Abstract
Next generation sequencing using platforms such as Illumina MiSeq provides a deeper insight into the structure and function of bacterioplankton communities in coastal ecosystems compared to traditional molecular techniques such as clone library approach which incorporates Sanger sequencing. In this study, structure of bacterioplankton communities was investigated from two stations of Sundarbans mangrove ecoregion using both Sanger and Illumina MiSeq sequencing approaches. The Illumina MiSeq data is available under the BioProject ID PRJNA35180 and Sanger sequencing data under accession numbers KX014101-KX014140 (Stn1) and KX014372-KX014410 (Stn3). Proteobacteria-, Firmicutes- and Bacteroidetes-like sequences retrieved from both approaches appeared to be abundant in the studied ecosystem. The Illumina MiSeq data (2.1 GB) provided a deeper insight into the structure of bacterioplankton communities and revealed the presence of bacterial phyla such as Actinobacteria, Cyanobacteria, Tenericutes, Verrucomicrobia which were not recovered based on Sanger sequencing. A comparative analysis of bacterioplankton communities from both stations highlighted the presence of genera that appear in both stations and genera that occur exclusively in either station. However, both the Sanger sequencing and Illumina MiSeq data were coherent at broader taxonomic levels. Pseudomonas, Devosia, Hyphomonas and Erythrobacter-like sequences were the abundant bacterial genera found in the studied ecosystem. Both the sequencing methods showed broad coherence although as expected the Illumina MiSeq data helped identify rarer bacterioplankton groups and also showed the presence of unassigned OTUs indicating possible presence of novel bacterioplankton from the studied mangrove ecosystem.
Keywords: Bacterioplankton, Mangrove ecosystem, Sanger sequencing, Illumina MiSeq sequencing
| Specifications | |
|---|---|
| Organism | Sundarbans bacterioplankton metagenome |
| Sex | Not applicable |
| Sequencer or array type | Illumina MiSeq |
| Data format raw data | Fastq file |
| Experimental factors | Environmental sample |
| Experimental features | 16S rRNA metagenome sequencing |
| Analysis using | QIIME, MEGAN5 |
| Consent | Not applicable |
| Sample source location | Water, estuary, Sundarbans, India |
Bacterioplankton play key roles in biogeochemical cycling through the microbial loop in marine environment including coastal ecosystems [1]. The composition and distribution patterns of bacterioplankton communities have been surveyed across various coastal ecosystems such as the Columbia estuary [8], Pearl estuary [12] and Delaware Bay [3] to understand their role in ecosystem processes. However, not much is known in terms of bacterioplankton community structure from mangrove ecosystems globally [7], [9], [11]. Sundarbans, the world's largest contiguous mangrove ecoregion, provides a unique set up to investigate and understand the structure and functional significance of bacterioplankton communities. Seasonal variation in surface water temperature, heavy local precipitation during monsoon, continuous flow of freshwater from Ganga-Brahmaputra-Meghna riverine systems, diurnal intrusion of saline water from Bay of Bengal and dynamicity in dissolved nutrients could act as stressors for bacterioplankton communities of Sundarbans. We analysed the bacterioplankton communities by constructing 16S rRNA clone libraries and subsequent sequencing of individual clones by Sanger sequencing method from extracted environmental DNA from two stations representing the Sundarbans Biological Observatory Time Series (SBOTS). High-throughput sequencing using Illumina MiSeq approach was then undertaken from the same set of environmental DNA to obtain a deeper insight into bacterioplankton community structure. This study was undertaken in SBOTS located in Sagar Island, the largest island of the Indian Sundarbans. Two spatially separated stations designated as Station 1 (Stn1; 21° 44′ 44.4″ N, 88° 08′ 49.5″ E) and Station 3 (Stn3; 21° 40′ 40.6″ N, 88° 09′ 19.2″ E) as part of SBOTS were selected for this study. One litre of surface water sample was collected from each station in July 2014 following standard published protocol [5]. Collected samples were immediately fixed with molecular grade alcohol and transferred to the laboratory. Biomass was concentrated by filtering water samples through a 0.22 μm nitrocellulose filter paper (Pall, USA) using standard methodology [5]. The filters were immediately stored at − 20 °C until further downstream processing. Extraction of environmental DNA (eDNA) pool was undertaken from each filter following published protocol [2]. Clone libraries comprising of forty clones from each library was generated from both the stations as part of an ongoing study spanning from June to December 2014 (Ghosh and Bhadury, 2016, in prep.). In this study, data representing 40 clones from each station only for the month of July 2014 has been discussed. Since maximum heterogeneity in bacterioplankton communities was observed in the month of July 2014 based on clone library data (Sanger sequencing) therefore in order to get a deeper resolution of their community structure, the extracted eDNA from each station for the same month was also sequenced using Illumina MiSeq platform. The V3-V4 hypervariable region of ~ 460 bp was amplified using Pro340F (5′-CCTACGGGNBGCASCAG-3′) and Pro805R (5′-GACTACNVGGGTATCTAATCC-3′) primers for Illumina sequencing. The generated sequences were first quality filtered using QIIME [4]. After quality filtration and trimming of adapter sequences of the raw reads, the clean sequences were clustered into operational taxonomic units (OTUs) at 97% similarity cut off. Taxonomic affiliation was generated using default parameters in the QIIME pipeline and cross validated by Blast2go tool [6]. Phylogram was generated in MEGAN5 using the taxonomic assignment files obtained from QIIME [10]. This helped to compare the abundance of sequences recovered from Stn1 and Stn3 at the phylum, class, order, family and genus levels. Previous generated clone library data was then used to check the congruency between the two sequencing methods.
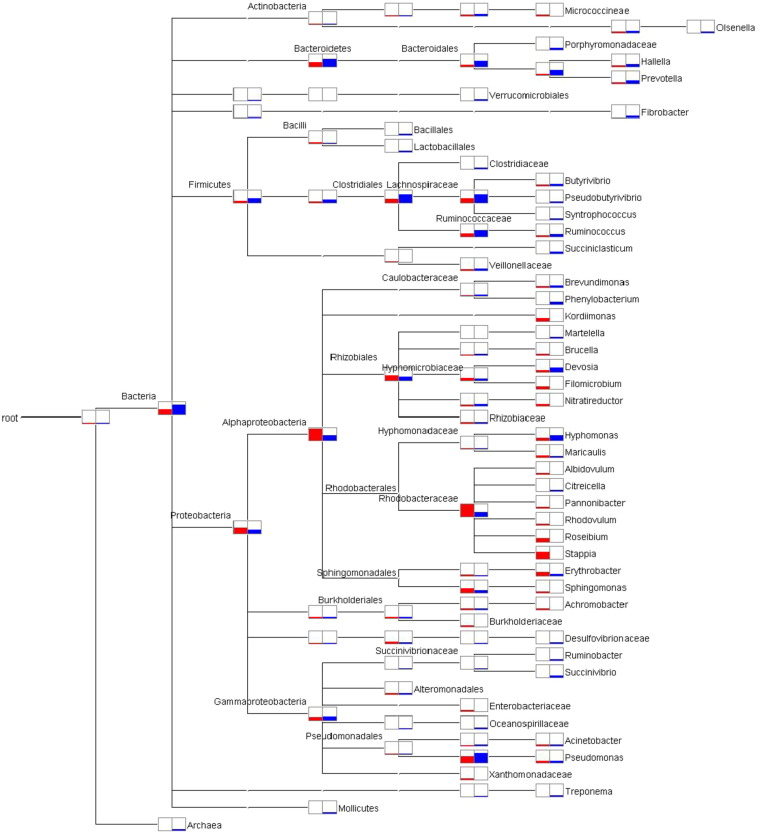
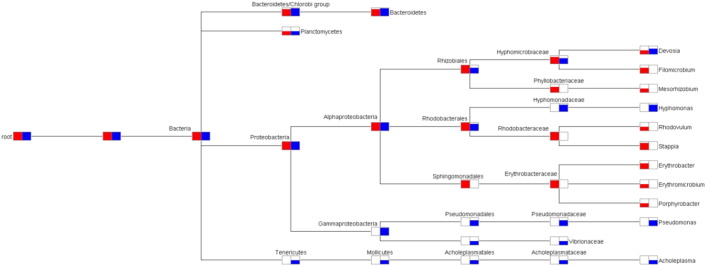
Approximately 888 MB data with 1,777,715 pair-end reads was generated from Stn1 and 1.24 GB data with 2,499,148 pair-end reads was generated from Stn3. The Illumina MiSeq data were grouped in 28,490 and 30,474 OTUs compared to 21 and 22 OTUs generated from clone libraries from Stn1 and Stn3 respectively. The bacterial communities at Stn1 appear to be dominated by Proteobacteria-like sequences (24,168 OTUs, abundance 94.05%), followed by Firmicutes-like sequences (2107 OTUs, abundance 1.84%) and Bacteroidetes-like sequences (1004 OTUs, abundance 1.77%). In the corresponding Stn1 clone library, Proteobacteria (18 OTUs) and Bacteroidetes-like sequences (2 OTUs) were encountered; however Firmicutes-like sequences were not encountered in clone library of Stn1. In Stn3, 11,773 OTUs (abundance 68.29%) and 8 OTUs of Proteobacteria-like sequences, 9824 OTUs (abundance 18.38%) and 0 OTUs of Firmicutes-like sequences and 5996 OTUs (abundance 9.22%) and 4 OTUs of Bacteroidetes-like sequences were recovered from the Illumina MiSeq data and clone library data respectively. A total of 31 phyla from Stn1 and 39 phyla from Stn3 were recovered from the Illumina MiSeq data. Upon further taxonomic classification, at the family level, abundant (in terms of number of sequences) families in Stn1 were found to be Hyphomicrobiaceae- (45.23%), Rhodobacteraceae- (14.08%), Pseudomonadaceae- (10.05%), Erythrobacteraceae- (7.55%) and Kordimonadaceae- (3.77%). These families were also found in Stn1_Jul clone library but number of sequence representation was low. Similarly, in Stn3 there was abundance (in terms of number of sequences) of Pseudomonadaceae-(29.15), Hyphomicrobiaceae- (11.11%), Hyphomonadaceae- (10.81) and Ruminococcaceae-(6%) -like sequences in both the Illumina and clone library data. Furthermore, apart from the abundant taxa, Illumina sequencing gave us a deeper insight into structure of bacterioplankton communities of SBOTS. Sequences from Stn1 could be affiliated to Acidobacteria- (0.03%), Actinobacteria- (0.55%), Chloroflexi- (0.017%), Cyanobacteria- (0.04%), Fibrobacteres- (0.03%), Spirochaetes- (0.03%), Tenericutes- (0.017%) and Verrucomicrobia-like sequences (0.017%). From Stn3, sequences could be affiliated to the phyla TM7- (0.66%), Actinobacteria- (0.55%), OD1- (0.5%), Planctomycetes- (0.25%), Fibrobacteres- (0.18%), Spirochaetes- (0.17%), Tenericutes- (0.14%), Synergistetes- (0.13%), WPS-2-(0.04%), Elusimicrobia- (0.03%), Cyanobacteria- (0.03%), Verrucomicrobia- (0.02%), Chloroflexi- (0.02%), Gemmatimonadetes- (0.01%) and SR1 (0.01%). Even though these two stations are interconnected in terms of freshwater and saline water mixing, higher number of bacterial phyla was encountered in Stn3 indicating higher diversity. To visualize the difference in bacterioplankton community structure at different taxonomic levels between the two stations, a phylogram was constructed. The comparative analysis of abundance of bacterial taxa and Archaea retrieved from Illumina MiSeq sequencing of Stn1 and Stn3 has been shown in Fig. 1(a). Higher abundance of Proteobacteria-like sequences was encountered in Stn1 whereas Bacteroidetes-like sequences were higher in abundance in Stn3. This trend was reflected in both the datasets generated by Sanger sequencing and Illumina MiSeq sequencing. The comparative analysis of bacterial taxa retrieved from the clone library followed by Sanger sequencing is shown in Fig. 1(b). The congruency between the two sequencing methods was reflected at all taxonomic levels. For example, at the class level, higher abundance of Rhizobiales-like sequences was observed in Stn1 compared to Stn3. At the family level, Stn1 appears to harbour a higher abundance of Hyphomicrobiaceae- and Rhodobacteraceae-like sequences compared to Stn3 whereas the number of Pseudomonadaceae-like sequences retrieved from Stn3 is higher than Stn1. At the genus level, higher number of Filomicrobium-, Rhodovulum-, Stappia-, Erythrobacter-like sequences was retrieved in Stn1 compared to higher number of Devosia- and Hyphomonas-like sequences in Stn3. The generated clone libraries of forty clones for each time point representing each station appeared to give a broader accurate snapshot of abundant bacterioplankton taxa in Sundarbans mangrove ecoregion. The generated Illumina MiSeq data showed congruency with the broad findings obtained from clone libraries. Given that Illumina sequencing is more in-depth, therefore the data provided a deeper insight into the structure of bacterioplankton communities and highlighted the presence of other bacterial phyla such as Actinobacteria- and Verrucomicrobia-like sequences. Moreover, as shown in Fig. 1(a), there was clear differentiation between bacterial genera that were encountered in both stations or exclusively recovered from only one station. For example, Hallela-, Prevotella-, Butyrivibrio-, Ruminococcus-, Brevundimonas-, Hyphomonas-, Maricaulis-, Erythrobacter-, Acinetobacter- and Pseudomonas-like sequences were retrieved from both the studied stations. Sequences affiliated to genera such as Micrococcineae-, Brucella-, Rhodovulum-, Roseibium-, Sphingomonas-, Achromobacter-like sequences were found exclusively in Stn1. Bacterial genera such as Ruminobacter- and Succinovibrio-like sequences were found only in Stn3. Interestingly, 975 OTUs from Stn1 and 305 OTUs from Stn3 obtained from Illumina data could not be assigned to any particular phylum highlighting the possible presence of novel bacterioplankton in this ecosystem with potentially unknown functions.
Fig. 1.
(a): Phylogram indicating abundance (number of sequences) of 16S rRNA sequences recovered from Stn1 (in red colour) and Stn3 (in blue colour) by Illumina MiSeq sequencing. Taxa showing differential distribution between the two stations have been shown.
(b): Phylogram indicating the abundance (number of sequences) of 16S rRNA sequences recovered from Stn1 (in red colour) and Stn3 (in blue colour) from the clone libraries followed by Sanger sequencing. Taxa showing differential distribution between the two stations have been shown.
The analysis of bacterioplankton communities from the world's largest contiguous mangrove ecosystem indicated the presence of numerous phyla such as Firmicutes-, Bacteroidetes-, Actinobacter-, in addition to overwhelming abundance of Proteobacteria-like sequences. Moreover, there was clear difference in terms of composition of bacterioplankton taxa irrespective of the close proximity of both stations. The two sequencing methods i.e. Sanger sequencing and Illumina MiSeq sequencing employed in this study showed broader congruency in terms of structure of bacterioplankton taxa but there was deeper resolution in terms of understanding the structure of bacterioplankton communities based on Illumina MiSeq sequencing.
Nucleotide sequence accession numbers
All sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under the accession numbers SRR4858370 and SRR4858369.
Conflict of interest
The authors declare they have no competing interests.
Acknowledgement
Anwesha Ghosh is the recipient of IISER Kolkata Integrated Ph.D. Fellowship. This work is supported by ARF grant awarded to Punyasloke Bhadury from IISER Kolkata. The authors would like to thank Somnath Halder for assistance during capillary DNA sequencing.
References
- 1.Azam F., Fenchel T., Field J.G., Gray J.S., Meyer-Reil L.A., Thingstad F. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 1983;10:257–263. [Google Scholar]
- 2.Bostrӧm K.H., Simu K., Hagstrӧm A., Riemann L. Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods. 2004;2:365–373. [Google Scholar]
- 3.Campbell B.J., Kirchman D.L. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J. 2013;7:210–220. doi: 10.1038/ismej.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choudhury A.K., Das M., Phillip P., Bhadury P. An assessment of the implications of seasonal precipitation and anthropogenic influences on a mangrove ecosystem using phytoplankton as proxies. Estuaries Coast. 2015;38(3):854–872. [Google Scholar]
- 6.Conesa A., Götz S., Garcίa-Gόmez J.M., Terol J., Talόn M., Robles M. Blast2Go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 7.Dann L.M., Mitchell J.G., Speck P.G., Newton K., Jeffries T., Paterson J. Virio- and bacterioplankton microscale distributions at the sediment-water interface. PLoS One. 2014;9(7):e102805. doi: 10.1371/journal.pone.0102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortunato C.S., Crump B.C. Bacterioplankton community variation across river to ocean environmental gradients. Microb. Ecol. 2011;62:374–382. doi: 10.1007/s00248-011-9805-z. [DOI] [PubMed] [Google Scholar]
- 9.Healey M.J., Moll R.A., Diall C.O. Abundance and distribution of bacterioplankton in the Gambia river, West Africa. Microb. Ecol. 1988;16(3):291–310. doi: 10.1007/BF02011701. [DOI] [PubMed] [Google Scholar]
- 10.Huson D.H., Mitra S., Ruscheweyh H.J., Weber N., Schuster S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21(9):1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silveira C.B., Vieira R.P., Cardoso A.M., Paranhos R., Albano R.M., Martins O.B. Influence of salinity on bacterioplankton communities from the Brazilian rain forest to the coastal Atlantic Ocean. PLoS One. 2011;6(3):e17789. doi: 10.1371/journal.pone.0017789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Jiao N., Cottrell M., Kirchman D. Contribution of major bacterial groups to bacterial biomass production along a salinity gradient in the South China Sea. Aquat. Microb. Ecol. 2006;43:233–241. [Google Scholar]