Abstract
TFAP2C (transcription factor-activating enhancer-binding protein 2C) expression has been positively correlated with poor prognosis in patients with certain types of cancer, but the mechanisms underlying TFAP2C-mediated tumorigenesis in non-small-cell lung cancer (NSCLC) are still unknown. We previously performed a microarray analysis to identify TFAP2C regulation genes, and TGFBR1 (transforming growth factor-β receptor type 1) was found to be upregulated by TFAP2C. We observed that TFAP2C or TGFBR1 overexpression led to oncogenic properties, such as cell viability, proliferation and cell cycle progression. TGFBR1 upregulation induced by TFAP2C also promoted cell motility and migration, leading to malignant development. We also found that PAK1 (p21 protein (Cdc42/Rac)-activated kinase 1) signaling was involved in TFAP2C/TGFBR1-induced tumorigenesis. These results were confirmed by an in vivo xenograft model and patient tissue samples. This study shows that TFAP2C promoted tumor progression by upregulation of TGFBR1 and consequent activation of PAK1 signaling.
Introduction
Elucidating the molecular mechanisms underlying lung tumorigenesis is imperative for efficient treatment, prevention and early diagnosis of lung cancer. Non-small-cell lung cancer (NSCLC) accounts for ~80% of lung cancer cases, and is associated with tumor aggressiveness and poor prognosis. Accumulation of several mutations in genes encoding oncogenes and tumor suppressor genes is known to be a major factor in NSCLC tumorigenesis. Many studies have reported that EGFR and KRAS are mutated in ~20–30% of NSCLC patients.1 To date, EGFR-targeting strategies, such as pharmacological inhibition, have been tested. However, the outcomes of these trials have not been satisfactory because of the occurrence of chemoresistance and activation of other survival signaling pathways to compensate for the EGFR inhibition.2, 3 Therefore, it is essential to elucidate other signaling mechanisms of NSCLC tumorigenesis to enhance the therapeutic efficacy.
TFAP2C (transcription factor-activating enhancer-binding protein 2C) is a member of the transcription factor AP-2 family and regulates the expression of multiple genes. TFAP2C has GC-rich binding activity and controls the transcription of a large number of genes, leading to differentiation, proliferation and cell death.4, 5 TFAP2C has been reported to regulate cancer-associated gene expression.6, 7 Several breast cancer patients showed gene amplification of TFAP2C,8 indicating that TFAP2C has a significant role in cancer development. However, little is known about the role of TFAP2C in lung tumorigenesis.
Transforming growth factor-β (TGF-β) receptor type 1 (TGFBR1) is a receptor for TGF-β ligands. When TGF-β ligands are present, they first bind to TGF-β receptor type 2 (TGFBR2), and TGFBR1 is recruited to TGFBR2 and activated.9, 10 TGFBR1 consequently activates Smad signaling and nuclear localization of the Smad complex to regulate gene transcription. In addition to the canonical Smad pathway, TGFBR1 can activate other types of signaling via noncanonical pathways, including the PAK1 (p21 protein (Cdc42/Rac)-activated kinase 1), MAPK (mitogen-activated protein kinase), PI3K (phosphatidylinositol-3-kinase) and AKT (a serine/threonine kinase) pathways.11, 12, 13, 14 Following stimulation with TGF-β ligands, cells undergo malignant transformation, metastatic conversion, angiogenesis and cancer progression through the interplay and cross-talk of TGFBR1 signal mediators.15, 16, 17
In this study, we found that TFAP2C-dependent expression of TGFBR1 was associated with cell proliferation and cell cycle progression. Interestingly, TGFBR1 upregulation led to epithelial–mesenchymal transition (EMT) induction through activation of PAK1 signaling in NSCLC cells. Our results identified a novel mechanism of TFAP2C-mediated lung tumorigenesis.
Materials and methods
Chemicals, antibodies and reagents
Antibodies specific for TFAP2C, TGFBR1, tubulin, E-cadherin, vimentin, fibronectin, Snail, p-Snail, Smad3 and p-Samd3 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies specific for PAK1, p-PAK1, MEK, p-MEK1, ERK and p-ERK were obtained from Cell Signaling Technology (Beverly, MA, USA). Cell culture media (RPMI-1640 and Dulbecco's modified Eagle's medium), fetal bovine serum (FBS), glutamine, penicillin and streptomycin were acquired from Gibco (Grand Island, NY, USA). Control small interfering RNA (siRNA) and siRNAs specific for TFAP2C #1 and TGFBR1 (ON-TARGETplus SMARTpool) were obtained from Dharmacon (Chicago, IL, USA) and siRNA specific for TFAP2C #2 (AccuTarget siRNA) was obtained from Bioneer (Daejeon, Republic of Korea).
Cell lines, cell culture and irradiation
The human NSCLC cell lines, NCI-H292 and NCI-H838, and the human normal lung cell lines, MRC5 and WI-26 VA4, were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA), authenticated and maintained in early passages, no more than 6 months after receipt from ATCC. NCI-H292, NCI-H838, MRC5 or WI-26 VA4 cells were grown in RPMI-1640 or Dulbecco's modified Eagle's medium supplemented with 10% FBS, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin at 37 °C in a 5% CO2/95% air atmosphere. Cells were exposed to a single dose of γ-rays using a Gamma Cell 40 Exactor (Nordion International, Kanata, ON, Canada) at a dose of 0.81 Gy min−1. Flasks containing control cells were placed in the irradiation chamber, but not exposed to radiation.
Real-time quantitative RT-PCR
mRNA levels were determined by real-time quantitative RT-PCR (qRT-PCR), as described previously.18 After the indicated treatments were applied to the cells, including siRNA treatment and/or gene overexpression, total RNA was prepared using TRIzol (Gibco, Grand Island, NY, USA). cDNA was generated using an MMLV reverse transcription system (Promega, Madison, WI, USA) according to the manufacturer's protocols. Aliquots of a master mix containing all reaction components and primers (Table 1) were dispensed into a real-time PCR plate (Applied Biosystems, Foster City, CA, USA). All PCR reagents were obtained from a SYBR Green Core Reagent Kit (Applied Biosystems). Gene mRNA levels were measured in triplicate in the reaction plate. qRT-PCR was performed using a Applied Biosystems-7900 HT qRT-PCR Instrument (Applied Biosystems). PCR was performed over 40 cycles of 15 s at 95 °C and 1 min at 60 °C, after which samples were subjected to thermal denaturation. Gene expression levels were normalized to GAPDH mRNA levels using the 2−ΔΔCT method.19 To simplify data presentation, relative expression values were multiplied by 102.
Table 1. Primers for determining expression levels of TFAP2C, TGFBR1 and EMT-related genes.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| TFAP2C | 5′-ACAGGATCCATGTTGTGGAAAATAACCGAT-3′ | 5′-ATACTCGAGTTTCCTGTGTTTCTCCATTTT-3′ |
| TGFBR1 | 5′-TCAGCTCTGGTTGGTGTCAG-3′ | 5′-ATGTGAAGATGGGCAAGACC-3′ |
| GAPDH | 5′-ATGACATCAAGAAGGTGGTG-3′ | 5′-CATACCAGGAAATGAGCTTG-3′ |
| CDH1 | 5′-GGATTGCAAATTCCTGCCATTC-3′ | 5′-AACGTTGTCCCGGGTGTCA-3′ |
| FN1 | 5′-TGACCTTTTCTGGCTCGTCT-3′ | 5′-GTTCAGCACAAAGGGCTCTC-3′ |
| VIM | 5′-GACAATGCGTCTCTGGCACGTCTT-3′ | 5′-TCCTCCGCCTCCTGCAGGTTCTT-3′ |
Abbreviations: CDH1, E-cadherin; EMT, epithelial–mesenchymal transition; FN1, fibronectin; TFAP2C, transcription factor-activating enhancer-binding protein 2C; TGFBR1, transforming growth factor-β receptor type 1; VIM, vimentin.
Western blot analysis
After the experimental treatments, western blotting was performed as described previously.20 Whole-cell lysates were prepared using RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 25 mM NaF, 1 mM dithiothreitol, 20 mM EGTA, 1 mM Na3VO4, 0.3 mM phenylmethanesulfonyl fluoride and 5 U ml−1 aprotinin) and protein concentrations in the lysates were determined using a BioRad Protein Assay Kit (BioRad Laboratories, Hercules, CA, USA). Protein samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane that was subsequently blocked with 5% skim milk in TBST (10 mM Tris, 100 mM NaCl and 0.1% Tween-20) for 1 h at room temperature. Next, membranes were probed with specific primary antibodies and peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Blots were visualized using an ECL Detection System (Roche Applied Science, Indianapolis, IN, USA), and densitometric analysis was performed using the Scion Image software (Scion Corporation, Frederick, MD, USA).
Colony-forming assay
A colony-forming assay was performed as described previously.21 Briefly, cells were plated at a density of 400 cells per well in 35 mm dishes. After 24 h, the cells were treated with the indicated siRNAs or genes and subsequently grown for 14 days. Next, cells were fixed with 10% methanol/10% acetic acid and then stained with 1% crystal violet. Colonies containing more than 50 cells were identified using densitometry software and scored as survivors.
Cell viability assay
Cell viability was assessed by measuring ATP levels using a CellTiter-Glo Assay Kit (Promega) according to the manufacturer's protocols. Briefly, treated cells (104 cells per ml) in 100 μl of culture medium were transferred to a 96-well microplate. Subsequently, 100 μl of CellTiter-Glo reagent (Promega) was added to each well. After the contents of the wells were gently mixed at 300–500 r.p.m. for 2 min, the plate was incubated at room temperature for 10 min. The luminescence of each sample was measured using a Glomax Multidetection System (Promega).
Cell cycle analysis
After each treatment, the cells were washed with phosphate-buffered saline (pH 7.4), collected by trypsinization, fixed with ice-cold 70% ethanol, washed with phosphate-buffered saline and stained with 10 mg ml−1 propidium iodide (Sigma, St Louis, MO, USA) containing 100 mg ml−1 RNase (Sigma) for 30 min at 37 °C. Cell cycle analysis was performed using a FACSVerse Flow Cytometer (BD Biosciences, San Jose, CA, USA). At least 10 000 cells were analyzed per sample and cell cycle distributions were obtained using BD FACSuite software (BD Biosciences).
Cell assay (3D culture) and immunofluorescence staining
Three-dimensional (3D) acini cultures and immunofluorescence staining were conducted as described previously22 to detect morphologic changes in the epithelial acini. Briefly, Matrigel (BD Biosciences) was thawed overnight at 4 °C and mixed well using prechilled pipette tips before use. Cells were cultured on eight-well chambered glass slides (Nunc, Naperville, IL, USA) containing 100% growth factor-reduced Matrigel at 37 °C for at least 30 min. Separately, NCI-H292 cells were harvested, counted and resuspended as single-cell suspensions of 25 000 cells per ml in RPMI medium. Aliquots (200 μl) of these cell suspensions were mixed with 200 μl of RPMI medium containing 4% Matrigel. These samples were placed in the wells of glass slides. The cells were then incubated and attached in a 37 °C, 5% CO2 incubator for 3 days. For transfection, media were exchanged for 400 μl of RPMI containing 4% Matrigel with the indicated siRNA and/or gene constructs, after which cells were incubated for 24 h. Cells were then exposed to a single dose of γ-rays (4 Gy), and further incubated for 24 h. For immunofluorescence staining studies, cells and acini were fixed with 2% paraformaldehyde for 20 min, permeabilized in 0.5% Triton X-100 for 10 min, washed three times with phosphate-buffered saline and blocked in immunofluorescence buffer (phosphate-buffered saline, 0.1% bovine serum albumin, 0.2% Triton X-100 and 0.05% Tween-20) containing 10% FBS at 37 °C for 30 min. Cells and acini were stained with primary antibody against tubulin overnight at 4 °C and then washed three times with immunofluorescence buffer. After being incubated with DyLight 488-conjugated secondary antibodies (Thermo Scientific, Rockford, IL, USA) and counterstained with 4′,6-diamidino-2-phenylindole, slides were mounted with VECTASHIELD Hard-Set Mounting Medium (Vector Laboratories, Burlingame, CA, USA). Fluorescence images were visualized using an Olympus IX71 fluorescence microscope (Olympus Optical Co Ltd, Tokyo, Japan).
Wound-healing assay
The effects of TFAP2C and TGFBR1 on cell motility were determined using a wound-healing assay, as described previously.23 After cells with the indicated treatments had reached 70% confluency in RPMI-1640 medium supplemented with 1% FBS, the monolayers were scratched using a 200 μl pipette tip. Cells were then further incubated for 24 or 48 h. Photomicrographs were taken at × 100 using an Olympus IX71 inverted microscope (Olympus Optical Co Ltd).
Transwell cell migration assay
The effects of TFAP2C and TGFBR1 on cell migration were investigated using Transwell cell migration assays, as described previously.24 Briefly, cells (1 × 104 in serum-free RPMI-1640 medium) subjected to the indicated treatments for 72 h were seeded into the upper chambers of a 24-well Transwell chamber (Corning, Corning, NY, USA) fitted with a 5 μm pore-size insert. The lower chamber was filled with RPMI-1640 medium containing 2% FBS. After 6 h, the upper membrane surface was wiped with a cotton swab to remove cells that had not migrated to the lower chamber. Cells that had migrated to the lower membrane surface were fixed with 4% paraformaldehyde, stained with hematoxylin and counted. Migration indices were calculated and normalized to the number of untreated cells that had migrated. The results are expressed as fold increases in migration compared with non-treated controls as determined by relative numbers of cells in a randomly selected field in experiments conducted in triplicate.
Tumor xenografts in nude mice
The effects of TFAP2C and TGFBR1 on tumor development were analyzed using a xenograft mouse model as described previously.25 Six-week-old male BALB/c athymic nude mice (Central Lab Animals, Seoul, South Korea) were used for the in vivo experiments. The animal protocols were approved by the Institutional Animal Care and Use Committee of Pusan National University (Busan, South Korea), and performed in accordance with the provisions of the NIH Guide for the Care and Use of Laboratory Animals. Mice were housed individually or in groups of up to five in sterile cages. Animals were maintained in animal care facilities in a temperature-regulated room (23±1 °C) under a 12 h light–dark cycle and quarantined for 1 week before the study. The animals were fed water and a standard mouse chow diet ad libitum. The mice (n=5 per group) were injected with NCI-H292 cells transfected with scrambled shRNA, TFAP2C shRNA, TGFBR1 shRNA and LV-TGFBR1 in a flank and tumors were allowed to develop. To produce sufficient lentiviral particles to transfect NSCLC cells, we used the Lenti-vpak Packaging Kit (Origene, Rockville, MD, USA). Tumor length (L) and width (W) were measured using a caliper, and tumor volumes were calculated using the formula (L × W2)/2. At the end of the treatment period, animals were killed and the tumors were used for biochemical studies.
Patient tissue samples
Specimens and data used in this study, which included frozen tissue samples from six NSCLC patients and paired normal adjacent tissues (Table 2), were provided by the Biobank of Pusan National University Hospital (Busan, South Korea) and a member of the Korea Biobank Network. The use of these archival tissues for gene expression analysis and immunohistochemistry was approved by the Ethics Committee of Pusan National University.
Table 2. Patient characteristics.
| Characteristics | No. of patients (n=6) |
|---|---|
| Sex | |
| Male | 3 |
| Female | 3 |
| Age (year) | |
| Median | 63 |
| Range | 55–78 |
| Histology | |
| Adenocarcinoma | 3 |
| Squamous cell carcinoma | 3 |
Immunohistochemistry
Expression levels of TFAP2C, TGFBR1, PAK1 and pPAK1 in lung cancer patients were analyzed by immunohistochemistry (IHC) as described previously.26 Frozen patient tumor/normal lung tissues were fixed in formalin, dehydrated and embedded in paraffin blocks, which were then sectioned at 4 μm. The sections were subsequently incubated in 3% hydrogen peroxide/methanol and then in 0.25% pepsin (Dako, Carpinteria, CA, USA) to retrieve the antigens. Next, the samples were blocked in a blocking solution (Dako), incubated at 4 °C overnight with primary antibodies, washed with TBST and incubated with polymer-horse radish peroxidase-conjugated secondary antibody (Dako). A 3,3′-diaminobenzidine substrate chromogen system (Dako) was then used to detect antibody binding, and stained sections were examined under an Olympus IX71 inverted microscope (Olympus Optical Co. Ltd).
Statistical analysis
All numeric data are presented as the mean±s.e.m. of at least three independent experiments. Experimental results were analyzed by Student's t-test, and one-way analysis of variance (ANOVA) followed by Tukey's honestly significant difference test. Prism 5 software (GraphPad Software, San Diego, CA, USA) was used to conduct the statistical analysis, and P-values <0.05 were considered significant.
Results
TFAP2C induces TGFBR1 expression in NSCLC cells
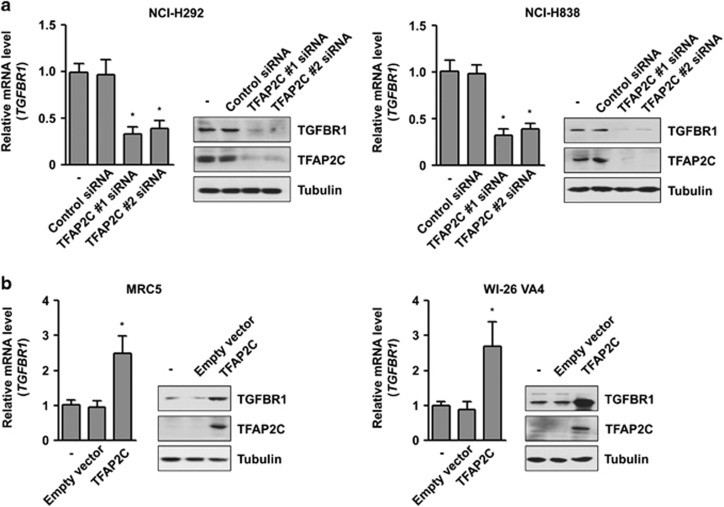
We previously investigated the tumorigenic role of TFAP2C in lung cancer, and the microarray data for transcriptional targets of TFAP2C were deposited in the Gene Expression Omnibus database (GEO Series accession number GSE79228; unpublished data). Among various genes, we focused on TGFBR1 as a TFAP2C target gene involved in lung tumorigenesis and malignancy based on the oncogenic properties of TGFBR1.15, 16, 17 To confirm the microarray data, we measured the expression levels of TGFBR1 in the NSCLC cell lines NCI-H292 and NCI-H838. After TFAP2C knockdown by treatment with TFAP2C-specific siRNA, both the mRNA and protein levels of TGFBR1 were decreased in NCI-H292 and NCI-H838 cells (Figure 1a). To confirm the relationship between TFAP2C and TGFBR1 in normal cells, we measured the expression levels of TGFBR1 in MRC5 and WI-26 VA4 cells, which are normal lung fibroblasts. When TFAP2C was overexpressed by transient transfection in MRC5 cells, the mRNA and protein levels of TGFBR1 were increased (Figure 1b). These results suggested that the expression of TGFBR1 is regulated by TFAP2C in NSCLC cells. NCI-H292 and MRC5 cells were used for subsequent experiments.
Figure 1.
TGFBR1 (transforming growth factor-β receptor type 1) was upregulated by TFAP2C (transcription factor-activating enhancer-binding protein 2C) in non-small-cell lung cancer (NSCLC) cells. (a) The effects of TFAP2C knockdown by treatment of TFAP2C small interfering RNA (siRNA) (#1 and #2) on the expression of TGFBR1 mRNA and protein in NCI-H292 and NCI-H838 cells were analyzed by real-time quantitative RT-PCR (qRT-PCR) and western blotting, respectively. Based on these results, TFAP2C siRNA no 1 was used for the subsequent experiments. (b) The effects of TFAP2C upregulation on the expression of TGFBR1 mRNA and protein in MRC5 and WI-26 VA4 cells were analyzed by real-time qRT-PCR and western blotting, respectively. *P<0.05 compared with control cells. Error bars represent±s.e.m. (n=3); Student's t-test.
TGFBR1 upregulation by TFAP2C promotes NSCLC tumorigenesis
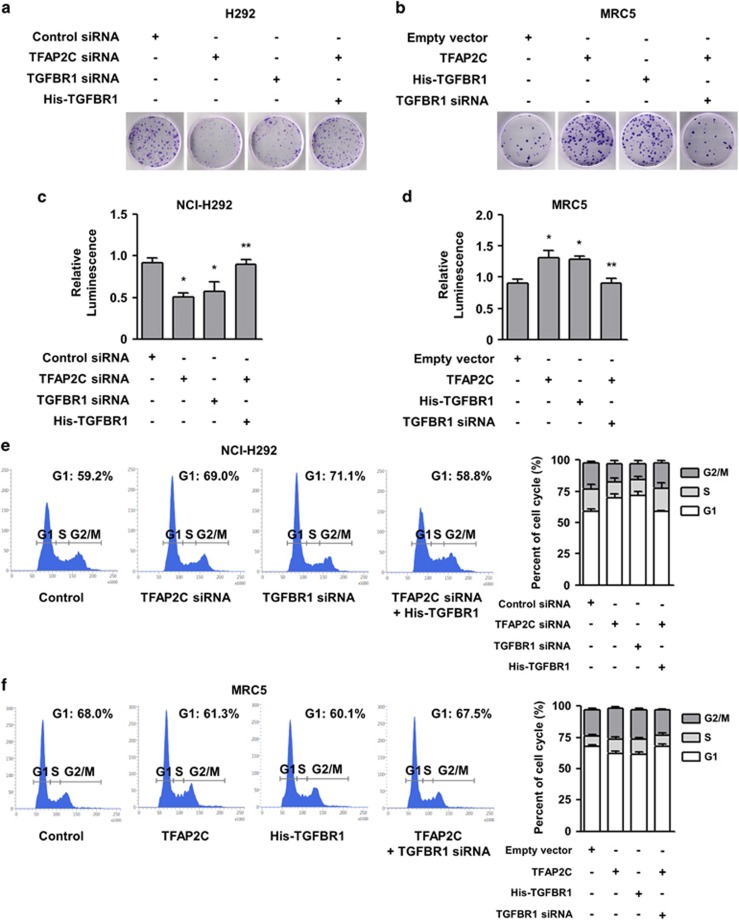
We next investigated whether TGFBR1 expression contributes to the proliferation of NSCLC cells in lung tumorigenesis. We conducted a colony-forming assay to observe the long-term effects of TGFBR1 knockdown on NSCLC cell proliferation. TFAP2C or TGFBR1 knockdown significantly suppressed the colony-forming ability of NCI-H292 cells (Figure 2a). We observed that the decreased colony numbers of cells with TFAP2C knockdown could be substantially recovered by overexpression of TGFBR1. In addition, ectopic expression of TFAP2C or TGFBR1 increased the number of colonies in MRC5 cells, and the colony number increase of cells overexpressing TFAP2C could be significantly reduced by further treatment with TGFBR1 siRNA (Figure 2b). To determine the short-term effects of TGFBR1 knockdown on NSCLC cell viability and proliferation, we conducted a cell viability assay that measured ATP levels produced in live cells.27 TFAP2C or TGFBR1 knockdown in NCI-H292 cells reduced ATP levels, indicating decreased cell viability (Figure 2c). However, the decreased viability of NCI-H292 cells treated with TFAP2C siRNA was rescued by further overexpression of TGFBR1, which is consistent with the results shown in Figure 2a. In MRC5 cells, we also observed that the overexpression of TFAP2C or TGFBR1 increased the ATP levels and that the increased viability of TFAP2C-overexpressing cells was further decreased by TGFBR1 knockdown (Figure 2d). We next determined whether TGFBR1-mediated regulation of cell viability and proliferation influenced cell cycle progression, which is a tumorigenic feature. As shown in Figure 2e, cell cycle arrest in the G1 phase was detected in NCI-H292 cells treated with TFAP2C or TGFBR1 siRNA (69.0% or 71.1%, respectively) compared with mock-transfected cells (59.2%). The G1 arrest induced by TFAP2C knockdown was inhibited by further overexpression of TGFBR1 (58.8%). In MRC5 cells, the G1 arrest was reduced in cells overexpressing TFAP2C or TGFBR1 (61.3% or 60.1%, respectively) compared with mock-transfected cells (68.0%), and further treatment with TGFBR1 siRNA resulted in increased G1 arrest in cells overexpressing TFAP2C (67.5%) (Figure 2f). Overall, these findings demonstrated that increased expression of TGFBR1 induced by TFAP2C is positively correlated with tumorigenic properties, including cell cycle progression and increased cell viability and proliferation in the lungs.
Figure 2.
TFAP2C (transcription factor-activating enhancer-binding protein 2C)-induced TGFBR1 (transforming growth factor-β receptor type 1) upregulation promotes non-small-cell lung cancer (NSCLC) tumorigenesis. (a) The effects of TFAP2C or TGFBR1 knockdown on the proliferation of NCI-H292 cells were measured by a colony-forming assay. (b) The effects of TFAP2C or TGFBR1 overexpression on the proliferation of MRC5 cells were measured by a colony-forming assay. (c) The effects of TFAP2C or TGFBR1 knockdown on the viability of NCI-H292 cells were measured by a cell viability assay. *P<0.05 compared with control cells; **P<0.05 compared with cells treated with TFAP2C small interfering RNA (siRNA). (d) The effects of TFAP2C or TGFBR1 overexpression on the viability of MRC5 cells were measured by a cell viability assay. *P<0.05 compared with control cells; **P<0.05 compared with cells transfected with TFAP2C. (e) The effects of TFAP2C or TGFBR1 knockdown on cell cycle progression of NCI-H292 cells were determined by cell cycle analysis. (f) The effects of TFAP2C or TGFBR1 overexpression on cell cycle progression of MRC5 cells were determined by cell cycle analysis. Error bars represent±s.e.m. (n=3); one-way analysis of variance (ANOVA), Tukey's honestly significant difference test.
TGFBR1 upregulation by TFAP2C promotes EMT of NSCLC cells
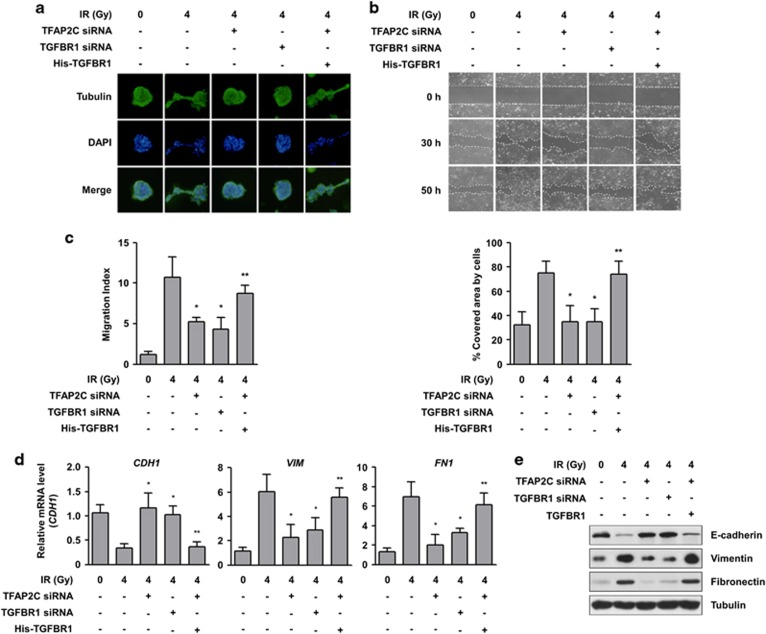
As TGFBR1 is positively correlated with invasion and metastasis in tumor development,16 we next determined whether TFAP2C-induced TGFBR1 expression promotes cell motility, EMT and metastatic conversion in NSCLC cells. To assess the effects of TFAP2C and TGFBR1 on morphological changes in NCI-H292 cells leading to cancer cell motility, we conducted a 3D culture assay. Cells treated with radiation, a potent EMT inducer,28 showed significantly increased numbers of acini invading the 3D Matrigel matrix, and the radiation-induced morphological changes were substantially minimized by treatment with TFAP2C or TGFBR1 siRNA to levels similar to those of non-irradiated control cells (Figure 3a). The morphological changes in TFAP2C-depleting NCI-H292 cells in response to radiation were increased by further ectopic expression of TGFBR1. Moreover, we observed that cells treated with TFAP2C or TGFBR1 siRNA and then exposed to radiation showed decreased motility compared with control cells treated with radiation alone based on a wound healing assay and a Transwell cell migration assay (Figures 3b and c). The decreased cell migration of NCI-H292 cells treated with TFAP2C siRNA was further recovered by TGFBR1 overexpression. Consistent with these results, treatment with TFAP2C or TGFBR1 siRNA suppressed radiation-induced EMT via induction of E-cadherin expression (an epithelial marker) and reduction of vimentin and fibronectin expression (two mesenchymal markers) at both the mRNA and protein levels (Figures 3d and e). When overexpressing TGFBR1, NCI-H292 cells treated with TFAP2C siRNA showed suppression of E-cadherin expression and induction of vimentin and fibronectin expression, leading to EMT. Thus, these results indicate that TFAP2C-induced TGFBR1 expression is positively involved in metastatic conversion in NSCLC cells.
Figure 3.
TFAP2C (transcription factor-activating enhancer-binding protein 2C)-induced TGFBR1 (transforming growth factor-β receptor type 1) upregulation promotes epithelial–mesenchymal transition (EMT) of non-small-cell lung cancer (NSCLC) cells. (a) A three-dimensional (3D) culture model was used to assess the effects of TFAP2C or TGFBR1 knockdown on the morphological changes of NCI-H292 cells. After treatment, the cells were exposed to 4 Gy of radiation, grown in 3D cultures, permeabilized and stained for tubulin (green) and nuclei (blue). (b) The effects of TFAP2C knockdown on the radiation-induced migration of NCI-H292 cells were assessed by a wound-healing assay. Representative images of each group at different time points are shown (top). The graphs show the percentage of the area covered by cells at a specific time point (30 h) from four randomly selected images (bottom). (c) Inhibitory effects of TFAP2C knockdown on the radiation-induced migration of NCI-H292 cells were assessed by a Transwell migration assay. (d) The effects of TFAP2C or TGFBR1 knockdown on the mRNA expression of CDH1, FN1 and VIM in NCI-H292 cells were measured by real-time quantitative RT-PCR (qRT-PCR). (e) Inhibitory effects of TFAP2C knockdown on the protein expression of E-cadherin, fibronectin and vimentin in NCI-H292 and NCI-H838 cells were measured by western blotting. *P<0.05 compared with cells treated with radiation alone; **P<0.05 compared with cells treated with radiation and TFAP2C small interfering RNA (siRNA). Error bars represent±s.e.m. (n=3); one-way analysis of variance (ANOVA), Tukey's honestly significant difference test.
PAK1 signaling activated by TGFBR1 upregulation is involved in NSCLC tumorigenesis
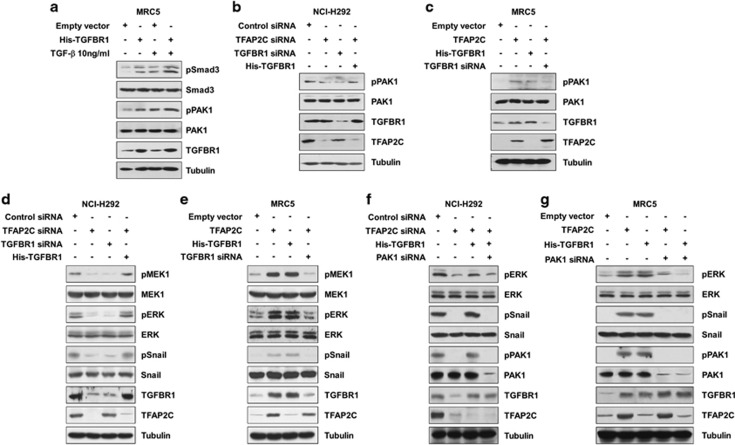
As TFAP2C knockdown decreased expression of TGFBR1, but not expression of TGFBR2 or TGF-β ligands in our microarray data, we determined whether there was a relationship between expression of TGFBR1 and activation of TGFBR1 signaling. Regardless of the TGF-β1 treatment, TGFBR1 overexpression resulted in the phosphorylation of its downstream targets (Figure 4a). These findings indicated that TGFBR1 could sufficiently transduce its signaling via overexpression regardless of the amounts of its ligands. Thus, we hypothesized that noncanonical pathways activated by increased TGFBR1 may be involved in lung tumorigenesis. Among various proteins affected by TGFBR1, PAK1 has been reported to have a critical role in tumor development and malignant progression.11, 29 To determine whether PAK1 is activated in response to TFAP2C-induced TGFBR1 expression, we measured the expression levels of PAK1 and its phosphorylation on the Ser144 residue, which is responsible for maximal kinase activity of PAK1. We observed that PAK1 phosphorylation of the Ser144 residue in NCI-H292 cells was reduced by treatment with TFAP2C or TGFBR1 siRNA and that the reduced phosphorylation of PAK1 by TFAP2C siRNA treatment was recovered by further overexpression of TGFBR1 up to the basal levels observed in non-treated control cells (Figure 4b). In MRC5 cells, PAK1 phosphorylation was induced by ectopic expression of TFAP2C or TGFBR1, and the increased PAK1 phosphorylation in cells overexpressing TFAP2C was reduced by further treatment with TGFBR1 siRNA (Figure 4c). The MEK1/ERK and Snail pathways are well-known downstream effectors of PAK1 that promote tumor development and EMT induction.30, 31 We observed that phosphorylation of MEK1, ERK and Snail in NCI-H292 cells decreased in response to TGFBR1 knockdown, whereas the decreased phosphorylation of these proteins in TFAP2C-depleted cells was rescued by ectopic expression of TGFBR1 (Figure 4d). In MRC5 cells, TFAP2C or TGFBR1 overexpression led to increased phosphorylation of MEK1, ERK and Snail, but these increases were reduced by further knockdown of TGFBR1 (Figure 4e). The involvement of PAK1 in the regulation of ERK and Snail phosphorylation in response to TFAP2C or TGFBR1 manipulation was confirmed by PAK1 knockdown (Figures 4f and g). Taken together, our results suggested that the MAPK and Snail pathways activated by TGFBR1-PAK1 signaling are critically involved in acquisition of tumorigenic properties.
Figure 4.
PAK1 (p21 protein (Cdc42/Rac)-activated kinase 1) signaling is activated by TFAP2C (transcription factor-activating enhancer-binding protein 2C)-induced TGFBR1 (transforming growth factor-β receptor type 1) overexpression. (a) The effects of TGFBR1 overexpression or TGF-β1 treatment on phosphorylation of TGFBR1 downstream targets were analyzed by western blotting. (b) The effects of TFAP2C or TGFBR1 knockdown on the expression and phosphorylation of PAK1 in NCI-H292 cells were analyzed by western blotting. (c) The effects of TFAP2C or TGFBR1 overexpression on the expression and phosphorylation of PAK1 in MRC5 cells were analyzed by western blotting. (d) The effects of TFAP2C or TGFBR1 knockdown on the PAK1 downstream pathways in NCI-H292 cells were analyzed by western blotting. (e) The effects of TFAP2C or TGFBR1 overexpression on the PAK1 downstream pathways in MRC5 cells were analyzed by western blotting. (f) The effects of PAK1 knockdown on the TFAP2C and TGFBR1 signaling in NCI-H292 cells were analyzed by western blotting. (g) The effects of PAK1 knockdown on the signaling mediated by TFAP2C or TGFBR1 overexpression in MRC5 cells were analyzed by western blotting.
Effects of TFAP2C and TGFBR1 in lung tumorigenesis in vivo
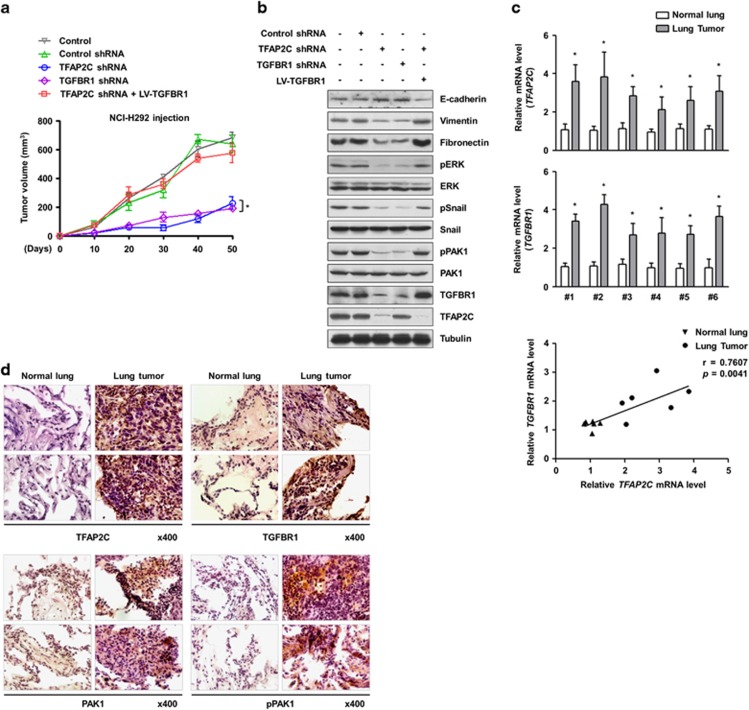
To examine the oncogenic effects of TFAP2C and TGFBR1 in vivo, we conducted mouse xenograft experiments. NCI-H292 cells were injected subcutaneously into the flanks of nude mice (n=5 per group) and the volumes of the generated tumors were measured for 50 days. Depletion of TFAP2C or TGFBR1 resulted in a significant reduction of tumor growth (Figure 5a). We confirmed that the volume of TFAP2C-suppressing tumors was markedly increased by overexpressing TGFBR1 in NCI-H292 cells. Consistent with in vitro studies using NSCLC cells and normal lung cells, we observed increased E-cadherin and decreased vimentin, fibronectin, PAK1 phosphorylation, ERK phosphorylation and Snail phosphorylation in xenografted tumors derived from TFAP2C- or TGFBR1-knockdown cells compared with the control groups. The effects induced by TFAP2C knockdown were rescued by further overexpression of TGFBR1 (Figure 5b). In addition, we found that the mRNA levels of TFAP2C and TGFBR1 were substantially increased in the tumor tissues of six NSCLC patients compared with their normal counterparts (Figure 5c). Pearson's correlation scatter plot showed a positive correlation between TFAP2C and TGFBR1 expression (Pearson's coefficient, r=0.7607; P=0.0041). IHC analyses of the phosphorylation levels of PAK1 and the protein levels of TFAP2C and TGFBR1 confirmed our in vitro and in vivo data (Figures 5d). Taken together, our findings demonstrated the importance of TFAP2C and TGFBR1 in lung tumorigenesis and malignant progression.
Figure 5.
The tumorigenic effects of TFAP2C (transcription factor-activating enhancer-binding protein 2C) and TGFBR1 (transforming growth factor-β receptor type 1) in vivo. (a) The effects of TFAP2C or TGFBR1 knockdown on in vivo tumorigenesis in a xenograft mouse model. Error bars represent±s.e.m. (n=5 per group); *P<0.05 compared with tumor tissues from mice transplanted with cells suppressing TFAP2C and overexpressing TGFBR1; one-way analysis of variance (ANOVA), Tukey's honestly significant difference test. (b) The protein levels of TFAP2C and TGFBR1 in tumor tissue lysates from xenograft mice were measured by western blotting. (c) The mRNA levels of TFAP2C and TGFBR1 in tumor tissues of lung cancer patients were measured by real-time quantitative RT-PCR (qRT-PCR) (top). Error bars represent±s.e.m. (n=3); *P<0.05 compared with normal lung tissues; Student's t-test. Pearson's correlation scatter plot of the fold change in the levels of TFAP2C and TGFBR1 mRNA in normal and tumor tissues of lung cancer patients is shown (bottom). (d) The protein levels of TFAP2C and TGFBR1 and the phosphorylation levels of PAK1 (p21 protein (Cdc42/Rac)-activated kinase 1) in tumor tissues of lung cancer patients were measured by immunohistochemistry (IHC) analysis.
Discussion
In this study, we showed that TFAP2C and TGFBR1 contribute to lung tumorigenesis. The results of this study indicated that TFAP2C has a critical role in lung cancer development and activation of TGFBR1-PAK1 signaling. We first demonstrated that TFAP2C upregulates TGFBR1, which is responsible for PAK1-mediated oncogenic signaling. Our data showed that increased TGFBR1 mediated by TFAP2C results in enhanced proliferative capacity and cell cycle progression in normal lung cells and NSCLC cells. We also found that TFAP2C-mediated upregulation of TGFBR1 increased tumor malignancy and aggressiveness, including EMT induction and increased migration in NSCLC cells. Furthermore, high levels of TFAP2C and TGFBR1 mRNA and protein were observed in lung cancer patients. Taken together, our results indicate that TFAP2C and TGFBR1 significantly contribute to oncogenic signaling to promote lung cancer.
In addition to our previous microarray profiling, the data sets (TCGA, Nature 2014; TCGA, Provisional) in the cBioportal database (http://www.cbioportal.org)32, 33 showed genetic amplification of TFAP2C. However, TGFBR1 was not amplified and TGFBR1 expression was not correlated with TFAP2C upregulation (Pearson's coefficient, r=−0.145). To investigate this discrepancy, we additionally analyzed the Oncomine database (http://www.oncomine.org). According to the data sets in the Oncomine database, the Garber data set34 and the Beer data set35 showed TFAP2C and TGFBR1 overexpression in squamous cell lung carcinoma (2.561- and 1.510-fold, respectively) and in lung adenocarcinoma (1.741- and 1.961-fold, respectively) compared with their normal counterparts, respectively. The information from the Oncomine database showed a positive trend for association between TFAP2C and TGFBR1 expression in lung cancer and supported the results of our microarray analysis that indicated TGFBR1 downregulation in response to TFAP2C knockdown in NSCLCs. Nevertheless, we could not eliminate the possibility of indirect involvement of TFAP2C in TGFBR1 expression via activation of crosstalk signaling or miRNAs, which should be clarified by further molecular studies.
The binding of TGF-β ligands to TGFBR2 results in recruitment and activation of TGFBR1, leading to the activation of a canonical Smad pathway.10 In our microarray data deposited as GSE79228 in the GEO database, TFAP2C knockdown in NSCLC cells decreased the expression of TGFBR1 but not that of TGFBR2 or TGF-β ligands. We also found that overexpression of TGFBR1 itself resulted in phosphorylation of its downstream targets compared with non-overexpression of TGFBR1 when the cells were treated with the same concentrations of its ligand. Among the various noncanonical pathways downstream of TGFBR1, we focused on PAK1 as a TGFBR1 downstream signal mediator because PAK1 has an important role in oncogenic signaling, including cell survival, proliferation and cell motility, through activation of the MAPK and Snail pathways.30, 31 To interact with TGFBR1, PAK1 must be transported from the cytoplasm to the plasma membrane via PAK1-adaptor proteins. A systemic analysis revealed that TGFBR1 under TGF-β stimuli interacted with PAK1 through several PAK1-binding proteins, including α-PIX, oxidative stress-response kinase-1 and occludin.11 Among these PAK1-binding proteins, α-PIX was found to be significantly downregulated by TFAP2C knockdown in our microarray analysis. Although further molecular analysis is needed, our findings suggested an increase in PAK1 recruitment to the plasma membrane by TFAP2C-induced α-PIX, which consequently increased the interaction of PAK1 with TFAP2C-induced TGFBR1 and led to the activation of oncogenic signaling.
In tumor microenvironments, several cytokines and secreted proteins enhance tumor malignancy and aggressiveness. TGF-β ligands secreted from both tumor cells and nearby normal stromal cells are major factors that induce EMT of tumor cells. Multiple studies have reported that increased secretion of TGF-β ligands in a paracrine or autocrine manner resulted in inhibition of apoptosis coupled with induction of cell survival, invasion and metastasis against chemotherapy.15, 16, 17 In the present study, we observed that TGFBR1-PAK1 signaling could be significantly increased by TGFBR1 overexpression with basal levels of its ligands and that TFAP2C-induced TGFBR1 led to oncogenic signaling with EMT induction through PAK1-activated MAPK and Snail pathways in NSCLC cells. These findings suggest that TFAP2C and TGFBR1 could be positively associated with NSCLC malignancy, including acquisition of tumor aggressiveness and metastatic potential.
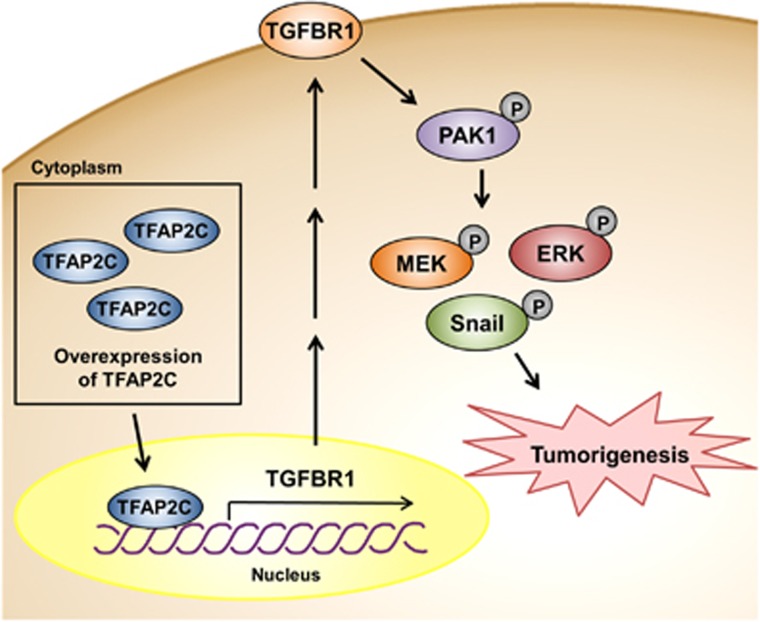
Little is known about the molecular mechanisms of TFAP2C expression-promoted tumorigenesis in NSCLC. The results of this study showed that TFAP2C is a crucial regulator of tumor development via TGFBR1 upregulation. We report for the first time that TGFBR1 expression induced by TFAP2C could significantly activate the oncogenic signaling of PAK1-activated MAPK and Snail pathways regardless of the levels of its ligands (Figure 6). We propose that TFAP2C and TGFBR1 are promising oncogenic factors in NSCLC and that pharmacological inhibition of TGFBR1 could increase the efficiency of NSCLC therapy.
Figure 6.
Schematic diagram showing an oncogenic role of TFAP2C (transcription factor-activating enhancer-binding protein 2C) in lung tumorigenesis through activation of TGFBR1 (transforming growth factor-β receptor type 1)/PAK1 (p21 protein (Cdc42/Rac)-activated kinase 1) signaling. The TFAP2C-mediated increase in TGFBR1 expression induces phosphorylation of PAK1. Increased PAK1 phosphorylation promotes cell cycle progression, proliferation and epithelial–mesenchymal transition (EMT) through activation of the mitogen-activated protein kinase (MAPK) and Snail pathways, resulting in lung tumorigenesis and malignant development.
Acknowledgments
This work was supported by a 2-Year Research Grant of Pusan National University.
The authors declare no conflict of interest.
References
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015; 16: e342–e351. [DOI] [PubMed] [Google Scholar]
- Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer—is it becoming a reality? Nat Rev Clin Oncol 2010; 7: 401–414. [DOI] [PubMed] [Google Scholar]
- Komurov K, Tseng JT, Muller M, Seviour EG, Moss TJ, Yang L et al. The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Mol Syst Biol 2012; 8: 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol 2005; 6: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene 2000; 260: 1–12. [DOI] [PubMed] [Google Scholar]
- Ailan H, Xiangwen X, Daolong R, Lu G, Xiaofeng D, Xi Q et al. Identification of target genes of transcription factor activator protein 2 gamma in breast cancer cells. BMC Cancer 2009; 9: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer 2010; 49: 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JA, Bosher JM, Skinner A, Sheer D, Williams T, Hurst HC. Chromosomal mapping of the human and mouse homologues of two new members of the AP-2 family of transcription factors. Genomics 1996; 35: 262–264. [DOI] [PubMed] [Google Scholar]
- Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett 2006; 580: 2811–2820. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003; 113: 685–700. [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science 2005; 307: 1621–1625. [DOI] [PubMed] [Google Scholar]
- Vogelmann R, Nguyen-Tat MD, Giehl K, Adler G, Wedlich D, Menke A. TGFbeta-induced downregulation of E-cadherin-based cell-cell adhesion depends on PI3-kinase and PTEN. J Cell Sci 2005; 118: 4901–4912. [DOI] [PubMed] [Google Scholar]
- Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res 2012; 347: 11–20. [DOI] [PubMed] [Google Scholar]
- Gomes LR, Terra LF, Wailemann RA, Labriola L, Sogayar MC. TGF-beta1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer 2012; 12: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet 2001; 29: 117–129. [DOI] [PubMed] [Google Scholar]
- Miyazono K. Transforming growth factor-beta signaling in epithelial–mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B 2009; 85: 314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman GJ. Switching TGFbeta from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev 2011; 21: 93–99. [DOI] [PubMed] [Google Scholar]
- Kwon T, Youn H, Son B, Kim D, Seong KM, Park S et al. DANGER is involved in high glucose-induced radioresistance through inhibiting DAPK-mediated anoikis in non-small cell lung cancer. Oncotarget 2016; 7: 7193–7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Kim W, Youn H, Kwon T, Kang J, Kim E, Son B et al. PIM1 kinase inhibitors induce radiosensitization in non-small cell lung cancer cells. Pharmacol Res 2013; 70: 90–101. [DOI] [PubMed] [Google Scholar]
- Kang J, Kim W, Kwon T, Youn H, Kim JS, Youn B. Plasminogen activator inhibitor-1 enhances radioresistance and aggressiveness of non-small cell lung cancer cells. Oncotarget 2016; 7: 23961–23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Youn H, Kwon T, Son B, Kang J, Yang HJ et al. PAK1 tyrosine phosphorylation is required to induce epithelial-mesenchymal transition and radioresistance in lung cancer cells. Cancer Res 2014; 74: 5520–5531. [DOI] [PubMed] [Google Scholar]
- Youn H, Son B, Kim W, Jun SY, Lee JS, Lee JM et al. Dissociation of MIF-rpS3 complex and sequential NF-kappaB activation is involved in IR-induced metastatic conversion of NSCLC. J Cell Biochem 2015; 116: 2504–2516. [DOI] [PubMed] [Google Scholar]
- Son B, Jun SY, Seo H, Youn H, Yang HJ, Kim W et al. Inhibitory effect of traditional oriental medicine-derived monoamine oxidase B inhibitor on radioresistance of non-small cell lung cancer. Sci Rep 2016; 6: 21986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Youn H, Kang C, Youn B. Inflammation-induced radioresistance is mediated by ROS-dependent inactivation of protein phosphatase 1 in non-small cell lung cancer cells. Apoptosis 2015; 20: 1242–1252. [DOI] [PubMed] [Google Scholar]
- Kim W, Kim E, Yang HJ, Kwon T, Han S, Lee S et al. Inhibition of hedgehog signalling attenuates UVB-induced skin photoageing. Exp Dermatol 2015; 24: 611–617. [DOI] [PubMed] [Google Scholar]
- Petty RD, Sutherland LA, Hunter EM, Cree IA. Comparison of MTT and ATP-based assays for the measurement of viable cell number. J Biolumin Chemilumin 1995; 10: 29–34. [DOI] [PubMed] [Google Scholar]
- Kang J, Kim E, Kim W, Seong KM, Youn H, Kim JW et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem 2013; 288: 27343–27357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi F, Lu J, Phan R, Lewis M, Trinidad K, Aljilani A et al. Significance of KRAS/PAK1/Crk pathway in non-small cell lung cancer oncogenesis. BMC Cancer 2015; 15: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res 2005; 65: 3179–3184. [DOI] [PubMed] [Google Scholar]
- Zhang X, Mao H, Chen JY, Wen S, Li D, Ye M et al. Increased expression of microRNA-221 inhibits PAK1 in endothelial progenitor cells and impairs its function via c-Raf/MEK/ERK pathway. Biochem Biophys Res Commun 2013; 431: 404–408. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA 2001; 98: 13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002; 8: 816–824. [DOI] [PubMed] [Google Scholar]