Abstract
Motivation
Measuring differential gene expression is a common task in the analysis of RNA-Seq data. To identify differentially expressed genes between two samples, it is crucial to normalize the datasets. While multiple normalization methods are available, all of them are based on certain assumptions that may or may not be suitable for the type of data they are applied on. Researchers therefore need to select an adequate normalization strategy for each RNA-Seq experiment. This selection includes exploration of different normalization methods as well as their comparison. Methods that agree with each other most likely represent realistic assumptions under the particular experimental conditions.
Results
We developed the NVT package, which provides a fast and simple way to analyze and evaluate multiple normalization methods via visualization and representation of correlation values, based on a user-defined set of uniformly expressed genes.
Availability
The R package is freely available under https://github.com/Edert/NVT
1. Introduction
High throughput sequencing of RNA or cDNA (RNA-Seq) had an enormous impact on basic and clinical research since its introduction in the 2000s (Wang et al., 2009; Wilhelm and Landry, 2009). Independently of the used sequencing technology, the vast majority of research projects attempting to measure global expression levels of features (genes, exons, small RNAs or non-coding RNAs) compare expression values of multiple samples that represent different biological states. Importantly, any such differential expression (DE) analysis requires normalized data. This means that all non-biological influence, such as potential effects of sample preparation or sequencing efficiency, has to be removed to make the data comparable in between different experiments. To balance sequencing depths several methods use scaling factors (Dillies et al., 2013) (Table S1): Total count (TC), Median (ME), Upper quartile (UQ), Trimmed mean of M-values (TMM) and the relative log expression method implemented in DESeq (DESeq). Both TMM and DESeq operate under the assumption that most of the genes are not differentially expressed. Normalization methods without scaling factors are (Dillies et al., 2013) (Table S1): Quantile (Q), Reads per kilobase per million mapped reads (RPKM) and normalization by a defined gene set (G). Thus, two main concepts for data normalization in RNA-seq applications exist. While TMM and DESeq mainly consider differential library size, other normalization methods account for the distribution adjustment of read counts (TC, UQ, ME, Q, RPKM). Normalization based on RNA spike-ins (Lovén et al., 2012) makes other methods obsolete but this requires the RNA spike-in which had to be planned and applied previous to the sequencing. All the previously described methods are based on specific assumptions. Thus, identifying the method(s) for which these assumptions agree with the specific experimental setting represents a significant challenge, an exception is quantro (Hicks and Irizarry, 2012) which gives recommendations on when to use Q normalization or not. For example: if an experiment compares gene expression levels of healthy vs. rapidly growing tumor cells, the assumptions of non-differentially expressed genes or equal amounts of mRNA might not apply. The decision to utilize a certain normalization method can therefore have an enormous impact on the entire downstream analysis. Also, conclusions drawn from the enrichment of differentially expressed genes with respect to functional categories might be severely affected. As RNA-Seq experiments have become popular and powerful research tools in many areas of biology and medicine, also non-specialists need to be able to explore and compare different normalization methods to select the most appropriate one. To assist researchers in these tasks we present the normalization visualization tool (NVT). NVT is a fast and simple way to visually and quantitatively assess the normalization strategy including a set of user-defined genes. This set should consist of genes that do not change their relative expression levels in the particular study, this requires preliminary knowledge or experimental data (e.g. from quantitative PCR measurements).
2. Methods and implementation
NVT is an easy to use and freely available R package that provides visualization and evaluation of 10 different normalization methods for the comparison of two RNA-Seq data samples provided by the user (use case and detailed description in vignette, see supplement). It works with raw expression values per feature, may it be genes, exons or short RNA, originating from RNA-Seq datasets. The expression data of two samples has to be provided as a table of features and their respective number of mapped reads. NVT includes the normalization methods TC, ME, TMM, UQ, the upper quartile implementation from the NOISeq (Tarazona et al., 2011) package (UQ2), Q, RPKM, RPM, TPM, DESeq and G. If required for comparison reasons, also no normalization (N) can be applied. For some of these methods (RPKM and TPM), in addition to the expression values per feature, the respective gene length is also required as input. It can be provided as list or directly uploaded from a gff or gtf annotation-file via the GenomicRanges (Lawrence et al., 2013) and rtracklayer (Lawrence et al., 2009) packages. NVT allows to compare and evaluate normalization methods based on genes that are expected to be equally expressed in both samples. The members of this set of control genes can be visualized and used for normalization (by using "G" as normalization parameter). The different normalization methods are evaluated via a plot function and a function which calculates correlation based on the control gene set. The correlation can serve as a main criterion for the evaluation of the performance of any normalization method implemented in NVT. Available correlation functions are the Pearson-correlation coefficient, the root-mean-square-deviation (RMSD) and the mean-absolute-error (MEA). The normalization methods can be applied individually or all methods can be applied in one step and the resulting correlation values are presented in a ranked list. If required, the normalized expression per feature can also be extracted. The basic plot function generates a scatter-plot of the normalized expression data of two RNA-Seq samples. Based on the selected control gene set, whose members are highlighted in the scatter-plot, a linear model is calculated and plotted as a red line (the linear model can also be retrieved via the respective function). If the control genes are stably expressed, the red line will overlap with the gray dashed diagonal line. The advanced plot function requires ggplot2 (Wickham et al., 2009) for additional density bars. This function is illustrated for human gene expression data from the airway package (Himes et al., 2014) (Figure 1). In addition, NVT also offers the possibility to compare different normalization methods in between replicates. If two biological replicates are compared, all data points (including the defined gene set) would ideally reside on the diagonal, indicated by the dashed gray line in the scatter plot. The nearer a data point is located to the diagonal line, the better its correlation of this particular feature is in the two samples.
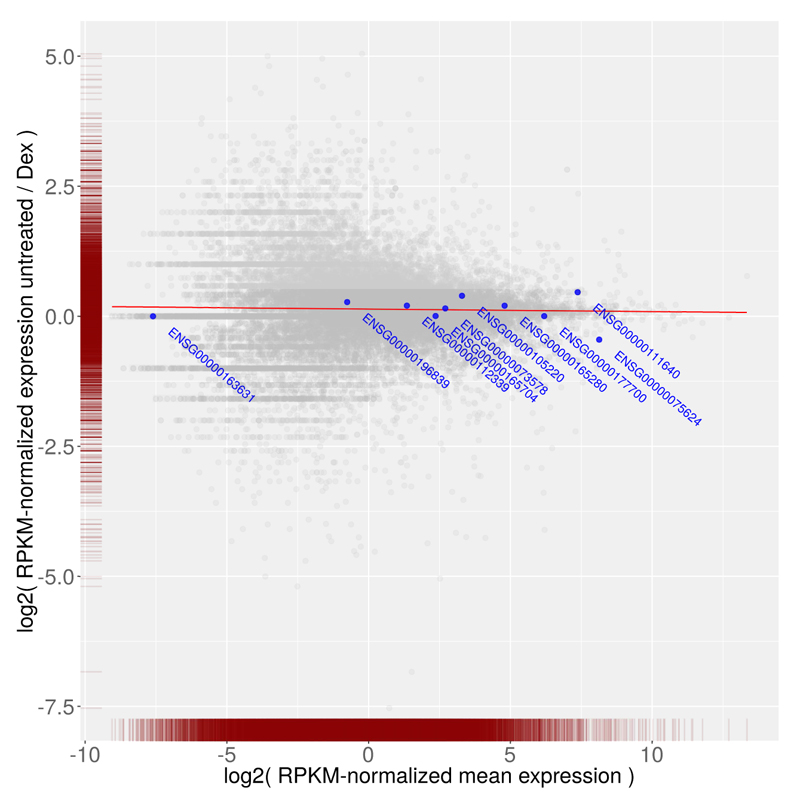
Fig. 1.
MA-plot of RPKM normalized gene expression of N61311 cells untreated vs. treated with the anti-inflammatory drug dexamethasone (Dex). The expression levels per gene are represented as gray dots, the user defined set of control genes (GAPDH, ALB, ACTB, HPRT1, ADA, POLR2L, VCP, GPI, HBS1L and SDHA) are shown in blue and the linear model calculated based on expression of the control gene set is depicted as red line.
3. Conclusions
The appropriate assumption(s) for correct normalization of RNA-Seq data critically depend on the nature of the particular experimental setup. NVT is a simple and fast tool to evaluate the normalization strategy for any given RNA-Seq data set. The visual comparison of normalization methods to a user-defined gene set of control genes in NVT is an efficient and intuitive way to assess the performance of different normalization methods. NVT generates publication-ready figures and also provides correlation measures. The package thereby facilitates the documentation of methodological decisions for RNA-Seq experiments. NVT is hosted on https://github.com, using its infrastructure for maintenance and bug tracking, updates and release of future versions. After assessing the demands of users, possible improvements and additional functions will be: the implementation of additional normalization methods and the possibility to include custom normalized data.
Supplementary Material
Supplementary information: Supplementary data are available at Bioinformatics online.
Funding
T.E. and F.G. were supported by the starting grant "ONCOMECHAML" from the European Research Council (ERC).
Footnotes
Conflict of Interest: none declared.
References
- Dillies M-A, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- Hicks S, Irizarry R. quantro: a data-driven approach to guide the choice of an appropriate normalization method. Genome Biology. 2015;16:117. doi: 10.1186/s13059-015-0679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, Whitaker RM, Duan Q, Lasky-Su J, Nikolos C, et al. RNA-Seq Transcriptome Profiling Identifies CRISPLD2 as a Glucocorticoid Responsive Gene that Modulates Cytokine Function in Airway Smooth Muscle Cells. PLoS ONE. 2014;9:e99625. doi: 10.1371/journal.pone.0099625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Revisiting Global Gene Expression Analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Ggplot2: elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- Wilhelm BT, Landry J-R. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.