Abstract
By changing the relative abundance of generated antigenic peptides through alterations in the proteolytic activity, interferon (IFN)-γ-induced immunoproteasomes influence the outcome of CD8+ cytotoxic T lymphocyte responses. In the present study, we investigated the effects of hepatitis C virus (HCV) infection on IFN-γ-induced immunoproteasome expression using a HCV infection cell culture system. We found that, although IFN-γ induced the transcriptional expression of mRNAs encoding the β1i/LMP2, β2i/MECL-1 and β5i/LMP7 immunoproteasome subunits, the formation of immunoproteasomes was significantly suppressed in HCV-infected cells. This finding indicated that immunoproteasome induction was impaired at the translational or posttranslational level by HCV infection. Gene silencing studies showed that the suppression of immunoproteasome induction is essentially dependent on protein kinase R (PKR). Indeed, the generation of a strictly immunoproteasome-dependent cytotoxic T lymphocyte epitope was impaired in in vitro processing experiments using isolated 20S proteasomes from HCV-infected cells and was restored by the silencing of PKR expression. In conclusion, our data point to a novel mechanism of immune regulation by HCV that affects the antigen-processing machinery through the PKR-mediated suppression of immunoproteasome induction in infected cells.
Introduction
The clearance of viral infection is dependent on vigorous CD8+ cytotoxic T lymphocyte (CTL) responses, which must be tightly regulated to prevent immune-mediated host tissue damage. Virus-infected cells are recognized and destroyed by specific CTLs that bind to virus-derived peptide epitopes associated with cell surface major histocompatibility complex (MHC) class I molecules.1, 2 Most of these antigenic peptides, which are usually 8–10 amino-acid residues in length, are generated by the 30S proteasome complex, which is the central proteolytic machinery of the ubiquitin-proteasome-system.3, 4
The 30S complex is composed of the 20S proteasome proteolytic core complex and two associated 19S regulatory particles.4, 5 The 20S complex is arranged as four staggered rings, each containing seven non-identical subunits. The outer rings contain the α subunits (α1–α7), which form the ‘gates' through which substrates enter and products are released.5 Each of the two inner rings contains the β subunits (β1–β7), three of which (β1, β2 and β5) harbor the six active sites.5
Type I and II interferons (IFNs), which are major cytokines in viral infection, induce the expression of the immunosubunits (i-subunits) β1i/LMP2, β2i/MECL-1 and β5i/LMP7 in non-immune cells and the assembly of the so called immunoproteasomes (i-proteasomes).5, 6 In addition, i-proteasomes are constitutively expressed in hematopoietic/immune cells, such as dendritic cells.7, 8 Because of the altered proteolytic activity, i-proteasomes have been shown to exhibit altered frequencies in cleavage site usage. This affects the relative abundance of the generated antigenic peptides, which in turn can influence the quality of the peptide-specific CD8+ CTL response.9 For example, the generation of the hepatitis B virus (HBV) core 141–151 epitope has been shown to be strongly influenced by the structural presence of the i-subunit β5i/LMP7.10 Additionally, it has been shown that β1i/LMP2- or β5i/LMP7-deficient mice are unable to efficiently generate and present some CD8+ CTL epitopes11, 12, 13 while the CD8+ CTL response was barely affected in β1i/LMP2- or β5i/LMP7-deficient mice infected with lymphocytic choriomeningitis virus.14, 15 Recent reports demonstrated that quantitative changes in the epitope generation of i-subunit-deficient mice result in alterations of the immunodominance hierarchy and the T-cell repertoire in a murine influenza infection model.16 Another study using mice completely lacking i-proteasomes indicated that the peptide repertoire presented by dendritic cells in the lymphoid organs differed from that presented by wild-type dendritic cells by 50%.17 In addition to affecting the outcome of the CTL response, i-proteasomes also possess an important proteostatic function in preserving cell viability under conditions of IFN-induced oxidative stress.18, 19 For example, in a murine model of coxsackievirus infection, i-proteasomes were shown to protect mice against oxidant protein damage in the injured myocardium.20
Hepatitis C virus (HCV) is one of the most common causes of chronic liver disease. Although some patients successfully clear the virus after acute HCV infection, most patients fail to eliminate the virus and develop chronic persistent infection accompanied by inflammatory liver injury.21 The outcome of HCV infection is determined by virus-specific cellular immune responses.22, 23, 24, 25 Indeed, patients who control their HCV infection have broad CD8+ T-cell responses with higher functional avidity, whereas CD8+ T-cell responses are impaired in patients with persistent HCV infection.23, 24, 25, 26
HCV evades host immune responses through various mechanisms, leading to chronic persistent infection.27 However, little is known regarding the effects of HCV infection on the epitope-processing machinery, which is essential for the recognition of infected cells by CTLs. In the present study, we investigated the i-proteasome induction in HCV-infected cells. We demonstrate that i-proteasome induction is suppressed and that protein kinase R (PKR) is responsible for the suppression of i-proteasome induction in HCV-infected cells.
Materials and Methods
Cell culture
Huh-7.5, a human hepatoma cell line, was obtained from Apath (Brooklyn, NY, USA) and maintained in Dulbecco's modified Eagle's medium (Welgene, Daegu, Korea) containing 10% fetal bovine serum (Welgene), 4.5 g l−1 glucose, 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA), and 4 mM L-glutamine in 5% CO2 at 37 °C.
HCV preparation, infection and IFN-γ treatment
The JFH-1 strain (genotype 2a) of HCV was produced by transfecting Huh-7.5 cells with in vitro-transcribed RNA from a plasmid encoding the full JFH-1 HCV genome (provided by Apath), as previously described.28 After HCV RNA transfection, the cell culture supernatant was used to infect naive Huh-7.5 cells. Subsequently, the cell culture supernatant with the highest HCV RNA titer was taken and further concentrated with WelProt virus concentration reagent (Welgene) to prepare the HCV cell culture (HCVcc) stock.
For the HCV infection experiments, naive Huh-7.5 cells were infected with HCVcc stock at a multiplicity of infection (MOI) of 0.01 in culture dishes. At 3 days after infection, the cells were subcultured for IFN-γ treatment. Two days after being subcultured, the cells were treated with 10 ng ml−1 of IFN-γ for 24 or 48 h, and i-proteasome induction was examined.
Immunoblot analysis
Cell lysates were prepared using RIPA buffer (Thermo Scientific, Rockford, IL, USA) supplemented with protease inhibitors and phosphatase inhibitors and were quantified by a bicinchoninic assay (Pierce, Rockford, IL, USA). Then 10 μg of cell lysate was electrophoresed through a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Richmond, CA, USA). Specific proteins were detected using mouse anti-HCV core (Thermo Scientific), rabbit anti-β1i (Abcam, Cambridge, MA, USA), goat anti-β2i (Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse anti-β5i (Santa Cruz Biotechnology), rabbit anti-α4, rabbit anti-β5 (Thermo Scientific), mouse anti-tubulin (Sigma-Aldrich, St Louis, MO, USA), rabbit anti-PKR (Santa Cruz Biotechnology), rabbit anti-phospho-PKR (Cell Signaling, Beverly, MA, USA) or anti-IFNGR1 (Abcam) antibodies. In addition, proteasome antibodies from a laboratory stock were used. After overnight incubation with primary antibodies at 4 °C, protein bands were visualized by horseradish peroxidase-conjugated anti-mouse Ig (BD Biosciences, San Jose, CA, USA), anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) or anti-goat IgG (Santa Cruz Biotechnology) antibodies and enhanced chemiluminescence. The density of the resulting protein bands was analyzed using the ImageJ densitometry software (National Institutes of Health, Bethesda, MD, USA).
Flow cytometry
HCV-infected cells were fixed and permeabilized using FoxP3 staining buffer (eBioscience, San Diego, CA, USA). The intracellular HCV core protein was stained with anti-HCV core IgG1 to detect HCV-infected cells. After secondary staining with allophycocyanin-conjugated goat anti-mouse Ig (BD Biosciences), the cells were analyzed using an LSR II instrument (BD Biosciences) and the FlowJo software (TreeStar, Ashland, OR, USA).
Purification of the 20S proteasome complex
Frozen cells were lysed in lysis buffer (20 mM Tris, pH 7.2, 1 mM EDTA, 1 mM sodium azide, 1 mM dithiothreitol, 50 mM NaCl, 0.1% NP-40) and homogenized. The lysates were centrifuged, the supernatant was applied onto DEAE-Sephacel (GE Healthcare, Little Chalfont, UK) and unbound proteins were removed by washing. Proteasome complexes were eluted with 400 mM NaCl in TEAD buffer (20 mM Tris, pH 7.2, 1 mM EDTA, 1 mM sodium azide, 1 mM dithiothreitol, 400 mM NaCl) and were subsequently concentrated by ammonium sulfate precipitation. Protein-containing fractions were separated by ultracentrifugation (285 000 g for 16 h) using a SW40Ti rotor (Beckman Coulter, Brea, CA, USA). Subsequently, fractions containing proteasomes were pooled, applied to a Mono Q column (GE Healthcare), and eluted with a gradient of 100–1000 mM NaCl in TEAD. The purity of the eluted proteasomes was assessed by Coomassie brilliant blue-stained SDS-PAGE.
Two-dimensional gel electrophoresis
To separate the subunits of the 20S proteasome complex, isoelectric focusing by carrier ampholytes was combined with SDS-PAGE. Proteasomes were applied to a carrier ampholyte isoelectric focusing gel. In the second dimension, proteins were loaded onto SDS-PAGE and stained with Coomassie brilliant blue G-250. Subsequently, protein identification was performed as described earlier.29 Liquid chromatography–tandem mass spectrometric analysis was carried out on a LTQ Orbitrap Velos Pro (Thermo Scientific, San Jose, CA, USA) mass spectrometer, and raw data were processed toward protein identification utilizing PEAKS Studio V.7.0 (Bioinformatics Solutions, Waterloo, Ontario, Canada).
Quantitative real-time PCR
Total RNA was extracted from cell pellets using a Ribospin Kit (GeneAll Biotechnology, Seoul, Korea). Subsequently, cDNA was synthesized using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA, USA). TaqMan real-time PCR was performed on a CFX96 real-time PCR detection machine (Bio-Rad Laboratories) using a Kapa Probe Fast qPCR Kit (Kapa Biosystems, Woburn, MA, USA) and TaqMan probes (Applied Biosystems). The expression level of each gene was normalized to the mRNA levels of β-actin, which was used as an endogenous control.
Short-hairpin RNA (shRNA) lentiviruses
A validated clone of the shRNA bacterial glycerol stock with lentiviral constructs expressing PKR shRNA was obtained from Sigma-Aldrich. The lentiviral vector was transfected into 293TN cells (System Biosciences, Mountain View, CA, USA) with Lipofectamine 2000 transfection reagent (Invitrogen), and lentiviral particles were harvested from the transfected cells after 48 h. Huh-7.5 cells were transduced with PKR shRNA lentiviruses, and stable cell lines were established by 4 weeks of antibiotic selection using 2 μg ml−1 of puromycin (Sigma-Aldrich).
In vitro peptide digestion assays
Twenty micrograms of the HBV core 131–162, 32-mer polypeptide and 3 μg of purified proteasome complexes were incubated in 300 μl TEAD buffer at 37 °C for the indicated time (37 μl per time point). The reaction was terminated by the addition of trifluoroacetic acid. The digested products were separated by reversed-phase chromatography on a 1 mm RP column (Beta Basic-18, 100 × 1 mm, 3 μm, 150 Å, Thermo Scientific). Peptides were detected online with electrospray ionization–mass spectrometry (DECA XP MAX iontrap instrument; Thermo Scientific). The kinetics of the identified peaks in time-dependent processing experiments (signal intensity versus time of digestion) was analyzed using the LCQuan software version 2.5 (Thermo Scientific).
Statistical analysis
The statistical significance of the data was analyzed by Mann–Whitney U-test or paired t-test using GraphPad Prism version 5.01 (GraphPad Software, San Diego, CA, USA). A P-value of <0.05 was considered statistically significant.
Results
Induction of i-proteasomes by IFN-γ is suppressed in HCV-infected cells
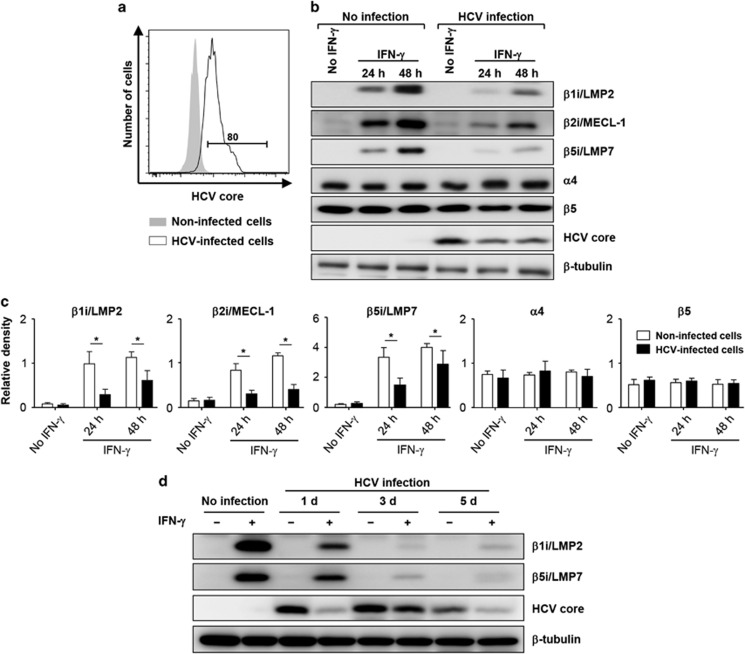
To examine the effects of HCV infection on i-proteasome induction, we used the in vitro JFH-1 HCVcc infection system, which reflects the entire viral life cycle from entry to virion release. HCV infection was confirmed by the intracellular staining of the core antigen (Figure 1a). Initially, Huh7.5 cells were treated with IFN-γ to induce the formation of i-proteasomes. IFN-γ increased the expression levels of the i-proteasome subunits β1i/LMP2, β2i/MECL-1 and β5i/LMP7 in non-infected cells but did not alter the standard proteasome subunit β5 expression and abundance of the non-replaceable subunit α4 (Figure 1b). In contrast, the IFN-γ-triggered induction of the i-proteasome subunits was significantly suppressed in HCV-infected cells (Figure 1b). The protein band densities were analyzed and are presented in Figure 1c. This effect was clearly observed as early as 3 days after HCV infection (Figure 1d).
Figure 1.
IFN-γ-induced i-subunit expression is suppressed in HCV-infected cells. (a–c) Huh-7.5 cells were infected with HCVcc at 0.01 MOI and cultured for 5 days. The infection rate was determined by anti-HCV core staining and flow cytometry (a).The infection rate was approximately 80%. HCV-infected or uninfected cells were treated with 10 ng ml−1 IFN-γ for the indicated times. The total cell lysates were analyzed by immunoblotting for i-subunits (β1i/LMP2, β2i/MECL-1 and β5i/LMP7), standard proteasome subunits (α4 and β5), the HCV core and tubulin (b). The protein band densities from three independent experiments are presented as a bar graph (mean±s.e.m.; c). *P<0.05. (d) Huh-7.5 cells were infected with HCVcc at 0.01 MOI, cultured for 1, 3 or 5 days and treated with 10 ng ml−1 IFN-γ for 24 h. The total cell lysates were analyzed by immunoblotting for i-subunits (β1i/LMP2 and β5i/LMP7), the HCV core and tubulin.
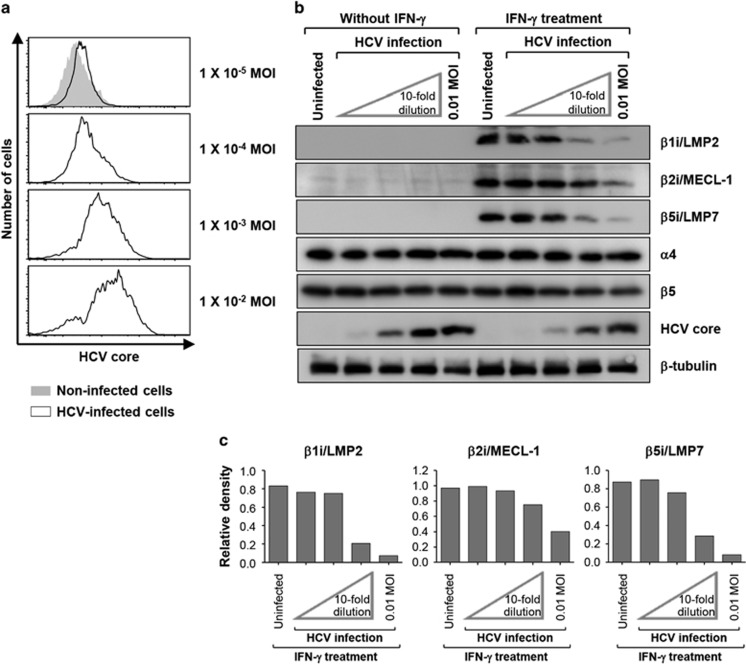
The suppressive effects of HCV infection on IFN-γ-mediated i-subunit induction were confirmed using different HCV infection rates. To produce HCV-infected cells with various infection rates, we infected Huh-7.5 cells with HCVcc at 10-fold dilution steps (Figure 2a). Protein expression analysis revealed that the induction of i-proteasome subunits was most severely suppressed in cells infected with the highest MOI (Figure 2b), whereas the suppressive effect of HCV on i-subunit induction was abrogated in cells infected with lower MOIs (Figure 2b). The protein band densities were analyzed and are presented in Figure 2c.
Figure 2.
HCV infection suppresses i-subunit expression in a dose-dependent manner. Huh-7.5 cells were infected with HCVcc at 10-fold dilution steps (MOIs from 10−2 to 10−5) and cultured for 5 days. (a) The infection rate was determined by anti-HCV core staining and flow cytometry. (b) Cells were treated with 10 ng ml−1 IFN-γ for 24 h. Total cell lysates were analyzed by immunoblotting for i-subunits (β1i/LMP2, β2i/MECL-1 and β5i/LMP7), standard proteasome subunits (α4 and β5), the HCV core and tubulin. (c) The protein band densities are presented as a bar graph.
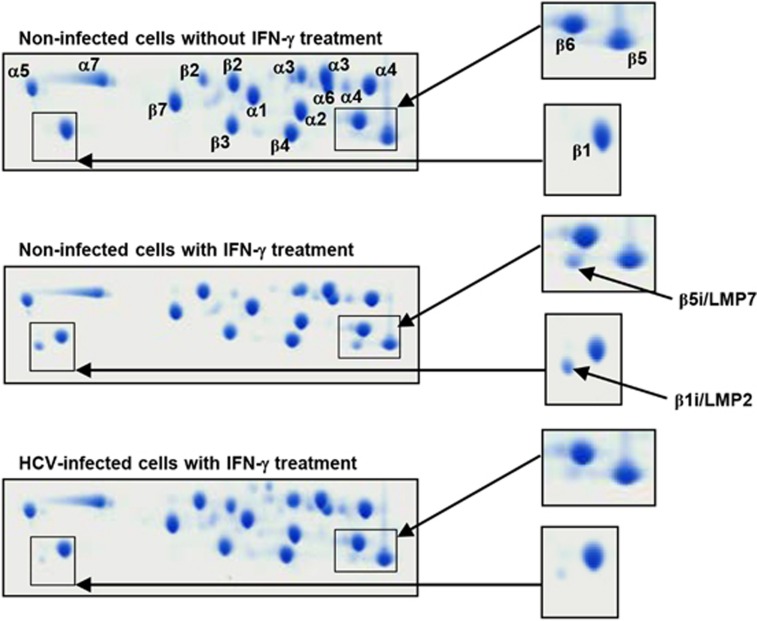
I-subunit incorporation into proteasome complexes is reduced in HCV-infected cells
To directly assess i-proteasome formation, we isolated 20S proteasomes from HCV-infected and IFN-γ-treated Huh-7.5 cells and analyzed their subunit composition by two-dimensional gel electrophoresis. As a control, 20S proteasome complexes were separated from non-infected Huh-7.5 cells either treated or not treated with IFN-γ. As shown in Figure 3, two-dimensional gel electrophoresis revealed the presence of both β1i/LMP2 and β5i/LMP7 in the IFN-γ-treated and non-infected cells, indicating that the i-subunits were incorporated into the 20S proteasome complex. In contrast, β1i/LMP2 and β5i/LMP7 could not be detected in the 20S proteasomes isolated from non-treated Huh-7.5 control cells (Figure 3). Interestingly, in agreement with the protein expression data shown in Figures 1 and 2, β1i/LMP2 and β5i/LMP7 incorporation was clearly reduced in the 20S proteasomes isolated from IFN-γ-treated, HCV-infected cells (Figure 3). This indicates that the IFN-γ-induced i-proteasome assembly was reduced in the HCV-infected cells.
Figure 3.
The subunit composition of isolated 20S proteasome complexes. Huh-7.5 cells were infected with HCVcc at 0.01 MOI and cultured for 5 days. The HCV-infected or uninfected cells were treated with 10 ng ml−1 IFN-γ for 48 h, and the 20S proteasome complexes were isolated from cell lysates. Purified 20S proteasomes were analyzed by two-dimensional gel electrophoresis and mass spectrometry.
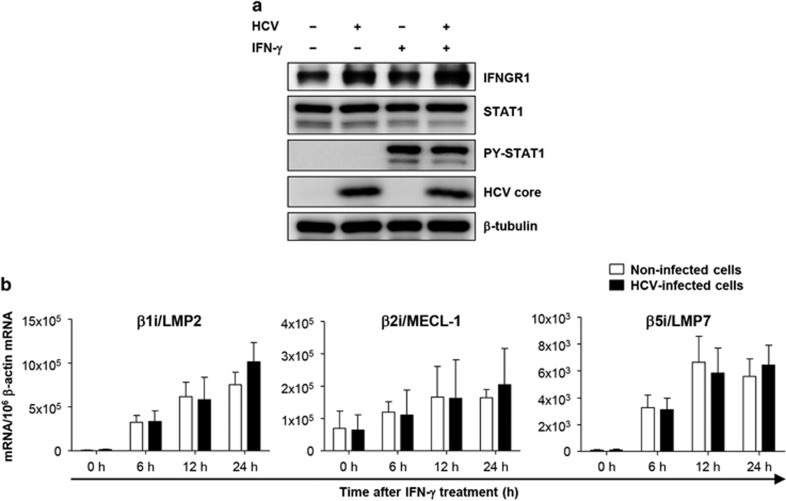
The induction of i-subunit mRNA expression is not suppressed in HCV-infected cells
To decipher the molecular mechanisms of interference with IFN-γ-mediated i-subunit induction in HCV-infected cells, we first examined the expression of IFN-γR in HCV-infected cells and found that IFNGR1 is well expressed in HCV-infected cells (Figure 4a). We also examined phosphorylation of signal transducer and activator of transcription factor 1 (STAT1) after IFN-γ treatment and found that STAT1 phosphorylation was not impaired in HCV-infected cells (Figure 4a). Consequently, we next studied the i-subunit expression at the mRNA level in non-infected and HCV-infected cells. Interestingly, the IFN-γ-induced mRNA expression of the i-subunits β1i/LMP2, β2i/MECL-1 and β5i/LMP7 did not differ between the uninfected cells and HCV-infected cells (Figure 4b). This suggests that HCV infection regulates the IFN-γ-induced i-proteasome expression at the translational or posttranslational level and not at the transcriptional level.
Figure 4.
Effect of HCV infection on IFN-γ signaling and i-subunit mRNA. Huh-7.5 cells were infected with HCVcc at an MOI of 0.01 and cultured for 5 days. (a) HCV-infected or uninfected cells were treated with 10 ng ml−1 IFN-γ for 30 mins, and the total cell lysates were analyzed by immunoblotting for IFNGR1, STAT1, phosphorylated STAT1, the HCV core and tubulin. (b) HCV-infected or uninfected cells were treated with 10 ng ml−1 IFN-γ for the indicated times, and the total RNA was then purified from the cells. After cDNA synthesis, the mRNA levels of i-proteasome subunits were quantified by TaqMan real-time PCR. The data from five independent experiments are presented as a bar graph (mean±s.e.m.).
The IFN-γ-mediated induction of i-proteasomes is impaired by PKR in HCV-infected cells
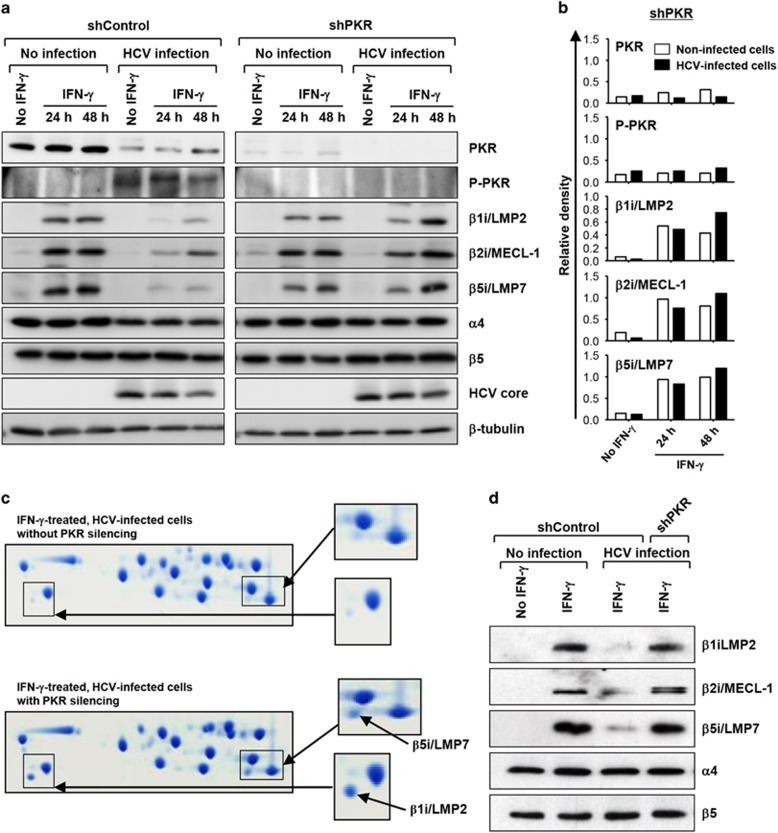
In HCV-infected cells, the translation of antiviral IFN-stimulated genes is suppressed by PKR phosphorylation and subsequent eukaryotic initiation factor 2α (eIF2α) phosphorylation.30 In addition, we recently reported that IFN-induced MHC class I expression is attenuated in HCV-infected cells by the same mechanism and that effector functions of CD8+ T cells are ultimately decreased.31 We therefore hypothesized that the suppressive effects of HCV on i-proteasome induction were mediated by PKR. To test this hypothesis, we established PKR-silenced cell lines and tested whether PKR silencing had an effect on the IFN-γ-induced i-proteasome expression in HCV-infected cells. We first confirmed that phosphorylation of eIF2α is abrogated in PKR-silenced cells (Supplementary Figure S1) and found that HCV RNA titer was not strikingly affected by PKR silencing (Supplementary Figure S2). Interestingly, in contrast to the data obtained for the HCV-infected Huh-7.5 cells expressing PKR, immunoblot analyses of HCV-infected and PKR-silenced cell lysates demonstrated that IFN-γ-mediated i-subunit induction was not suppressed (Figure 5a). The protein band densities from PKR-silenced cell lysates were analyzed and are presented in Figure 5b. We next examined the subunit composition of isolated 20S proteasome complexes. Two-dimensional gel electrophoresis showed that PKR silencing reconstituted the incorporation of β1i/LMP2 and β5i/LMP7 into 20S proteasomes in IFN-γ-treated, HCV-infected cells (Figure 5c). These data were corroborated by immunoblot analyses of the isolated 20S proteasomes for the i-subunits β1i/LMP2, β2i/MECL-1 and β5i/LMP7 (Figure 5d). Our data thus show that PKR is responsible for the suppression of IFN-γ-triggered i-proteasome induction in HCV-infected cells.
Figure 5.
I-proteasome induction in PKR-silenced cells. PKR-silenced Huh-7.5 cell lines were established by transduction with shRNA lentiviruses. Control cells (shControl) and PKR-silenced cells (shPKR) were infected with HCVcc at an MOI of 0.01 and cultured for 5 days. (a) Cells were treated with 10 ng ml−1 IFN-γ for the indicated times. Cell lysates were analyzed by immunoblotting for PKR, phosphorylated PKR, i-subunits (β1i/LMP2, β2i/MECL-1 and β5i/LMP7), standard proteasome subunits (α4 and β5), the HCV core and tubulin. (b) The protein band densities from PKR-silenced cell lysates are presented as a bar graph. (c and d) The cells were treated with 10 ng ml−1 IFN-γ for 48 h, and the 20S proteasome complexes were isolated from the cell lysates. Separated 20S proteasomes were analyzed by two-dimensional gel electrophoresis and Coomassie blue staining. β1i/LMP2 and β5i/LMP7 are indicated in the insets to the right (c). The isolated 20S proteasome complexes were analyzed by immunoblotting for i-subunits (β1i/LMP2, β2i/MECL-1 and β5i/LMP7) and standard proteasome subunits (α4 and β5) (d).
Impaired induction of i-proteasomes affects peptide processing in HCV-infected cells
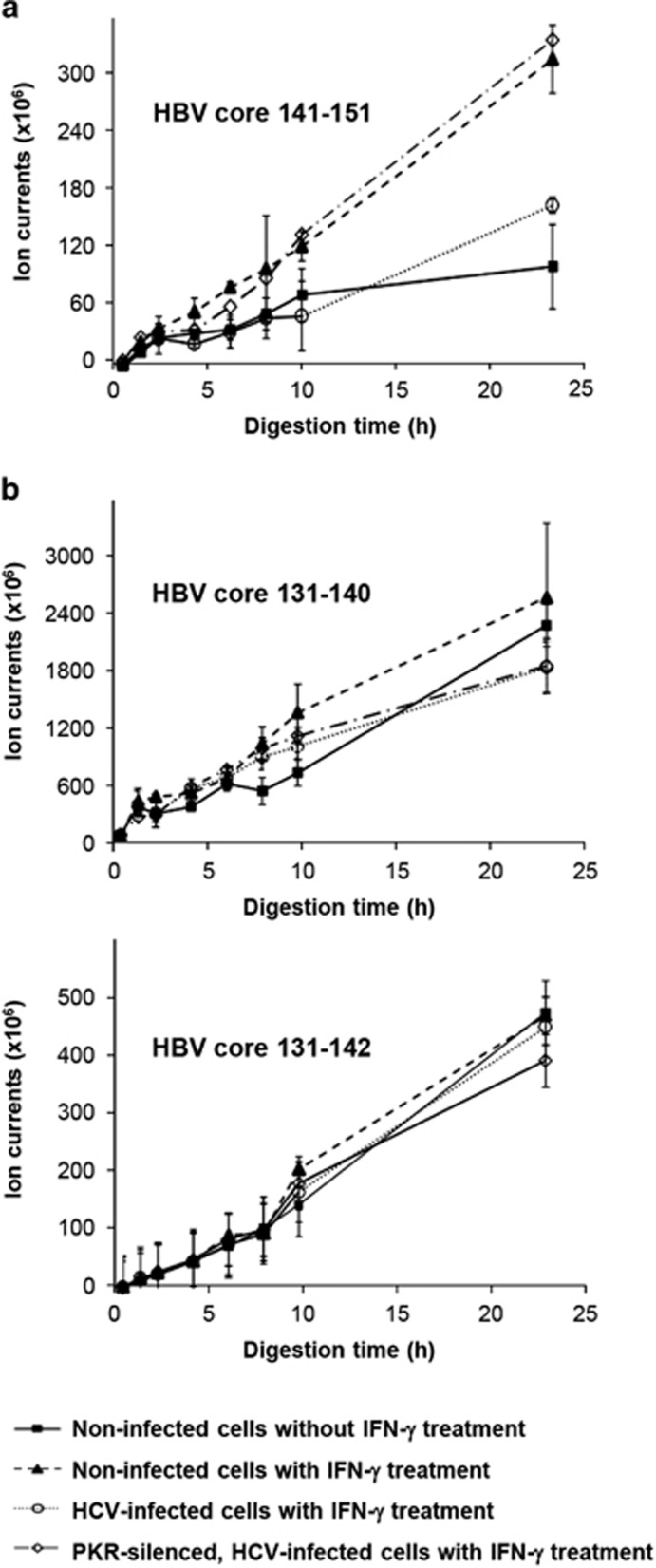
To investigate the function of the different 20S proteasomes, we performed in vitro processing experiments analyzing the proteasomal capacity to generate an epitope peptide from a polypeptide precursor by mass spectrometry. Because an i-proteasome-dependent epitope has not yet been identified in HCV proteins, we investigated the generation of the HBV core 141–151 peptide, known to be strongly dependent on the presence of the i-subunit β5i/LMP7, from the substrate peptide HBV core 131–162.10 As control, we determined the abundance of the i-proteasome-independent peptides HBV core 131–140 and HBV core 131–142 among the cleavage products. The 20S proteasomes isolated from the IFN-γ-treated Huh-7.5 cells generated the HBV core 141–151 peptide, whereas those from cells without IFN-γ treatment processed only low epitope amounts (Figure 6a). Importantly, and as expected, the 20S proteasomes from the IFN-γ-treated, HCV-infected cells displayed a strongly diminished level of HBV core 141–151 epitope generation, which was comparable to that of non-treated Huh-7.5 control cells. In striking contrast, the HBV core 141–151 epitope was generated by 20S proteasomes derived from IFN-γ-treated, HCV-infected cells when PKR was silenced (Figure 6a). As shown in Figure 6b, the generation of the i-proteasome-independent peptides HBV core 131–140 and HBV core 131–142 was not regulated by IFN-γ treatment or HCV infection. Collectively, our data point to a decisive role of PKR in the HCV-mediated reduction of IFN-γ-induced i-proteasome expression. The data suggest that PKR inhibits the generation of i-proteasome-dependent epitopes or epitopes that are favorably generated by the i-proteasomes in infected cells.
Figure 6.
Generation of an i-proteasome-dependent epitope by the isolated 20S proteasomes. The cells were infected with HCVcc at an MOI of 0.01 and were cultured for 5 days. HCV-infected or uninfected cells were treated with 10 ng ml−1 IFN-γ for 48 h, and 20S proteasome complexes were isolated from cell lysates. Purified 20S proteasome complexes were subsequently incubated with the precursor substrate HBV core 131–162 polypeptide for the indicated time periods. In vitro digests were analyzed by HPLC and mass spectrometry for the presence of the β5i/LMP7-dependent HBV core 141–151 epitope (a) and the β5i/LMP7-independent HBV core 131–140 and 131–142 peptides (b).
Discussion
In the present study, we investigated the effects of HCV infection on IFN-γ-induced i-proteasome expression using an in vitro HCVcc infection system. We found that HCV infection prevented the induction of the i-proteasome subunits β1i/LMP2, β2i/MECL-1 and β5i/LMP7 at the protein level in a PKR-dependent manner. Furthermore, we demonstrated i-proteasome suppression by studying not only the subunit composition but also the function of the 20S proteasome complexes isolated from HCV-infected cells. Previously, we showed that type I and III IFNs are produced from HCV-infected cells32 and that i-proteasomes can be induced by type I IFNs.33 In future research, it should be examined whether type I IFN-induced i-proteasome expression is also impaired by the PKR-dependent mechanism in HCV-infected cells.
To combat viral infections, the expression of i-proteasomes triggered by type I or type II IFNs, the generation of a virus-derived peptide repertoire and the subsequent induction of viral antigen-specific CD8+ CTLs is of major importance. On one hand, a vigorous CTL response is beneficial because it allows the infected cells to be destroyed. On the other hand, the release of IFNs by activated lymphocytes results in the induction of oxidative stress. In particular, newly synthesized proteins are sensitive to oxygen radicals, and the exposure of cellular proteins to such radicals leads to oxidative protein damage, the accumulation of proteins and the destruction of both infected and non-infected cells.18
Viral interference with the i-proteasome function has been shown to occur either on the transcriptional level or through the direct interaction of viral proteins with the i-subunits. In both human and mouse cytomegalovirus-infected fibroblasts, the transcription and protein expression of the i-proteasome subunits are inhibited, thus preventing their incorporation into nascent proteasome complexes.34 This mechanism was suggested to lead to changes in the CTL epitope repertoire during the effector phase of the immune response against cytomegalovirus infection.34 Direct interference with the proteasome complex has been reported for the HIV-1 Tat protein, which interacts with six β subunits of the 20S proteasome, as well as with the β2i/MECL-1 and β5i/LMP7 i-subunits.35 In addition, HCV NS3 directly binds to β5i/LMP7.36 Both mechanisms result in impaired proteasomal proteolytic activity.35, 36
In the present study, IFN-γ-induced i-proteasomes were examined as important players in the antigen processing of cytoplasmic proteins. However, the expression of other components of the antigen-processing machinery is also increased by IFN-γ. These antigen-processing proteins include proteasome activator 28, transporters associated with antigen processing, and endoplasmic reticulum aminopeptidases.5, 6, 37, 38, 39 Given that IFN-γ-induced i-proteasome expression is suppressed by a PKR-mediated mechanism in HCV-infected cells, the protein expression of other antigen-processing machinery components might also be suppressed by the same mechanism in HCV-infected cells. Indeed, we confirmed this hypothesis by showing that IFN-γ-induced proteasome activator 28 expression was suppressed in HCV-infected cells (data not shown).
PKR phosphorylation, which is induced by double-stranded RNA, leads to eIF2α phosphorylation and ultimately suppresses new protein translation. In HCV infection, this pathway was shown to be responsible for the impaired expression of antiviral IFN-stimulated gene proteins.30 This is considered to be one of the major mechanisms of HCV immune evasion and has been shown to be dependent on the presence of a specific polymorphism within the HCV strains.40 We demonstrated that this mechanism operates to suppress IFN-induced MHC class I molecule expression31 and i-proteasome induction in HCV-infected cells. This indicates that the PKR-eIF2α pathway regulates not only antiviral innate immune responses but also CD8+ CTL responses and non-immune functions, such as the maintenance of protein homeostasis under conditions of IFN-induced oxidative stress. This further suggests that the pathway can be targeted for anti-HCV drug development. Recently, it was shown that cyclophilin inhibitors reduce PKR phosphorylation and restore IFN-stimulated gene protein expression in HCV-infected cells.41 It is thus worthwhile to investigate whether cyclophilin inhibitors restore i-proteasome induction in HCV-infected cells.
In the present study, we could not study directly the function of CD8+ CTLs specific to i-proteasome-dependent HCV epitopes because i-proteasome-dependent epitopes have not yet been identified in HCV proteins. Instead, we examined the generation of the HBV core 141–151 peptide, a strictly i-proteasome-dependent CTL epitope. Previously, it was demonstrated that the generation of the HBV core 141–151 peptide determines the activity of the CTL response.10 Moreover, the generation of the HBV core 141–151 peptide has been used as a functional evidence of the i-proteasome induction.33
In summary, we have shown that i-proteasome induction, assembly and function is suppressed in HCV-infected cells in a PKR-dependent manner. We propose the existence of an HCV immune-regulation mechanism that affects the antigen-processing machinery and the processing of cytoplasmic viral proteins.
Acknowledgments
This work was supported by a grant from the Korean Health Technology R&D Project through the Ministry of Health and Welfare of the Republic of Korea (HI13C1263). This work was also supported by National Research Foundation grants (NRF-2012-M3C1A1-048860 and NRF-2014R1A2A1A10053662) and by the Korea Advanced Institute of Science and Technology (KAIST) Future Systems Healthcare Project, which is funded by the Ministry of Science, ICT and Future Planning of Korea. The work was further supported by grants from the Deutsche Forschungsgemeinschaft (CRC854) to U Seifert and M Naumann.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Supplementary Material
References
- Klein J, Sato A. The HLA system. First of two parts. N Engl J Med 2000; 343: 702–709. [DOI] [PubMed] [Google Scholar]
- Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics 1995; 41: 178–228. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994; 78: 761–771. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82: 373–428. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2001; 2: 179–187. [DOI] [PubMed] [Google Scholar]
- Van den Eynde BJ, Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr Opin Immunol 2001; 13: 147–153. [DOI] [PubMed] [Google Scholar]
- Tanaka K. Role of proteasomes modified by interferon-gamma in antigen processing. J Leukoc Biol 1994; 56: 571–575. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol Rev 1998; 163: 161–176. [DOI] [PubMed] [Google Scholar]
- Mishto M, Liepe J, Textoris-Taube K, Keller C, Henklein P, Weberruß M et al. Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur J Immunol 2014; 44: 3508–3521. [DOI] [PubMed] [Google Scholar]
- Sijts AJ, Ruppert T, Rehermann B, Schmidt M, Koszinowski U, Kloetzel PM. Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J Exp Med 2000; 191: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kaer L, Ashton-Rickardt PG, Eichelberger M, Gaczynska M, Nagashima K, Rock KL et al. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity 1994; 1: 533–541. [DOI] [PubMed] [Google Scholar]
- Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med 2001; 193: 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling HJ, Swat W, Laplace C, Kühn R, Rajewsky K, Müller U et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science 1994; 265: 1234–1237. [DOI] [PubMed] [Google Scholar]
- Nussbaum AK, Rodriguez-Carreno MP, Benning N, Botten J, Whitton JL. Immunoproteasome-deficient mice mount largely normal CD8+ T cell responses to lymphocytic choriomeningitis virus infection and DNA vaccination. J Immunol 2005; 175: 1153–1160. [DOI] [PubMed] [Google Scholar]
- Basler M, Youhnovski N, Van Den Broek M, Przybylski M, Groettrup M. Immunoproteasomes down-regulate presentation of a subdominant T cell epitope from lymphocytic choriomeningitis virus. J Immunol 2004; 173: 3925–3934. [DOI] [PubMed] [Google Scholar]
- Zanker D, Waithman J, Yewdell JW, Chen W. Mixed proteasomes function to increase viral peptide diversity and broaden antiviral CD8+ T cell responses. J Immunol 2013; 191: 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol 2012; 13: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schröter F et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 2010; 142: 613–624. [DOI] [PubMed] [Google Scholar]
- Ebstein F, Voigt A, Lange N, Warnatsch A, Schröter F, Prozorovski T et al. Immunoproteasomes are important for proteostasis in immune responses. Cell 2013; 152: 935–937. [DOI] [PubMed] [Google Scholar]
- Opitz E, Koch A, Klingel K, Schmidt F, Prokop S, Rahnefeld A et al. Impairment of immunoproteasome function by β5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog 2011; 7: e1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med 2001; 345: 41–52. [DOI] [PubMed] [Google Scholar]
- Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 2005; 436: 946–952. [DOI] [PubMed] [Google Scholar]
- Cerny A, Chisari FV. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 1999; 30: 595–601. [DOI] [PubMed] [Google Scholar]
- Francavilla V, Accapezzato D, De Salvo M, Rawson P, Cosimi O, Lipp M et al. Subversion of effector CD8+ T cell differentiation in acute hepatitis C virus infection: exploring the immunological mechanisms. Eur J Immunol 2004; 34: 427–437. [DOI] [PubMed] [Google Scholar]
- Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med 2000; 191: 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisii C, Tempestilli M, Agrati C, Poccia F, Tocci G, Longo MA et al. Accumulation of dysfunctional effector CD8+ T cells in the liver of patients with chronic HCV infection. J Hepatol 2006; 44: 475–483. [DOI] [PubMed] [Google Scholar]
- Park SH, Rehermann B. Immune responses to HCV and other hepatitis viruses. Immunity 2014; 40: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Shin EC. Colorimetric focus-forming assay with automated focus counting by image analysis for quantification of infectious hepatitis C virions. PLoS ONE 2012; 7: e43960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach N, Herrmann U, Kähne T, Schicknick H, Pielot R, Naumann M et al. Differential effects of dopamine signalling on long-term memory formation and consolidation in rodent brain. Proteome Sci 2015; 13: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 2009; 6: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Sung PS, Park SH, Yoon S, Chang DY, Kim S et al. Hepatitis C virus attenuates interferon-induced major histocompatibility complex class I expression and decreases CD8+ T cell effector functions. Gastroenterology 2014; 146: 1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung PS, Cheon H, Cho CH, Hong SH, Park DY, Seo HI et al. Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc Natl Acad Sci USA 2015; 112: 10443–10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EC, Seifert U, Kato T, Rice CM, Feinstone SM, Kloetzel PM et al. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J Clin Invest 2006; 116: 3006–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Zimmermann A, Basler M, Groettrup M, Hengel H. A cytomegalovirus inhibitor of gamma interferon signaling controls immunoproteasome induction. J Virol 2004; 78: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apcher GS, Heink S, Zantopf D, Kloetzel PM, Schmid HP, Mayer RJ et al. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Lett 2003; 553: 200–204. [DOI] [PubMed] [Google Scholar]
- Khu YL, Tan YJ, Lim SG, Hong W, Goh PY. Hepatitis C virus non-structural protein NS3 interacts with LMP7, a component of the immunoproteasome, and affects its proteasome activity. Biochem.J 2004; 384: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabathuler R, Reid G, Kolaitis G, Driscoll J, Jefferies WA. Comparison of cell lines deficient in antigen presentation reveals a functional role for TAP-1 alone in antigen processing. J Exp Med 1994; 180: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol 2002; 3: 1169–1176. [DOI] [PubMed] [Google Scholar]
- York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol 2002; 3: 1177–1184. [DOI] [PubMed] [Google Scholar]
- Tasaka-Fujita M, Sugiyama N, Kang W, Masaski T, Murayama A, Yamada N et al. Amino acid polymorphisms in hepatitis C virus core affect infectious virus production and major histocompatibility complex class I molecule expression. Sci Rep 2015; 5: 13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daito T, Watashi K, Sluder A, Ohashi H, Nakajima S, Borroto-Esoda K et al. Cyclophilin inhibitors reduce phosphorylation of RNA-dependent protein kinase to restore expression of IFN-stimulated genes in HCV-infected cells. Gastroenterology 2014; 147: 463–472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.