78 phosphoglycerate mutase,79 PDK,80 and PKM2,36, 38) lipid synthesis/fatty acid metabolism targets (ATP citrate lyase,81 fatty acid synthase,82 monoglyceride lipase83 and carnitine palmitoyltransferase 1(CPT1),84) and the PPP proteins (glucose-6-phosphate dehydrogenase,63 transaldolase and transketolase).85 Several other glycolytic enzymes and transporters, including PFKFB3,32 GAPDH,86 LDH-A,87 GLUT1 and GLUT488, 89 and monocarboxylate transporter 4 (MCT4), may become candidates for anticancer therapy.90
Table 1. Representative inhibitors of metabolic enzyme and oncogenes that entered clinical trials (from https://clinicaltrials.gov/).
| Compound name | Protocol number | Status | Study completed or last updated year |
|---|---|---|---|
| AMPK activator | |||
| Metformin | NCT01433913 | Phase II completed | 2015 |
| NCT01941953 | Phase II completed | 2015 | |
| NCT01210911 | Phase II completed | 2010 | |
| GLUTs inhibitor | |||
| 2-DG | NCT00633087 | Phase I/II terminated | 2011 |
| NCT00096707 | Phase I | 2008 | |
| HIF1 inhibitor | |||
| EZN-2968, Antisense Oligonucleotide Inhibitor | NCT01120288 | Phase I completed | 2014 |
| RO7070179, HIF1A mRNA antagonist | NCT01251926 | Phase I completed | 2014 |
| PX-478 | NCT02564614 | Phase I recruiting | 2016 |
| BAY87–2243 | NCT00522652 | Phase I completed | 2010 |
| NCT01297530 | Phase I terminated | 2012 | |
| IDH inhibitor | |||
| AG-120, AG-221 | NCT02677922 | Phase I/II recruiting | |
| NCT02073994 | AG-120, phase I recruiting | ||
| NCT02074839 | AG-120, phase I recruiting | ||
| NCT02632708 | Phase I recruiting | ||
| NCT01915498 | Phase I active, not recruiting | ||
| NCT02577406 | AG-221, phase III recruiting | ||
| LDH inhibitor | |||
| Gossypol(AT-101) | NCT00848016 | Phase II completed & results | 2014 |
| NCT00540722 | Phase II completed | 2013 | |
| NCT00666666 | Phase II completed | 2013 | |
| MCT1 inhibitor | |||
| AZD3965 | NCT01791595 | Phase I recruiting | 2016 |
| NCT01278615 | Phase II terminated | ||
| PDK inhibitor | |||
| Dichloroacetate | NCT01029925 | Phase II terminated | 2013 |
| NCT00540176 | Phase II completed | 2014 | |
| PKM2 inhibitor | |||
| TLN-232 | NCT00422786 | Phase II completed | 2008 |
| Myc inhibitor | |||
| Quarfloxin | NCT00780663 | Phase II completed | 2010 |
| CX-3543 | NCT00955786 | Phase I completed | 2008 |
Although performed in vitro or at preclinical stages, successful modulation of oxidative stress by targeting cancer cell metabolism has been reported as follows. Inhibition of CPT1 with etomoxir impairs NADPH production and promotes oxidative stress-induced cell death in human glioblastoma cells.91 Inhibition of PDKs could upregulate the activity of the pyruvate dehydrogenase complex and rectify the balance between the demand and supply of oxygen, which could lead to cancer cell death.80 Inhibition of glycolysis and PPP with the disruption of thioredoxin systems showed selective cancer targeting in pancreatic and breast cancer cells.92 Inhibition of LDH-A with FX11 impaired malignant progression of lymphoma and pancreatic xenografts by inducing oxidative stress.87 Inhibition of glycolysis and PPP through 2-deoxy-d glucose and 6-aminonicotinamide induced oxidative stress and sensitized cancer cells to radiotherapy.93 Deprivation of glutamine through the inhibition of GLS1 decreases the GSH level to alter the redox balance and increase chemosensitivity in cancer cells.92, 94 There are other reports that inhibition of the redox pathway can effectively eradicate cancer cells. NOV-002 and canfosfamide targeting S-glutathionylation, PX-12 targeting thioredoxin and arsenic derivatives with unknown mechanisms demonstrated anti-cancer activity by increasing oxidative stress.95
Have our attempts to correct genetic or epigenetic changes reversed metabolic reprogramming in cancer cells, and does the metabolic reprogramming affect responses to anti-oncogene targeted therapies? The answers appear to be ‘No' and ‘Yes', respectively. Recent reports show that rewiring of metabolic circuits in cancer cells results in resistance to several oncogene-targeted therapies and that the combined use of glycolytic inhibitors is somewhat successful in avoiding the resistance.12, 96, 97, 98, 99, 100, 101 Importantly, these findings indicate that altered glycolytic metabolism can be both an oncogenesis and treatment-resistance mechanism and that simple reversion of genetic or epigenetic causes responsible for the metabolic reprogramming may not be effective in cancer cells that have been metabolically fully rewired during the progression of tumorigenesis. The metabolic changes that occur in cancer cells were previously considered secondary to the transformation process and also completely different from non-transformed normal cells. However, studies have revealed that changes in cellular metabolism and epigenetic and genetic changes in malignant transformation are intertwined and cannot be dissociated from other facets of malignant transformation.102, 103 To add complexity, a recent report that circulating cancer cells increase mitochondrial biogenesis and oxidative phosphorylation, without slowing the glycolysis rate, to increase viability and invasiveness104 indicates that cancer cells can adapt to ATP demand by enhancing oxidative phosphorylation and that the balance between mitochondrial productivity and detoxification of ROS can be easily shifted based on various cellular needs. Therefore, methods to correct glycolysis-shifted metabolic reprogramming in localized primary or metastatic cancer may be effective but may also affect the viability and invasive properties of circulating cancer cells with increased oxidative phosphorylation (the metabolic therapy may reduce localized cancer burden at the cost of increasing the incidence of metastasis).
Targeting metabolic synergy between cancer and stromal cells in the tumor microenvironment
Put simply, the tumor microenvironment is comprised of cancer and stromal cells such as cancer associated fibroblasts (CAFs), immune cells and blood vessels.105 Cancer aggressiveness is driven by metabolic synergy between cancer and stromal cells.106 Although cancer research has previously focused on cancer cells, the role of stromal cells in cancer is becoming a new central focus. Metabolic synergy, which induces the efficient utilization of catabolites by cancer cells, results from differential expression of catabolite transporters in stromal and cancer cells.107 For example, cancer and stromal cells secrete lactate, and extracellular lactate directly increases ROS levels in neutrophils, CD34+ vasculogenic stem cells, and cancer cells.108, 109
Microenvironments of cancer tissues are found to produce ROS levels that are higher than normal intracellular ROS levels.22, 110, 111 It has been determined that cancer-associated metabolic alterations (Warburg effect) are not a strictly uniform feature of malignant cells. They differ across distinct cancers and are found even in non-transformed cells in tumor microenvironments.112, 113 Furthermore, non-transformed stromal, endothelial, and immune cells outnumber their neoplastic counterparts in cancer.5, 114 Because rapid cell proliferation requires accelerated production of basic cellular building blocks for assembling new cells, differences in metabolism between cancer cells and non-transformed stromal and endothelial cells together can fuel cancer growth by lactate shuttling, maximally producing substrates for biosynthesis.89, 107, 115 The provision of lactate to oxidative cancer cells by lactate shuttling or by other unknown mechanisms may compensate for the lack of intracellular lactate and anti-oxidative resources in cancer cells.116 The tumor microenvironment strongly affects the metabolic status of cancer cells, and the supporting role of CAFs in the metabolic reprogramming of cancer cells is being emphasized.8, 17 Cancer cells induce aerobic glycolysis in neighboring fibroblasts by providing a hypoxic ROS-rich microenvironment.107, 117 Induced fibroblasts differentiate to myofibroblasts and upregulate MCT4 to secrete lactate and pyruvate that transforms normal stroma to ultimately help cancer cells grow (reverse Warburg effect). Lactate uptake by cancer cells can increase the surrounding stromal pH and can protect cells from harsh acidic microenvironments. Lactate shuttling between normoxic and hypoxic cancer cells or adjacent stromal cells induces a switch from glycolysis to oxidative phosphorylation and increases the generation of intracellular ROS.89 However, it also increases the survival of cancer cells by increasing anti-apoptotic mechanisms.118 These findings imply that metabolic adaptations in tumor microenvironments may obscure the direct tumoricidal effect of high-level ROS easily observable in in vitro culture systems. Alterations in oncogenes and cancer suppressor genes direct cellular metabolism to satisfy the biosynthetic demands for continuous cancer cell proliferation. However, CAFs are considered to be free from the alterations observed in oncogenes or tumor suppressors, and it is known that the induction of metabolic reprogramming in CAFs is driven by hypoxic status of the microenvironment.17, 119 Stromal cells such as endothelial cells and other cancer-associated cells are also considered free of genetic changes and have an important role in ROS metabolism within the tumor microenvironment. Functional changes in their mitochondria induced by cancer cells can be considered reversible.120 Thus, modulation of ROS in stromal cells may be more effective for both preventing myofibroblastic differentiation of fibroblasts into CAFs and reducing stabilization of HIF1-α and the resulting increase of GLUT1, GLUT3, MCT4 and PKM2 than in cancer cells where genetic changes are primarily and permanently responsible for the irreversible metabolic reprogramming. It has been shown that PDGF signaling is increased by ROS-induced oxidative inactivation of protein tyrosine phosphatases.121, 122 These findings indicate that PDGF signaling induced by ROS may at least be responsible for the proliferation of a PDGFR-β+ population of CAFs and that modulation of PDGF signaling may become a potential target in the future.
Targeting metabolic changes in blood vessels
Angiogenesis is a hallmark of hypoxic cancer mass, and intensive studies have been performed to target cancer angiogenesis. Chemical inhibition of vascular endothelial growth factor (VEGF) signaling is the only clinically approved anti-angiogenesis-based strategy, but resistance from genetic mutations is acquired within months after treatment initiation, seriously limiting its benefit.123, 124, 125 The endothelium is a single layer of endothelial cells (ECs) lining the blood vessel lumen and is in direct contact with blood.126 Surprisingly, ECs predominantly rely on glycolysis rather than oxidative metabolism for ATP production.127, 128 Over 80% of ATP is produced from conversion of glucose into lactate in the physiological state. Less than 1% of pyruvate from glycolysis enters mitochondria for oxidative metabolism and subsequent ATP production.127 However, ECs can also switch to oxidative metabolism of glucose, amino acids and fatty acids.129, 130 This is strikingly similar to the aerobic glycolysis observed in cancer cells, and compounds targeting cancer cellular metabolic reprogramming show similar effects in cancer cells and ECs.127, 131, 132 Currently, it is not clear whether metabolic phenotypes in ECs are modulated by cancer cells or CAFs.
Importantly, the switch to an angiogenic phenotype (as occurs in cancer) is even more metabolically demanding and mediated by an increase in aerobic glycolysis in EC metabolism.128 Stabilization of HIF-1α can also induce pro-angiogenic signaling pathways such as NF-κB, IL-8 and VEGFR2, leading to increased angiogenesis.133, 134 LDH-B is upregulated in the endothelium, and VEGF signaling increases glycolytic flux by inducing expression of GLUT1 and PFKFB3.127 Indeed, inhibition with 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one (3PO) or EC-specific gene knockdown of PFKFB3 inhibits cancer growth in vivo by inhibiting glycolytic flux.135 Recently, 3PO has been shown to reduce pathological angiogenesis in a variety of disease models.128, 132, 135
Targeting endothelial MCT1 can reduce the import of lactate released from hypoxic cancer cells and stromal cells in cancer microenvironments (lactate shuttling), which can stabilize HIF-1α and increase angiogenesis through the NF-κB/IL-8 pathway.134 Inhibition of MCT1 suppresses angiogenesis and reduces cancer growth in mice.133, 134 These observations are encouraging for validation in clinical trials. It should be noted that the therapeutic strategy of targeting glucose metabolism in ECs to inhibit cancer angiogenesis is very recent, but evidence suggests that it is possible.
Cellular response to modulation of ROS is difficult to predict
There are conflicting reports about the potency of cellular anti-oxidant systems and the efficiency of ROS targeting cancer therapies and even an increased risk of cancer in a long-term trial of redox modulation.3, 15, 110, 111, 136, 137, 138 Furthermore, the lack of any comprehensive study of the redox status of various cancer cell types critically limits the validation of speculation about the contribution of redox shifts to phenotypes across a broad spectrum of cancer cell types.139 Consequently, reports have confusingly shown that ROS can be cytotoxic and also tumorigenic, can activate or inhibit telomerase activity and that it can also contribute to metabolic reprogramming, invasion and metastasis of cancer cells.19 Notably, the related therapeutic effects at the bedside appear disappointingly controversial, and neither increasing nor decreasing levels of ROS show uniform therapeutic effects.140
ROS have dual functions, implying antagonism between different types and levels of ROS (Figure 1).137 Low levels of ROS can activate various signaling pathways to stimulate cell proliferation, survival and even extend the lifespans of C. elegans, invertebrates and mice.141, 142, 143 Because excess levels of ROS irreversibly damage cellular macromolecular components and result in cell death, both the mitochondrial competence and detoxification of ROS are known to be critical for cancer cell viability.9 ROS can also mediate motility and invasive properties of cancer cells, contribute to extracellular matrix remodeling, increase neo-angiogenesis and induce the metabolic reprogramming of both cancer and stromal cells.17 To detoxify ROS, cells express ROS scavenging enzymes.11 The mitochondrial manganese superoxide dismutase (MnSOD or SOD2), the expression of which is induced upon accumulation of ROS in hypoxic cancer cells, converts superoxide to the less reactive hydrogen peroxide. Hydrogen peroxide is broken down into water and dioxygen by enzymatic and non-enzymatic anti-oxidants. The balance between the production and elimination of ROS leads to redox homeostasis. Other important defensive mechanisms include peroxiredoxin, NADPH-dependent thioredoxin and glutathione/glutaredoxin systems.14 For detailed descriptions of ROS features, readers are referred to other excellent reviews.11, 16, 110, 111, 139, 144
It has been generally speculated that an anti-ROS strategy can prevent tumorigenesis by suppressing oncogenic functions of ROS and that increasing ROS will preferentially eradicate vulnerable cancer cells.11 Indeed, disruption of ROS maintenance and the resulting redox imbalance in cancer cells has been regarded as an important principle in cancer therapeutics, including chemotherapy because cancer cells are generally considered to be more vulnerable to disruption of redox balance and mitochondrial function than those of normal cells.110, 145, 146, 147
However, previous speculation that cancer cells are already flooded with high levels of ROS and that reducing ROS scavenging activity to increase ROS toxicity will uniformly eradicate cancer cells, appears to be an oversimplification. Cancer cells develop heightened anti-oxidant systems to survive in a high oxidative stress environment,21, 22 and this is regarded as an important drug resistance mechanism. A heightened anti-oxidant system is thought to be the reason that Ras- and Myc-driven cancer cells are among the most difficult to treat.23 Although numerous mechanisms are responsible for cancer resistance to chemotherapy, glycolytic phenotypes also count as a resistance mechanism. Increased glucose consumption supports cell proliferation and even enhances anti-oxidant capacity by activating the PPP pathway and keeping pyruvate away from mitochondrial oxidation to avoid the generation of excess ROS, implying that metabolic reprogramming can inherently increase anti-oxidant capacity that favors survival of cancer cells against high levels of ROS.3 Indeed, stabilization of HIF1-α, cysteine oxidation of PKM2 by increased ROS, activation of hexokinase 2 and major NADPH-producing PPP, de novo serine metabolism and NAD+ production from conversion of pyruvate into lactate by LDH, all upregulated along with the Warburg effect, can also presumably downregulate cytotoxic ROS.40, 50, 148, 149, 150 Furthermore, export of increased lactate leads to an acidic tumor microenvironment, which accounts for the resistance to many chemotherapy drugs from decreased uptake of the drugs and increased NADPH and GSH levels to maintain redox status.12, 151, 152 These mechanisms can potentially reduce the efficiency of chemotherapies expected to exploit vulnerabilities of cancer cells with presumably high ROS levels.
Recent findings that regulation of cellular signaling by redox homeostasis is highly sophisticated and that modulation of metabolism and ROS may produce conflicting results depending on whether cancer cells are resident in a tumor microenvironment or circulating in blood also emphasize that benefits from the regulation of ROS are not easily predictable.104 Nuclear respiratory factor 2 (Nrf2), a potent regulator of redox homeostasis, regulates ROS levels in normal cells by increasing expression of anti-oxidant genes. However, the activating mutations of Nrf2 or treatment of cancer cells with anti-oxidants can not only reduce ROS levels but also activate oncogenic activities.153 Furthermore, another report that treatment with antioxidants, N-acetylcysteine and vitamin E increases cancer cell proliferation by reducing ROS, DNA damage, and even p53 expression in mouse and human lung tumor cells138 again emphasizes that consequences from the modulation of ROS are hard to predict.
Influence of oxidative stress on metabolic enzymes
For predicting cellular response to ROS, it is important to understand how changes in ROS levels and redox imbalance can affect macromolecules in the progression of cancer. A significant body of evidence supports a crucial role for ROS in cellular functions and shows that ROS can interact with and modify biological macromolecules including DNA, lipids and proteins.19 However, surprisingly little is known about the cellular targets of ROS or how redox signaling is integrated in the oncogenic response.154 ROS and/or redox imbalance-sensitive PTMs of metabolic enzymes, oncogenes, tumor-suppressor transcription factors and signaling molecules may play considerable roles in metabolic reprogramming. The PTMs on these proteins identified to be modulated by redox changes in cancer cells are phosphorylation, nitration, cysteine oxidation, glutathionylation, acetylation, methylation and SUMOylation (Figure 3),19, 155, 156, 157, 158, 159 and studies showed acetylation of most enzymes functioning in glycolysis and the Krebs cycle.160, 161 Oxidative stress induced with menadione affects the activities of core metabolic enzymes, such as PDH, IDH, fumarase, GAPDH, malic enzyme and citrate synthase, as well as levels of various metabolites from glycolysis and the Krebs cycle.162 Hypoxia induces O-GlcNAcylation at serine 529 of PFK1 and inhibits PFK1 activity.163 The S529 glycosylation of PFK1 redirects glucose flux toward oxidative PPP, leading to increased NADPH production and concentration of glutathione. It has long been known that ROS can activate Akt.164, 165 Akt can phosphorylate serine 466 of PFK2, leading to activation of PFK1 by the PFK2 product F2,6BP.166 Furthermore, ROS can induce oxidation of cysteine 358 of PKM2 and result in decreased PKM2 activity.40 Importantly, it has been hypothesized that SUMOylation of key metabolic enzymes and glucose transporters may shift cellular metabolic strategies toward increased flux through the glycolytic pathway during periods of hypoxic stress.167, 168, 169 Mitochondrial aconitase catalyzes the reversible conversion of citrate to isocitrate. Oxidative stress causes various PTMs on aconitase resulting in functional impairment, and subcellular localization of the enzyme is shifted from mitochondria to the cytosol, resulting in slowing of the Krebs cycle.170
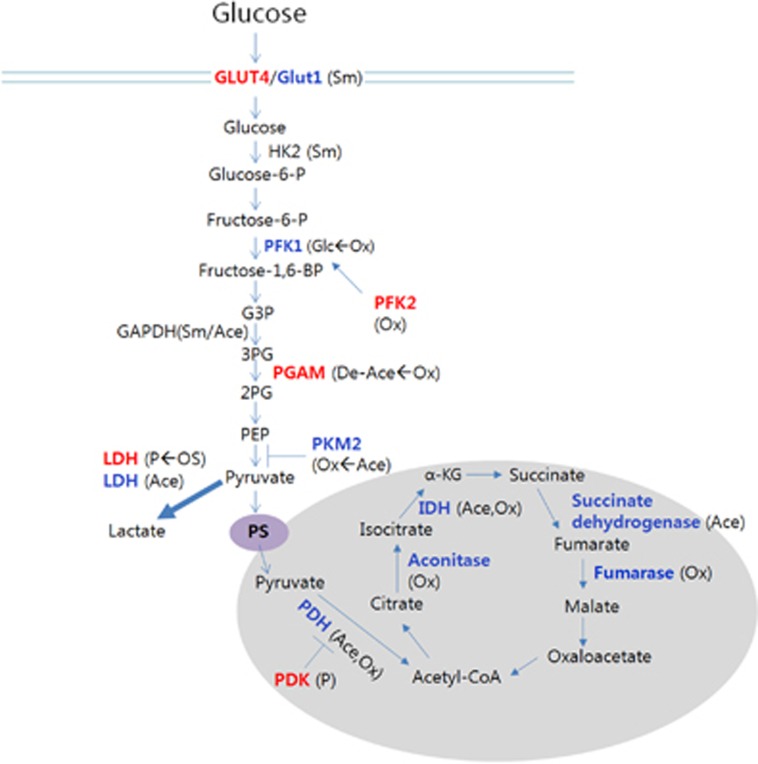
Figure 3.
PTMs affecting enzymatic activities in glucose metabolism. Enzyme activities that are increased or decreased upon specific PTMs are shown in red and blue, respectively. We use arrows to indicate if any specific inducer is known for a PTM (inducer→resultant PTM). Whether sumoylation increases enzyme activities of HK2 and GAPDH is not clear; however, induced sumoylation promotes glycolysis, and the two enzymes are found to be SUMOylated(167). Ace, acetylation; De-Ace, deacetylation; Glc, glycosylation; OS, oxidative stress; Ox, oxidation; P, phosphorylation; PGAM, phosphoglycerate mutase; Sm, SUMOylation.
Increased ROS levels can reduce oxidative phosphorylation via various mechanisms, including stabilization of HIF1-α, and cysteine modifications, including S-glutathionylation of complex I proteins, and affect cellular signaling from inactivation of phosphatases, including PTP1b, PTEN and MAPK phosphatases, favoring aerobic glycolysis and survival.171, 172, 173, 174 Furthermore, ROS-dependent activation of receptor tyrosine kinases175, 176 and stabilization of HIF1-α by ligand-induced activation of multiple receptor tyrosine kinases177 have been shown. Thus, oxidative stress induced by chemotherapeutic interventions can aggravate metabolic reprogramming patterns in cancer cells by stabilization of HIF1-α and also possibly by PTMs, such as acetylation, and oxidation of metabolic enzymes, such as PKM2.
Transient oxidation of thiols in protein tyrosine phosphatases (PTPs) and other structurally related phosphatases, such as the tumor suppressor PTEN, leads to their reversible inactivation by the formation of either an intramolecular disulfide bridge or a sulfenyl-amide bond.173, 178, 179 Conversely, oxidation of some non-receptor protein tyrosine kinases leads to their activation, either by direct SH modification or indirectly by concomitant inhibition of PTPs that guides sustained activation of protein tyrosine kinases. PDK1 phosphorylates and inactivates PDH and consequently the pyruvate dehydrogenase complex.180 It is known that PDK1 is commonly tyrosine phosphorylated in human cancers by diverse oncogenic tyrosine kinases.181 Although it is known that oncogenic tyrosine kinases, including fibroblast growth factor receptor 1 (FGFR1), BCR-ABL and FLT3-ITD, can phosphorylate diverse metabolic enzymes such as PKM2, LDH-A and PDK1,159 whether and how oxidative stress can affect the phosphorylation of these enzymes are still not clear. Interestingly, direct oxidation of cysteine 488 of FGFR1 decreases its activity.182 FGFR1 phosphorylates tyrosine 10 and 83 of LDH-A and enhances LDH-A enzyme activity to promote the Warburg effect and tumor growth by regulating NADH/NAD(+) redox homeostasis.183 FGFR1 can also inhibit PKM2 by direct phosphorylation of PKM2 tyrosine 105.184
All of these findings suggest that changes in ROS levels can affect metabolic rewiring directly or indirectly and also that ROS modulation of metabolic enzymes can be another potential mechanism of resistance to chemotherapeutic approaches, which are not easily observable in vitro. Thus, the current strategy of cancer therapy, based on the long-held speculation of supposedly higher ROS levels in cancer cells, aimed at finding a possible therapeutic window that can be used to eradicate cancer cells rather than normal non-transformed cells appears to warrant further studies. The caveat is that the studies cited that form the basis of our understanding about metabolic reprogramming and cellular response to oxidative stress have been acquired primarily from cancer cell lines rather than from intact cancer tissues.185 Considering our ignorance regarding the role of ROS and anti-oxidant systems in different tumor microenvironments, the lack of reliable tools to evaluate in vivo levels of ROS, and the heterogeneity of tumor microenvironments comprised of cancer, stromal, endothelial, immune and even circulating cancer cells,105 the general administration of oxidative stress inducers for cancer therapy should be weighed carefully.
Conclusions
In conclusion, for the successful implementation of cancer therapy, potential therapeutic targets will need to be identified based on their roles in cancer metabolism coupled with cancer-specific expression/isoforms, potential mutations and oncogenic control mechanisms as well as based on the prediction of whether functional reversal of specific metabolic pathways will counteract the effects of ROS modulation and vice versa. The parameters that we need to clarify may include cancer type and staging, tissue type in which the cancer originated (the metabolic profile of tumors depends not only on the type of genetic lesion but also on the tissue in which the mutation arises186), duration and types of metabolic or ROS modulation, further genetic changes acquired from treatment, composition of the microenvironment, degree of metabolic synergy, the current lack of biomarkers to represent in vivo ROS levels, incomplete and confusing understanding of the role of ROS in cancer progression, in vivo validation of prior in vitro findings, identification of cellular targets of ROS and the competence of mitochondrial function in cancer cells.
A breakthrough will likely come from the advent of a technology that can detect in vivo redox status across a wide range of cancer cells at different stages as well as other non-transformed cells in the tumor microenvironment and from the development of small molecules that can locally target specific biochemical nodes. We expect the feasibility of combined approaches targeting ROS and metabolism for successful cancer therapy to become an exciting topic of study.
Acknowledgments
We apologize for not being able to cite the work of colleagues in many other important related studies because of space restrictions. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2014R1A1A1004682 and 2014R1A2A1A11049526).
The authors declare no conflict of interest.
References
- Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927; 8: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tan M, Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett 2015; 356: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA. Is cancer a metabolic disease? Am J Pathol 2014; 184: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel J. Targeting tumour metabolism. Nat Rev Drug Discov 2010; 9: 503–504. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Vander Heiden MG, Kroemer G. Metabolic targets for cancer therapy. Nat Rev Drug Discov 2013; 12: 829–846. [DOI] [PubMed] [Google Scholar]
- Jones NP, Schulze A. Targeting cancer metabolism–-aiming at a tumour's sweet-spot. Drug Discov Today 2012; 17: 232–241. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012; 491: 364–373. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011; 11: 325–337. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012; 12: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009; 8: 579–591. [DOI] [PubMed] [Google Scholar]
- Granja S, Pinheiro C, Reis RM, Martinho O, Baltazar F. Glucose addiction in cancer therapy: advances and drawbacks. Curr Drug Metab 2015; 16: 221–242. [DOI] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer 2013; 12: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis 2016; 7: e2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 2009; 417: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyev AY, Kushnareva YE, Starkov AA. Mitochondrial metabolism of reactive oxygen species. Biochemistry 2005; 70: 200–214. [DOI] [PubMed] [Google Scholar]
- Costa A, Scholer-Dahirel A, Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin Cancer Biol 2014; 25: 23–32. [DOI] [PubMed] [Google Scholar]
- Fiaschi T, Chiarugi P. Oxidative stress, tumor microenvironment, and metabolic reprogramming: a diabolic liaison. Int J Cell Biol 2012; 2012: 762825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafani M, Sansone L, Limana F, Arcangeli T, De Santis E, Polese M et al. The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid Med Cell Longev 2016; 2016: 3907147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchler MJ, Domann FE. Redox regulation of the epigenetic landscape in cancer: a role for metabolic reprogramming in remodeling the epigenome. Free Radical Biol Med 2012; 53: 2178–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Zhang H, Ma XF, Zhou X, Gan L, Liu YY et al. Isoliquiritigenin enhances radiosensitivity of HepG2 cells via disturbance of redox status. Cell Biol Biophys 2013; 65: 433–444. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11: 85–95. [DOI] [PubMed] [Google Scholar]
- Watson J. Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol 2013; 3: 120144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson TC, Zhong W, Oberley TD. Redox imbalance and biochemical changes in cancer. Cancer Res 2013; 73: 6118–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiswing L, Zhong W, Oberley TD. Increasing discordant antioxidant protein levels and enzymatic activities contribute to increasing redox imbalance observed during human prostate cancer progression. Free Radical Biol Med 2014; 67: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper ME, Antoniou A, Villalobos-Menuey E, Russo A, Trauger R, Vendemelio M et al. Characterization of a novel metabolic strategy used by drug-resistant tumor cells. FASEB J 2002; 16: 1550–1557. [DOI] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011; 17: 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan MM, Chi JT. Alternative fuels for cancer cells. Cancer J 2015; 21: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest 2013; 123: 3678–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A, Clem BF, Imbert-Fernandez Y, Ozcan SC, Peker S, O'Neal J et al. 6-Phosphofructo-2-kinase (PFKFB3) promotes cell cycle progression and suppresses apoptosis via Cdk1-mediated phosphorylation of p27. Cell Death Dis 2014; 5: e1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem BF, O'Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA 2nd et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther 2013; 12: 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 2013; 23: 316–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Jeon HM, Ju MK, Kim CH, Yoon G, Han SI et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res 2012; 72: 3607–3617. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010; 463: 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008; 452: 230–233. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol 2012; 8: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 2008; 452: 181–186. [DOI] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell 2011; 42: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011; 334: 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem Biol 2012; 19: 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci 2013; 13: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. NeuroOncology 2009; 11: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012; 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 2006; 3: 177–185. [DOI] [PubMed] [Google Scholar]
- Kluza J, Corazao-Rozas P, Touil Y, Jendoubi M, Maire C, Guerreschi P et al. Inactivation of the HIF-1alpha/PDK3 signaling axis drives melanoma toward mitochondrial oxidative metabolism and potentiates the therapeutic activity of pro-oxidants. Cancer Res 2012; 72: 5035–5047. [DOI] [PubMed] [Google Scholar]
- Lu CW, Lin SC, Chen KF, Lai YY, Tsai SJ. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J Biol Chem 2008; 283: 28106–28114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhu Y, Morinibu A, Kobayashi M, Shinomiya K, Itasaka S et al. HIF-1-mediated metabolic reprogramming reduces ROS levels and facilitates the metastatic colonization of cancers in lungs. Sci Rep 2014; 4: 3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonanez-Organis JG, Rodriguez-Armenta M, Leal-Rubio B, Peregrino-Uriarte AB, Gomez-Jimenez S, Yepiz-Plascencia G. Alternative splicing generates two lactate dehydrogenase subunits differentially expressed during hypoxia via HIF-1 in the shrimp Litopenaeus vannamei. Biochimie 2012; 94: 1250–1260. [DOI] [PubMed] [Google Scholar]
- Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J Biol Chem 1995; 270: 21021–21027. [DOI] [PubMed] [Google Scholar]
- Ullah MS, Davies AJ, Halestrap AP. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J Biol Chem 2006; 281: 9030–9037. [DOI] [PubMed] [Google Scholar]
- Pelicano H, Lu W, Zhou Y, Zhang W, Chen Z, Hu Y et al. Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14-mediated mechanism. Cancer Res 2009; 69: 2375–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello D, Balsa E, Acosta-Iborra B, Fuertes-Yebra E, Elorza A, Ordonez A et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1alpha decreases oxygen consumption by inhibiting complex I activity. Cell Metab 2011; 14: 768–779. [DOI] [PubMed] [Google Scholar]
- Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C et al. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol 2007; 27: 755–761. [DOI] [PubMed] [Google Scholar]
- Gorlach A, Diebold I, Schini-Kerth VB, Berchner-Pfannschmidt U, Roth U, Brandes RP et al. Thrombin activates the hypoxia-inducible factor-1 signaling pathway in vascular smooth muscle cells: Role of the p22(phox)-containing NADPH oxidase. Circ Res 2001; 89: 47–54. [DOI] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab 2013; 17: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun 2013; 4: 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O et al. p53 regulates mitochondrial respiration. Science 2006; 312: 1650–1653. [DOI] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res 2004; 64: 2627–2633. [DOI] [PubMed] [Google Scholar]
- Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res 2012; 72: 560–567. [DOI] [PubMed] [Google Scholar]
- Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol 2011; 13: 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimeni M, Voena C, Kopecka J, Riganti C, Pescarmona G, Bosia A et al. Modulation of doxorubicin resistance by the glucose-6-phosphate dehydrogenase activity. Biochem J 2011; 439: 141–149. [DOI] [PubMed] [Google Scholar]
- Won KY, Lim SJ, Kim GY, Kim YW, Han SA, Song JY et al. Regulatory role of p53 in cancer metabolism via SCO2 and TIGAR in human breast cancer. Hum Pathol 2012; 43: 221–228. [DOI] [PubMed] [Google Scholar]
- Dang CV. The interplay between MYC and HIF in the Warburg effect. Ernst Schering Found Symp Proc 2007, 35–53. [DOI] [PubMed]
- Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life 2013; 65: 904–910. [DOI] [PubMed] [Google Scholar]
- Shen L, O'Shea JM, Kaadige MR, Cunha S, Wilde BR, Cohen AL et al. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci USA 2015; 112: 5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 2011; 35: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CW, Ayer DE. An extended Myc network contributes to glucose homeostasis in cancer and diabetes. Front Biosci (Landmark Ed) 2011; 16: 2206–2223. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 2011; 10: 671–684. [DOI] [PubMed] [Google Scholar]
- Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer 2010; 10: 267–277. [DOI] [PubMed] [Google Scholar]
- Sborov DW, Haverkos BM, Harris PJ. Investigational cancer drugs targeting cell metabolism in clinical development. Expert Opin Investig Drugs 2015; 24: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L, Buhler L, Icard P, Lincet H, Steyaert JM. Metabolic treatment of cancer: intermediate results of a prospective case series. Anticancer Res 2014; 34: 973–980. [PubMed] [Google Scholar]
- Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene 2006; 25: 4633–4646. [DOI] [PubMed] [Google Scholar]
- Pedersen PL. Warburg, me and hexokinase 2: multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the ‘Warburg Effect', i.e., elevated glycolysis in the presence of oxygen. J Bioenerg Biomembr 2007; 39: 211–222. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Dai J, Jung Y, Wei CL, Wang Y et al. A glycolytic mechanism regulating an angiogenic switch in prostate cancer. Cancer Res 2007; 67: 149–159. [DOI] [PubMed] [Google Scholar]
- Zieker D, Konigsrainer I, Tritschler I, Loffler M, Beckert S, Traub F et al. Phosphoglycerate kinase 1 a promoting enzyme for peritoneal dissemination in gastric cancer. Int J Cancer 2010; 126: 1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 2010; 329: 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang SL, Hu X, Tam KY. Targeting tumor metabolism for cancer treatment: is pyruvate dehydrogenase kinases (PDKs) a viable anticancer target? Int J Biol Sci 2015; 11: 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 2005; 24: 6314–6322. [DOI] [PubMed] [Google Scholar]
- Chirala SS, Wakil SJ. Structure and function of animal fatty acid synthase. Lipids 2004; 39: 1045–1053. [DOI] [PubMed] [Google Scholar]
- Labar G, Wouters J, Lambert DM. A review on the monoacylglycerol lipase: at the interface between fat and endocannabinoid signalling. Curr Med Chem 2010; 17: 2588–2607. [DOI] [PubMed] [Google Scholar]
- Casals N, Zammit V, Herrero L, Fado R, Rodriguez-Rodriguez R, Serra D. Carnitine palmitoyltransferase 1C: From cognition to cancer. Prog Lipid Res 2016; 61: 134–148. [DOI] [PubMed] [Google Scholar]
- Langbein S, Zerilli M, Zur Hausen A, Staiger W, Rensch-Boschert K, Lukan N et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Brit J Cancer 2006; 94: 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, Larochette N et al. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene 2007; 26: 2606–2620. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA 2010; 107: 2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H et al. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther 2012; 11: 1672–1682. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest 2013; 123: 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Xie N, Banerjee S, Cui H, Fu M, Thannickal VJ et al. The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J Biol Chem 2015; 290: 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta 2011; 1807: 726–734. [DOI] [PubMed] [Google Scholar]
- Li J, Csibi A, Yang S, Hoffman GR, Li C, Zhang E et al. Synthetic lethality of combined glutaminase and Hsp90 inhibition in mTORC1-driven tumor cells. Proc Natl Acad Sci USA 2015; 112: E21–E29. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sharma PK, Bhardwaj R, Dwarakanath BS, Varshney R. Metabolic oxidative stress induced by a combination of 2-DG and 6-AN enhances radiation damage selectively in malignant cells via non-coordinated expression of antioxidant enzymes. Cancer Lett 2010; 295: 154–166. [DOI] [PubMed] [Google Scholar]
- Izaki S, Goto H, Yokota S. Increased chemosensitivity and elevated reactive oxygen species are mediated by glutathione reduction in glutamine deprived neuroblastoma cells. J Cancer Res Clin Oncol 2008; 134: 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero AJ, Jassem J. Cellular redox pathways as a therapeutic target in the treatment of cancer. Drugs 2011; 71: 1385–1396. [DOI] [PubMed] [Google Scholar]
- Klawitter J, Kominsky DJ, Brown JL, Klawitter J, Christians U, Leibfritz D et al. Metabolic characteristics of imatinib resistance in chronic myeloid leukaemia cells. Br J Pharmacol 2009; 158: 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluza J, Jendoubi M, Ballot C, Dammak A, Jonneaux A, Idziorek T et al. Exploiting mitochondrial dysfunction for effective elimination of imatinib-resistant leukemic cells. PLoS ONE 2011; 6: e21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Marignac VL, Smith S, Toban N, Bazile M, Aloyz R. Resistance to Dasatinib in primary chronic lymphocytic leukemia lymphocytes involves AMPK-mediated energetic re-programming. Oncotarget 2013; 4: 2550–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumarola C, Caffarra C, La Monica S, Galetti M, Alfieri RR, Cavazzoni A et al. Effects of sorafenib on energy metabolism in breast cancer cells: role of AMPK-mTORC1 signaling. Breast Cancer Res Treat 2013; 141: 67–78. [DOI] [PubMed] [Google Scholar]
- Lu H, Li X, Luo Z, Liu J, Fan Z. Cetuximab reverses the Warburg effect by inhibiting HIF-1-regulated LDH-A. Mol Cancer Ther 2013; 12: 2187–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwich J, Orr WS, Ng CY, Spence Y, Morton C, Davidoff AM. HIF-1 alpha activation mediates resistance to anti-angiogenic therapy in neuroblastoma xenografts. J Pediatr Surg 2013; 48: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 2012; 13: 270–276. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab 2011; 14: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, O'Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 2014; 16: 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Bae JS. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediat Inflamm 2016; 2016: 6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotgia F, Martinez-Outschoorn UE, Lisanti MP. Cancer metabolism: new validated targets for drug discovery. Oncotarget 2013; 4: 1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin Oncol 2014; 41: 195–216. [DOI] [PubMed] [Google Scholar]
- Echigoya Y, Morita S, Itou T, Sakai T. Effects of extracellular lactate on production of reactive oxygen species by equine polymorphonuclear leukocytes in vitro. Am J Vet Res 2012; 73: 1290–1298. [DOI] [PubMed] [Google Scholar]
- Milovanova TN, Bhopale VM, Sorokina EM, Moore JS, Hunt TK, Hauer-Jensen M et al. Lactate stimulates vasculogenic stem cells via the thioredoxin system and engages an autocrine activation loop involving hypoxia-inducible factor 1. Mol Cell Biol 2008; 28: 6248–6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L, Chuang CC, Wu S, Zuo L. Reactive oxygen species in redox cancer therapy. Cancer Lett 2015; 367: 18–25. [DOI] [PubMed] [Google Scholar]
- Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res 2010; 44: 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Rathmell JC. The metabolic life and times of a T-cell. Immunol Rev 2010; 236: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman BJ, Dang CV. Normal and cancer cell metabolism: lymphocytes and lymphoma. FEBS J 2012; 279: 2598–2609. [DOI] [PubMed] [Google Scholar]
- Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res 2010; 316: 1324–1331. [DOI] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 2008; 118: 3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hee VF, Perez-Escuredo J, Cacace A, Copetti T, Sonveaux P. Lactate does not activate NF-kappaB in oxidative tumor cells. Front Pharmacol 2015; 6: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlides S, Whitaker-Menezes D, Castello-Cros R, Flomenberg N, Witkiewicz AK, Frank PG et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle 2009; 8: 3984–4001. [DOI] [PubMed] [Google Scholar]
- Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol 2008; 18: 165–173. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Li SH, Park HS, Park JW, Lee B, Chun YS. Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J BiolChem 2011; 286: 6963–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Trachootham D, Liu J, Chen G, Pelicano H, Garcia-Prieto C et al. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat Cell Biol 2012; 14: 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Rovira II, Finkel T. Oxidants painting the cysteine chapel: redox regulation of PTPs. Dev Cell 2002; 2: 251–252. [DOI] [PubMed] [Google Scholar]
- Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 1995; 270: 296–299. [DOI] [PubMed] [Google Scholar]
- Hayman SR, Leung N, Grande JP, Garovic VD. VEGF inhibition, hypertension, and renal toxicity. Curr Oncol Rep 2012; 14: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inai T, Mancuso M, Hashizume H, Baffert F, Haskell A, Baluk P et al. Inhibition of vascular endothelial growth factor (VEGF) signaling in cancer causes loss of endothelial fenestrations, regression of tumor vessels, and appearance of basement membrane ghosts. Am J Pathol 2004; 165: 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti J, Loges S, Dimmeler S, Carmeliet P. Recent molecular discoveries in angiogenesis and antiangiogenic therapies in cancer. J Clin Invest 2013; 123: 3190–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Introduction to cerebral cavernous malformation: a brief review. BMB Rep 2016; 49: 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 2013; 154: 651–663. [DOI] [PubMed] [Google Scholar]
- Goveia J, Stapor P, Carmeliet P. Principles of targeting endothelial cell metabolism to treat angiogenesis and endothelial cell dysfunction in disease. EMBO Mol Med 2014; 6: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med 2010; 48: 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel A, Jarmuszkiewicz W. Aerobic metabolism and reactive oxygen species in endothelial cells. Postepy Biochem 2013; 59: 386–394. [PubMed] [Google Scholar]
- Del Bufalo D, Trisciuoglio D, Scarsella M, D'Amati G, Candiloro A, Iervolino A et al. Lonidamine causes inhibition of angiogenesis-related endothelial cell functions. Neoplasia 2004; 6: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoors S, De Bock K, Cantelmo AR, Georgiadou M, Ghesquiere B, Cauwenberghs S et al. Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metab 2014; 19: 37–48. [DOI] [PubMed] [Google Scholar]
- Sonveaux P, Copetti T, De Saedeleer CJ, Vegran F, Verrax J, Kennedy KM et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS ONE 2012; 7: e33418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-kappaB/IL-8 pathway that drives tumor angiogenesis. Cancer Res 2011; 71: 2550–2560. [DOI] [PubMed] [Google Scholar]
- Xu Y, An X, Guo X, Habtetsion TG, Wang Y, Xu X et al. Endothelial PFKFB3 plays a critical role in angiogenesis. Arterioscler Thromb Vasc Biol 2014; 34: 1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayed SM, Mahmoud AA, El Sawy SA, Abdelaal EA, Fouad AM, Yousif RS et al. Warburg effect increases steady-state ROS condition in cancer cells through decreasing their antioxidant capacities (anticancer effects of 3-bromopyruvate through antagonizing Warburg effect). Med Hypotheses 2013; 81: 866–870. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 2013; 12: 931–947. [DOI] [PubMed] [Google Scholar]
- Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Sci Transl Med 2014; 6: 221ra15. [DOI] [PubMed] [Google Scholar]
- Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat Rev Cancer 2014; 14: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EA, Thompson IM Jr, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ et al. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 2011; 306: 1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab 2007; 6: 280–293. [DOI] [PubMed] [Google Scholar]
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med 2011; 51: 327–336. [DOI] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp Gerontol 2010; 45: 410–418. [DOI] [PubMed] [Google Scholar]
- Glasauer A, Chandel NS. Targeting antioxidants for cancer therapy. Biochem Pharmacol 2014; 92: 90–101. [DOI] [PubMed] [Google Scholar]
- Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov 2010; 9: 447–464. [DOI] [PubMed] [Google Scholar]
- Song IS, Jeong JY, Jeong SH, Kim HK, Ko KS, Rhee BD et al. Mitochondria as therapeutic targets for cancer stem cells. World J Stem Cells 2015; 7: 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YC, Ou DL, Hsu C, Lin KL, Chang CY, Lin CY et al. Activating oxidative phosphorylation by a pyruvate dehydrogenase kinase inhibitor overcomes sorafenib resistance of hepatocellular carcinoma. Br J Cancer 2013; 108: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros S, Schulze A. Glycolysis back in the limelight: systemic targeting of HK2 blocks tumor growth. Cancer Discov 2013; 3: 1105–1107. [DOI] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 2014; 510: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 2014; 55: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Milito A, Fais S. Tumor acidity, chemoresistance and proton pump inhibitors. Future Oncol 2005; 1: 779–786. [DOI] [PubMed] [Google Scholar]
- Backos DS, Franklin CC, Reigan P. The role of glutathione in brain tumor drug resistance. Biochem Pharmacol 2012; 83: 1005–1012. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011; 475: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin B, Tonks NK. PTP1B: mediating ROS signaling to silence genes. Mol Cell Oncol 2015; 2: e975633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak DW, Weerapana E. Cysteine-mediated redox signalling in the mitochondria. Mol Biosyst 2015; 11: 678–697. [DOI] [PubMed] [Google Scholar]
- Miki H, Funato Y. Regulation of intracellular signalling through cysteine oxidation by reactive oxygen species. J Biochem 2012; 151: 255–261. [DOI] [PubMed] [Google Scholar]
- Lizama-Manibusan B, McLaughlin B. Redox modification of proteins as essential mediators of CNS autophagy and mitophagy. FEBS Lett 2013; 587: 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Dalle-Donne I. Redox proteomics. Antioxid Redox Signal 2012; 17: 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Chen J. Post-translational modifications and the Warburg effect. Oncogene 2014; 33: 4279–4285. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010; 327: 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Li FL, Cheng ZL, Lei QY. Impact of acetylation on tumor metabolism. Mol Cell Oncol 2014; 1: e963452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Matthes A, Koszior S, Lehmann M, Araujo WL, Bock R et al. Alteration of mitochondrial protein complexes in relation to metabolic regulation under short-term oxidative stress in Arabidopsis seedlings. Phytochemistry 2011; 72: 1081–1091. [DOI] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd et al. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012; 337: 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushio-Fukai M, Alexander RW, Akers M, Yin Q, Fujio Y, Walsh K et al. Reactive oxygen species mediate the activation of Akt/protein kinase B by angiotensin II in vascular smooth muscle cells. J Biol Chem 1999; 274: 22699–22704. [DOI] [PubMed] [Google Scholar]
- Okoh VO, Felty Q, Parkash J, Poppiti R, Roy D. Reactive oxygen species via redox signaling to PI3K/AKT pathway contribute to the malignant growth of 4-hydroxy estradiol-transformed mammary epithelial cells. PLoS ONE 2013; 8: e54206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem 1997; 272: 17269–17275. [DOI] [PubMed] [Google Scholar]
- Agbor TA, Cheong A, Comerford KM, Scholz CC, Bruning U, Clarke A et al. Small ubiquitin-related modifier (SUMO)-1 promotes glycolysis in hypoxia. J Biol Chem 2011; 286: 4718–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgino F, de Robertis O, Laviola L, Montrone C, Perrini S, McCowen KC et al. The sentrin-conjugating enzyme mUbc9 interacts with GLUT4 and GLUT1 glucose transporters and regulates transporter levels in skeletal muscle cells. Proc Natl Acad Sci USA 2000; 97: 1125–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti VS, Vergarajauregui S, Pulido D, Sandoval IV. The insulin-sensitive glucose transporter, GLUT4, interacts physically with Daxx. Two proteins with capacity to bind Ubc9 and conjugated to SUMO1. J Biol Chem 2002; 277: 19783–19791. [DOI] [PubMed] [Google Scholar]
- Lushchak OV, Piroddi M, Galli F, Lushchak VI. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep 2014; 19: 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux RJ, Jin X, Willmore WG. Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biol 2014; 2: 123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go YM, Chandler JD, Jones DP. The cysteine proteome. Free Radic Biol Med 2015; 84: 227–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell 2005; 121: 667–670. [DOI] [PubMed] [Google Scholar]
- Yang M, Haase AD, Huang FK, Coulis G, Rivera KD, Dickinson BC et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Mol Cell 2014; 55: 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank GD, Mifune M, Inagami T, Ohba M, Sasaki T, Higashiyama S et al. Distinct mechanisms of receptor and nonreceptor tyrosine kinase activation by reactive oxygen species in vascular smooth muscle cells: role of metalloprotease and protein kinase C-delta. Mol Cell Biol 2003; 23: 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi MZ, Azar ZM, Srivastava AK. Role of receptor and nonreceptor protein tyrosine kinases in H2O2-induced PKB and ERK1/2 signaling. Cell Biochem Biophys 2007; 47: 1–10. [DOI] [PubMed] [Google Scholar]
- Nilsson MB, Zage PE, Zeng L, Xu L, Cascone T, Wu HK et al. Multiple receptor tyrosine kinases regulate HIF-1alpha and HIF-2alpha in normoxia and hypoxia in neuroblastoma: implications for antiangiogenic mechanisms of multikinase inhibitors. Oncogene 2010; 29: 2938–2949. [DOI] [PubMed] [Google Scholar]
- Chiarugi P. PTPs versus PTKs: the redox side of the coin. Free Radic Res 2005; 39: 353–364. [DOI] [PubMed] [Google Scholar]
- Ostman A, Frijhoff J, Sandin A, Bohmer FD. Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem 2011; 150: 345–356. [DOI] [PubMed] [Google Scholar]
- Patel MS, Korotchkina LG. Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases. Exp Mol Med 2001; 33: 191–197. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell 2011; 44: 864–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble DJ, Sun G. Direct and specific inactivation of protein tyrosine kinases in the Src and FGFR families by reversible cysteine oxidation. Proc Natl Acad USA 2009; 106: 5070–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hitosugi T, Chung TW, Xie J, Ge Q, Gu TL et al. Tyrosine phosphorylation of lactate dehydrogenase A is important for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol 2011; 31: 4938–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2009; 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016; 2: e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 2012; 15: 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]