Abstract
DNA damaging agents are a constant threat to genomes in both the nucleus and the mitochondria. To combat this threat, a suite of DNA repair pathways cooperate to repair numerous types of DNA damage. If left unrepaired, these damages can result in the accumulation of mutations which can lead to deleterious consequences including cancer and neurodegenerative disorders. The base excision repair (BER) pathway is highly conserved from bacteria to humans and is primarily responsible for the removal and subsequent repair of toxic and mutagenic oxidative DNA lesions. Although the biochemical steps that occur in the BER pathway have been well defined, little is known about how the BER machinery is regulated. The budding yeast, Saccharomyces cerevisiae is a powerful model system to biochemically and genetically dissect BER. BER is initiated by DNA N-glycosylases, such as S. cerevisiae Ntg1. Previous work demonstrates that Ntg1 is post-translationally modified by SUMO in response to oxidative DNA damage suggesting that this modification could modulate the function of Ntg1. In this study, we mapped the specific sites of SUMO modification within Ntg1 and identified the enzymes responsible for sumoylating/desumoylating Ntg1. Using a non-sumoylatable version of Ntg1, ntg1ΔSUMO, we performed an initial assessment of the functional impact of Ntg1 SUMO modification in the cellular response to DNA damage. Finally, we demonstrate that, similar to Ntg1, the human homologue of Ntg1, NTHL1, can also be SUMO-modified in response to oxidative stress. Our results suggest that SUMO modification of BER proteins could be a conserved mechanism to coordinate cellular responses to DNA damage.
Keywords: Base Excision Repair (BER), Ntg1, NTHL1, SUMO, Sumoylation
Introduction
Genomes in both the nucleus and mitochondria are constantly exposed to various exogenous and endogenous DNA damaging agents (1). A suite of DNA repair pathways cooperate to ensure the efficient repair of numerous types of DNA damage that result from such exposures (2, 3). Oxidative DNA damage, caused by numerous sources including cellular metabolism (4, 5) and exogenous factors (6), is one of the most common forms of DNA damage. Estimates suggest that 90,000 oxidative lesions and 200,000 apurinic/apyrimidinic (AP) sites are generated per human cell per day (7–9). Unrepaired lesions can result in the accumulation of mutations which can trigger deleterious consequences including cancer and neurodegenerative disorders (1–3, 7, 8, 10–16). The base excision repair (BER) pathway is primarily responsible for the removal and repair of toxic and mutagenic oxidative DNA damage (3, 17–19). Numerous studies have defined in detail the biochemical steps that occur in the BER pathway (3, 20), but little is known about how the BER machinery is regulated (21).
BER is initiated by the recognition and hydrolysis of a damaged base by a DNA N-glycosylase leaving an AP site (3, 20, 22, 23). The AP site is then further processed to create a nick in the DNA backbone (3, 20, 22, 23). Subsequent steps create a single-strand break that is then filled by a specialized DNA polymerase and sealed by ligase (3, 20, 22, 23). These steps must occur in a sequential manner ensuring that AP sites and single strand breaks are properly managed to allow repair at the initial site of DNA damage without causing collateral damage via accumulation of BER intermediates (3, 20, 22, 23). The human NTHL1 protein, which is a bifunctional Endonuclease III–like N-glycosylase/AP lyase, is responsible for initiating repair of a wide array of oxidative lesions (21, 24–26). As the initiating factor in the BER pathway (3, 21, 22), NTHL1 must be regulated to ensure that repair is rapid, but also regulated to prevent the accumulation of toxic and mutagenic AP sites and single strand breaks that are the products of NTHL1 enzymatic activity (21, 24–26). N-glycosylase regulation could occur through a number of distinct mechanisms including modulating protein levels, protein localization, protein-protein interactions, and post-translational modifications (27–35).
Recent discoveries highlight the importance of N-glycosylase regulation in cancer (36, 37). Several studies identified mutations in the NTHL1 gene in a recently characterized cancer predisposition syndrome (38–40). These heterozygous loss-of-function mutations in NTHL1 predispose patients to colorectal cancer and other forms of cancer (38–40). Altered NTHL1 function can also result in mislocalization/accumulation of the protein in the cytoplasm of cancer cells in a subset of gastric tumors (36). These studies provide evidence that proper function of NTHL1 is critical to maintain genomic integrity and cellular homeostasis.
Much of the work that has contributed to our knowledge of DNA repair mechanisms has exploited the budding yeast S. cerevisiae as DNA repair pathways are conserved through evolution (41). Recent studies of the S. cerevisiae orthologues of NTHL1, Ntg1 and Ntg2, reveal that these proteins are post-translationally modified by the Small Ubiquitin-like MOdifier, SUMO (24, 42). The Ntg1 protein is modified in response to DNA damage (24, 42). Sumoylation has the potential to function in a number of regulatory roles including modulating protein-protein interactions and protein activity (27–35). One well-characterized example of SUMO-mediated regulation of the BER pathway is the human thymine DNA glycosylase (TDG), where sumoylation of TDG triggers a conformational change which alters the DNA binding pocket of the enzyme to influence enzyme turnover (43–45). This conformational change in TDG decreases the affinity of TDG for DNA leading to an increase in the off rate and hence an increase in the catalytic efficiency (turnover) of TDG (43, 44). Similarly, sumoylation could also modulate the function of Ntg1; however, the impact of SUMO modification on Ntg1 function has not yet been explored.
Critical to defining the functional role of SUMO modification of Ntg1 is identifying the SUMO modified sites within Ntg1. In this study, we identify the enzymes that mediate/regulate sumoylation of Ntg1. We also map the SUMO-modified sites on Ntg1 and perform an initial assessment of the functional importance of sumoylation of Ntg1. In addition, we demonstrate that, similar to Ntg1, human NTHL1 can also be SUMO-modified in response to oxidative stress. Our results suggest that SUMO modification of BER proteins could represent an evolutionarily conserved mechanism by which cells respond to oxidative DNA damage.
Materials and Methods
3.1 Strains, Plasmids, and Media
All haploid S. cerevisiae strains and plasmids used in this study are listed in Table 1. S. cerevisiae cells were cultured at 25°C, 30°C, or 37°C in YPD medium (1% yeast extract, 2% peptone, 2% dextrose, 0.005% adenine sulfate, and 2% agar for plates) or SD medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% dextrose, 0.5% adenine sulfate, and 2% agar for plates). In order to introduce plasmids, cells were transformed by a modified lithium acetate method (46).
Table 1.
Strains and Plasmids Used in this Study
| Strain or Plasmid | Description | References |
|---|---|---|
| DSC0295 |
MATahis3Δ1 leu2Δ0 met 15Δ0 ura3Δ0; Tet-Off C-terminally TAP-tagged Ntg1 |
(24) |
| YSC1178-7499106 (DSC0297) |
MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0; C- terminally TAP-tagged Ntg1 |
Open Biosystems |
| BY4147 (DSC0313) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| DSC0470 |
MATa
ntg1::hphMX4, his7-1, lys2Δ5'::LEU- lys2Δ3', ade5-1, trp1-289, ura3-52 |
This study |
| EJY341 (DSC0527) |
MATa
trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 [cir°] |
(99) |
| EJY342 (DSC0528) |
MATa
trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 siz1Δ::LEU2 [cir°] |
(99) |
| EJY343 (DSC0529) |
MATa
trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 siz2Δ::TRP1 [cir°] |
(99) |
| EJY344 (DSC0530) |
MATa
trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 siz1Δ::LEU2 siz2Δ::TRP1 [cir°] |
(99) |
| MHY1488 (DSC0534) | MATa ulp1Δ::HIS3 LEU2::ulp1-333 | (58) |
| EJY447 (DSC0535) |
MATa
trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 ulp2Δ::kanMX [cir°] |
(60) |
| GBY5 (DSC0536) | MATa smt3-allR::TRP1 | (55) |
| DSC0537 |
MATa
ntg1::hphMX4, his7-1, lys2Δ5'::LEU- lys2Δ3', ade5-1, trp1-289, ura3-52, pD0436 |
This study |
| DSC0538 |
MATa
trp1-Δ1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 [cir°], pD0436 |
This study |
| DSC0539 | MATa smt3-allR::TRP1, pD0436 | This study |
| DSC0540 | MATa ulp1Δ::HIS3 LEU2::ulp1-333, pD0436 | This study |
| hDNP19 |
MATa/MATα rad1::kanMX/RAD1 ntg1::hphMX4/NTG1 ntg2::BSD/NTG2 apn1::TRP1/APN1 DSF1::URA3/DSF1 his7- 1/his7-1 lys2Δ5′::LEU-lys2Δ3′/lys2Δ5′::LEU- lys2Δ3′ ade5-1/ade5-1 trp1-289/trp1-289 ura3- 52/ura3-52 |
(19) |
| DSC0367 |
MATa his7-1 lys2Δ5′::LEU-lys2Δ3′ ade5-1 trp1- 289 ura3-52 |
(51) |
| DSC0369 |
MATa ntg1::hphMX4 rad1::kanMX ntg2::BSD apn1::TRP1 his7-1 lys2Δ5′::LEU-lys2Δ3′ ade5-1 trp1-289 ura3-52 |
(51) |
| DSC0371 |
MATa rad1::kanMX ntg2::BSD apn1::TRP1 his7-1 lys2Δ5′::LEU-lys2Δ3′ ade5-1 trp1-289 ura3-52 |
(51) |
| DSC0561 |
MATa ntg1k20,38,376,388,396R rad1::kanMX ntg2::BSD apn1::TRP1 DSF1::URA3 his7-1 lys2Δ5′::LEU-lys2Δ3′ ade5-1 trp1-289 ura3-52 |
This study |
| DSC0549 |
MATa ntg1k396R rad1::kanMX ntg2::BSD apn1::TRP1 DSF1::URA3 his7-1 lys2Δ5′::LEU- lys2Δ3′ ade5-1 trp1-289 ura3-52 |
This study |
| DSC0551 |
MATa ntg1k20,38R rad1::kanMX ntg2::BSD apn1::TRP1 DSF1::URA3 his7-1 lys2Δ5′::LEU- lys2Δ3′ ade5-1 trp1-289 ura3-52 |
This study |
| DSC0558 |
MATa ntg1k20,38,376,388R rad1::kanMX ntg2::BSD apn1::TRP1 DSF1::URA3 his7-1 lys2Δ5′::LEU- lys2Δ3′ ade5-1 trp1-289 ura3-52 |
This study |
| DSC0561 |
MATa ntg1k20,38,376,388,396R rad1::kanMX ntg2::BSD apn1::TRP1 DSF1::URA3 his7-1 lys2Δ5′::LEU-lys2Δ3′ ade5-1 trp1-289 ura3-52 |
This study |
| DSC0555 |
MATa ntg1k376,388,396R rad1::kanMX ntg2::BSD apn1::TRP1 DSF1::URA3 his7-1 lys2Δ5′::LEU- lys2Δ3′ ade5-1 trp1-289 ura3-52 |
This study |
| pD0390 | pET-15b His6-NTG1 | (51) |
| pD0394 | pET -15b His6-NTG1Δcat | (51) |
| pD0493 | pET -15b His6-NTG1(K->R)5 | This study |
| pD0436 | Tet-Off NTG1-TAP, CEN, URA3, ampR | This study |
| pD0437 | Tet-Off ntg1K20R-TAP, CEN, URA3, ampR | This study |
| pD0438 | Tet-Off ntg1K38R-TAP, CEN, URA3, ampR | This study |
| pD0444 | Tet-Off ntg1K376R-TAP, CEN, URA3, ampR | This study |
| pD0445 | Tet-Off ntg1K388R-TAP, CEN, URA3, ampR | This study |
| pD0446 | Tet-Off ntg1K396R-TAP, CEN, URA3, ampR | This study |
| pD0447 | Tet-Off ntg1K20,38R-TAP, CEN, URA3, ampR | This study |
| pD0448 | Tet-Off ntg1K20,38,376R-TAP, CEN, URA3, ampR | This study |
| pD0449 | Tet-Off ntg1K20,38,396R-TAP, CEN, URA3, ampR | This study |
| pD0450 | Tet-Off ntg1K20,38,376,388R-TAP, CEN, URA3, ampR |
This study |
| pD0451 | Tet-Off ntg1K20,38,376,388,396R-TAP, CEN, URA3, ampR |
This study |
| pD0452 | Tet-Off ntg1K376,388,396R-TAP, CEN, URA3, ampR | This study |
A centromeric vector (CEN, URA3), pRS316 (47) was employed as the backbone for the generation of a construct expressing C-terminally tagged Ntg1-TAP fusion protein (pD0436). The insert was amplified using the primers listed in Table 2 and inserted at the NotI restriction site of pRS316 (47). The insert includes the tetracycline repressible promoter (Tet-Off) and the C-terminally tagged NTG1-TAP fusion from the DSC0295 strain (24). The S. cerevisiae haploid deletion mutant ntg1Δ (DSC0470) generated by dissection of tetrads derived from heterozygous diploid hDNP19 (19), and the SUMO pathway mutant collection (E3 ligase mutant strains, siz1Δ, siz2Δ, and siz1Δ/siz2Δ and desumoylase mutant stains ulp1-ts and ulp2Δ) were utilized to assess the level of sumoylated wildtype and mutant Ntg1 (19, 48, 49). All lysine to arginine amino acid substitutions (Supplemental Figure 1C) were created by site-directed mutagenesis performed using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) with the primers listed in Table 2. The resulting plasmids were sequenced to ensure the introduction of the desired mutation and the absence of any additional mutations.
Table 2.
Plasmid Construction Primers
| Primer Purpose |
Primer Name |
Sequence (5' - 3') |
|---|---|---|
| pD0436 | tetNtg1Cla- F1 |
GAATCGATTGCAGTTTCATTTGATGCTCGATGAG |
| His- Ntg1Cla- R1 |
GAATCGATGTATTCTGGGCCTCCATGTCGC | |
| K20R | K20R2-F | CAATTCTGAGGAAAAGACCGCTGGTAAGGACTGAAACTGG |
| K20R2-R | CCAGTTTCAGTCCTTACCAGCGGTCTTTTCCTCAGAATTG | |
| K38R | K38R-F | GGACCAAAATCAGACAAGAAGAGGTTGTCCCTCAACCCGTG |
| K38R-R | CACGGGTTGAGGGACAACCTCTTCTTGTCTGATTTTGGTCC | |
| K157R | K157R-F | GATGCTATCATCGCAAACAAGAGATGAAGTTACCGCAATGGC |
| K157R-R | GCCATTGCGGTAACTTCATCTCTTGTTTGCGATGATAGCATC | |
| K194R | K194R-F | CCGTTTTACAAATCAATGAGACCAGATTAGACGAATTGATTCATTCAG |
| K194R-R | CTGAATGAATCAATTCGTCTAATCTGGTCTCATTGATTTGTAAAACGG | |
| K255R | K255R-F | CATTACAAAAGGCATGGGGCAGGATTGAAGGTATCTGCGTTGACG |
| K255R-R | CGTCAACGCAGATACCTTCAATCCTGCCCCATGCCTTTTGTAATG | |
| K359R | K359R-F | GCAAAATATCATGAGTTATCCAAAGTGGGTGAGATACCTGGAAGG |
| K359R-R | CCTTCCAGGTATCTCACCCACTTTGGATAACTCATGATATTTTGC | |
| K364R | K364R-F | TACCTGGAAGGAAGACGTGAACTGAACGTGGAGGCGG |
| K364R-R | CCGCCTCCACGTTCAGTTCACGTCTTCCTTCCAGGTA | |
| K376R | K376R-F | CGTGGAGGCGGAAATCAATGTTAGACACGAGGAGAAAACAG |
| K376R-R | CTGTTTTCTCCTCGTGTCTAACATTGATTTCCGCCTCCACG | |
| K388R | K388R-F | CGAGGAGAAAACAGTTGAAGAAACTATGGTCAGACTGGAAAATG |
| K388R-R | CATTTTCCAGTCTGACCATAGTTTCTTCAACTGTTTTCTCCTCG | |
| K396R | K396R-F | GGAAAATGATATTTCTGTTAGAGTAGAGGACGGTCGACGG |
| K396R-R | CCGTCGACCGTCCTCTACTCTAACAGAAATATCATTTTCC | |
| NTHL1 | NTHL1- Flag-F |
ACACTGGCGGCCGTTACTAGTGGATCCT |
| NTHL1- Flag-R |
ACGACTCACTATAGGGAGACCCAAGCTT |
To express recombinant Ntg1, the NTG1 open reading frame was cloned into pET-15b (Invitrogen) to generate N-terminal His6 epitope tagged His6-Ntg1 (pD0390) (Table 1). Site-directed mutagenesis of His6-NTG1 was performed at lysines 20, 38, 376, 388, and 396 (lysines to arginines) to create a nonsumoylatable Ntg1 (ntg1ΔSUMO), His6-Ntg1ΔSUMO (pD0493), and at lysine 243 (lysine to glutamine), His6-Ntg1Δcat (pD0394) (Table 1). Expression vectors were transformed into DE3 cells.
Site-directed mutagenesis at the endogenous NTG1 locus of the wildtype (DSC0367) parent was performed via delitto perfetto protocol (50) to generate ntg1K20,38,376,388,396R. The resulting variants were then crossed with haploid BER-Nucleotide Excision Repair- (NER-) mutants to create diploids which were then dissected to identify cells with each Ntg1 variant BER*/NER- strain (DSC0367, DSC0369, DSC0371, DSC0561).
NTHL1 was cloned from the RG214598 plasmid (Origene) using the NTHL1-Flag primer pair for the addition of the Flag-tag and cloned into the pcDNA3.1 (+) vector using the HindIII and BamHI sites.
3.2 Exposure to DNA Damaging Agents
S. cerevisiae cells were grown in 5–35 mL YPD or SD -URA media to either 2 × 107 or 1 × 108 cells/mL, centrifuged, and washed with water. Cells were then resuspended in 5–35 mL water, YPD, or plated onto YPD agar plates containing the appropriate agent: 20 mM hydrogen peroxide (Sigma); or 0.005–0.3% methyl methanesulfonate (MMS) (Sigma). Cells were exposed to agents for 1–2 hours as indicated at 30°C or 37°C.
3.3 Immunoblotting Ntg1
The steady-state level of each Ntg1-TAP fusion protein variant was assessed by immunoblotting whole cell lysates with the rabbit polyclonal anti-TAP antibody (1:3,333 dilution, Open Biosystems) to determine the relative level of differentially modified Ntg1 products. An anti-3-phosphoglycerate (PGK) antibody (1:10,000 dilution; Invitrogen) was used as a control determine the relative level of protein lysate loaded into each lane.
The analysis of immunoblots was performed utilizing the ECL Plex immunoblotting detection system (Amersham), the Typhoon Trio variable mode imager (GE Healthcare), and the ImageQuant TL software package (GE Healthcare). To quantify the percentage of modified Ntg1-TAP, the ratio of modified Ntg1 bands to total Ntg1 signal (including modified and unmodified) was determined for wildtype Ntg1 and each lysine to arginine amino acid substitution variant of Ntg1. Previous work demonstrates that modified Ntg1 contains at least one covalently linked SUMO and the size of higher bands is consistent with multiple SUMO additions(24). Standard error of the mean was calculated for each. The two-sample Student’s t-test was employed to test for significance (α=0.05).
3.4 Cultured cell lines and cell culture
HT29 colon adenocarcinoma cells were cultured in McCoy’s 5A modified media (Corning) and supplemented with 10% FBS, penicillin and streptomycin. Cultured cells were passaged every 3–4 days, or upon 80% confluency.
3.5 NTHL1-Flag immunoprecipitation
HT29 colon adenocarcinoma cells were seeded at a density of 1×106 cells in 100 cm2 dishes. Transfection of the NTHL1-Flag construct or empty Flag vector was performed using Lipofectamine3000 (Invitrogen) and a final concentration of 10 µg plasmid/dish. The hydrogen peroxide incubation was performed with a final concentration of 125 µM hydrogen peroxide in sterile PBS for 15 minutes at 37°C. All cells were lysed in NP40 buffer (50 mM Tris pH 8.0, 100 mM NaCl, 32 mM NaF, 0.5% NP40 detergent) supplemented with protease and phosphatase inhibitors (Thermo Scientific), and SENP (de-SUMOylase) SUMO-2 aldehyde inhibitors (Enzo Life Sciences). Antibodies for Flag (Rabbit, 2368; Cell Signaling) or IgG (mouse, ab77118; abcam) were conjugated to Protein G Dynabeads (10007D; Life Sciences) for 2 hours prior to adding lysates. For each sample, 500 µg of total protein was added to the beads and rotated overnight at 4°C. Beads were washed three times in NP40 buffer for 5 minutes each. NTHL1-Flag was eluted from the beads using a 3X Flag® peptide (Sigma) for 2 hours at a working concentration of 100 µg/mL per the manufacturer’s instructions.
Following Flag peptide elution, samples were added to Laemmli buffer (50% glycerol, 10% SDS, 100 mM Tris, pH 6.8), boiled for 5 minutes at 95°C, and loaded on 4–12% Bis-Tris gels (Invitrogen). Proteins were transferred onto a polyvinylidene fluoride membrane and blocked with 5% ECL prime (GE Healthcare) in 0.1% PBST for 1 hour at room temperature. Blots were incubated in primary antibodies overnight at 4°C. All washes were performed in 0.1% PBST at room temperature, and the corresponding horseradish peroxidase-conjugated secondary antibodies were added for 1 hour. Antigen-antibody complexes were detected using Supersignal™ west pico chemiluminescent substrate kit (Thermo Scientific). Antibodies used for western blotting were: NTHL1 (mouse, cat # MAB2675; R&D Systems) and SUMO-2/3 (rabbit, made in Nicholas Seyfried lab, Emory University).
3.6 Structural modeling
The Protein Homology/analogY Recognition Engine version 2.0 (Phyre2) server was used to generate a model of Ntg1 based on its E. coli Endonuclease III homolog (PDB ID: 2ABK). The N-terminal and C-terminal domains do not share homology with E. coli Endonuclease III but align to other bacterial endonucleases. The N-terminal domain aligns to the restriction endonuclease BsaWI (PDB ID: 4ZSF) and the C-terminal domain aligns to the endonuclease BglII (PDB ID: 1DFM). PyMOL Molecular Graphics System, Version 1.8 Schrödinger LLC was used to model these structures.
3.7 Overexpression and purification of the recombinant Ntg1 variants for in vitro DNA strand scission assay
To assess the functional consequences of changing five lysines (K20,K38,K376,K388,K396) to arginine within Ntg1, we expressed and purified recombinant protein containing these five amino acid substitutions. We designated this recombinant protein ntg1(K->R)5. As controls, we employed wildtype Ntg1 and a catalytic mutant of Ntg1 (lysine 243 to glutamine) which we term ntg1Δcat. Recombinant Ntg1 was purified as previously described (51). Briefly, Escherichia coli BL21 (DE3) cells containing each variant His6-Ntg1 plasmids were grown to an OD600 of 0.5–1.0 and expression induced for 4 hours at 25°C. Cells were lysed via sonication and the supernatant was applied to Ni-NTA agarose beads purification (Qiagen) to crudely purify the His6-Ntg1 variants. Crude lysate was eluted through a gravity flow column (BIORAD) and dialyzed. Crudely purified His6-Ntg1 variants were further purified to apparent homogeneity by fast protein liquid chromatography.
3.8 Preparation of oligonucleotide and DNA strand scission assay
To assess the functional consequences of changing five lysines (K20,K38,K376,K388,K396) to arginine within Ntg1, we employed an in vitro strand scission assay. An oligonucleotide containing dihydrouracil (DHU) at position 13 (DHU-31mer) was purchased from Midland Certified Reagent Company (Midland, TX, USA). A complementary strand containing a guanine opposite the DHU position was obtained from Eurofins MWG/Operon (Huntsville, AL, USA). The DHU-31mer was 5’-end-labeled with [γ-32P] ATP (Amersham) and T4 polynucleotide kinase (Promega) prior to annealing to the complementary strand (24). Single-stranded DHU-31mer was annealed in a 1:1.6 molar ratio to the appropriate complementary strand, heated to 80°C for 10 minutes and cooled slowly to room temperature.
The AP lyase activity of purified Ntg1 variants (Ntg1, ntg1(K->R)5, and ntg1Δcat) was assayed as previously described (51). Briefly, DNA strand scission assays were carried out in a standard reaction buffer (20 mL) containing 100 mM KCl, 10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 50 fmol of labeled DNA substrate and 20 fmol of Ntg1 protein. Reactions were performed at 37°C for 15 minutes and then stopped by the addition of 10 µL of loading buffer (90% formamide, 1mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue) followed by heating at 95°C for 5 minutes. Reaction products were then resolved on a denaturing-urea polyacrylamide gel (15%) and analyzed with a Typhoon Trio variable mode imager (GE Healthcare).
3.9 Functional analysis of Ntg1 in vivo
To test the sensitivity and the biological function of the Ntg1 complete sumoylation null mutant (ntg1K20,38,376,388,396R), which we term ntg1ΔSUMO (DSC0561), to DNA damaging agents, a serial dilution and spotting assay was employed. Each strain was grown at 30°C to an OD600 of 0.3 – 0.6 in YPD, washed in 5 mL of water, and then diluted to 2×107 cells/mL in water. Five-fold serial dilutions of cells were then plated onto plates containing only YPD or YPD with 0.005% MMS. Plates were incubated at 30°C and then analyzed for sensitivity at days 2 and 4.
Growth kinetics experiments were carried out using S. cerevisiae cells that express each Ntg1 variant encoded at the endogenous NTG1 locus in a DNA repair compromised background (DSC0367, DSC0369, DSC0371, and DSC0561). The growth kinetics of four independently isolated ntg1ΔSUMO variants (DSC0561) and four wildtype Ntg1 (DSC0371), all in a DNA repair compromised background, were tested by analyzing growth curves. Ntg1 sumoylation mutants were grown to saturation over 2 days at 30°C in YPD. Cell concentrations were normalized by OD600, and then samples were diluted to an OD600 of 0.05 in 150 µL of YPD medium containing 0, 0.005, or 0.010% MMS and added to the wells of a 96-well microtiter plate. Cell samples were loaded in duplicate, were grown at 30°C with shaking, and absorbance at OD600 was measured every 30 minutes for 48 hours in an ELX808 Ultra microplate reader with KCjunior software (Bio-Tek Instruments, Inc.). The samples for each genotype and duplicate were averaged for every time point and differences between the two genotypes was analyzed by Students t-test.
Results
4.1 Genetic analysis of the Ntg1 sumoylation pathway
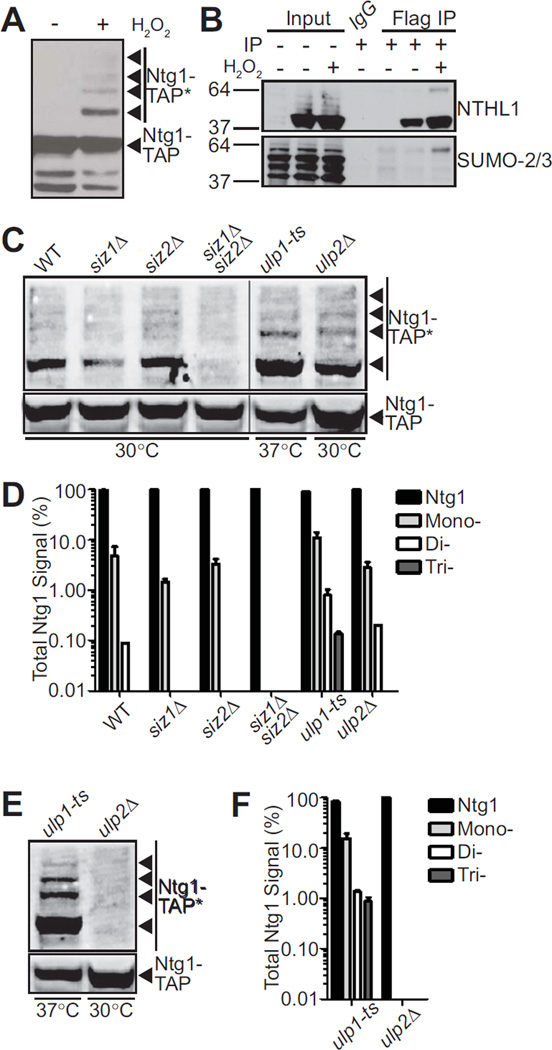
Our group has previously shown that in response to cellular exposure to hydrogen peroxide, Ntg1 is post-translationally modified by SUMO (24). As shown in Figure 1A, several Ntg1 bands are detected upon exposure to hydrogen peroxide suggesting that Ntg1 could be modified by multiple post-translational modifications, including the possibility for addition of multiple SUMO moieties. To determine whether SUMO modification of BER proteins that initiate repair is conserved, we tested whether we could detect sumoylation of human NTHL1. For this experiment, we transfected HT29 colon adenocarcinoma cells with NTHL1-Flag or an empty Flag vector. Cells were treated with hydrogen peroxide and immunoprecipitated with anti-Flag or with IgG as a control. Total lysate and bound fractions were subjected to immunoblotting to detect NTHL1 and SUMO. As shown in the top panel of Figure 1B, we detect NTHL1 in the input and NTHL1 is enriched in the bound fraction, as expected. In samples from cells treated with hydrogen peroxide, a higher molecular weight band of NTHL1 appears, suggesting a post-translational modification. Consistent with SUMO modification, a band of the same molecular weight is recognized by an anti-SUMO-2/3 antibody. The extent of modification of NTHL1 is greatly increased in response to hydrogen peroxide exposure (Figure 1B, bottom). We do not detect NTHL1 or SUMO-2/3 in the control IgG immunoprecipitation. Thus, both S. cerevisiae Ntg1/Ntg2 (24) and human NTHL1 can be modified by SUMO.
Figure 1. Sumoylation of Ntg1 is conserved and mediated by Siz1/2.
A. Wildtype S. cerevisiae cells expressing Ntg1-TAP were exposed to 0 (−) or 20 mM (+) H2O2 for 1 hour at 30°C. Cells were pelleted, lysed, and immunoblotted to detect TAP-tagged Ntg1. Bands corresponding to post-translationally modified Ntg1 including SUMO-modified Ntg1 (24) are indicated by Ntg1-TAP*. B. Colon adenocarcinoma cells (HT29) were transfected with NTHL1-Flag or empty Flag vector and treated with 0 (−) or 125 µM (+) H2O2 for 15 minutes at 37°C. Cells were lysed, immunoprecipitated with Flag antibodies and both the Input and Flag IP fractions were subjected to immunoblotting. An IgG bead alone immunoprecipitation was included as a control. The blot was probed with NTHL1 and SUMO-2/3 antibodies as indicated. C. Wildtype (WT), siz1Δ, siz2Δ, siz1Δsiz2Δ, ulp1-ts, or ulp2Δ cells were transformed with a plasmid expressing Ntg1-TAP. Cells were (C) exposed to 20 mM hydrogen peroxide or (E) not treated. Cells were incubated at 30°C except ulp1-ts cells which were shifted to the non-permissive temperature of 37°C. Each sample was lysed, immunoblotted, and bands were quantified. Nonadjacent lanes in the same image are separated by a black line. D. The data from (C) were quantitated. The total amount of Ntg1-TAP including unmodified and modified Ntg1-TAP was set to 100% (Ntg1) and the fraction of signal present in bands (Total Ntg1 Signal %) corresponding to the size consistent with Mono-, Di-, and Tri-sumoylation is plotted on a log scale. Results shown are the average of two independent experiments. Error bars represent SEM. E. To examine sumoylation of Ntg1 in the absence of oxidative damage, ulp1-ts and ulp2Δ cells expressing Ntg1-TAP were analyzed to detect any modified Ntg1 species (Ntg1-TAP*). F. The data from (E) were quantitated. The total amount of Ntg1-TAP including unmodified and modified Ntg1-TAP was set to 100% (Ntg1) and the fraction of signal present in bands (Total Ntg1 Signal %) corresponding to the size consistent with Mono-, Di-, and Tri-sumoylation is plotted on a log scale. Results shown are the average of two independent experiments. Error bars represent SEM.
Sumoylation involves a series of conjugations that, in S. cerevisiae, are catalyzed by the E1 (Uba2/Aos1 heterodimer), the E2 (Ubc9), and one of four E3 (Siz1, Siz2, Mms21, and Zip3) ligases (52–57). These enzymes catalyze the attachment of the SUMO protein to a substrate lysine residue through formation of an isopeptide bond (52–57). Sumoylation is a dynamic process that is readily reversible by SUMO proteases, which in S. cerevisiae are Ulp1 and Ulp2 (58, 59).
To define the pathway by which Ntg1 is SUMO modified, we examined S. cerevisiae cells lacking the E3 ligases, Siz1 and Siz2, as well as Siz1/Siz2 double deletion cells (53, 60). We first examined Ntg1 sumoylation in these siz1Δ and siz2Δ mutant cells in response to hydrogen peroxide exposure (Figure 1C, D). In the siz1Δ cells, we detected reduced levels of Ntg1 sumoylation (1.5%) compared to wildtype control cells (4.7%) (Figure 1D). The level of Ntg1 sumoylation in the siz2Δ cells was largely unchanged (3.2%) as compared to wildtype. In the siz1Δsiz2Δ double mutant cells, we could not detect Ntg1 sumoylation (Figure 1C, D). These results demonstrate that Siz1 is the primary E3 ligase responsible for hydrogen peroxide–induced sumoylation of Ntg1 while Siz2 could play a minor role in Ntg1 sumoylation.
We next examined Ntg1 sumoylation in cells defective for the SUMO proteases, Ulp1 and Ulp2. The ULP1 gene is essential so we employed a temperature sensitive mutant, ulp1-1 (ulp1-ts) (61), and shifted cells to 37°C to inactivate Ulp1. The levels of hydrogen peroxide–induced Ntg1 sumoylation in the ulp1-ts mutant were significantly higher than in the wildtype control cells. The ulp1-ts mutant cells displayed 10.9% monosumoylated Ntg1, contrasting with 4.7% monosumoylated Ntg1 detected in the wildtype cells (Figure 1C, D). In contrast to the ulp1-ts cells, ulp2Δ cells exhibit no detectable change in sumoylation in response to hydrogen peroxide exposure when compared to wildtype (Figure 1D). These results suggest that Ulp1 serves as the primary de-sumoylase for Ntg1.
Sumoylation is a dynamic process where only a very small percent of sumoylated product is present at any given time (35). In fact, in wildtype cells, without exogenous exposure to reactive oxygen species (ROS) or oxidative stress, we do not detect modification of Ntg1 (Figure 1A). To determine whether Ntg1 is modified only in response to hydrogen peroxide or is endogenously modified at low levels, we examined Ntg1 sumoylation in the ulp1-ts cells and the ulp2Δ mutant cells in the absence of any treatment. Loss of Ulp1 function resulted in a dramatic increase in both monosumoylated and multi-modified Ntg1 compared to wildtype (Figure 1E, F). In contrast, loss of Ulp2 had no impact on Ntg1 sumoylation levels. These data indicate that Ntg1 can be sumoylated in the absence of exogenous stress.
4.2 Identification of Ntg1 sumoylation sites
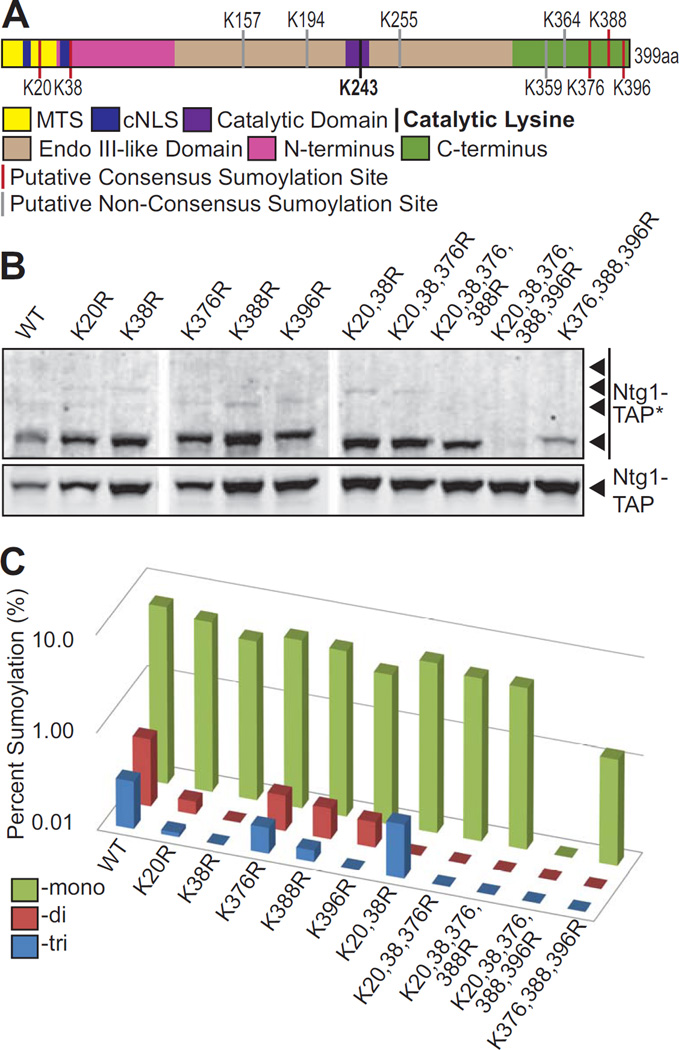
Sumoylation occurs on lysine residues, typically within SUMO consensus sequences (62, 63). More than two-thirds of known SUMO substrates contain at least one consensus sumoylation motif Ψ-K-x-D/E (where Ψ is a hydrophobic residue, K is the lysine conjugated to SUMO, x is any amino acid, and D/E is an acidic residue) (62, 63). We used freely available search engines to identify predicted sumoylation sites in both NTHL1 and Ntg1 (Supplemental Figure 1A, B). Prediction software identified multiple candidate sumoylation sites in NTHL1 (Supplemental Figure 1A). To identify candidate sumoylation sites in Ntg1, we used a combination of five SUMO prediction programs: SUMOsp 1.0 (64), SUMOsp 2.0/GPS-SUMO (65, 66), SUMOplot (http://www.abgent.com/sumoplot), SUMOpre (67), and PCI-SUMO (68) This analysis identified five putative consensus sumoylation sites (K20, K38, K376, K388, K396) within Ntg1 (Figure 2A and Supplemental Figure 1B). Five putative non-consensus sumoylation sites were also identified (Figure 2A and Supplemental Figure 1B). Consensus and non-consensus motifs of identified putative sumoylation sites and prediction scores are shown in Supplemental Figure 1B.
Figure 2. Identification of sumoylation sites in Ntg1.
A. A domain schematic of Ntg1 is shown with the following functional motifs/domains indicated: The Mitochondrial Targeting Sequence (MTS) in yellow, the classical Nuclear Localization Signal (cNLS) in dark blue, the Catalytic Domain in purple. The Catalytic Lysine, K243, is depicted as a black bar. The central region of Ntg1 that is homologous to E. coli Endonuclease III is shown in tan (amino acids 95–335) while the non-conserved N- and C-terminal domains are indicted in magenta (amino acids 1–94) and green (amino acids 336–399), respectively. Putative Consensus Sumoylation Sites are shown as red bars and Putative Non-Consensus Sumoylation Sites are shown as grey bars. B. A series of Ntg1 variants with candidate SUMO modification sites altered from lysine to arginine were generated and expressed in temperature sensitive ulp1 cells. Cells were treated with 20 mM hydrogen peroxide for 1 hour at 30°C, lysed, and immunoblotted to detected Ntg1-TAP and modified Ntg1-TAP (Ntg1-TAP*). Nonadjacent lanes in the same image are separated by white space. C. Results from (B) were quantitated. For each Ntg1 variant, the percent of total Ntg1-TAP signal present in the band corresponding to the size of Mono-, Di-, and Tri-sumoylation (indicated as Percent Sumoylation) is plotted on a log scale.
We initially tested for SUMO modification within these sites on Ntg1 via mass spectrometry. However, when we analyzed the bacterially expressed Ntg1 through mass spectrometry the peptides containing the putative SUMO modification sites were not detected. As an alternative approach, to determine which of these putative sites are sumoylated and to generate a form of Ntg1 that cannot be sumoylated, we performed site-directed mutagenesis to create conservative amino acid substitutions of the ten putative sumoylation site lysines to arginines. These substitutions were made in order of predicted site strength for all single sites (Supplemental Figure 1C). In total, we created 25 single and combination lysine to arginine substitutions beginning with a single substitution and proceeding with double, triple, etc. substitutions (Supplemental Figure 1C). We then analyzed the sumoylation status of all the resulting variants of Ntg1 in response to hydrogen peroxide.
The single lysine to arginine substitutions were tested first and the results showed that all Ntg1 variants containing single lysine to arginine substitutions can still be sumoylated (Figure 2B). Quantification of the single substitution data showed that one substitution examined (K396) results in a detectable decrease in the amount of monosumoylated Ntg1, suggesting that K396 could be the primary site of monosumoylation (Figure 2C). The finding that no single lysine to arginine substitution leads to a complete loss of SUMO modification supports our earlier results suggesting that Ntg1 is sumoylated at multiple lysines simultaneously. Thus, multiple substitutions are required to produce a variant that cannot be sumoylated. Single substitution of the five putative non-consensus sumoylation sites (K157, 194, 255, 359, 364) did not alter levels of Ntg1 sumoylation, indicating that these sites are not essential for Ntg1 sumoylation. Next, we tested a series of combinations of lysine to arginine Ntg1 variants for sumoylation. The double and triple mutant proteins involving the N- and C-termini, Ntg1K20,38R-TAP and Ntg1K20,38,376R-TAP, can both still be sumoylated (Figure 2B, C). The quadruple mutant protein, Ntg1K20,38,376,388R-TAP, shows only a single sumoylated species; while an additional K396 to arginine substitution, leads to the complete loss of all detectable SUMO-modification of Ntg1 (ntg1ΔSUMO). For the collection of variants, changes in the levels of Ntg1 sumoylation were quantified and are presented in Table 3. These results demonstrate that Ntg1 is sumoylated at any of five consensus sumoylation sites and that all five sites must be simultaneously changed to arginine to generate an Ntg1 variant that cannot be modified by SUMO. Supplemental Figure 1C shows all of the combinations of Ntg1 variants generated and summarizes the total sumoylation loss. Thus, we have identified the five lysine residues within Ntg1 that can be sumoylated.
Table 3.
Quantification of Mono-, Di-, Tri- Sumoylation of Ntg1 Variants
| Potential Sumo Modification |
Fraction of Modified Ntg1 (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | K20R | K38R | K376R | K388R | K396R | K20, 38R |
K20,38, 376R |
K20,38, 376, 388R |
K20,38, 376,388, 396R |
K376, 388, 396R |
|
| Ntg1- Tri |
0.31 | 0.11 | 0.02 | 0.18 | 0.13 | 0.00 | 0.36 | 0.02 | 0.00 | 0.00 | 0.00 |
| Ntg1- Di |
0.49 | 0.14 | 0.00 | 0.23 | 0.21 | 0.18 | 0.00 | 0.05 | 0.00 | 0.00 | 0.00 |
| Ntg1- Mono |
6.52 | 5.53 | 4.27 | 5.30 | 4.98 | 3.45 | 5.50 | 4.66 | 4.51 | 0.04 | 1.24 |
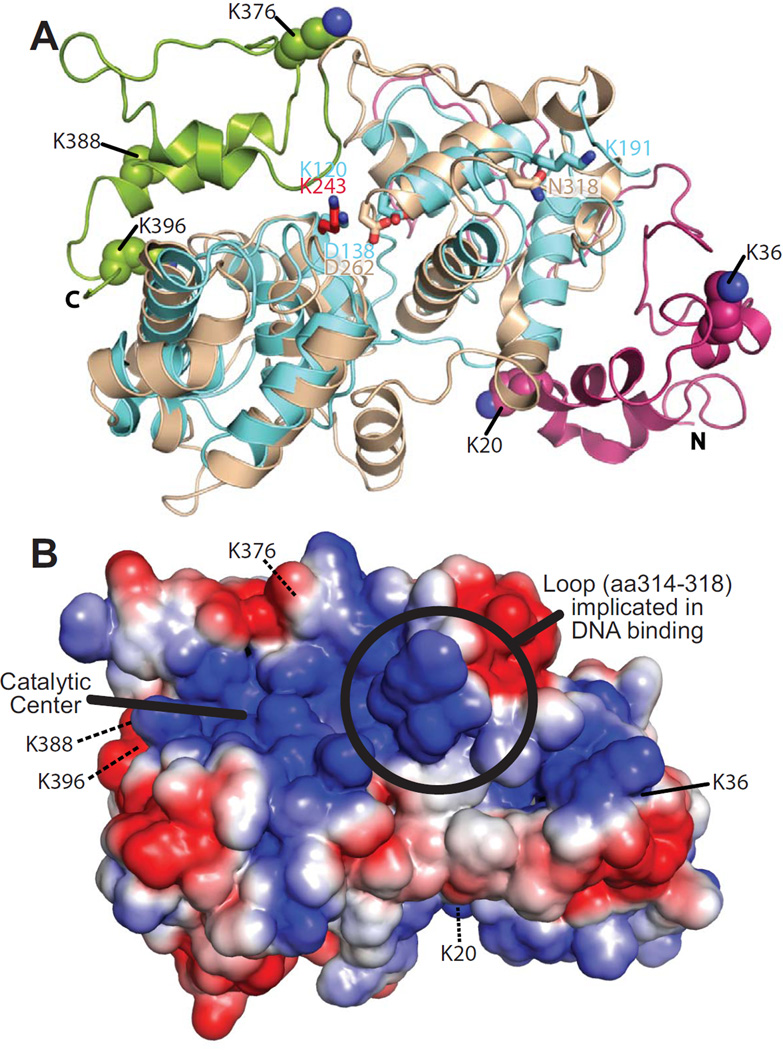
The lysines within Ntg1 that can be sumoylated reside in the N- and C-terminal domains that are specific to the eukaryotic enzyme and outside of the 307 residues which comprise the evolutionarily-conserved catalytic core with homology to the bacterial Endonuclease III protein (69). To provide insight into the location of these lysines within the three-dimensional structure of Ntg1, we generated a homology model of S. cerevisiae Ntg1 (Figure 3A) using the Protein Homology/analogY Recognition Engine version 2.0 (Phyre2) (70). The predicted model (Figure 3A) is based on the structure of the E. coli Endonuclease III protein (PDB ID: 2ABK). Tan regions display the high confidence (90%) homology mapping of the region of Ntg1 (amino acids 95–335) with homology to Endonuclease III (Figure 2A). The magenta and green regions correspond to the N-terminal (amino acids 1–94) and C-terminal (amino acids 335–399) domains, respectively (Figure 2A). Although the N- and C-terminal domains of Ntg1 do not align to Endonuclease III, they are modeled based on homology to other endonucleases. The N-terminal domain aligns to the restriction endonuclease BsaWI (PDB ID: 4ZSF) and the C-terminal domain aligns to the endonuclease BglII (PDB ID: 1DFM). Based on our homology model (Figure 3A), the five lysines that we defined as SUMO modification sites are all surface exposed, consistent with being accessible for modification.
Figure 3. Homlogy model of Ntg1.
A. A homology model of Ntg1 shown as a ribbon diagram was generated as described in Materials and Methods. The model is overlaid on the E. coli Ntg1 homologue, Endonuclease III, structure (cyan, PDB ID: 2ABK). The Ntg1 catalytic domain (amino acids 95–335; tan), N-terminal domain (amino acids 1–94; magenta), C-terminal domain (amino acids 336–399; green), catalytic amino acid of Ntg1 (K243, red) and Endonuclease III (K120, blue) are shown in addition to Endonuclease III amino acids D138, important for catalysis, and K191, implicated in DNA binding (69), and the corresponding amino acids in Ntg1 (D262 and N318, respectively). The five consensus sumoylation sites (K20, K38, K376, K388, and K396) are shown as balls and indicated by the labeling. B. An electrostatic model of Ntg1 is shown based on the homology model. Positive and negative residues are colored in blue and red, respectively. White indicates neutral residues. The loop containing residues 314–318, indicated by a circle, has been implicated in DNA binding by Endo III (69). The catalytic center is indicated by a bold black line and the five consensus sumoylation sites are labeled and indicated by black lines. Residues 20, 376, 388, and 396 are located on the back face of the model and are indicated by black dotted lines.
As sumoylation influences DNA binding and turnover of TDG (43–45), we analyzed the proximity of the sumoylation sites to the DNA binding and catalytic centers in our Ntg1 model. The structure of sumoylated-TDG (PDB ID: 1WYW) shows the close proximity of the SUMO modification to the DNA binding and catalytic site of TDG (71), illustrating why sumoylation of TGD might influence TGD catalysis. To assess whether sumoylation could impact DNA binding or catalysis by Ntg1, we superimposed our model of Ntg1 with the structure of Endonuclease III (Figure 3A). Previous work implicated K120 and D138 in catalysis and K191 in DNA binding of Endonuclease III (Figure 3A) (69). We identified the analogous amino acids in our model of Ntg1 (Figure 3A, B). Loop residues in Endo III corresponding to residues 314–318 in Ntg1 are important for DNA binding (Figure 3B). Both the catalytic residues and the DNA binding loop in Ntg1 are distant from the sumoylated lysines and extend from the opposite face of the protein. This analysis suggests that sumoylation is unlikely to directly influence Ntg1-mediated catalysis.
4.3 In vitro functional analysis of the nonsumoylatable Ntg1 variant, ntg1K20,38,376,388,396R (ntg1(K->R)5)
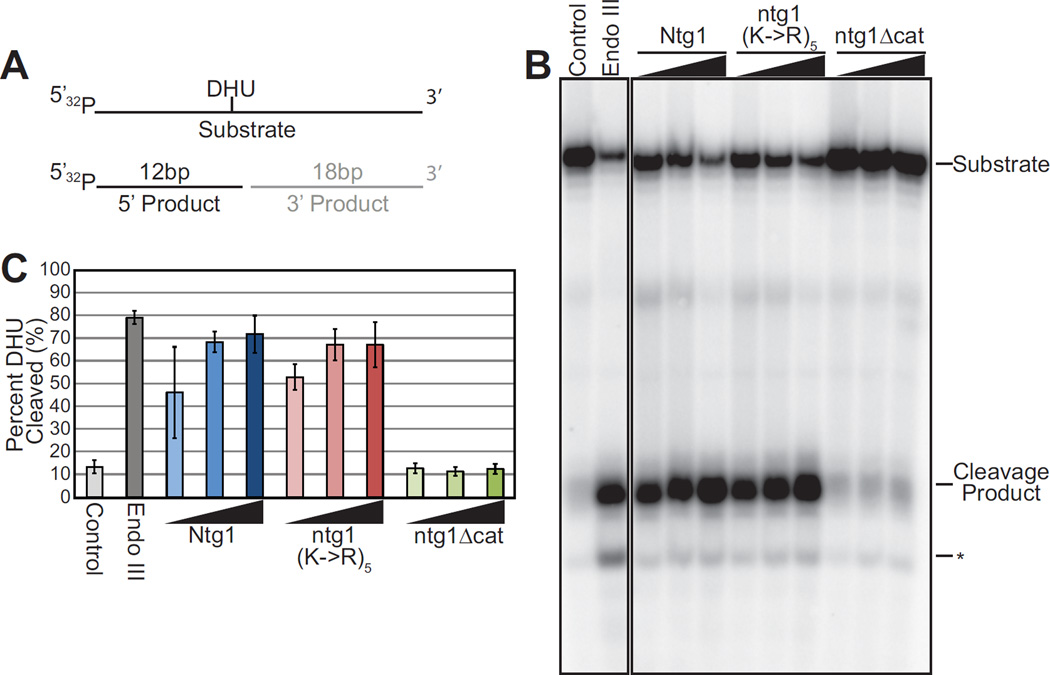
Although changing a lysine residue to an arginine residue conserves the charge and size of the amino acid, such modest changes could induce a conformational change potentially impacting function. To address whether the conservative substitution of the five lysines that constitute the Ntg1 sumoylation sites (K20, K38, K376, K388, K396) impacts the catalytic activity of Ntg1 in vitro, we employed an in vitro oligonucleotide cleavage assay to compare the enzymatic activity of wildtype Ntg1 to ntg1K20,38,376,388,396R, which we designate ntg1(K->R)5. The oligonucleotide substrate contains dihydrouracil (DHU) which is an Ntg1 substrate (72). As a control, we employed a catalytically inactive Ntg1 (ntg1Δcat) variant created by changing the catalytic lysine at position 243 to glutamine (73). We incubated purified recombinant His6-Ntg1 variants with the oligonucleotide containing the Ntg1 substrate and detected Ntg1 enzymatic activity as cleavage of the oligonucleotide at the position of the DHU (26). As shown in the cleavage assay presented in Figure 4B, His6-ntg1(K->R)5 shows enzymatic activity comparable to wildtype His6-Ntg1 whereas a catalytically inactive form of Ntg1, His6-ntg1Δcat, did not cleave the substrate. We quantitated the results of three independent cleavage experiments (Figure 4C). These results confirm that there is no difference in the activity of ntg1(K->R)5 compared to wildtype Ntg1 in this assay. These results indicate that changing the five SUMO modification sites from lysine to arginine does not alter the enzymatic activity of Ntg1.
Figure 4. Functional analysis of Ntg1 variant.
A. A schematic of the substrate employed for the in vitro cleavage assay, which contains dihydrouracil (DHU) embedded in a 31mer oligo, illustrating the substrate and expected products of the cleavage reaction is shown. B. Recombinant E. coli Endonuclease III (Endo III), and Ntg1 variants, His6-Ntg1, His6-ntg1(K->R)5, catalytically inactive ntg1 (His6-ntg1Δcat), were employed for the in vitro cleavage assay. Increasing amounts of recombinant protein (5–50 ng) were added to radioactively-labeled substrate. Oligonucleotide Cleavage Products were electrophoresed and subjected to phosphorimager analysis. The Control lane shows the substrate with no added protein. The positive control is addition of 50 ng of E. coli Endo III. The position of the labeled product generated by cleavage (Cleavage Product) is indicated. Random degradation product is indicated by an asterisk (*). Nonadjacent lanes in the same image are separated by black lines. Results shown in (B) are representative of three independent experiments. C. Quantification of Cleavage Product generated for each Ntg1 variant from three independent experiments. Results are shown as Percent DHU Cleaved. Error bars represent standard deviation in the data.
4.4 Functional analysis of Ntg1 in vivo
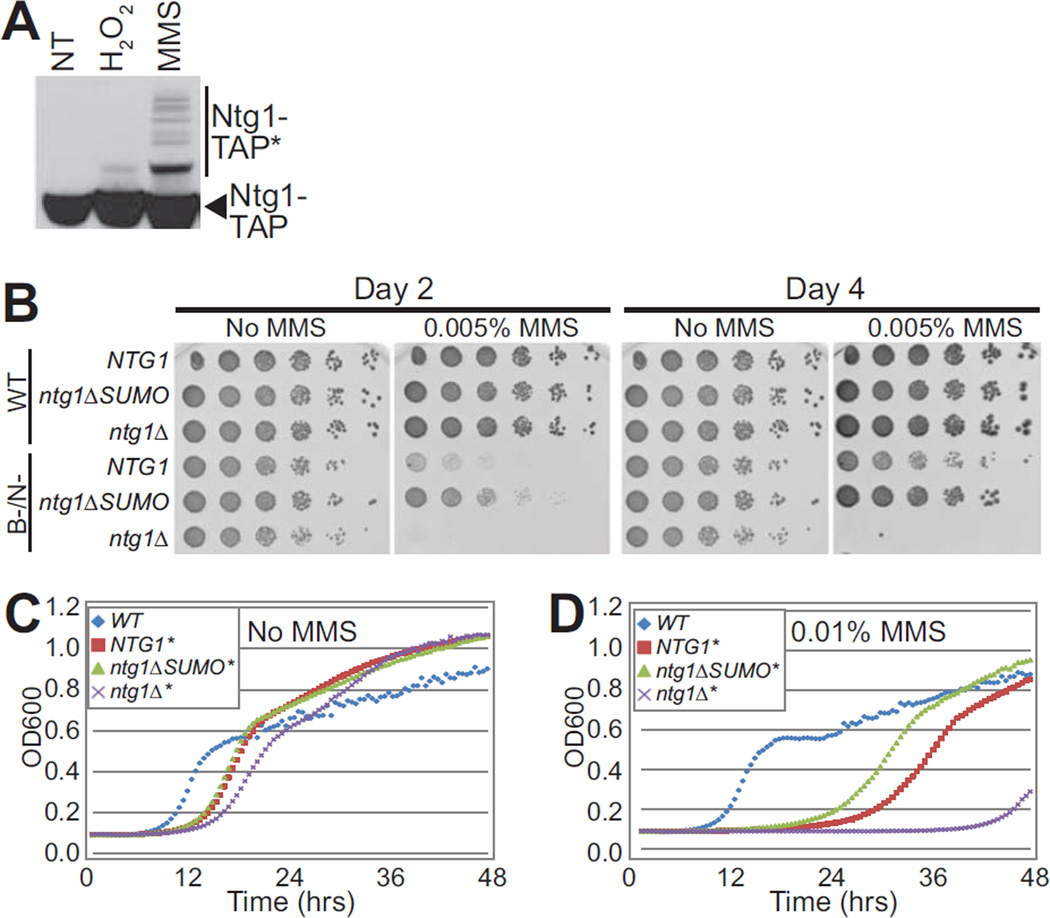
Previous studies showed that Ntg1 is SUMO–modified in response to treatment with hydrogen peroxide (24). To assess whether other types of DNA damage can induce Ntg1 sumoylation, cells were exposed to methyl methanesulfonate (MMS), which induces alkylating DNA damage (74, 75). As shown in Figure 5A, Ntg1 is sumoylated in response to treatment with MMS. We exploited this observation to examine how cells that express an Ntg1 variant that cannot be modified by SUMO (ntg1K20,38, 376,388,396R), which we designate ntg1ΔSUMO, respond to DNA damage. For these experiments, Ntg1 or ntg1ΔSUMO was expressed either in base excision and nucleotide excision repair (NER)-proficient wildtype cells (WT) or repair-deficient (ntg1Δntg2Δapn1Δrad1Δ) cells, which lack BER and NER (B-/N-) (19, 76). Cells were exposed to MMS and the growth characteristics of these cells expressing wildtype Ntg1 were compared to those expressing ntg1ΔSUMO. We then examined growth in the absence or presence of MMS (Figure 5B). Growth was analyzed at days 2 and 4 following serial dilution and spotting on plates. As expected (76), the repair proficient (WT) cells grew well under all conditions tested, regardless of which Ntg1 variant was expressed. In contrast, the repair-deficient cells display slow growth in the presence of MMS even with wildtype NTG1. The repair-deficient ntg1Δ cells were extremely sensitive to MMS (Figure 5B). Surprisingly, the ntg1ΔSUMO cells were less sensitive to MMS compared to cells with wildtype NTG1. This result suggests that sumoylation of Ntg1 could be important for coordinating DNA repair with cell cycle progression or DNA damage response.
Figure 5. Functional analysis of ntg1ΔSUMO in DNA damage pathways.
A. Wildtype cells expressing Ntg1-TAP were exposed to hydrogen peroxide (H2O2), methyl methanesulfonate (MMS), or were not treated (NT) and lysed. Lysate was subjected to immunoblotting to detect Ntg1-TAP and modified forms of Ntg1-TAP (Ntg1-TAP*). B. Cells with either a full complement of wildtype (WT) DNA repair pathways or deficient in both base excision repair and nucleotide excision repair (B-/N-) were employed. As described in Materials and Methods, the genotype for B-/N- cells (DSC0369) is ntg1Δntg2Δapn1Δrad1Δ. Both the WT and B-/N- cells were engineered to express ntg1ΔSUMO and compared to cells expressing NTG1 or lacking Ntg1 (ntg1Δ). Cultures were 5-fold serially diluted and spotted onto rich media or rich media containing 0.005% MMS and incubated at 30°C for 4 days. Pictures were taken at Day 2 and Day 4. C/D. The same samples as shown in (B) with either intact DNA repair pathways (WT) (blue diamond) or deficient in base excision repair and nucleotide excision repair (B-/N-), denoted by an *, contain wildtype NTG1 (red square), or ntg1ΔSUMO (green triangle), or lack Ntg1 (ntg1Δ) (purple X) at the endogenous NTG1 locus. The genotype for B-/N- is ntg1Δntg2Δapn1Δrad1Δ (DSC0369). Cells were grown in liquid culture with No MMS (C) or with 0.01% MMS (D) for 48 hours. OD600 readings were taken every 30 minutes and plotted vs time. Results shown in (B, C, and D) are representative of at least three independent experiments.
To further examine the growth of the ntg1ΔSUMO cells following treatment with MMS, growth curves were generated for wildtype, repair-deficient cells, Ntg1 in repair-deficient cells, and ntg1ΔSUMO in repair-deficient cells grown in YPD with and without MMS (Figure 5C, D). The results indicate that repair-deficient cells that express ntg1ΔSUMO emerge from lag-phase earlier than repair-deficient cells expressing wildtype Ntg1 (Figure 5D). As expected, repair-deficient cells expressing either Ntg1 or ntg1ΔSUMO grew equally well in the absence of MMS (Figure 5C). These data further suggest that the sumoylation of Ntg1 plays a role in coordinating the growth arrest that occurs in response to DNA damage.
Discussion
We report here that SUMO modification is a conserved post-translational modification of S. cerevisiae Ntg1 and the human orthologue, NTHL1. In S. cerevisiae, we identified the two SUMO ligases, Siz1 and Siz2, and the desumoylase, Ulp1, critical for reversible regulation of this modification. We mapped the sites of SUMO modification in Ntg1 and created an Ntg1 that cannot be SUMO modified. Our preliminary analysis of this non-sumoylatable form of Ntg1 reveals that SUMO modification may be important for proper cellular response to DNA damage.
We identified Ulp1 as the primary desumoylase for Ntg1 with little impact of Ulp2. As the primary role of Ulp2 is to remove SUMO from poly(SUMO) chains (55), and we detect no change in SUMO modification of Ntg1 in ulp2Δ mutant cells (Figure 1C, D), we speculate that Ntg1 could be modified by multiple independent SUMOs rather than a single chain of multiple SUMOs. This model is consistent with our finding that five different lysine residues in Ntg1 can be modified by SUMO. While we cannot rule out the possibility that the Ntg1-TAP used in this study is also modified by other post-translational modifications, the band shifts are consistent with the molecular size and charge of multiple SUMO molecules. Consistent with a possible role for additional post-translational modifications, mass spec analysis reveals that Ntg1 serine 71 is phosphorylated (77). Regardless, the data presented here in combination with our previous publication (24) show that Ntg1 is SUMO modified by at least one SUMO molecule and that there are at least five lysines on Ntg1 that can be SUMO modified. These SUMO molecules could coordinate other post-translational modifications.
Our data show that both hydrogen peroxide and MMS can induce sumoylation of Ntg1 (24). Like hydrogen peroxide, treatment with MMS can cause oxidative stress and generate ROS (78). Therefore, we cannot yet clearly distinguish whether hydrogen peroxide and MMS trigger sumoylation of Ntg1 through the same or distinct mechanisms. Further work will be required to determine how the sumoylation machinery responds to DNA damage and/or oxidative stress. Regulation could occur through activation of SUMO E3 ligases or through inhibition of the Ulp1 desumoylase. Further analysis will be required to dissect this mechanism.
Sumoylation can influence numerous functions of a protein including catalytic activity, localization, stability, and/or protein-protein interactions (79). As sumoylation plays a role in regulating the binding capabilities of TDG by modulating the interaction with DNA, sumoylation could also impact the DNA binding ability of Ntg1. However, based on homology modeling and mapping of the sumoylation sites, our model suggests that sumoylation at any of the five sites we identified likely does not directly interfere with the DNA binding to Ntg1. Consistent with this result, none of the Ntg1 lysine to arginine substitutions altered catalytic activity of the recombinant protein in vitro (Figure 4). With respect to localization, Ntg1 sumoylation at K20 and K38 on Ntg1 are within or just adjacent to the consensus organelle targeting localization sequences. The N-terminus of Ntg1 contains a mitochondrial targeting sequence (MTS) at amino acids 1–26 and a classical bipartite nuclear localization signal (cNLS) at amino acids 14–17 and 31–37 (51). The proximity of the sumoylation sites, specifically K20 and K38, to these localization signals suggests a potential role for sumoylation in regulating subcellular localization of Ntg1. In fact, our previous biochemical fractionation studies showed that sumoylated Ntg1 is detected only in the nucleus (24). Consistent with a conserved regulatory model, human NTHL1 also contains putative SUMO sites at K56 and K60 that overlap a predicted cNLS at amino acids 56–60 (Supplemental Figure 1A). Another possible function of sumoylation is to modulate proteinprotein interactions (79). Little is known about the interacting partners of Ntg1. One high-throughput yeast two-hybrid study identified two DNA damage response proteins, Rad59 and Rfc2, as physical interactors of Ntg1 (80). Rad59 is involved in double strand break repair (81), and Rfc2 is part of the ATPase clamp loader for the proliferating cell nuclear antigen (PCNA) processivity factor for DNA polymerases (82). As both of these proteins are implicated in DNA damage response, they could mediate crosstalk between BER and the DNA damage response pathway. A critical next step in understanding the functional impact of sumoylation on Ntg1 is to identify SUMO-dependent interacting proteins.
Our data (Figure 5B, C, D) show cells expressing ntg1ΔSUMO display more rapid growth compared to cells expressing wildtype Ntg1 in a DNA repair deficient background in response to MMS. Alkylation damage induced by MMS can be mutagenic and lead to cytotoxic blockage of replication forks (75, 83, 84). One possibility is that sumoylation of Ntg1 is required for proper checkpoint activation or maintenance. Cell cycle checkpoints are activated by sensor proteins, such as Rad9 (85), detecting an increase in DNA damage and initiating a signal cascade that ultimately leads to activation of Rad53, the protein kinase responsible for cell cycle arrest (86, 87). Activation of Rad53 is critical for stabilization of replication forks and activating the DNA repair pathway (81). Interestingly, improper activation of Rad53 results in an increased resistance to MMS via engagement of translesion synthesis (TLS) (88). Further investigation of this potential connection between the DNA checkpoint protein, Rad53, and the BER protein, Ntg1, could reveal a novel DNA damage response activator.
A number of studies have identified roles for SUMO in modulating DNA repair (89–98). The work presented here suggests SUMO-mediated regulation could extend to the evolutionarily conserved BER pathway. Indeed regulation of the initial step of the BER pathway could be crucial to ensure genome integrity.
Supplementary Material
Highlights.
Base excision repair (BER) proteins can be modified by SUMO
Both S. cerevisiae Ntg1 and human NTHL1 can be sumoylated
Five lysines are identified as the sites of SUMO modification in Ntg1
An Ntg1 variant that cannot be modified by SUMO is generated
Cells where Ntg1 cannot be modified by SUMO fail to respond properly to DNA damage
Acknowledgments
We would like to thank Christine Dunham for her help with the generation of the protein homology model. We would also like to acknowledge members of the Corbett and Doetsch laboratories for helpful discussions and advice.
This study was supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Funding Sources
This work was supported by the National Institutes of General Medical Sciences [RO1 GM05872816; T32 GM008490 22; and F31 GM115178 01] and the National Institutes of Health [NIH ES011163].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA damage and repair: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10(5):891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- 2.Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85(11):1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Boesch P, Weber-Lotfi F, Ibrahim N, Tarasenko V, Cosset A, Paulus F, et al. DNA repair in organelles: Pathways, organization, regulation, relevance in disease and aging. Biochim Biophys Acta. 2011;1813(1):186–200. doi: 10.1016/j.bbamcr.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutation research. 2011;711(1–2):193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Pan Y. Mitochondria, reactive oxygen species, and chronological aging: a message from yeast. Experimental gerontology. 2011;46(11):847–852. doi: 10.1016/j.exger.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 6.de Gruijl FR, van Kranen HJ, Mullenders LH. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J Photochem Photobiol B. 2001;63(1–3):19–27. doi: 10.1016/s1011-1344(01)00199-3. [DOI] [PubMed] [Google Scholar]
- 7.Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2'-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990;87(12):4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59(11):2522–2526. [PubMed] [Google Scholar]
- 9.Salmon TB, Evert BA, Song B, Doetsch PW. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32(12):3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefl S, Nishi H, Petukh M, Panchenko AR, Alexov E. Molecular mechanisms of disease-causing missense mutations. J Mol Biol. 2013;425(21):3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evert BA, Salmon TB, Song B, Jingjing L, Siede W, Doetsch PW. Spontaneous DNA damage in Saccharomyces cerevisiae elicits phenotypic properties similar to cancer cells. J Biol Chem. 2004;279(21):22585–22594. doi: 10.1074/jbc.M400468200. [DOI] [PubMed] [Google Scholar]
- 12.Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272(32):19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 13.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 14.Wei YH. Oxidative stress and mitochondrial DNA mutations in human aging. Proc Soc Exp Biol Med. 1998;217(1):53–63. doi: 10.3181/00379727-217-44205. [DOI] [PubMed] [Google Scholar]
- 15.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 16.Lenaz G. Role of mitochondria in oxidative stress and ageing. Biochim Biophys Acta. 1998;1366(1–2):53–67. doi: 10.1016/s0005-2728(98)00120-0. [DOI] [PubMed] [Google Scholar]
- 17.Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, et al. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem. 2009;284(14):9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5(2):89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Degtyareva NP, Chen L, Mieczkowski P, Petes TD, Doetsch PW. Chronic oxidative DNA damage due to DNA repair defects causes chromosomal instability in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28(17):5432–5445. doi: 10.1128/MCB.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sentürker S, Auffret van der Kemp P, You HJ, Doetsch PW, Dizdaroglu M, Boiteux S. Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26(23):5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swartzlander DB, Bauer NC, Corbett AH, Doetsch PW. Chapter 5 – Regulation of base excision repair in eukaryotes by dynamic localization strategies. In: Doetsch PW, editor. Mechanisms of DNA Repair. Progress in Molecular Biology and Translational Science. Vol. 110. Oxford, UK: Academic Press; 2012. pp. 93–121. [DOI] [PubMed] [Google Scholar]
- 22.Nilsen H, Krokan HE. Base excision repair in a network of defence and tolerance. Carcinogenesis. 2001;22(7):987–998. doi: 10.1093/carcin/22.7.987. [DOI] [PubMed] [Google Scholar]
- 23.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451(1–2):39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths LM, Swartzlander D, Meadows KL, Wilkinson KD, Corbett AH, Doetsch PW. Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Mol Cell Biol. 2009;29(3):794–807. doi: 10.1128/MCB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda S, Kohmoto T, Tabata R, Seki Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA Repair (Amst) 2002;1(10):847–854. doi: 10.1016/s1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- 26.You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Sentürker S, et al. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38(35):11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 27.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316(2):113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 28.Cremona CA, Sarangi P, Zhao X. Sumoylation and the DNA damage response. Biomolecules. 2012;2(3):376–388. doi: 10.3390/biom2030376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartek J, Hodny Z. SUMO boosts the DNA damage response barrier against cancer. Cancer Cell. 2010;17(1):9–11. doi: 10.1016/j.ccr.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 30.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 31.Dery U, Masson JY. Twists and turns in the function of DNA damage signaling and repair proteins by post-translational modifications. DNA Repair (Amst) 2007;6(5):561–577. doi: 10.1016/j.dnarep.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ET. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 2011;585(18):2891–2896. doi: 10.1016/j.febslet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: The molecular choreography. Mutation Research/Reviews in Mutation Research. 2012;751(2):158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Kirkin V, Dikic I. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol. 2007;19(2):199–205. doi: 10.1016/j.ceb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458(7237):461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 36.Goto M, Shinmura K, Igarashi H, Kobayashi M, Konno H, Yamada H, et al. Altered expression of the human base excision repair gene NTH1 in gastric cancer. Carcinogenesis. 2009;30(8):1345–1352. doi: 10.1093/carcin/bgp108. [DOI] [PubMed] [Google Scholar]
- 37.Koketsu S, Watanabe T, Nagawa H. Expression of DNA repair protein: MYH, NTH1, and MTH1 in colorectal cancer. Hepatogastroenterology. 2004;51(57):638–642. [PubMed] [Google Scholar]
- 38.Short E, Thomas LE, Hurley J, Jose S, Sampson JR. Inherited predisposition to colorectal cancer: towards a more complete picture. J Med Genet. 2015;52(12):791–796. doi: 10.1136/jmedgenet-2015-103298. [DOI] [PubMed] [Google Scholar]
- 39.Rivera B, Castellsague E, Bah I, van Kempen LC, Foulkes WD. Biallelic NTHL1 Mutations in a Woman with Multiple Primary Tumors. The New England journal of medicine. 2015;373(20):1985–1986. doi: 10.1056/NEJMc1506878. [DOI] [PubMed] [Google Scholar]
- 40.Weren RD, Ligtenberg MJ, Kets CM, de Voer RM, Verwiel ET, Spruijt L, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nature genetics. 2015;47(6):668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 41.Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015;43(21):10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280(6):4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 43.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO JOURNAL. 2002;21(6):1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smet-Nocca C, Wieruszeski JM, Leger H, Eilebrecht S, Benecke A. SUMO-1 regulates the conformational dynamics of thymine-DNA glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem. 2011;12:4. doi: 10.1186/1471-2091-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15(7):616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 46.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. Embo J. 1998;17(17):5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang CH, Wei W, Liu L. Regulation of DNA repair by S-nitrosylation. Biochim Biophys Acta. 2012;1820(6):730–735. doi: 10.1016/j.bbagen.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 51.Swartzlander DB, Griffiths LM, Lee J, Degtyareva NP, Doetsch PW, Corbett AH. Regulation of base excision repair: Ntg1 nuclear and mitochondrial dynamic localization in response to genotoxic stress. Nucleic Acids Res. 2010;38(12):3963–3974. doi: 10.1093/nar/gkq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276(24):21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 53.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106(6):735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 54.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272(43):26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 55.Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. The Journal of biological chemistry. 2003;278(45):44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 56.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276(38):35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 57.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20(15):2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li SJ, Hochstrasser M. A new protease required for cell-cycle progression in yeast. Nature. 1999;398(6724):246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 59.Li SJ, Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol. 2000;20(7):2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwienhorst I, Johnson ES, Dohmen RJ. SUMO conjugation and deconjugation. Molecular & general genetics : MGG. 2000;263(5):771–786. doi: 10.1007/s004380000254. [DOI] [PubMed] [Google Scholar]
- 61.Soustelle C, Vernis L, Freon K, Reynaud-Angelin A, Chanet R, Fabre F, et al. A new Saccharomyces cerevisiae strain with a mutant Smt3-deconjugating Ulp1 protein is affected in DNA replication and requires Srs2 and homologous recombination for its viability. Molecular and cellular biology. 2004;24(12):5130–5143. doi: 10.1128/MCB.24.12.5130-5143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5(12):2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 64.Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–W257. doi: 10.1093/nar/gkl207. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, et al. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9(12):3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–W330. doi: 10.1093/nar/gku383. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu J, He Y, Qiang B, Yuan J, Peng X, Pan XM. A novel method for high accuracy sumoylation site prediction from protein sequences. BMC Bioinformatics. 2008;9:8. doi: 10.1186/1471-2105-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Green JR, Dmochowski GM, Golshani A. Can Med Biol Eng Conf. Vancouver, BC: 2006. Prediction of protein sumoylation sites via parallel cascade identification. [Google Scholar]
- 69.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14(16):4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435(7044):979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 72.Dizdaroglu M, Bauche C, Rodriguez H, Laval J. Novel substrates of Escherichia coli nth protein and its kinetics for excision of modified bases from DNA damaged by free radicals. Biochemistry. 2000;39(18):5586–5592. doi: 10.1021/bi9927787. [DOI] [PubMed] [Google Scholar]
- 73.Augeri L, Lee Y-M, Barton AB, Doetsch PW. Purification, characterization, gene cloning, and expression of Saccharomyces cerevisiae redoxyendonuclease, a homolog of Escherichia coli Endonuclease III. Biochemistry. 1997;36(4):721–729. doi: 10.1021/bi9625511. [DOI] [PubMed] [Google Scholar]
- 74.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3(1):1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 75.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6(4):429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 76.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(4):2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rowe LA, Degtyareva N, Doetsch PW. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic Biol Med. 2008;45(8):1167–1177. doi: 10.1016/j.freeradbiomed.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jentsch S, Muller S. Regulatory Functions of Ubiquitin and SUMO in DNA Repair Pathways. Subcell Biochem. 2010;54:184–194. doi: 10.1007/978-1-4419-6676-6_15. [DOI] [PubMed] [Google Scholar]
- 80.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 81.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & development. 1994;8(6):652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 82.Noskov V, Maki S, Kawasaki Y, Leem SH, Ono B, Araki H, et al. The RFC2 gene encoding a subunit of replication factor C of Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22(9):1527–1535. doi: 10.1093/nar/22.9.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boiteux S, Laval J. Mutagenesis by alkylating agents: coding properties for DNA polymerase of poly (dC) template containing 3-methylcytosine. Biochimie. 1982;64(8–9):637–641. doi: 10.1016/s0300-9084(82)80103-x. [DOI] [PubMed] [Google Scholar]
- 84.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutation research. 1985;150(1–2):77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 85.Prakash L. Lack of chemically induced mutation in repair-deficient mutants of yeast. Genetics. 1974;78(4):1101–1118. doi: 10.1093/genetics/78.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell. 2002;9(5):1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- 87.Toh GW, Lowndes NF. Role of the Saccharomyces cerevisiae Rad9 protein in sensing and responding to DNA damage. Biochemical Society transactions. 2003;31(Pt 1):242–246. doi: 10.1042/bst0310242. [DOI] [PubMed] [Google Scholar]
- 88.Conde F, Ontoso D, Acosta I, Gallego-Sanchez A, Bueno A, San-Segundo PA. Regulation of tolerance to DNA alkylating damage by Dot1 and Rad53 in Saccharomyces cerevisiae. DNA Repair (Amst) 2010;9(10):1038–1049. doi: 10.1016/j.dnarep.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Sarangi P, Bartosova Z, Altmannova V, Holland C, Chavdarova M, Lee SE, et al. Sumoylation of the Rad1 nuclease promotes DNA repair and regulates its DNA association. Nucleic Acids Res. 2014;42(10):6393–6404. doi: 10.1093/nar/gku300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hang LE, Lopez CR, Liu X, Williams JM, Chung I, Wei L, et al. Regulation of Ku-DNA association by Yku70 C-terminal tail and SUMO modification. J Biol Chem. 2014;289(15):10308–10317. doi: 10.1074/jbc.M113.526178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, Damborsky J, et al. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 2010;38(14):4708–4721. doi: 10.1093/nar/gkq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21(6):1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8(11):1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 94.Steinacher R, Schar P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr Biol. 2005;15(7):616–623. doi: 10.1016/j.cub.2005.02.054. [DOI] [PubMed] [Google Scholar]
- 95.Bergink S, Ammon T, Kern M, Schermelleh L, Leonhardt H, Jentsch S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51–Rad52 interaction. Nat Cell Biol. 2013;15(5):526–532. doi: 10.1038/ncb2729. [DOI] [PubMed] [Google Scholar]
- 96.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19(1):123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 97.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436(7049):428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 98.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9(8):923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 99.Chen XL, Reindle A, Johnson ES. Misregulation of 2 microm circle copy number in a SUMO pathway mutant. Molecular and cellular biology. 2005;25(10):4311–4320. doi: 10.1128/MCB.25.10.4311-4320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web References
- http://www.abgent.com/sumoplot Date Last used: 8/4/16.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.