Abstract
Campylobacter jejuni, the most common cause of bacterial diarrhoeal disease, is normally helical. However, it can also adopt straight rod, elongated helical and coccoid forms. Studying how helical morphology is generated, and how it switches between its different forms, is an important objective for understanding this pathogen. Here, we aimed to determine the genetic factors involved in generating the helical shape of Campylobacter. A C. jejuni transposon (Tn) mutant library was screened for non-helical mutants with inconsistent results. Whole genome sequence variation and morphological trends within this Tn library, and in various C. jejuni wild type strains, were compared and correlated to detect genomic elements associated with helical and rod morphologies. All rod-shaped C. jejuni Tn mutants and all rod-shaped laboratory, clinical and environmental C. jejuni and Campylobacter coli contained genetic changes within the pgp1 or pgp2 genes, which encode peptidoglycan modifying enzymes. We therefore confirm the importance of Pgp1 and Pgp2 in the maintenance of helical shape and extended this to a wide range of C. jejuni and C. coli isolates. Genome sequence analysis revealed variation in the sequence and length of homopolymeric tracts found within these genes, providing a potential mechanism of phase variation of cell shape.
Campylobacter spp. are the most common cause of bacterial food-borne diarrhoeal disease worldwide1. A major route of human infection is through the consumption of contaminated meats, especially chicken, and milk or through faecal-oral transmission2,3,4,5,6. A fundamental property of most Campylobacter spp. is that its cells are helical, and that these can change their shape to become rod or coccoid-shaped7. Helical cell shape is relatively rare within bacterial species and is thought to confer various colonisation and pathogenic advantages for Campylobacter jejuni and Campylobacter coli, including improved translocation across mucus membranes by facilitating a ‘corkscrew’-like motility7,8,9 and a loss of helical cell shape is linked to attenuated chick colonisation10.
To better understand how helical-shaped Campylobacter spp. maintain their cell shape and thus any associated colonisation, pathogenic or other advantage, it is first important to identify the genes and proteins involved. While the C. jejuni genome encodes cytoskeleton regulatory elements MreB, RodA and FtsZ, important for maintaining rod morphology in many bacteria11,12,13,14,15,16, these proteins alone are not sufficient to generate helical and curved bacterial morphologies11,17,18. Mechanisms for generating helical and curved morphologies have been identified in other bacterial species, including the proteins encoded by the genes ccrp59, ccrp1143, csd1, csd2, csd3 and ccmA in Helicobacter pylori19,20 and creS in Caulobacter crescentus21. So far, the only genes known to be involved in determination of the helical cell shape of C. jejuni are pgp1 and pgp2, and their protein products are peptidoglycan (PG) peptidases that are important for PG synthesis9,10,22. When either of the pgp1 or pgp2 genes were mutated, the muropeptide profile radically changes and helical cell shape could not be maintained9,10.
Pgp1 is a DL-carboxypeptidase which cleaves monomeric PG tripeptides to dipeptides9. Deletion or over-expression of pgp1 results in rod-shaped morphologies. Thus, the proper ratio of monomeric tripeptides to dipeptides may be required for proper shape determination9. Loss of pgp1 affects pathogenic attributes such as chicken colonisation, biofilm formation, motility and activation of host inflammatory mediators9. Pgp2 is an LD-carboxypeptidase cleaving PG tetrapeptides to tripeptides, which provide the substrate for the DL-carboxypeptidase Pgp110. Unlike pgp1, pgp2 is not restricted to helical and vibroid bacteria and is conserved in both Gram negative and Gram positive bacteria10.
In this study, we aimed to identify novel genetic determinants of C. jejuni helical cell shape. To do this, a C. jejuni signature-tagged transposon mutant (STM) library23 was screened by light microscopy for altered cell morphologies. Of 1,933 STMs screened, 132 revealed changes in cell shape, including rod and short helical morphologies. However, transposon (Tn) insertion sites could not account for these cell shape differences. Instead, whole genome sequencing (WGS) was used to investigate genomic variation between STMs. In addition, we found a direct correlation between cell and colony morphologies, enabling the isolation and sequencing of rod-shaped bacteria from wild-type (WT) populations. All sequenced rod-shaped C. jejuni isolates contained a single nucleotide polymorphism (SNP) or small insertion or deletion (INDEL) in the genes encoding the peptidoglycan remodelling enzymes Pgp1 or Pgp2.
Results and Discussion
Identification and isolation of rod-shaped C. jejuni mutants
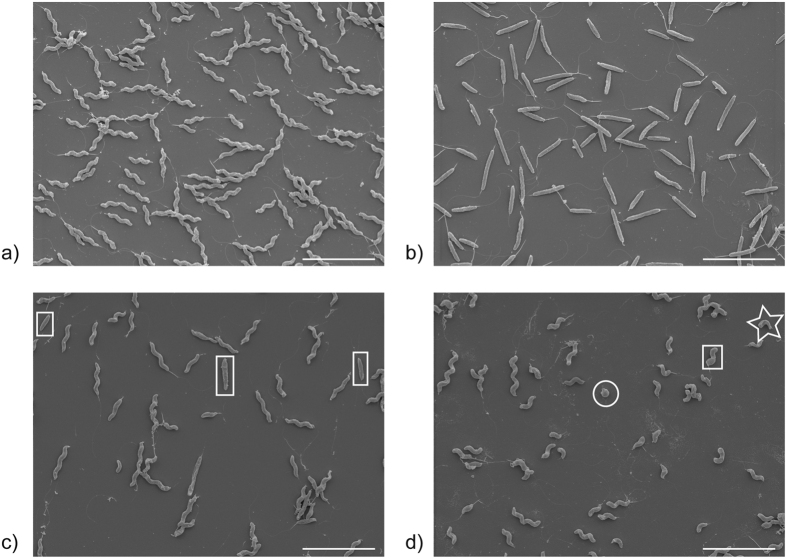
Tn mutant libraries are a useful tool for screening a range of phenotypes derived from random mutation. We screened a pre-existing STM Tn library constructed in C. jejuni strain M123 by light microscopy, with the aim of identifying bacteria with altered cell morphologies. STMs analysed in the screen were derived from 45 different signature tags, with each tag comprising 10–50 STMs. Of 1,933 Tn mutants cultured and screened individually, 89 were exclusively rod-shaped, 22 had mixed rod and helical cell morphologies and 25 showed an exclusively “short-helical” morphology, which included a mixture of helical, S-shaped, crescent and coccoid forms (Fig. 1).
Figure 1. Scanning electron micrographs of observed C. jejuni morphologies.
(a) helical, (b) rod, (c) rod-and-helical and (d) short helical C. jejuni M1 isolates. Rod bacteria in (c) are boxed. Short helical cultures (d) include S-shaped (box), crescent (star) and/or coccoid (circle) bacteria. Scale bars represent 5 μm.
The STM Tn mutants screened were generated as 1,933 independent and random mutation events but, the observed morphologies had a seemingly non-random distribution within the library: 109 of the 136 non-helical Tn mutants belonged to only seven signature tags (Fig. S1). We used linker PCR and plasmid rescue to determine the chromosomal location of the Tn insertion sites within 49 non-helical Tn mutants, and found that the pattern of non-helical morphologies did not correlate clearly with particular Tn insertion sites, suggesting that the observed cell morphologies were not the result of the primary Tn mutation event. To investigate this in more detail, we decided to test some of the genes mutated in the Tn library for their role in cell shape determination. We chose three genes for this initial analysis that were similar to genes from other bacteria that are involved in cell shape determination. These were CJM1_0159 which encodes a predicted coiled-coil region also seen in crescentin, a protein responsible for curvature in C. crescentus21, and CJM1_0631 and dtpT which encode large, transmembrane di-/tripeptide transporters that we predicted were either important for cell wall integrity or transported substrates necessary for PG synthesis. We mutated these genes in C. jejuni by allelic-replacement and showed that this did not consistently result in rod morphology in the WT background. Taken together, the data indicated that the Tn mutation event was not responsible for rod shape cells in the library and that therefore another source of variation may be responsible for the loss of helical cell shape.
Without a clear pattern of Tn insertion sites to account for the different morphologies within the library, we hypothesised that there was another source of variation within the non-helical STMs. Since STM morphologies were maintained throughout the cell cycle and across multiple generations, we further hypothesised this was due to a genetic mutation, as opposed to differences in genetic regulation.
Pulsed-field gel electrophoresis (PFGE), a highly discriminatory technique used for distinguishing bacterial strains24,25 was performed to investigate whether major chromosomal rearrangements could account for the difference between rod-shaped STMs and the WT. No major chromosomal rearrangements could be detected by pulse-field gel electrophoresis.
In lieu of major chromosomal rearrangement, we performed WGS to search for polymorphisms that might explain the phenotypes. In total, 66 rod-shaped Tn mutants, 6 short-helical shaped Tn mutants and 15 Tn mutants showing mixed rod and helical morphologies were selected and sequenced alongside the WT parent and 6 helical Tn mutants as controls.
Importantly, we observed that, although the WT M1 strain is characterised by helical morphology, our WT M1 laboratory strain (believed to have been passaged only twice since original isolation) contained a mixture of helical and rod-shaped cells. This finding is consistent with the discovery of a proportion of rod-shaped bacteria within the predominantly helical targeted deletion strains already mentioned. Following this discovery, we sought to isolate pure populations of the helical and rod-shaped bacteria and noticed a clear association between cell and colony morphologies. Helical bacteria formed shiny, rounded colonies on MH agar whereas rod-shaped bacteria formed dull, flat and often larger colonies (Fig. 2). This association was confirmed by light microscopy for over 150 colonies. We hypothesise that the different colony morphologies may be due to a discrepancy in the packing of helical versus rod cells within the colony26, as observed in cell shape mutants of other bacterial species18. We used colony purification to isolate helical and rod-shaped populations from C. jejuni strains M1, NCTC11168, 81116 and 81–176 (Fig. 3), as well as from the isogenic M1 mutant CJM1_0159. A selection of helical and rod-shaped WT isolates from these four C. jejuni strains was included in the WGS analysis. We have summarized the string of methods applied and conclusions made in the process of the STM analysis, as described above, in Fig. S2.
Figure 2. Helical and rod C. jejuni have distinct colony morphologies.
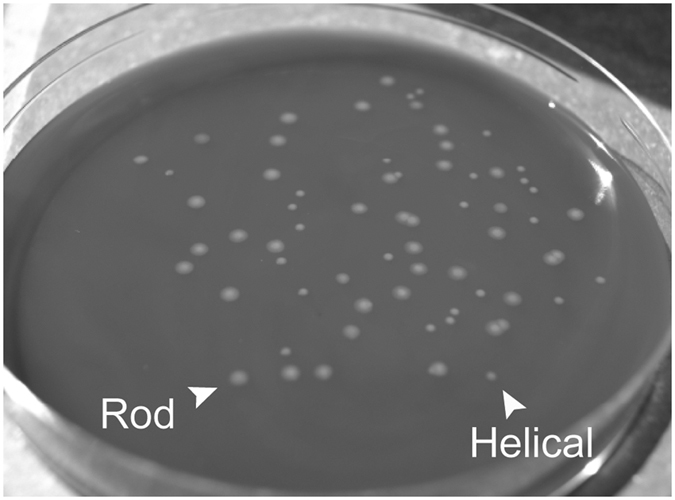
Colony growth of a mixed population of helical and rod C. jejuni M1 WT bacteria. The plate shows the, flattened and often larger colonies of rod bacteria and the shiny and rounded colonies of helical bacteria.
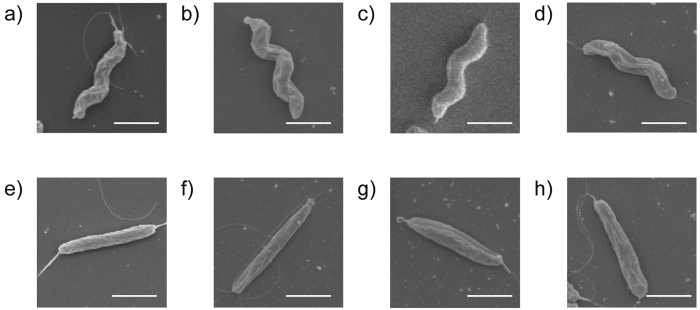
Figure 3. Scanning electron micrographs of helical and rod WT C. jejuni isolates.
Helical isolates from C. jejuni strains (a) M1, (b) 81116, (c) 81–176 and (d) NCTC11168 and rod isolates from (e) M1, (f) 81116, (g) 81–176 and (h) NCTC11168. Scale bars represent ~1 μm.
To determine whether C. jejuni morphological changes could be observed during in vitro growth, we performed serial passages of single colonies on MH agar and screened each passage for cell shape mutants. Specifically, we identified rod-shaped bacteria in passages of helical bacteria, and for helical-shaped bacteria in passages of rod bacteria. We screened over 40,000 colonies from four passages of both helical and rod WT lineages and identified four rod-shaped mutants from the helical lineages. These rod-shaped mutants were from the first passage of isolate H2 (H2_P1_R1, where H2 = helical isolate 2, P1 = first passage and R1 = rod 1), the second passage of isolate H5 (H5_P2_R1 and H5_P2_R2) and the fourth passage of isolate H5 (H5_P4_R1). No helical-shaped revertants from the rod lineages were identified. The four rod-shaped clones, each with a helical isolate from the same lineage and passage number as a control, were analysed by WGS. This screen for cell shape variants was also used to determine the rate of helical-to-rod morphological change in C. jejuni M1, which we calculated as a frequency of 7.7 × 10−4, or 7.25 × 10−5 phenotypic variants/division (Table S1).
Genome sequenced C. jejuni isolates were genetically distinct
The genome sequence data from 133 helical and rod Tn mutants and WT isolates were analysed for the presence of single nucleotide polymorphisms (SNPs) or small insertions and deletions (INDELs) (Table S2) and changes in the number of bases in documented phase variable regions (PVRs) (Table S3).
Phase variation (PV) is a common source of genetic and phenotypic variation in many bacteria, including C. jejuni27,28,29. PV enables a stochastic ‘on’ and ‘off’ switching of genes in bacteria, providing population diversity that may promote immune evasion or survival in varying environmental conditions26 or if the bacterium inhabits more than one niche30. Regions of the bacterial genome that are particularly prone to these reversible mutations are known as PVRs and include simple sequence repeats, inverted repeats, gene duplications and methylation sites. Homopolymeric tracts (HTs) are highly susceptible to slipped-strand mispairings, which alter the length of the tracts and generate frameshift mutations during DNA replication and repair26,31. The frameshift mutations cause a change to the downstream amino acid sequence and almost always result in premature stop codons. In this way, PVRs are able to randomly switch genes ‘on’ and ‘off’ and stochastically regulate gene expression26. Although these mutations are heritable, they are also often reversible. This is usually evident by a variety of HT lengths of a PVR within a population27,28 or observed directly by bacterial passage32,33. The rate of PV is generally greater than that of spontaneous mutation and is typically calculated to 10−4 mutations/division or more26,28,34. Rates of PV can differ depending on environmental conditions and/or whether the genotypic switch is from ‘on-to-off’ or ‘off-to-on’ and these rates can vary up to 10-fold28,34,35,36,37.
The propensity of C. jejuni to undergo PV29 may be exacerbated by the absence of a functional mismatch repair (MMR) system in this species38. MMR systems have been found to protect bacterial genomes from slipped-strand mispairing39,40,41 and it has been postulated that the lack of this repair mechanism in C. jejuni makes its genome more susceptible to PV38. Indeed, MMR mutants in other species show increased mutation rates42,43. Alternatively, higher rates of PV in C. jejuni could be due to inaccuracies of the C. jejuni DNA polymerase, as has also been suggested for H. pylori44.
In C. jejuni, PVRs have been identified in capsule, LOS and flagellin glycosylation loci as well as a range of loci of unknown function4,27,28,45. Most identified PVRs in C. jejuni are within polyG:C tracts of seven or more nucleotides in length26,27,45. However, PV in C. jejuni has also been observed in polyA:T tracts and in HTs as short as two nucleotides27,32, demonstrating that this phenomenon is not limited by nucleotide or tract length. However, investigations into variable polyA:T tracts are limited by their density within the C. jejuni genome, which is roughly 70% AT rich27. Considering all these complexities of PV analysis, it is reasonable to hypothesise that PV in C. jejuni may occur in more regions than currently recorded.
The WGS demonstrated the fluidity and susceptibility to mutation of the C. jejuni genome. There were numerous SNPs and INDELs that were distinct from the GenBank reference genome of the same strain (CP001900.1). Interestingly, genetic differences were observed in isolates from the same laboratory stock, the same isolates following in vitro passage and from isolates subjected to genetic manipulation. Many mutations identified throughout the C. jejuni genomes fell within HTs of a range of lengths and nucleotide sequence. As previously described, slipped strand mispairing at runs of Cs or Gs has been described in C. jejuni27. Although rarely classified as a mechanism of PV in Campylobacter, we present evidence here that runs of As or Ts are also susceptible to length variation, suggesting that these regions are particularly susceptible to mutation and that PV in Campylobacter should be re-evaluated. However, whether these mutations occur naturally within C. jejuni populations, due to an error prone DNA polymerase, a lack of DNA repair mechanisms and/or the instability of HTs, or whether the mutations emerge from stresses induced by the laboratory environment and genetic manipulation remains to be determined. For example, differences within the STM library could be derived from differences in growth or transformation micro-environments while the STM library was being made. Collectively, the presence of these mutations warns against assumptions that isolates within any C. jejuni population are genetically identical (also addressed in refs 46, 47, 48, 49) or that any site-directed genotypic changes to C. jejuni are responsible for observed phenotypic changes. To overcome this uncertainty during phenotypic characterisation, the precedent should be maintained that C. jejuni isolates should be assessed by genome sequencing, by appropriate complementation analyses of mutants, and/or by performing laboratory manipulations and physiological assays on a panel of isolates.
Mutations in pgp1 or pgp2 were present in all rod-shaped C. jejuni laboratory isolates
Every rod-shaped Tn mutant from the M1 STM library and every rod-shaped isolate from WT strains M1, 81116, 81–176 and NCTC11168 had a SNP or INDEL within the PG peptidase-encoding genes pgp1 or pgp2 (Fig. 4. and Table 1). Rod-shaped bacteria isolated from the mixed rod-and-helical Tn mutants also contained a SNP or INDEL within pgp1 or pgp2 (as determined by PCR and Sanger sequencing) (Fig. 4. and Table 1). No mutations in either of these genes were observed in any of the helical isolates. This strict correlation between the rod-shaped morphology and mutations in these genes was confirmed by site-directed mutagenesis (SDM) of a selection of rod STMs and rod WT isolates. SDM was performed to lock one of the variable homopolymeric tracts in pgp1 and pgp2 into its ‘on’ length. The rod STM isolate adopted a helical morphology after the incorporation of pgp1_ON (Fig. S3) and all three tested rod STMs adopted a helical morphology after the incorporation of pgp2_ON (Fig. S4).
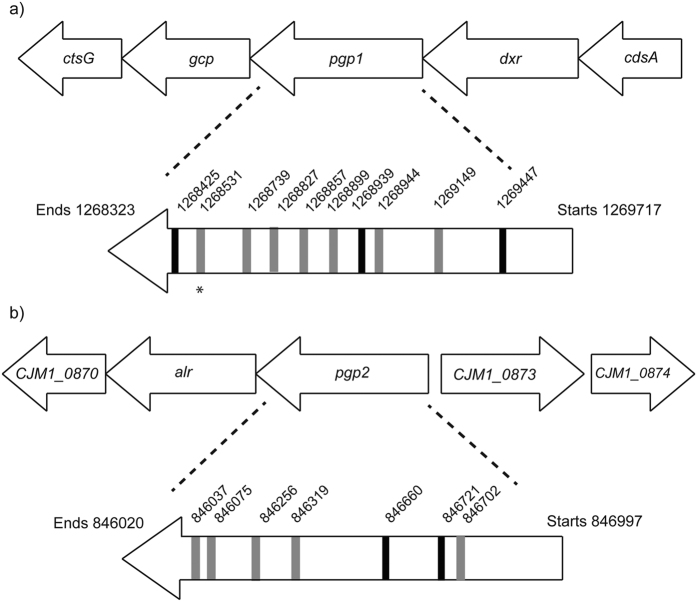
Figure 4. Mutations observed in pgp1 and pgp2.
Base locations of mutations in (a) pgp1 and (b) pgp2 are in reference to the C. jejuni M1 genome (CP001900.1)4. Black horizontal bars indicate SNPs; grey horizontal bars indicate INDELs. All INDELs are located in homopolymeric tracts. Asterisk (*) below pgp1 indicates the 8-A tract, in which mutations were observed in more than one C. jejuni strain.
Table 1. Mutations detected in pgp1 and pgp2 in rod-shaped C. jejuni isolates.
| Base location of change in M1 | Nucleotide change | Amino acid change | C. jejuni isolates with mutation |
|---|---|---|---|
| 1269447 (pgp1) | C > T | R91C | 51-40, 51-43, 54-40, 54-43, 55-49 |
| 1269149 (pgp1) | 5 A > 4 A | Stop at 205 | 11168_R1, 11168_R2, 11168_R3 |
| 1268944 (pgp1) | 4 A > 5 A | Stop at 261 | 19-36 R°, H5_P2_R1 |
| 1268939 (pgp1) | T > A | V260D | 17-19 R° |
| 1268899 (pgp1) | 5 T > 4 T | Stop at 281 | 5-45 R*, 17-48 R* |
| 1268857 (pgp1) | 2 T > T | Stop at 295 | 38-14 R* |
| 1268827 (pgp1) | 3 A > 2 A | Stop at 299 | 17-14 R* |
| 1268739 (pgp1) | 3 A > 2 A | Stop at 334 | 17-49* |
| 1268531 (pgp1) | 8 A > 7 A | Stop at 403 | Δ0159_R, 19-34, 17-38 R°, 18-26 R*, R1, R2, R3, R4, R5, R6, R7, 81116_R2, 81116_R3, 81-176_R2, 81-176_R3, R’1, R’2, R’3, R’4, R’5, R’6, R’7, R’8, R’9, R’10, R’11, R’12, R’13, R’14, R’15, R’16, R’17, R’18, R’19 m R’20, R’21, R’22, R’23, R’24, R’25, R’26, R’27, R’28, R’29, R’30, R’31, R’32, R’33, R’34, R’35, R’36, R’37, R’38, R’39, R’40, R’41, R’42, R’43, R’44, R’45, R’46, R’47, R’48, R’49, R’51, R’52, R’53, R’54, R’55, R’56, R’57, R’58, R’59, R’60, R’61, R’62, R’63, R’64, R’65, R’66, R’67, R’68, R’69, R’70, R’71, R’72, R’73, R’74, R’75, R’76, R’77, R’78, R’79, R’80, R’81, R’83, R’84, R’85, R’86, R’87, R’89, R’90, R’91, R’92, R’93, R’94, R’95, R’96, R’97, R’98, R’99, R’100, R’101, R’102, R’103, R’104, R’105, R’106, R’107, R’108, R’109, R’110, R’111 |
| 8 A > 9 A | Stop at 408 | 17-34 Ro, 81116_R1 | |
| 1268425 (pgp1) | ATA > A | Stop at 438 | 43-4 R* |
| 846702 (pgp2) | 2 G > 4 G | Stop at 101 | R’50, R’82, R’88 |
| 846721 (pgp2) | C > T | Q93Stop | 22-12 |
| 846660 (pgp2) | C > T | P113L | H5_P2_R2 |
| 846319 (pgp2) | 3 A > 2 A | Stop at 234 | H5_P4_R1 |
| 846256 (pgp2) | 4 A > 3 A | Stop at 247 | 50-1, 50-2, 50-3, 50-6, 50-9, 50-10, 50-18, 50-19, 50-20, 50-34, 51-6, 51-9, 51-31, 51-35, 51-39, 51-41, 51-44, 53-1, 53-9, 53-10, 53-14, 53-17, 53-23, 53-23, 53-26, 53-27, 53-34, 53-35, 53-42, 54-6, 54-26, 54-35, 54-37, 54-44, 54-45, 54-48, 54-49, 54-50, 55-28, 55-29, 55-33, 55-34, 55-38, 55-40, 55-41, 55-42, 55-50, 57-32, 57-33, 57-34, 57-36, 57-37, 57-38, 57-39, 57-40, 57-41 R, 55-35 R |
| 846075 (pgp2) | 4 T > 5 T | Stop at 312 | H2_P1_R1 |
| 846037 (pgp2) | 4 T > 3 T | Stop at 322 | 62-5 R* |
| 846702 (pgp2) | 2 G > 4 G | Stop at 101 | R’50, R’82, R’88 |
The base location, nucleotide change and predicted effect on translation of each mutation in pgp1 and pgp2 are listed alongside the rod-shaped C. jejuni isolates in which each mutation was observed. Isolates include the rod M1 Tn mutants, rod isolates from rod-and-helical M1 Tn mutants (labelled ‘R’), the rod-shaped targeted deletion strain ΔCJM1_0159::cat (abbreviated Δ0159_R), rod-shaped isolates from the WT M1, 81116, 81-176 and NCTC11168 strains, rod-shaped mutants from serial passages of helical-shaped M1 isolates (i.e., H2_P1_R1, where H2 = helical isolate 2, P1 = first passage and R1 = rod 1) and rod-shaped mutants from a colony screen of helical-shaped M1cam. Base locations for all strains are in reference to the C. jejuni M1 GenBank genome (CP001900.1). The full-length Pgp1 protein is 464 aa and Pgp2 is 325 aa. All isolates were analysed by Illumina next generation sequencing unless noted; *analysed by Sanger sequencing, °analysed by Sanger sequencing and WGS.
Mutations in pgp1 and pgp2 were in a wide array of base locations (Fig. 4) and their predicted effects on translation included single amino acid changes and 4 bp truncations – all of which were sufficient to cause a predicted loss of function of these proteins. Function was tested by analysis of the cell wall muropeptides of rod-shaped and helical isolates, using High-Performance Liquid Chromatography (HPLC) (Fig. S5, and Table S4). These analyses demonstrated that rod-shaped pgp1 mutants in both the M1 and 81116 backgrounds have similar muropeptide profiles, and that these are distinct from the muropeptide profiles from their respective helical WT cells. Likewise, muropeptide profiles of rod-shaped pgp2 mutants in the M1 and 81116 backgrounds were similar to each other and distinct from WT cells. The muropeptide compositions are also similar to those of pgp1 mutants, pgp2 mutants or WT in the 81–176 background9,10, except that the putative dipeptide (peak 3) and putative acetylated tetra-tetrapeptide (peak 12) fractions are greater in the 81116 and M1 pgp1 mutants than in the 81–176 pgp1 mutant. These profiles also expand our understanding of muropeptide compositions of helical versus rod-shaped bacterial species.
Combined, these data demonstrate that many different mutations can affect the functionality of Pgp1 and Pgp2. Interestingly, the most frequent site of mutation within pgp1 was an HT consisting of 8 As towards the 3′ end of the gene. This HT had 7 or 9 As in many of the rod-shaped isolates in the M1, 81116 and 81–176 backgrounds, resulting in a truncation that is predicted to remove approximately 14% of the WT protein. The rate of morphological change is greater than that of the spontaneous mutation (Table S1), so we speculated that there may be phase variable control of cell shape.
To further investigate this hypothesis, we conducted another experiment looking for rod-shaped bacteria in passages of helical bacteria, and for helical-shaped bacteria in passages of rod bacteria. In this experiment, the starting helical isolate was M1cam (Table S5) and the rod isolate was M1 ‘R2’ containing the HT 7A (Table S5). 112 spontaneous rod derivatives were identified from 375,000 screened colonies. Over 525,000 colonies were screened looking for helical derivatives from the rod-shaped starting bacteria without detection of changes in colony morphology. Next we performed WGS on genomic DNA obtained from each of these colonies and mapped the reads to the M1cam (CP012149) and identified the SNPs and INDELS. We discovered that the helical to rod isolates R’50, R’82 and R’88 all had normal CJM1cam_0872 (pgp1), and all had a frameshift caused by a 2-base insertion in the same position in CJM1cam_1302 (pgp2) (Fig. 4, Table 1). All other helical to rod isolates had normal CJM1cam_1302 (pgp2), and all had a frameshift caused by a single base deletion in the same position in CJM1cam_0872 (pgp1) (Fig. 4, Table 1). This screen was also used to determine the rate of helical-to-rod morphological change in C. jejuni M1, under these experimental conditions, which we calculated as a frequency of 2.1 × 10−4 (Table S1). This frequency is similar to the frequency that we calculated with a different experimental procedure (Table S1), and the differences are likely to be due to differences in the experimental procedure, and the estimation of the number of colonies screened.
These data, combined with the data from the earlier experiments, demonstrate that the helical-to-rod morphological switch occurs repeatedly in standard laboratory growth conditions. It remains possible that the rod-to-helical switch is also occurring but at a lower rate. Alternatively, different environmental pressures may be necessary to observe the rod-to-helical switch.
Clinical and environmental C. jejuni and C. coli isolates contain length variants of the 8-A tract in pgp1
Next, we examined whether HT length polymorphisms in pgp1 and pgp2 also existed in a range of C. jejuni isolates from various environmental and clinical sources. We investigated the presence and allelic variances of pgp1 and pgp2 in 859 genomes of C. jejuni and C. coli. The genomes were from a wide range of isolates: 192 from clinical, agricultural and wild bird sources48, 319 from multiple stages of poultry processing, including farms, abattoirs and retail chicken meat50 and 348 from clinical human cases51. Analysis of these genomes revealed that all isolates contained pgp1 and pgp2, suggesting that these genes are core to C. jejuni and C. coli. Furthermore, although no length variation was detected at the 4-A HT in pgp2 (where variation had been observed in the C. jejuni Tn mutant library), 29 genomes (3% of isolates) contained length variation at the 8-A HT in pgp1 (Table 2).
Table 2. Prevalence of various lengths of poly-A tract in the pgp1 allele of Campylobacter spp. from various isolation sources.
| Poly-A tract lengtha | C. jejuni | C. coli | Total number of genomes | ||||
|---|---|---|---|---|---|---|---|
| Farm chicken | Clinical cases | Otherb | Farm chicken | Clinical cases | Otherb | ||
| 9 nt | 1 | 18 | 0 | 0 | 1 | 0 | 20 |
| 8 nt | 248 | 345 | 64 | 90 | 59 | 24 | 830 |
| 7 nt | 2 | 2 | 0 | 1 | 1 | 3 | 9 |
aLength of poly-A at position 1,180 of the pgp1 gene.
bOther sources of isolation comprise wild birds, cattle, ducks, environmental water and unknown sources.
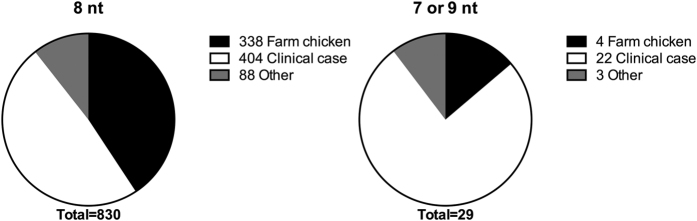
The morphology of a selection of these C. jejuni and C. coli isolates was investigated by light microscopy. All isolates with 7-A or 9-A length polymorphisms at the 8-A HT in pgp1 were rod-shaped. Furthermore, a large proportion of isolates with the 7-A or 9-A HTs were from clinical sources (Fig. 5). Additionally, a recent report22 described the complete genome sequence of C. jejuni RM1285, a rod-shaped morphological variant, an environmental isolate derived from the exudate of a commercial chicken breast acquired from a retail store. RM1285 was reported to have a single A deletion at the 1,187th nucleotide of pgp1, that resulted in a 62-amino acid truncation of the Pgp1 protein22. We have identified that the A deletion occurs in the 8-A HT described above. Together, these data suggest that HT length variation in pgp1 may not simply be an artefact of laboratory conditions or manipulation and can affect the cell shape of C. jejuni and C. coli in the host and natural environment. However, it should be noted that none of our isolates, and we presume RM128522, were screened directly from the primary source and each isolate had undergone a passage in the laboratory, therefore we cannot rule out the possibility that the change in the HT occurred during this growth step.
Figure 5. Prevalence of poly-A tract lengths in the pgp1 allele of Campylobacter spp. isolated from various sources.
The two distributions show a significant discrepancy (p = 0.0083; χ2 for goodness of fit test; χ2 = 9.571, d.f. = 2).
Conclusions
Cell shape is a defining characteristic of bacterial species and members of a particular species normally retain a constant morphology for generation after generation. Yet functional questions pertaining to how bacteria have particular shapes, and what evolutionary advantage being a particular shape confers, have been largely neglected until recently.
In this study, an STM library screen was performed to identify random mutants of C. jejuni with morphological changes. None of the genes disrupted by Tns were found to be responsible for helical cell shape. Instead, through extensive genome sequence analysis, we found that the observed morphological changes in the C. jejuni M1 STM library were explained by background mutations (SNPs or INDELS) in either of the carboxypeptidase genes pgp1 or pgp2, independent of the Tn insertions.
Reviewing the method used to generate the STM library, there were three stages in which background mutations could have emerged. The first was the growth of the WT population used to extract the genomic DNA, subsequently used in in vitro transposition. The second stage was the growth of the WT population used for natural transformation with the transposed DNA. The third stage was the growth of the transformants, prior to making frozen stocks. In any of these stages, a random mutational event could have occurred and proliferated to generate a non-helical STM. Considering the density of rod STMs within certain tags, with many STMs containing the same genomic variation, it is likely the rod-causing mutations in signature tags 50, 51, 53, 54, 55 and 57 occurred in the first or second stage. Mutations generated in these early stages would allow a single change to propagate throughout an entire tag or group of tags generated on the same day.
The discovery that both pgp1 and pgp2 are present in every genome from a collection of laboratory, environmental, wild bird, poultry and clinical C. jejuni and C. coli isolates strongly suggests pgp1 and pgp2 are core to these two helical Campylobacter species. Furthermore, the novel discovery that the 8-A HT in pgp1 (AT PPVR3) is variable within this collection of C. jejuni and C. coli isolates, the recent study by Gunther et al.22, and the fact that indels in pgp1 consistently correlates to a rod morphology when lengthened or shortened, emphasises the role of stochastic, heritable mutation in regulating Campylobacter cell shape in a range of environmental and host conditions.
Together, our work demonstrates the susceptibility of C. jejuni to transforming into a rod-shaped bacterium. Helical cell shape is maintained by a delicate balance of PG lengths and crosslinking9,10, which can be disrupted by a single point mutation at numerous locations in the PG carboxypeptidase genes pgp1 or pgp2. Yet despite a propensity of C. jejuni to undergo mutations of these genes, the helical morphology remains the dominant form in laboratory and natural environments – reinforcing the importance of helical cell shape to the success of this bacterium. While the possibility of phase variation suggests that the rod C. jejuni morphology may have an advantage in certain environments or host associations, various observations from human sources3,52,53 and infection models54,55 suggest that the ability to cause human campylobacteriosis is most often dependent on a helical Campylobacter, a shape that confers the greatest motility (Fig. 6) and enables the colonisation of the cecal crypts.
Figure 6. Motility of helical and rod WT C. jejuni M1 isolates.
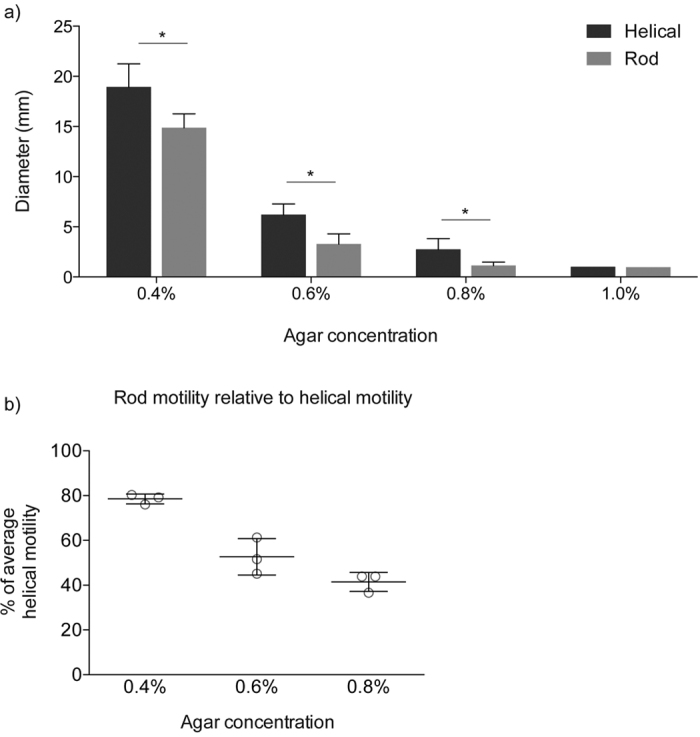
(a) Average motility of helical and rod WT C. jejuni M1 isolates in 0.4%, 0.6%, 0.8% and 1.0% (w/v) select agar. Motility for all isolates decreased with increasing agar concentration until isolates were effectively non-motile through 1.0% agar (all isolates measured 1 mm diameter, roughly equivalent to the original pipette stab). Statistical differences at each agar concentration were determined using a Mann-Whitney test (*p < 0.0001). Data shown is mean and SD (n = 15). (b) Relative motility of rod isolates compared to the average motility of helical isolates decreases with increasing agar concentrations. Data shown is mean and SD (n = 3). All helical (H2, H3, H4) and rod (R2, R3, R4) isolates were also analysed by genome sequencing.
The PG peptidases necessary for maintaining helical cell shape may offer useful antimicrobial targets to reduce the pathogenicity of C. jejuni or decrease its prevalence within the food chain. These targets are broad-spectrum in the sense that endo- and carboxypeptidases are common throughout the bacterial kingdom56,57, but many PG peptidases are redundant58,59 and therefore detrimental effects might be compensated in other species and it may also be possible to design narrow-spectrum options that specifically target Pgp1 or Pgp2. Intriguingly, Liu et al.60 recently reported a bacterial cell shape-determining inhibitor of H. pylori Csd4 (homologue of Pgp1 in C. jejuni) that causes significant cell straightening of H. pylori and a diminished, yet observable, effect on the morphology of C. jejuni.
Methods
Bacterial strains, media and growth conditions
C. jejuni strains were routinely cultured on Mueller Hinton (MH) agar (Oxoid) supplemented with 5% defibrinated horse blood (Oxoid) and 5 μg/ml trimethoprim (Tp). Defined and Tn mutants and complemented strains were selected on 10 μg/ml chloramphenicol (Cm) or 50 μg/ml kanamycin (Km), as appropriate. Campylobacter spp. bacterial cultures were grown in standard microaerophilic conditions (5% CO2, 5% H2, 85% N2, 5% O2) at 42 °C in a MACS VA500 variable atmosphere work station (Don Whitley Scientific). Electrocompetent Escherichia coli and C. jejuni used in cloning were prepared and transformed as previously described61. Bacterial strains and plasmids used in this study are detailed in Table S5.
Signature-tagged transposon mutant library
Generation of the C. jejuni M1 STM library was described in Grant et al.23. Briefly, mariner-based Tns containing a chloramphenicol acetyl-transferase (cat) cassette and a unique 40 bp DNA tag (‘signature tag’) were used in an in vitro transposition of WT C. jejuni M1 DNA. Transposed DNA was cloned into WT C. jejuni M1 by natural transformation. Chloramphenicol-resistant Tn mutant colonies were selected after 60–72 h growth on MH agar, subcultured on fresh agar and stored at −80 °C. Fifty mutants were recovered from each transformation. Mutants were labelled first by their signature tag (1–95). and then by a mutant number (1–50).
Colony purification by colony morphology
Agar or broth cultures of C. jejuni were serially diluted in MH broth, spread onto fresh MH agar and incubated in standard growth conditions. Colonies were inspected by eye after 24–48 h growth and either quantified or isolated based on colony morphology. Helical colonies displayed a shiny and rounded morphology, whereas rod colonies displayed a grey, flat and often larger morphology. Cell morphology was confirmed by transferring bacteria from a colony to a glass slide and viewing cells by light microscopy.
DNA sequencing
Sanger sequencing was performed by Source BioScience LifeSciences. WGS was performed at the Wellcome Trust Sanger Institute. Isolates were sequenced as multiplex libraries with 100 or 150 bp paired-end reads using next-generation Illumina HiSeq or MiSeq sequencing technology, respectively. For the sequencing of M1cam helical to rod isolates, Sequencing libraries were prepared using the NEBNext Ultra II DNA library prep kit (New England Biolabs). 250 ng DNA was sheared to 400 bp fragments in microTUBE screw-cap tubes in a M220 focused-ultrasonicator (Covaris). Following DNA library preparation, the library size was determined with a Bioanalyzer 2100 (Agilent), quantified using the Qubit dsDNA BR kit (Life Technologies), pooled in equal quantities, and analysed with the NEBNext library quant kit (New England Biolabs). The pooled library was subjected to 150 bp paired-end sequencing (Genomics core facility at Cancer Research UK). The read files were demultiplexed using the demuxFQ tool developed at Cancer Research UK. For WGS analysis, de novo draft assemblies of each isolate were created and sequencing reads were mapped to the reference genome for the relevant C. jejuni strain using the pipeline described in ref. 62. SNPs and INDELs were called using SAMtools mpileup63.
Analysis of homopolymeric tracts
The length of HTs in PVRs was assessed using the HT length of each sequencing read spanning a defined region. To be counted, the sequencing read must cover the entire region, as determined by the presence of nucleotides on either side of the HT. The distribution of HT lengths at each PVR was converted into a percentage of total reads and compiled based on whether the HT length was in or out of frame with the annotated gene sequence (‘on’ or ‘off’, respectively).
Recombinant DNA techniques
Standard methods were used for molecular cloning64. Chromosomal and plasmid DNA purification, DNA modification and ligations were performed using commercial kits according to the manufacturers’ instructions (QIAGEN, Thermo Scientific, New England Biolabs). DNA concentration was measured using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). PCR primers were purchased from Sigma (Sigma Aldrich). Thermal cycling was performed in a Gene Amp PCR System 9700 (PE Applied Biosystems) or T100T Thermal Cycler (Bio-Rad). Thermal cycling conditions were 96 °C for 2 min, then 30 cycles at 96 °C for 1 min, 55–60 °C for 1 min and 72 °C for 30 sec/kb, and finally an extension at 72 °C for 5 min.
Generation of C. jejuni defined gene deletion mutants, complemented strains and site-directed mutants
Targeted gene deletions of CJM1_0159, CJM1_0631 and dtpT were performed by exchanging the gene of interest with a cat cassette from pRY11165. The cat cassette was amplified with primers containing KpnI (dare008), BamHI (dare009), PstI (dare010) or SacI (dare011) restriction endonuclease (RE) target sites. Flanking regions of each deleted gene were amplified using upstream and downstream primers (0159_1 to 4; 0631_1 to 4; or dtpT_1 to 4) containing RE sites matched to the chosen cat cassette primers. PCR-amplified fragments were ligated to pUC19 prior to transformation into E. coli. Purified plasmid DNA was used to naturally transform C. jejuni. The correct genomic rearrangements in the resulting C. jejuni mutants were confirmed by PCR and sequencing using the primers 0159_ck1 to ck5; 0631_ck1 to ck4; or dtpT_ck1 and ck2, respectively. Primers used in this study are listed in Table S6.
Site-directed mutagenesis of pgp1 and pgp2 was performed by amplifying the gene of interest with primers incorporating nucleotide change (s) (pgp1_1 to 6 or pgp2_1 to 6, respectively). Using overlapping sequences, an antibiotic resistance cassette (amplified using primers pgp1_7 and 8 or pgp2_7 and 8, respectively) was incorporated between the mutated gene and the downstream region (amplified using primers pgp1_9 and 10 or pgp2_9 and 10). PCR-amplified fragments with overlapping ends were annealed using a Gibson Assembly® Cloning Kit (New England Biolabs) according to the manufacturer’s instructions and ligated into the pMiniT vector (New England Biolabs) prior to transformation of E. coli. Purified plasmid DNA was used to naturally transform C. jejuni. The correct genomic rearrangements in the resulting C. jejuni mutants were confirmed by PCR and sequencing using the primers pgp1_ck1 to ck3 and pgp2_ck1 to ck3. Bacterial strains and plasmids used in this study are detailed in Table S5. Primers used in this study are listed in Table S6.
Plasmid rescue
Plasmid rescue was used to assess the chromosomal location of Tns within C. jejuni M1 STMs. The technique was based on the method described in Grant et al.23 but was amended to make use of vector cloning. BglII-digested STM genomic DNA was ligated to BamHI-digested and dephosphorylated pUC19. Ligations were transformed into E. coli DH5α (Thermo Scientific) according to manufacturer’s instructions and transferred onto LB agar supplemented with chloramphenicol (Cm). Colonies present after O/N incubation were used to inoculate 5 ml LB broth with Cm. After O/N incubaction of LB cultures, plasmid DNA was extracted and analysed by Sanger sequencing using primers directed out of the Tn (AJG227 and CC1, Table S6).
Linker PCR
Linker PCR was an alternative method used to assess the chromosomal location of Tns within C. jejuni M1 STMs. Linker oligonucleotides (254 and 256, Table S6) were annealed in 1x annealing buffer (100 mM Tris (pH 8), 500 mM NaCl, 10 mM EDTA) at 95 °C for 3 min and cooled at 21 °C for 1 h. The annealed Linker oligonucleotides were ligated to RsaI-digested STM genomic DNA. Ligated Linker-STM DNA fragments were amplified in a PCR using a Tn-specific primer and a Linker-specific primer (CAT15 and 258, respectively, Table S6). The PCR product was resolved by gel electrophoresis and analysed by Sanger sequencing using primers directed out of the Tn (AJG227 and CC1, Table S6).
Pulsed-field gel electrophoresis
Major chromosomal patterns were compared between WT and STM C. jejuni M1 DNA using PFGE. The RE SacII was chosen for DNA fragmentation as SalI, SmaI and KpnI (also used for PFGE of Campylobacter species)24,25,66 all had recognition sequences within the Tn55. The PFGE method used was based on those described in Rivoal et al. and Ribot et al.24,25. WT and STM C. jejuni M1 agar cultures, washed and diluted in PBS to an OD600 nm of 0.6–0.8, were preserved and lysed within plugs made with 2% low-melt agarose (Promega) in TE pH 8. Genomic DNA within the agarose plugs was digested with SacII for 5 h at 21 °C, washed in 0.5x TBE and resolved by gel electrophoresis in 1% agarose in 0.5x TBE. Electrophoresis was performed according to manufacturer’s instructions at 6 V/cm for 20 h with a ramped pulse of 5–50 s using CHEF-DR® II Pulsed Field Electrophoresis Systems (Bio-Rad) and connected to a LTD 20 cooling system (Hybaid). Gels were stained with 0.3x SYBR® Safe (Invitrogen) and imaged on a GelDocTM XR + (Bio-Rad) with Image Lab 3.0 software (Bio-Rad).
Muropeptide analysis
Peptidoglycan (PG) purification and digestion protocols were adapted from those described in Glauner67, Li et al.68 and Frirdich et al.10. HPLC of purified and muramidase-digested C. jejuni PG was performed in the same manner and using the same instrumentation as described in Christie et al.69.
Calculations of mutation frequency and rate
The helical-to-rod switch observed in C. jejuni populations was quantified by mutation frequency and mutation rate. Mutation frequency, f, was calculated as the proportion of rod-shaped mutants, PR, that emerged during exponential growth of spiral isolates70. This value was incorporated into Drake’s calculation for a constant rate of mutation71:
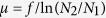 |
where μ is the mutation rate and N is the population size at time 1 or 2. We applied these equations to colony counts of helical and rod bacteria, plated on MH agar from liquid cultures of helical C. jejuni M1 undergoing exponential growth.
Light Microscopy
Light microscopy was performed on a Nikon Eclipse E200 light microscope under a 100x objective lens.
Scanning electron microscopy
To prepare samples for SEM, C. jejuni overnight (O/N) agar cultures were resuspended in 1 ml ddH2O and pelleted by centrifugation (9,300 × g) in microcentrifuge tubes. Bacteria were washed a total of four times before being fixed in 4% paraformaldehyde-1% glutaraldehyde in 0.1 M PBS. Fixed cells were stored at 4 °C prior to microscopy.
Motility assay
The motility of C. jejuni was quantified using motility agar made with 0.4%, 0.6%, 0.8% and 1.0% (w/v) select agar (Sigma) in MH broth. Motility agar was used to fill 6-well plates (7 ml of agar per well) 20 min prior to use. C. jejuni isolates were transferred via pipette tip from 12 h lawn growth (on MH agar plates) into each well of the motility agar. For each strain to be tested, three replicate 6-well plates were incubated for each motility agar concentration. Motility was measured as the diameter of the halo of motility after 12 h incubation.
Sequencing data
Genome sequencing data has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena), study accession PRJEB16677; ERS and ERR accession numbers are provided in Table S7.
Additional Information
How to cite this article: Esson, D. et al. Genomic variations leading to alterations in cell morphology of Campylobacter spp. Sci. Rep. 6, 38303; doi: 10.1038/srep38303 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Scanning Electron Microscopy was kindly performed by Dr Jeremy Skepper (Cambridge Advanced Imaging Centre). This work was funded by The Wellcome Trust through a PhD training studentship awarded to DE, and was supported by an Isaac Newton Trust/Wellcome Trust ISSF/University of Cambridge joint research grant awarded to AJG. SG and SPWDV were funded by BBSRC grant BB/K004514/1. AEM, SH, NRT and JP were supported by the Wellcome Trust grant number 098051. AEM was also supported by BBSRC grant BB/M014088/1. SKS was funded by Biotechnology and Biological Sciences Research Council grant BB/I02464X/1, Medical Research Council grant MR/L015080/1 and Wellcome Trust grant 088786/C/09/Z. GM was supported by a National Institute for Social Care and Health Research Fellowship (HF-14-13).
Footnotes
Author Contributions Conceived and designed the experiments D.E., E.S., N.R.T., J.P., D.J.M., A.J.G.; Performed the experiments: D.E., E.S., S.G., S.P.W.d.V., D.B., S.K.B., G.C. and A.J.G.; Analyzed the data: D.E., A.E.M., E.S., S.P.W.d.V., S.R.H., T.J.M., G.M., G.C., and A.J.G.; Wrote the paper: D.E. and A.J.G.; All authors reviewed the manuscript.
References
- Kirk M. D. et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS. Med. 12, e1001921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks In 2012. EFSA. Journal. 12, 3547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirrow M. B. Campylobacter enteritis: a “new” disease. Br. Med. J. 2, 9–11 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis C. et al. Genomic Characterization of Campylobacter jejuni Strain M1. PLoS. One. 5, e12253 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel M. E., Hayes S. F., Joens L. A. & Cieplak W. Jr. Characteristics of the internalization and intracellular survival of Campylobacter jejuni in human epithelial cell cultures. Microb. Pathog. 13, 357–370 (1992). [DOI] [PubMed] [Google Scholar]
- Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C. & Robinson R. A. 1985. A point-source outbreak of Campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152, 592–596 (1985). [DOI] [PubMed] [Google Scholar]
- Smibert R. M. The genus Campylobacter. Annu. Rev. Microbiol. 32, 673–709 (1978). [DOI] [PubMed] [Google Scholar]
- Alemka A. Corcionivoschi N. & Bourke B. Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. J. Gen. Microbiol. 134, 53–59 (2012). [DOI] [PubMed] [Google Scholar]
- Fridich E. et al. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS. Pathog. 8, e1002602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frirdich E. et al. Peptidoglycan LD-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J. Biol. Chem. 289, 8007–8018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge R. M., Divakaruni A. V. & Gober J. W. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51, 1321–1332 (2004). [DOI] [PubMed] [Google Scholar]
- Divakaruni A. V., Baida C., White C. L. & Gober J. W. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66, 174–188 (2007). [DOI] [PubMed] [Google Scholar]
- Shiomi D., Sakai M. & Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO. J. 27, 3081–3091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulmer D. M. et al. The bacterial cytoskeleton modulates motility, Type 3 secretion, and colonization in Salmonella. PLoS. Pathog. 8, e1002500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M., Doi M., Tamaki S., Park W., Nakajima-lijima S. & Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 167, 4935–4940 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., Joseleau-Petit D., Thevenet D., Bouloc P. & D’Ari R. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J. Bacteriol. 175, 6704–6710 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisah Alyahya S. et al. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl. Acad. Sci. USA. 106, 1239–1244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidner B. et al. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS. Pathog. 5, e1000669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro L. K. et al. Multiple peptidoglycan networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS. Pathog. 8, e1002603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro L. K. et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 141, 822–833 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N., Kuhn J. R. & Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 115, 705–713 (2003). [DOI] [PubMed] [Google Scholar]
- Gunther N. W., Bono J. L. & Needleman D. S. Complete genome sequence of Campylobacter jejuni RM1285, a rod-shaped morphological variant. Genome. Announc. 6, e01361–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A. J., Coward C., Jones M. A., Woodall C. A., Barrow P. A. & Maskell D. J. Signature-tagged transposon mutagenesis studies demonstrate the dynamic nature of cecal colonization of 2-week-old chickens by Campylobacter jejuni. Appl. Environ. Microbiol. 71, 8031–8041 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoal K., Ragimbeau C., Salvat G., Colin P. & Ermel G. Genomic diversity of Campylobacter coli and Campylobacter jejuni isolates recovered from free-range broiler farms and comparison with isolates of various origins. Appl. Environ. Microbiol. 71, 6216–6227 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribolt E. M., Fitzgerald C., Kubota K., Swaminathan B. & Barrett T. J. Rapid pulsed-field gel electrophoresis protocol for subtyping Campylobacter jejuni. J. Clin. Microbiol. 39, 1889–1894 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude M. W. & Baumler A. J. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17, 581–611 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J. et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 403, 665–668 (2000). [DOI] [PubMed] [Google Scholar]
- Bayliss C. D. et al. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic. Acids. Res. 40, 5876–5889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. M., Wagenaar J. A., Rigter A., Fearnley C., Newell D. G. & Duim B. Homonucleotide stretches in chromosomal DNA of Campylobacter jejuni display high frequency polymorphism as detected by direct PCR analysis. FEMS. Microbiol. Lett. 212, 77–85 (2002). [DOI] [PubMed] [Google Scholar]
- Inzana T. J. Identification of phase-variable genes that may contribute to nontypeable Haemophilus influenza nasopharyngeal colonization in humans contributes to our understanding of specific host-pathogen interactions. J. Infect. Dis. 208, 713–716 (2013). [DOI] [PubMed] [Google Scholar]
- Moxon E. R., Rainey P. B., Nowak M. A. & Kenski R. E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4, 24–33 (1994). [DOI] [PubMed] [Google Scholar]
- Hendrixson D. R. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol. Microbiol. 70, 519–536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. K. et al. Comparative variation within the genome of Campylobacter jejuni NCTC 11168 in human and murine hosts. PLoS. One. 9, e88229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A., Sakamoto C., Ghigo J.-M. & Beloin C. Did I pick the right colony? Pitfalls in the study of the regulation of phase variable antigen 43 adhesin. PLoS. One. 8, e73568 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennemann L. et al. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect. Immun. 80, 4364–4373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally D. K., Bogan J. A., Eisenstein B. I. & Blomfield I. C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K12: effects of temperature and media. J. Bacteriol. 175, 616–6193 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.-C. et al. A low-pH medium in vitro or the environment within a macrophage decreases the transcriptional levels of fimA, fimZ and lrp In Salmonella enterica serovar Typhimurium. J. Biosci. 38, 499–507 (2013). [DOI] [PubMed] [Google Scholar]
- Gaasbeek E. J. et al. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J. Bacteriol. 191, 3785–3793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci. C. et al. Hypermutation in pathogenic bacteria: frequent phase variation in Meningococci is a phenotypic trait of a specialized mutator biotype. Mol. Cell. 3, 435–445 (1999). [DOI] [PubMed] [Google Scholar]
- van der Broek D., Bloember G. V. & Lugtenberg B. The role of phenotypic variation in rhizosphere Pseudomonas bacteria. Environ. Microbiol. 7, 1686–1697 (2005). [DOI] [PubMed] [Google Scholar]
- Richardson A. R. & Stojilijkovi I. Mismatch repair and the regulation of phase variation in Neisseria meningitides. Mol. Microbiol. 40, 645–655 (2001). [DOI] [PubMed] [Google Scholar]
- Schaaper R. M. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268, 23762–23765 (1993). [PubMed] [Google Scholar]
- Martin P., Sun L., Hood, & Moxon E. R. Involvement of genes of genome maintenance in the regulation of phase variation frequencies In Neisseria meningitides. Microbiology. 150, 3001–3012 (2004). [DOI] [PubMed] [Google Scholar]
- Bjorkholm B., Sjolund M., Falk P. G., Berg O. G., Engstrand L. & Andersson D. I. Mutation frequency and biological cost of antibiotic resistance In Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 98, 14607–14612 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S. et al. Passage of Campylobacter jejuni through the chicken reservoir or mice promotes phase variation in contingency genes Cj0045 and Cj0170 that strongly associates with colonization and disease in a mouse model. Microbiology. 158, 1304–1316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson D. R. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61, 1646–1659 (2006). [DOI] [PubMed] [Google Scholar]
- Snipen L. et al. 2012. Analysis of evolutionary patterns of genes in Campylobacter jejuni and C. coli. Microb. Inform. Exp. 2, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric G. et al. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic Campylobacter. PLoS. One. 9, e92798 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S. P. et al. Motility defects in Campylobacter jejuni defined gene deletion mutants caused by second-site mutations. Microbiology. 161, 2316–2327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K. et al. Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. PeerJ. Preprints. 4, e2300v1 (2016). [DOI] [PubMed] [Google Scholar]
- Cody A. J. et al. Real-Time Genomic Epidemiological Evaluation of Human Campylobacter Isolates by Use of Whole-Genome Multilocus Sequence Typing. J. Clin. Microbiol. 51, 2526–2534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley H. van den Akker B. Giglio S. & Bentham R. The role of environmental reservoirs in human campylobacteriosis. Int. J. Environ. Res. Public. Health. 10, 5886–5907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R. E. Levine M. M. Clements M. L. Hughes T. P. & Blaser M. J. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157, 472–479 (1988). [DOI] [PubMed] [Google Scholar]
- Stahl M. et al. A novel mouse model of Campylobacter jejuni gastroenteritis reveals key pro-inflammatory and tissue protective roles for Toll-like receptor signalling during infection. PLoS. Pathog. 10, e1004264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., O’Rourkem J. L., Barrington P. J. & Trust T. J. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect. Immun. 51, 536–546 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F. & de Pedro M. A. Peptidoglycan plasticity in bacteria: emerging variability of the murein sacculus and their associated biological functions. Curr. Opin. Microbiol. 18, 46–53 (2014). [DOI] [PubMed] [Google Scholar]
- Frirdich E. & Gaynor E. C. Peptidoglycan hydrolases, bacterial shape, and pathogenesis. Curr. Opin. Microbiol. 16, 767–778 (2013). [DOI] [PubMed] [Google Scholar]
- Vollmer W., Joris B. Charlier P. & Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS. Microbiol. Rev. 32, 259–286 (2008). [DOI] [PubMed] [Google Scholar]
- van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 75, 636–663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. A bacterial cell shape-determining inhibitor. ACS. Chem. Biol. 11, 981–991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F. & Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic. Acids. Res. 16, 6127–6145 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. J. et al. Robust high throughput prokaryote de novo assembly and improvement pipeline for Illumina data. bioRxiv doi: 10.1101/052688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. & Russell D. W. 2001. Molecular Cloning. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory press.
- Yao R., Alm R. A., Trustm T. J. & Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 130, 127–130 (1993). [DOI] [PubMed] [Google Scholar]
- Hanninen M. L., Hakkinen M. & Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studies by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65, 2272–2275 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172, 451–464 (1988). [DOI] [PubMed] [Google Scholar]
- Li S.-Y., Holtje J.-V. & Young K. D. Comparison of high-performance liquid chromatography and fluorophore-assisted carbohydrate electrophoresis methods for analysing peptidoglycan composition of Escherichia coli. Anal. Biochem. 326, 1–12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G., Ustok F. L., Lu Q., Packman L. C. & Lowe C. R. Mutational analysis of Bacillus megaterium QM B1551 cortex-lytic enzymes. J. Bacteriol. 192, 5378–5389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A. & Foster P. L. Determining mutation rates in bacterial populations. Methods. 20, 4–17 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D. & Crow J. F. Rates of spontaneous mutation. Genetics. 148, 1667–1686 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.