Abstract
Bacterial endospore formers can produce spores that are resistant to many food processing conditions, including heat. Some spores may survive heating processes aimed at production of commercially sterile foods. Recently, it was shown that a spoVA operon, designated spoVA2mob, present on a Tn1546 transposon in Bacillus subtilis, leads to profoundly increased wet heat resistance of B. subtilis spores. Such Tn1546 transposon elements including the spoVA2mob operon were also found in several strains of Bacillus amyloliquefaciens and Bacillus licheniformis, and these strains were shown to produce spores with significantly higher resistances to wet heat than their counterparts lacking this transposon. In this study, the locations and compositions of Tn1546 transposons encompassing the spoVA2mob operons in B. amyloliquefaciens and B. licheniformis were analyzed. Introduction of these spoVA2mob operons into B. subtilis 168 (producing spores that are not highly heat resistant) rendered mutant 168 strains that produced high-level heat resistant spores, demonstrating that these elements in B. amyloliquefaciens and B. licheniformis are responsible for high level heat resistance of spores. Assessment of growth of the nine strains of each species between 5.2°C and 57.7°C showed some differences between strains, especially at lower temperatures, but all strains were able to grow at 57.7°C. Strains of B. amyloliquefaciens and B. licheniformis that contain the Tn1546 elements (and produce high-level heat resistant spores) grew at temperatures similar to those of their Tn1546-negative counterparts that produce low-level heat resistant spores. The findings presented in this study allow for detection of B. amyloliquefaciens and B. licheniformis strains that produce highly heat resistant spores in the food chain.
Keywords: spores, heat resistance, Tn1546 transposon, spoVA operon, genome analysis
Introduction
The ubiquitous presence of bacterial spore formers in nature can be largely attributed to their ability to produce endospores (spores) that can survive harsh environmental conditions (Nicholson et al., 2000; Setlow, 2006). Bacterial spores can enter the food chain from many different sources, for example via soil, dust, and biofilms (Heyndrickx, 2011). The intrinsic resistance properties of spores may result in survival during food processing, in which heating is one of the most commonly applied treatments to reduce bacterial loads. Such treatments put selective pressure on the microflora that is present, allowing for survival of those strains that produce spores with high heat resistance (Postollec et al., 2012). Surviving spores may germinate upon exposure to certain environmental triggers, and can subsequently resume vegetative growth, potentially resulting in food pathogenicity or food spoilage, depending on the species (Scheldeman et al., 2006; Wells-Bennik et al., 2016).
Spores of mesophilic species belonging to the B. subtilis group are commonly found in various food ingredients and food products. The B. subtilis group encompasses the species B. subtilis, B. amyloliquefaciens, B. licheniformis, B. vallismortis, B. mojavensis, B. atropheus, and B. sonorensis, which are phylogenetically close, yet distinguishable (Logan and Vos, 2009). These species can generally grow between temperatures of 30–50°C, with reported growth temperatures of B. licheniformis up to 58°C (Warth, 1978). The spores of B. subtilis, B. amyloliquefaciens and B. licheniformis are commonly found in various food ingredients and food products including cocoa, herbs, spices, bread, soups, milk, and milk powders (te Giffel et al., 1996; Oomes et al., 2007; Lima et al., 2011; Lücking et al., 2013; Miller et al., 2015). These species are for instance well-known contaminants of raw materials used in bread making (Rosenkvist and Hansen, 1995; Sorokulova et al., 2003), and the spores can potentially even survive the bread baking process (Valerio et al., 2015). After spore survival, germination, and outgrowth, vegetative cells of B. amyloliquefaciens, B. subtilis or B. licheniformis can result in spoiled food products. B. subtilis, for instance has been reported to be present in cocoa (Lima et al., 2011) leading to spoiled chocolate drinks, B. licheniformis may be present in milk and milk powders leading to spoilage of heat treated dairy products (Gopal et al., 2015), and B. amyloliquefaciens may spoil bread, resulting in ropy bread by degradation of starch and the formation of extracellular polysaccharides (Sorokulova et al., 2003; Valerio et al., 2012, 2015). Certain strains of B. licheniformis can produce a toxin, lichenisyn A, that can cause foodborne illness (Salkinoja-Salonen et al., 1999; Nieminen et al., 2007; Logan, 2012). Lichenisyn is a non-ribosomally synthesized lipo-peptide that is heat-stable (Konz et al., 1999). Due to the pathogenic potential of strains of B. licheniformis, it is critical to control these spores in the food chain (Madslien et al., 2013).
Notable differences have been observed with respect to the spore wet heat resistance properties of strains within the B. subtilis group (Kort et al., 2005; Oomes et al., 2007; Lima et al., 2011; Berendsen et al., 2015). Following a detailed analysis of the heat resistance of spores of 14 strains belonging to the B. subtilis group, strains could be divided in two groups based on spore heat resistance (Berendsen et al., 2015). For B. subtilis strains, it was recently demonstrated that spores with high-level heat resistance contain a Tn1546 transposon, encompassing a spoVA operon that is directly responsible for this phenotype (designated spoVA2mob, where mob indicates the presence on a mobile genetic element; Berendsen et al., 2016a). In addition, we observed high-level heat resistance of spores of B. licheniformis and B. amyloliquefaciens strains that carried the Tn1546 transposon; the spores of these strains showed heat resistance levels similar to as those of spores of B. subtilis strains with a Tn1546 transposon (Berendsen et al., 2015, 2016a).
In this study, we report the presence and composition of Tn1546 transposon homologs of B. subtilis which were found in strains of B. amyloliquefaciens and strains of B. licheniformis that produced highly heat resistant spores. This was performed by genome analysis or PCR detection. The spoVA2mob operons found in B. amyloliquefaciens and B. licheniformis were introduced into B. subtilis to assess their role in spore heat resistance. In addition, the growth temperatures of all B. amyloliquefaciens and B. licheniformis strains (nine each) with or without the Tn1546 transposons were assessed.
Materials and methods
Bacterial strains used in this study
The strains used in this study for genomic and phenotypic analyses are listed in Table 1. This included nine strains of B. amyloliquefaciens isolated from natural and food environments, nine strains of B. licheniformis from food environments, and two strains of B. subtilis. The genome sequences were available for the two B. subtilis strains, all B. amyloliquefaciens strains, and four B. licheniformis strains (Berendsen et al., 2016a,b; Krawczyk et al., 2016). For B. amyloliquefaciens strains B425 and B4140, the heat inactivation kinetics of spores were described previously (Berendsen et al., 2015) and for all other B. amyloliquefaciens and B. licheniformis strains, the heat inactivation kinetics of spores were previously determined (Berendsen et al., 2016a). Strains of B. subtilis 168 and B. subtilis B4146 were included in the genome analysis as reference strains, and the heat resistances of the spores of these strains were previously analyzed (Berendsen et al., 2015).
Table 1.
Strains used in this study.
| Strain no | Species | Description | Tn1546 | Genome sequence | References |
|---|---|---|---|---|---|
| B425 | B. amyloliquefaciens | Isolated from sterilized milk | + | LQYP00000000 | Berendsen et al., 2015; Krawczyk et al., 2016 |
| B4140 | B. amyloliquefaciens | Isolated from pizza | − | LQYO00000000 | Berendsen et al., 2015; Krawczyk et al., 2016 |
| 10A5 | B. amyloliquefaciens | Known as 10A5; NRRL B-14393, isolated from soil | −(PCR) | No | Priest et al., 1988 |
| FZB42 | B. amyloliquefaciens | Known as 10A6; FZB42, isolated from plant soil | − | NC_009725 | Chen et al., 2007 |
| 10A18 | B. amyloliquefaciens | Known as 10A18; CU8004 | −(PCR) | No | Zahler et al., 1987 |
| DSM7 | B. amyloliquefaciens | Known as DSM7, isolated from soil | + | FN597644 | Ruckert et al., 2011 |
| DSM1060 | B. amyloliquefaciens | Known as DSM1060 | −(PCR) | No | Priest et al., 1987 |
| 101 | B. amyloliquefaciens | Received as 101 | −(PCR) | No | Berendsen et al., 2016a |
| SB42 | B. amyloliquefaciens | Received as SB42 | −(PCR) | No | Berendsen et al., 2016a |
| B4089 | B. licheniformis | Known as E5/T12, isolated from pea soup | − | LKPM00000000 | Oomes et al., 2007 |
| B4090 | B. licheniformis | Known as T1, isolated from pea soup | + | LQYL00000000 | Oomes et al., 2007; Krawczyk et al., 2016 |
| B4091 | B. licheniformis | Known as T29, isolated from mushroom soup | − | LQYM00000000 | Oomes et al., 2007; Krawczyk et al., 2016 |
| B4092 | B. licheniformis | Isolated from buttermilk powder | + | LQYK00000000 | Krawczyk et al., 2016 |
| B4094 | B. licheniformis | Isolated from camomile tea | + | LKPN00000000 | Berendsen et al., 2016a |
| B4121 | B. licheniformis | Isolated from sateh pastry | − | LKPO00000000 | Berendsen et al., 2016a |
| B4123 | B. licheniformis | Isolated from sateh pastry | − | LKPP00000000 | Berendsen et al., 2016a |
| B4124 | B. licheniformis | Isolated from pancakes | − | LKPQ00000000 | Berendsen et al., 2016a |
| B4125 | B. licheniformis | Isolated from pancakes | − | LKPR00000000 | Berendsen et al., 2016a |
| B4062 | B. subtilis | Type strain 168 | − | NC_000964 | Kunst et al., 1997 |
| B4146 | B. subtilis | Isolated from curry sauce | + | NZ_JXHR01000000 | Berendsen et al., 2015, 2016a,b |
| 168-spoVA2mob (B4090) | B. subtilis | 168 amyE::spoVA2mob (B4090) specR | − | − | This study |
| 168-spoVA2mob(DSM7) | B. subtilis | 168 amyE::spoVA2mob (DSM7) specR | − | − | This study |
Genome analysis
Multiple sequence alignments were made for protein sequences of conserved genes that were present in single copy in all genomes using MUSCLE (Edgar, 2004). The core genome phylogenetic tree was constructed using PHYML (Guindon and Gascuel, 2003). To investigate the presence of the Tn1546 transposon and the encoded spoVA (designated spoVA2mob) operon that mediates high-level heat resistance, an orthology matrix was constructed using Ortho-MCL (Li et al., 2003) with the genomes of the four B. amyloliquefaciens strains, the nine B. licheniformis strains, B. subtilis B4146 as a strain that produces spores with high heat resistance, and B. subtilis 168 as a reference strain that produces spores with low-level heat resistance (Supplementary Data Sheet 1). The genomic organization of the Tn1546 transposon was visualized using the Artemis comparison tool (ACT), and using microbial genomic context viewer (MGcV) (Carver et al., 2005, 2012; Overmars et al., 2013). For the identified Tn1546 transposons, operon predictions were performed using FGENESB (http://www.softberry.com). Additionally, manual sequence comparisons and searches for pseudogenes were performed for all genes in the transposon.
Generic PCR primers were used, as described previously, for the detection of the Tn1546 encoded genes tnpA, spoVAC2mob and cls in the five strains of B. amyloliquefaciens for which no genome sequence information was available (i.e., strains 10A5, FZB42, 10A18, 101 and SB42; Berendsen et al., 2016a).
Comparison of heat inactivation kinetics of spores
For all strains the heat resistances of their spores (produced at 37°C) was previously determined using capillary tubes, at three different temperatures, using at least five time points, as described in Berendsen et al. (2015) and Berendsen et al. (2016a). For comparison of strain specific heat resistance of spores, the z-values (i.e., the increase in temperature required to achieve an additional log unit reduction) were calculated as described previously (van Asselt and Zwietering, 2006). Furthermore, the D-values (i.e., the time needed at that temperature to decrease the spore count 10-fold) reported previously, were used to calculate reference D-values (Dref) at the reference temperature of 110°C with the corresponding 95% prediction intervals, as presented in Supplementary Table 1 (van Asselt and Zwietering, 2006).
Cloning of the spoVA2mob operon
The spoVA2mob operon, including the predicted promotor region as present in strain B. licheniformis B4090 and B. amyloliquefaciens DSM7 was cloned into plasmid pDG1730, using a procedure as previously described (Berendsen et al., 2016a). The obtained constructs were transformed to B. subtilis 168 and integrated in the amyE locus as described in Berendsen et al. (2016a), to verify the role of this operon in increased heat resistance of spores. Spores were prepared for strains 168 amyE::spoVA2mob (DSM7), 168 amyE::spoVA2mob (B4090) and 168 as described above, and the heat resistance of spores of these strains was assessed by heating at 100°C for 1 h, followed by plating, incubation and enumeration of survivors.
Determination of the growth temperature
All strains were cultured from the −80°C glycerol stocks in brain heart infusion broth (BHI, Oxoid). For assessing growth at different temperatures, 24-well plates were filled with 1.5 ml of BHI agar and surface inoculated with 20 μl of turbid individual overnight cultures (>107 cfu mL−1, i.e., > 2 × 105 cells on the surface of the agar in each well). Growth or no growth on the agar surface was assessed on day 2, 6, 13, 23, and 30 at the following temperatures: 5.2, 11.5, 14.5, 30.5, 36.3, 45.9, 55.1, and 57.7°C. Growth was scored visually, based on the formation of colonies or a lawn on the agar surface in the wells. The day on which growth was first observed was noted. Two biological replicates were performed, and for each biological replicate, two technical replicates were performed.
Results
Genome mining for the Tn1546 transposon
For B. subtilis strains, it has been demonstrated that the presence of a Tn1546 transposon is responsible for high-level heat resistance of the spores (Berendsen et al., 2016a). The presence of the Tn1546 transposon was assessed in nine strains of B. amyloliquefaciens and nine strains of B. licheniformis. The genome sequences of all nine strains of B. licheniformis were available and in three of these strains the Tn1546 transposon was found (namely, strains B4090, B4092, and B4094; Figure 1). The genome sequences of four strains of B. amyloliquefaciens were available, and the Tn1546 transposon was found in two of these strains of B. amyloliquefaciens (namely in B425 and DSM7; Figure 1). The predicted protein for the transposase TnpA, which is part of the Tn1546 transposon, was found in orthologous group OG3133 for all of the strains that carry the Tn1546 transposon. Genome sequences were not available for the other five strains of B. amyloliquefaciens. PCR-based detection of the genes tnpA, spoVAC and cls (that are present on the transposon and very well conserved) using primers for these three target genes showed positive results for B. amyloliquefaciens strains B425 and DSM7 but did not reveal the transposon in the other five strains. In short, two out of nine strains of B. amyloliquefaciens and three out of nine strains of B. licheniformis contained the Tn1546 transposon.
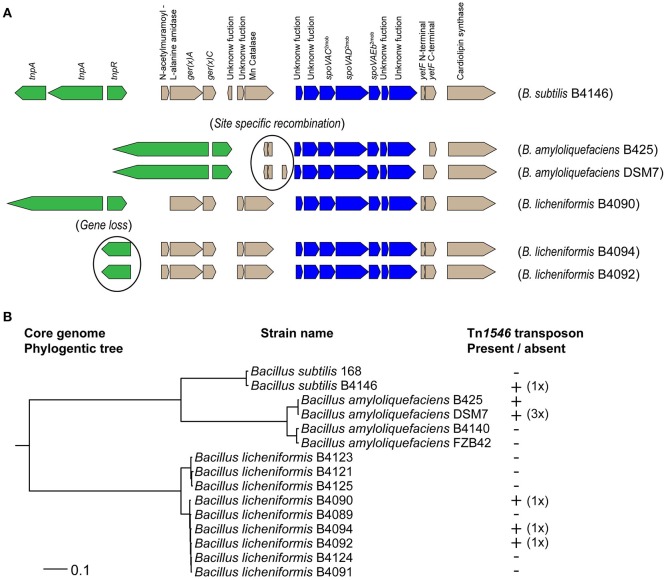
Figure 1.
(A) Overview of the Tn1546 transposons found in B. subtilis B4146, B. amyloliquefaciens B425 and DSM7, and B. licheniformis B4090, B4092, and B4090. The predicted gene functions are indicated for the transposon of B. subtilis B4146: a transposase gene (tnpA), a resolvase gene (res), an operon of N-acetylmuramoyl-L-alanine amidase, gerKA and ger(X)C (Operon 1), an operon of a gene with unknown function and a manganese catalase (Operon2), an operon of two genes with unknown functions, spoVAC, spoVAD, spoVAEb and two genes with unknown functions (Operon 3, spoVA2mob), a fragmented yetF gene (Gene 4), and a cardiolipin synthase gene (Gene 5). The transposons in B. amyloliquefaciens are smaller, probably due to site specific recombination events, whereby operon 1 and operon 2 were lost. In B. licheniformis B4092 and B4094 the resolvase gene was not present, which is most likely a result of gene loss. (B) Maximum likelihood phylogenetic tree based on core genome of single genes of 15 strains of the B. subtilis group. The presence and copy number of the Tn1546 transposon was indicated behind the corresponding strains. B. subtilis B4146, B. licheniformis B4090, B4092, and B4094 carry a single transposon. The B. amyloliquefaciens strain DSM7 carries three copies of the transposon, while for strain B425 the copy number could not be determined.
Heat resistance of spores is related to the presence of the Tn1546 transposon
The heat resistances of spores of B. amyloliquefaciens and B. licheniformis were assessed in relation to the presence or absence of the Tn1546 transposon (Supplementary Table 1). The heat resistance of spores was displayed as a reference decimal reduction time of spores per strain at the reference temperature of 110°C (-values) in relation to the presence or absence of the Tn1546 transposon (Supplementary Table 1).
Two strains of B. amyloliquefaciens, namely B425 and DSM7, contained the Tn1546 transposon. These strains produced spores that required significantly longer heating times (unpaired t-test p < 0.001) at 110°C, i.e., approximately 15 times, to achieve one decimal reduction, than spores of the other seven strains without this transposon. Strains B. licheniformis B4090, B4092, and B4094 contained the Tn1546 transposon, and the spores of these strains all required longer heating times (unpaired t-test p < 0.001) (2.5 times) to reach a decimal reduction than the spores of the six B. licheniformis strains that did not possess the Tn1546 transposon.
The spoVA2mob operon is responsible for increased heat resistance of spores
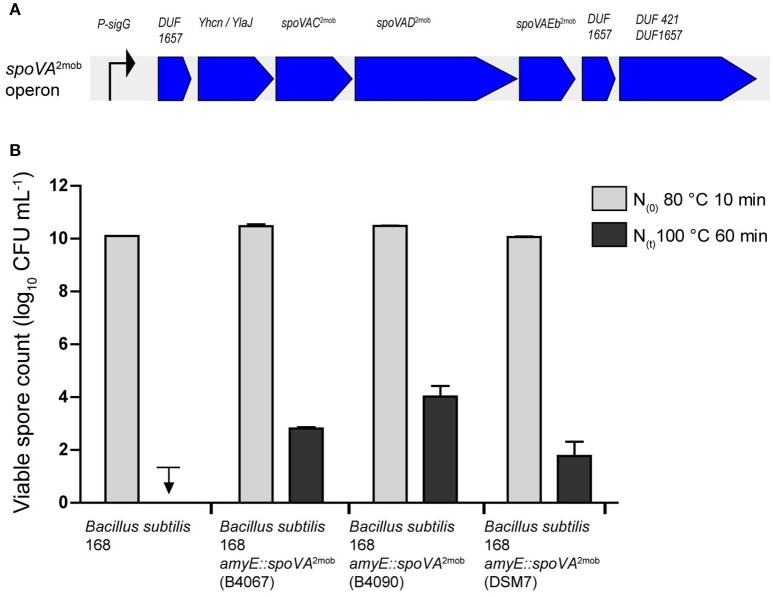
We previously showed that the introduction of the spoVA2mob operon originating from B. subtilis strain B4067 into laboratory strain B. subtilis 168 resulted in the formation of high-level heat resistant spores by this strain (Berendsen et al., 2016a). To establish whether the spoVA2mob operons present in the Tn1546 transposons of B. licheniformis B4090 and B. amyloliquefaciens DSM7 have a functional role, these operons were also introduced individually into B. subtilis 168. The B. subtilis 168 mutants carrying the spoVA2mob genes of B. licheniformis B4090 and the spoVA2mob genes of B. amyloliquefaciens produced spores with significantly higher heat resistances than the parent strains after heating at 100°C for 60 min (Figure 2). The spores of B. subtilis 168 were inactivated to a level below the detection limit, i.e., more than 8 log units reduction, indicative of a low level of heat resistance of spores. The spores of B. subtilis 168 amyE::spoVA2mob (containing the spoVA2mob operon of B. licheniformis B4090) and spores of B. subtilis 168 amyE::spoVA2mob (containing the spoVA2mob operon of B. amyloliquefaciens DSM7) showed survival of 4.0 log10 unit (±0.4), and 1.7 log10 unit (±0.5), respectively, indicating a high level of heat resistance of spores. The control strain B. subtilis 168 amyE::spoVA2mob producing high level heat resistant spores (containing the spoVA2mob operon of B. subtilis B4067) (Berendsen et al., 2016a), showed survival of 2.8 log10 unit (±0.05).
Figure 2.
(A) Overview of the spoVA2mob operon, as initially found in B. subtilis strain B4146. The spoVA2mob operon has a predicted sigma G binding site upstream of the first gene. The operon consists of seven genes: a gene of unknown function with a predicted DUF1657 domain, a gene of unknown function with a predicted YhcN/YlaJ domain, spoVAC, spoVAD, spoVAEb, a gene of unknown function with a predicted DUF1657 domain, and a gene of unknown function with a predicted DUF421 and a DUF1657 domain. (B) Spore counts after heating at 80°C for 10 min and at 100°C for 60 min for strains of B. subtilis 168, B. subtilis 168 amyE::spoVA2mob [B4067, data from Berendsen et al. (2016a)], B. subtilis 168 amyE::spoVA2mob (B4090), and B. subtilis 168 amyE::spoVA2mob (DSM7). The downward arrow indicates that the spores were inactivated below detection limit.
Detailed analysis of the Tn1546 transposon
The composition of the Tn1546 transposon in B. licheniformis strains B4090, B4092 and B4094 is shown in Figure 1A. In these strains, the Tn1546 transposon is highly similar to the one found in B. subtilis B4146 (Figure 1A). The transposons found in B. subtilis and B. licheniformis consist of genes that are required for transposition, and furthermore contain three operons and two single genes. The Tn1546 transposon found in the B. amyloliquefaciens strains DSM7 and B425 was smaller than the transposon found in B. subtilis and B. licheniformis. In both B. amyloliquefaciens strains, the first two operons were absent, possibly due to a site-specific recombination event, as a recombinase gene and a hypothetical gene were present at that genomic location.
The evolutionary relatedness of the different strains and species was visualized in a maximum likelihood core genome phylogenetic tree, based on concatenated protein sequences of conserved genes present in single copy in all genomes (Figure 1B). The species B. amyloliquefaciens, B. licheniformis and B. subtilis clustered in separate branches of the phylogenetic tree. For B. amyloliquefaciens, the strains with the Tn1546 transposon clustered together, whereas this was not the case for B. licheniformis strains carrying the Tn1546 transposon.
The genomic locations of the Tn1546 transposons were different for B. subtilis, B. amyloliquefaciens and B. licheniformis. In B. subtilis, the transposon was found at two genomic locations, namely inserted in yitF and between yxjA and yxjB (Berendsen et al., 2016a). In B. amyloliquefaciens, three Tn1546 transposons were found in strain DSM7 at three different genomic locations, namely between a gene encoding for a fructose-1,6-bisphophatase and a hypothetical gene, between two hypothetical genes, and between a hypothetical gene and rapK. For B. amyloliquefaciens strain B425, it was not possible to determine the genomic location(s) and copy number of the Tn1546 transposon, since contig breaks were present on both sides of the Tn1546 transposon. In B. licheniformis strains B4090, B4092, and B4094, a single Tn1546 transposon was found integrated in a gene that encodes a D-alanyl-D-alanine carboxypeptidase.
Detailed analysis revealed that some genes in the Tn1546 transposon were mutated and present as pseudogenes in the transposon of some strains. The genes tnpA and tnpR in the Tn1546 transposon (which are required for active transposition) were intact and present in B. amyloliquefaciens strains B425 and DSM7 and in B. licheniformis strain B4090.
Determination of vegetative growth at different temperatures
None of the 18 B. amyloliquefaciens and B. licheniformis strains were able to grow at 5.2°C within 30 days. At 11.5°C, six out of nine B. amyloliquefaciens strain and five out of nine B. licheniformis strains showed growth. At 14.5°C, all but one strain showed growth. All strains grew at the highest temperature tested (57.7°C), and at all other temperatures, growth of strains was observed except for strain 101 at 55.1°C. In the case of the nine B. amyloliquefaciens and nine B. licheniformis strains, high level heat resistance of spores due to the presence of the Tn1546 transposon was not correlated with the ability to grow at different temperatures (Table 2).
Table 2.
Determination of ability to grow at different temperatures for nine strains of B. amyloliquefaciens and nine strains of B. licheniformis.
| Species | Strain | Tn1546 transposon | Growth observed at temperaturea | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5.2°C | 11.5°C | 14.5°C | 30.5°C | 36.3°C | 45.9°C | 55.1°C | 57.7°C | |||
| B. amyloliquefaciens | 10A5 | No | − | +++ | +++ | ++++ | ++++ | ++++ | +++ | +++ |
| FZB42 | No | − | − | − | ++++ | ++++ | ++++ | ++++ | +++ | |
| 10A18 | No | − | − | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
| 101 | No | − | +++ | +++ | ++++ | ++++ | ++++ | − | +++ | |
| SB42 | No | − | ++ | ++++ | ++++ | ++++ | ++++ | +++ | +++ | |
| B4140 | No | − | +++ | ++++ | ++++ | ++++ | ++++ | ++ | ++++ | |
| DSM1060 | No | − | − | ++ | ++++ | ++++ | ++++ | +++ | ++++ | |
| DSM7 | Yes | − | ++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
| B425 | Yes | − | +++ | +++ | ++++ | ++++ | ++++ | ++ | ++++ | |
| B. licheniformis | B4089 | No | − | − | ++ | ++++ | ++++ | ++++ | ++++ | +++ |
| B4091 | No | − | − | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
| B4121 | No | − | − | +++ | ++++ | ++++ | ++++ | +++ | +++ | |
| B4123 | No | − | +++ | +++ | ++++ | ++++ | ++++ | ++++ | +++ | |
| B4124 | No | − | ++ | +++ | ++++ | ++++ | ++++ | ++++ | +++ | |
| B4125 | No | − | + | +++ | ++++ | ++++ | ++++ | +++ | +++ | |
| B4090 | Yes | − | + | ++ | ++++ | ++++ | ++++ | ++++ | +++ | |
| B4092 | Yes | − | +++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
| B4094 | Yes | − | − | ++ | ++++ | ++++ | ++++ | ++++ | +++ | |
Growth in: 2 days (++++), 6 days (+++), 13 days (++), 23 days (+), and no growth within 30 days(−).
Discussion
B. licheniformis strains B4090, B4092, and B4094 contained a single copy of the Tn1546 transposon with a single spoVA2mob operon. The heat resistances of spores of B. licheniformis with or without this operon were significantly different, but relatively modest. For B. amyloliquefaciens, spores of strains B425 and DSM7 showed comparable high-levels of heat resistance, which were significantly higher than those of the spores of other B. amyloliquefaciens strains. Strain DSM7 contains three Tn1546 transposable elements, and it is likely that strain B425 also contains multiple copies, however this remains to be established. The number of spoVA2mob operons have previously been found to correlate with the level of heat resistance of spores in B. subtilis; strains carrying three copies produced spores with the highest level of heat resistance (Berendsen et al., 2016a). For B. subtilis, it has been shown that the Tn1546 transposon was found at different locations in the genome, all leading to high-level heat resistance of spores (Berendsen et al., 2016a). It remains to be established whether the location of the insertion of the transposon in the genome of B. amyloliquefaciens and B. licheniformis plays a role in the level of heat resistance of spores.
The presence of intact tnpA and tnpR genes in B. amyloliquefaciens DSM7 and B425, and B. licheniformis B4090 suggests that active transposition of the Tn1546 element may be possible for these strains, although active transposition of the Tn1546 transposon is believed to require a plasmid intermediate, as has been described in Enterococcus faecium (Arthur et al., 1993). Interestingly, B. amyloliquefaciens DSM7, containing the intact tnpA and tnpR genes, contained three Tn1546 transposons. The encoded proteins required for transposition potentially allowed for internal transposition within the chromosome of strain DSM7. In B. subtilis strain B4146 and in B. licheniformis strains B4092 and B4094, the transposition genes are absent or not intact, suggesting that active transposition of the Tn1546 transposon is not likely to occur in these strains. This does not mean that the transposons cannot be transferred; transfer of genetic material including the Tn1546 transposon can be mediated by other transfer mechanisms, such as phage transduction, as described previously for B. subtilis (Berendsen et al., 2016a), or via the uptake of external DNA via natural competence (Kovacs et al., 2009).
The spoVA2mob operons present on the Tn1546 transposons differ from the spoVA operons (designated spoVA1) that are encoded on the chromosomes of B. subtilis (Tovar-Rojo et al., 2002), B. amyloliquefaciens and B. licheniformis. The SpoVA proteins encoded in the spoVA1 operon are required for uptake of DPA during the sporulation process, and sporulation cannot be completed upon deletion or disruption of these genes in B. subtilis 168 (Tovar-Rojo et al., 2002). In addition, the SpoVA proteins are involved in the release of Ca-DPA during the germination process (Vepachedu and Setlow, 2004, 2007), with the SpoVAC protein functioning as a mechano sensitive channel during germination (Velasquez et al., 2014). The SpoVAD protein has a binding pocket, whereby it can directly bind DPA (Li et al., 2012). Both the spoVA1 and the spoVA2mob operons contain genes encoding SpoVAC, SpoVAD and SpoVAEb, while the other genes in the operons are different. Presumably the presence of the spoVA2mob encoded proteins results in the increased uptake of DPA into the spore core, as was previously found for B. subtilis (Berendsen et al., 2016a).
It is known that environmental conditions during sporulation, such as temperature, matrix and medium composition, can influence the heat resistance of spores of the B. subtilis group (Cazemier et al., 2001; Melly et al., 2002; Rose et al., 2007). To allow for a direct comparison of the resistance properties of spores of the different strains, the applied sporulation conditions of all B. licheniformis and B. amyloliquefaciens strains were the same (Berendsen et al., 2015, 2016a). Factors that are known to influence spore heat resistance include the composition of the sporulation medium, and it is known that the addition of salts (Ca2+, Mn2+, Mg2+ and K+) results in higher resistances of spores to heat (Cazemier et al., 2001). Furthermore, the temperature during sporulation can influence the heat resistance of B. subtilis spores (Melly et al., 2002). In line with these findings, the heat resistances of spores of a B. licheniformis strain have been reported to be higher upon sporulation at 45°C, with a modeled optimum at 49°C, than at lower temperatures such as 20°C (Baril et al., 2012). Overall, the environmental conditions during sporulation, the presence or absence of genetic elements such as the spoVA2mob operon, and the storage conditions of spores will ultimately determine the heat resistance properties of spores. It is therefore conceivable that spores produced under laboratory conditions do not necessarily reach the same levels of heat resistance as spores found in foods (van Zuijlen et al., 2010; Lima et al., 2011).
It is generally assumed for bacterial spore formers that higher optimal growth temperatures of vegetative cells correlates positively with spore heat resistance (Nicholson et al., 2000). In this study, some differences were seen between strains with respect to their abilities to grow at different temperatures, but no consistent pattern was seen for strains that contain the Tn1546 element and produce high level heat resistant spores versus the ones that do not harbor the element and produce low-level heat resistant spores. All strains were able to grow at 57.7°C. All genes present on the Tn1546 transposon are under the control of sporulation-specific sigma factors K or G (Berendsen et al., 2016a) and are thus only expressed during late stages of sporulation. It is therefore in line with expectations that the genes on the Tn1546 transposon, which determine spore heat resistance, do not influence the ability of vegetative cells to grow at high temperatures as these genes are not expressed during vegetative growth.
Conclusions
Variation in heat resistance of spores exists between strains of different spore forming species but also within species (Oomes et al., 2007; Orsburn et al., 2008; Lima et al., 2011; Berendsen et al., 2015). In this study, a genomic analysis revealed the presence of Tn1546 transposons in two strains of B. amyloliquefaciens and in three strains of B. licheniformis. The presence of this transposon, containing the spoVA2mob operon, correlated with high-level heat resistance of spores. Strains producing low or high level heat resistant spores showed similar temperature ranges for growth. A functional role of the spoVA2mob operon in increasing the heat resistance of spores was demonstrated by cloning these operons in B. subtilis 168, resulting in spores with high-level heat resistance. Clearly, mere identification of the species of spores in food products does not provide information on the heat resistance levels of these spores. The knowledge obtained in this study on the role of the spoVA2mob operon in spore heat resistance can be used for specific detection of strains of the B. subtilis group that produce high-level heat resistant spores. Multiple DNA based methods can be used for the detection of such genetic elements, such as whole genome sequencing and specific PCR detection, among others (Caspers et al., 2011; Alkema et al., 2016). The ability to detect certain strains of B. subtilis group that have the ability to produce high level heat resistant spores can aid control of these sporeformers in the food chain.
Author contributions
EB and RK collected the data. EB, RK, AD, and JB analyzed the data. EB, OK, and MW wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Patrick Janssen for technical assistance. The authors have declared that no competing interests exist. The research was funded by TI Food and Nutrition, a public-private partnership on pre-competitive research in food and nutrition. The funding organization had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01912/full#supplementary-material
Orthology matrix constructed using Ortho-MCL with the genomes of the four B. amyloliquefaciens strains, the nine B. licheniformis strains, B. subtilis B4146 as a strain that produces spores with high heat resistance and B. subtilis 168 as a reference strain that produces spores with normal heat resistance.
Calculated reference D-value (Dref) at a reference temperature (Tref) of 110°C, and z-value, of spores of nine strains of B. amyloliquefaciens and spores of nine strains of B. licheniformis (Berendsen et al., 2015, 2016a). In these studies, all strains were sporulated on the same media using the same incubation, namely using Nutrient Agar (Difco, 23g/L) plates supplemented with CaCl2 (1 mM), KCl (13 mM), MgSO4 (1 mM) and MnSO4 (0.13 mM), with a pH of 7.0, and incubation for 7 days at 37°C to allow for sporulation (Berendsen et al., 2015). Subsequently spores were harvested from the plates and washed three times with sterile water (5000 g, 10 min, 4°C) (Berendsen et al., 2015).
References
- Alkema W., Boekhorst J., Wels M., van Hijum S. A. (2016). Microbial bioinformatics for food safety and production. Brief. Bioinform. 17, 283–292. 10.1093/bib/bbv034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Molinas C., Depardieu F., Courvalin P. (1993). Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175, 117–127. 10.1128/jb.175.1.117-127.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril E., Coroller L., Couvert O., Leguérinel I., Postollec F., Boulais C., et al. (2012). Modeling heat resistance of Bacillus weihenstephanensis and Bacillus licheniformis spores as function of sporulation temperature and pH. Food Microbiol. 30, 29–36. 10.1016/j.fm.2011.09.017 [DOI] [PubMed] [Google Scholar]
- Berendsen E. M., Boekhorst J., Kuipers O. P., Wells-Bennik M. H. J. (2016a). A mobile genetic element profoundly increases heat resistance of bacterial spores. ISME J. 10, 2633–2642. 10.1038/ismej.2016.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen E. M., Wells-Bennik M. H. J., Krawczyk A. O., de Jong A., van Heel A., Eijlander R. T., et al. (2016b). Draft genome sequences of 10 Bacillus subtilis strains that form spores with a high or low heat-resistance. Genome Announc. 4:e00124–16. 10.1128/genomeA.00124-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen E. M., Zwietering M. H., Kuipers O. P., Wells-Bennik M. H. (2015). Two distinct groups within the Bacillus subtilis group display significantly different spore heat resistance properties. Food Microbiol. 45(Pt A), 18–25. 10.1016/j.fm.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Carver T., Harris S. R., Berriman M., Parkhill J., McQuillan J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. 10.1093/bioinformatics/btr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. (2005). ACT: the artemis comparison tool. Bioinformatics 21, 3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Caspers M. P., Schuren F. H., van Zuijlen A. C., Brul S., Montijn R. C., Abee T., et al. (2011). A mixed-species microarray for identification of food spoilage bacilli. Food Microbiol. 28, 245–251. 10.1016/j.fm.2010.03.010 [DOI] [PubMed] [Google Scholar]
- Cazemier A. E., Wagenaars S. F., ter Steeg P. F. (2001). Effect of sporulation and recovery medium on the heat resistance and amount of injury of spores from spoilage bacilli. J. Appl. Microbiol. 90, 761–770. 10.1046/j.1365-2672.2001.01302.x [DOI] [PubMed] [Google Scholar]
- Chen X. H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K., Heinemeyer I., et al. (2007). Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. 10.1038/nbt1325 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal N., Hill C., Ross P. R., Beresford T. P., Fenelon M. A., Cotter P. D. (2015). The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 6:1418. 10.3389/fmicb.2015.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Heyndrickx M. (2011). The importance of endospore-forming bacteria originating from soil for contamination of industrial food processing. Appl. Environ. Soil Sci. 2011:11 10.1155/2011/561975 [DOI] [Google Scholar]
- Konz D., Doekel S., Marahiel M. A. (1999). Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol. 181, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort R., O'Brien A. C., van Stokkum I. H., Oomes S. J., Crielaard W., Hellingwerf K. J., et al. (2005). Assessment of heat resistance of bacterial spores from food product isolates by fluorescence monitoring of dipicolinic acid release. Appl. Environ. Microbiol. 71, 3556–3564. 10.1128/AEM.71.7.3556-3564.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs A. T., Smits W. K., Mironczuk A. M., Kuipers O. P. (2009). Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries. Environ. Microbiol. 11, 1911–1922. 10.1111/j.1462-2920.2009.01937.x [DOI] [PubMed] [Google Scholar]
- Krawczyk A. O., de Jong A., Holsappel S., Eijlander R. T., van Heel A. J., Berendsen E. M., et al. (2016). Genome sequences of 12 spore-forming Bacillus species, comprising Bacillus coagulans, Bacillus licheniformis, Bacillus amyloliquefaciens, Bacillus sporothermodurans, and Bacillus vallismortis, isolated from foods. Genome Announc. 4:e00103–16. 10.1128/genomeA.00103-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Ogasawara N., Moszer I., Albertini A. M., Alloni G., Azevedo V., et al. (1997). The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390, 249–256. 10.1038/36786 [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Jr., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Davis A., Korza G., Zhang P., Li Y. Q., Setlow B., et al. (2012). Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 194, 1875–1884. 10.1128/JB.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima L. J., Kamphuis H. J., Nout M. J., Zwietering M. H. (2011). Microbiota of cocoa powder with particular reference to aerobic thermoresistant spore-formers. Food Microbiol. 28, 573–582. 10.1016/j.fm.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Logan N. A. (2012). Bacillus and relatives in foodborne illness. J. Appl. Microbiol. 112, 417–429. 10.1111/j.1365-2672.2011.05204.x [DOI] [PubMed] [Google Scholar]
- Logan N. A., Vos P. (2009). Bergey's Manual of Systematic Bacteriology. New York, NY: Springer-Verlag. [Google Scholar]
- Lücking G., Stoeckel M., Atamer Z., Hinrichs J., Ehling-Schulz M. (2013). Characterization of aerobic spore-forming bacteria associated with industrial dairy processing environments and product spoilage. Int. J. Food Microbiol. 166, 270–279. 10.1016/j.ijfoodmicro.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Madslien E. H., Rønning H. T., Lindbäck T., Hassel B., Andersson M. A., Granum P. E. (2013). Lichenysin is produced by most Bacillus licheniformis strains. J. Appl. Microbiol. 115, 1068–1080. 10.1111/jam.12299 [DOI] [PubMed] [Google Scholar]
- Melly E., Genest P. C., Gilmore M. E., Little S., Popham D. L., Driks A., et al. (2002). Analysis of the properties of spores of Bacillus subtilis prepared at different temperatures. J. Appl. Microbiol. 92, 1105–1115. 10.1046/j.1365-2672.2002.01644.x [DOI] [PubMed] [Google Scholar]
- Miller R. A., Kent D. J., Watterson M. J., Boor K. J., Martin N. H., Wiedmann M. (2015). Spore populations among bulk tank raw milk and dairy powders are significantly different. J. Dairy Sci. 98, 8492–8504. 10.3168/jds.2015-9943 [DOI] [PubMed] [Google Scholar]
- Nicholson W. L., Munakata N., Horneck G., Melosh H. J., Setlow P. (2000). Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64, 548–572. 10.1128/MMBR.64.3.548-572.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen T., Rintaluoma N., Andersson M., Taimisto A. M., Ali-Vehmas T., Seppälä A., et al. (2007). Toxinogenic Bacillus pumilus and Bacillus licheniformis from mastitic milk. Vet. Microbiol. 124, 329–339. 10.1016/j.vetmic.2007.05.015 [DOI] [PubMed] [Google Scholar]
- Oomes S. J., van Zuijlen A. C., Hehenkamp J. O., Witsenboer H., van der Vossen J. M. B. M., Brul S. (2007). The characterisation of Bacillus spores occurring in the manufacturing of (low acid). canned products. Int. J. Food Microbiol. 120, 85–94. 10.1016/j.ijfoodmicro.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Orsburn B., Melville S. B., Popham D. L. (2008). Factors contributing to heat resistance of Clostridium perfringens endospores. Appl. Environ. Microbiol. 74, 3328–3335. 10.1128/AEM.02629-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmars L., Kerkhoven R., Siezen R. J., Francke C. (2013). MGcV: the microbial genomic context viewer for comparative genome analysis. BMC Genomics 14:209. 10.1186/1471-2164-14-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postollec F., Mathot A. G., Bernard M., Divanac'h M. L., Pavan S., Sohier D. (2012). Tracking spore-forming bacteria in food: from natural biodiversity to selection by processes. Int. J. Food Microbiol. 158, 1–8. 10.1016/j.ijfoodmicro.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Priest F. G., Goodfellow M., Shute L. A., Berkeley R. C. W. (1987). Bacillus amyloliquefaciens sp. nov., nom. rev. Int. J. Syst. Bacteriol. 37, 69–71. 10.1099/00207713-37-1-69 [DOI] [Google Scholar]
- Priest F. G., Goodfellow M., Todd C. (1988). A numerical classification of the genus Bacillus. J. Gen. Microbiol. 134, 1847–1882. 10.1099/00221287-134-7-1847 [DOI] [PubMed] [Google Scholar]
- Rose R., Setlow B., Monroe A., Mallozzi M., Driks A., Setlow P. (2007). Comparison of the properties of Bacillus subtilis spores made in liquid or on agar plates. J. Appl. Microbiol. 103, 691–699. 10.1111/j.1365-2672.2007.03297.x [DOI] [PubMed] [Google Scholar]
- Rosenkvist H., Hansen Å. (1995). Contamination profiles and characterisation of Bacillus species in wheat bread and raw materials for bread production. Int. J. Food Microbiol. 26, 353–363. 10.1016/0168-1605(94)00147-X [DOI] [PubMed] [Google Scholar]
- Ruckert C., Blom J., Chen X., Reva O., Borriss R. (2011). Genome sequence of B. amyloliquefaciens type strain DSM7(T) reveals differences to plant-associated B. amyloliquefaciens FZB42. J. Biotechnol. 155, 78–85. 10.1016/j.jbiotec.2011.01.006 [DOI] [PubMed] [Google Scholar]
- Salkinoja-Salonen M. S., Vuorio R., Andersson M. A., Kämpfer P., Andersson M. C., Honkanen-Buzalski T., et al. (1999). Toxigenic strains of Bacillus licheniformis related to food poisoning. Appl. Environ. Microbiol. 65, 4637–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheldeman P., Herman L., Foster S., Heyndrickx M. (2006). Bacillus sporothermodurans and other highly heat-resistant spore formers in milk. J. Appl. Microbiol. 101, 542–555. 10.1111/j.1365-2672.2006.02964.x [DOI] [PubMed] [Google Scholar]
- Setlow P. (2006). Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101, 514–525. 10.1111/j.1365-2672.2005.02736.x [DOI] [PubMed] [Google Scholar]
- Sorokulova I. B., Reva O. N., Smirnov V. V., Pinchuk I. V., Lapa S. V., Urdaci M. C. (2003). Genetic diversity and involvement in bread spoilage of Bacillus strains isolated from flour and ropy bread. Lett. Appl. Microbiol. 37, 169–173. 10.1046/j.1472-765X.2003.01372.x [DOI] [PubMed] [Google Scholar]
- te Giffel M. C., Beumer R. R., Leijendekkers S., Rombouts F. M. (1996). Incidence of Bacillus cereus and Bacillus subtilis in foods in the Netherlands. Food Microbiol. 13, 53–58. 10.1006/fmic.1996.0007 [DOI] [Google Scholar]
- Tovar-Rojo F., Chander M., Setlow B., Setlow P. (2002). The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J. Bacteriol. 184, 584–587. 10.1128/JB.184.2.584-587.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio F., De Bellis P., Di Biase M., Lonigro S. L., Giussani B., Visconti A., et al. (2012). Diversity of spore-forming bacteria and identification of Bacillus amyloliquefaciens as a species frequently associated with the ropy spoilage of bread. Int. J. Food Microbiol. 156, 278–285. 10.1016/j.ijfoodmicro.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Valerio F., Di Biase M., Huchet V., Desriac N., Lonigro S. L., Lavermicocca P., et al. (2015). Comparison of three Bacillus amyloliquefaciens strains growth behaviour and evaluation of the spoilage risk during bread shelf-life. Food Microbiol. 45(Pt A), 2–9. 10.1016/j.fm.2014.07.006 [DOI] [PubMed] [Google Scholar]
- van Asselt E. D., Zwietering M. H. (2006). A systematic approach to determine global thermal inactivation parameters for various food pathogens. Int. J. Food Microbiol. 107, 73–82. 10.1016/j.ijfoodmicro.2005.08.014 [DOI] [PubMed] [Google Scholar]
- van Zuijlen A., Periago P. M., Amézquita A., Palop A., Brul S., Fernández P. S. (2010). Characterization of Bacillus sporothermodurans IC4 spores; putative indicator microorganism for optimisation of thermal processes in food sterilisation. Food Res. Int. 43, 1895–1901. 10.1016/j.foodres.2009.11.011 [DOI] [Google Scholar]
- Velasquez J., Schuurman-Wolters G., Birkner J. P., Abee T., Poolman B. (2014). Bacillus subtilis spore protein SpoVAC functions as a mechanosensitive channel. Mol. Microbiol. 92, 813–823. 10.1111/mmi.12591 [DOI] [PubMed] [Google Scholar]
- Vepachedu V. R., Setlow P. (2004). Analysis of the germination of spores of Bacillus subtilis with temperature sensitive spo mutations in the spoVA operon. FEMS Microbiol. Lett. 239, 71–77. 10.1016/j.femsle.2004.08.022 [DOI] [PubMed] [Google Scholar]
- Vepachedu V. R., Setlow P. (2007). Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J. Bacteriol. 189, 1565–1572. 10.1128/JB.01613-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth A. D. (1978). Relationship between the heat resistance of spores and the optimum and maximum growth temperatures of Bacillus species. J. Bacteriol. 134, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells-Bennik M. H. J., Eijlander R. T., den Besten H. M. W., Berendsen E. M., Warda A. K., Krawczyk A. O., et al. (2016). Bacterial spores in food: survival, emergence, and outgrowth. Annu. Rev. Food Sci. Technol. 7, 457–482. 10.1146/annurev-food-041715-033144 [DOI] [PubMed] [Google Scholar]
- Zahler S. A., Korman R. Z., Thomas C., Fink P. S., Weiner M. P., Odebralski J. M. (1987). H2, a temperate bacteriophage isolated from Bacillus amyloliquefaciens strain H. J. Gen. Microbiol. 133, 2937–2944. 10.1099/00221287-133-10-2937 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Orthology matrix constructed using Ortho-MCL with the genomes of the four B. amyloliquefaciens strains, the nine B. licheniformis strains, B. subtilis B4146 as a strain that produces spores with high heat resistance and B. subtilis 168 as a reference strain that produces spores with normal heat resistance.
Calculated reference D-value (Dref) at a reference temperature (Tref) of 110°C, and z-value, of spores of nine strains of B. amyloliquefaciens and spores of nine strains of B. licheniformis (Berendsen et al., 2015, 2016a). In these studies, all strains were sporulated on the same media using the same incubation, namely using Nutrient Agar (Difco, 23g/L) plates supplemented with CaCl2 (1 mM), KCl (13 mM), MgSO4 (1 mM) and MnSO4 (0.13 mM), with a pH of 7.0, and incubation for 7 days at 37°C to allow for sporulation (Berendsen et al., 2015). Subsequently spores were harvested from the plates and washed three times with sterile water (5000 g, 10 min, 4°C) (Berendsen et al., 2015).