Abstract
Although bacteriophages are ubiquitous in various environments, their genetic diversity is primarily investigated in pelagic marine environments. Corresponding studies in terrestrial environments are few. In this study, we conducted the first survey of phage diversity in the paddy ecosystem by targeting a new viral biomarker gene, phoH. A total of 424 phoH sequences were obtained from four paddy waters generated from a pot experiment with different soils collected from open paddy fields in northeast China. The majority of phoH sequences in paddy waters were novel, with the highest identity of ≤70% with known phoH sequences. Four unique groups (Group α, Group β, Group γ and Group δ) and seven new subgroups (Group 2b, Group 3d, Group 3e, Group 6a, Group 6b, Group 6c and Group 6d) were formed exclusively with the clones from the paddy waters, suggesting novel phage phoH groups exist in the paddy ecosystem. Additionally, the distribution proportions of phoH clones in different groups varied among paddy water samples, suggesting the phage community in paddy fields is biogeographically distributed. Furthermore, non-metric multidimensional scaling analysis indicated that phage phoH assemblages in paddy waters were distinct from those in marine waters.
The paddy field is a unique agro-ecosystem in which flooding and drainage are repeated during the annual cycle of rice cultivation, which results in the alternation of aerobic and anoxic processes in the paddy field ecosystem. Thus, the paddy field ecosystem is considered to be a hotspot for studying microbial ecology and biochemical cycles1,2. A large body of literature addresses the microbial ecology of paddy fields, including total bacterial and fungal communities3, methanogenic archaea4, methanotrophic bacteria5, and ammonia-oxidizing bacteria and archaea6. Recently, research on viral ecology or phage ecology in paddy ecosystems has aroused much attention7,8,9,10,11,12,13,14,15. For instance, Nakayama et al.9 observed that the abundance of virus-like particles (VLPs) in paddy floodwaters during the rice cultivation period ranged from 5.6 × 106 to 1.2 × 109 VLPs·mL−1 with a mean of 1.5 × 108 VLPs·mL−1, which was greater than the number of viruses in oceans and estuaries16,17. Chae et al.18 isolated 34 phages infecting Xanthomonas oryzae pv. oryzae from paddy floodwaters and observed that using a phage mixture is an effective method to control the occurrence of rice bacterial leaf blight disease. Moreover, many novel phage sequences or specific phage groups were observed in paddy fields by analysing several biomarker genes12,13,19,20,21,22.
The phoH gene is a host-derived auxiliary metabolic gene (AMG) carried by some phages23. This gene belongs to the Pho regulon and regulates phosphate uptake and metabolism under conditions of low-phosphate and phosphate limitation24,25. Unlike popular biomarker genes (g23, g20, DNA pol and psbA), that are restricted to the analysis of the genetic diversity of specific morphological phages, phoH is carried by various morphological types of phages (including siphophages, myophages and podophages), phages having wide host range (including autotrophic hosts and heterotrophic hosts), and even viruses of autotrophic eukaryotes23. By targeting this gene, Goldsmith et al.23 designed the degenerate primers vPhoHf/vPhoHr and were the first to use phoH to examine marine phage diversity throughout a depth profile in the Sargasso Sea and worldwide oceans. They found that viral phoH sequences in marine waters were highly diverse, and they identified six novel groups of phoH sequences. Subsequently, they further investigated the viral community composition throughout the water column both in summer and in winter across three years at the Bermuda Atlantic Time-series Study site in the Sargasso Sea, and this study revealed that the distribution patterns of viral communities varied not only with depth but also with time26,27. Their findings indicated that phoH is an effective signature gene for examining phage diversity in marine environments.
In researching the genetic diversity of phages in paddy ecosystems, we have previously found that several degenerate primers used for investigating phage diversity in marine environments, such as MZIAbis/MZIA628, CPS1/CPS829, psbA-F/psbA-R30 and CP-DNAP-349F/CP-DNAP-533Ra/b31, were also suitable for studying phage diversity in paddy ecosystems. Our overall findings showed that the phage communities were significantly different between paddy and marine ecosystems. In this study, to further understand the phage communities in paddy ecosystems, we targeted the phoH gene by using the primers vPhoHf/vPhoHr with the goal of addressing the following questions: (i) Do phages carry the phoH gene in paddy ecosystems? (ii) If so, how diverse and novel are they compared with reported sequences? (iii) Are the phage community compositions similar or different among different paddy fields or between paddy and marine ecosystems?
Materials and Methods
Sample collection and processing
An incubation experiment was designed to survey phage phoH genes in paddy waters in northeast (NE) China. The reason for using an incubation experiment rather than sampling floodwater from open paddy fields was to ensure that the phages were actually generated from the paddy fields. Because paddy fields in NE China are occasionally irrigated with river water or underground water, inappropriate sampling times directly from the open fields might result in data that do not truly reflect phages normally present in paddy waters. In brief, approximately 20 kg of soil (0~10 cm depth) were collected from the paddy fields of Daan (45°36′N, 123°50′E), Suihua (46°43′N, 126°59′E), Mudanjiang (44°26′N, 129°29′E), and Yanjiagang (45°35′N, 126°20′E) (Table S1) in NE China on 9~13 May, 2014. Each paddy soil sample was subpackaged equally into two plastic containers with dimensions of 60 × 40 × 28 cm and incubated with autoclaved water. One week later, after basal nutrients of 0.4 g KCl, 1.0 g Ca3(PO4)2, 1.0 g (NH4)2SO4 per kilogram of soil were added to the soil for rice growth, we transplanted eleven rice seedlings (Oryza sativa L. ssp. japonica, cv. Daohuaxiang) into each container on 22 May, 2014. Hereafter, all the plastic containers were put outside on days without rain and in the greenhouse on rainy days, and the water layer was maintained 8 cm above soil by timely supplementation with autoclaved water.
Approximately 1 L of water samples from the two containers of each soil sample was collected on 22 June. After being centrifuged twice at 5,000 rpm for 30 min at 4 °C to remove soil particles, plankton and partial bacteria, the supernatant was sequentially filtered through 0.4-μm and 0.2-μm carbonate membrane filters (Nuclepore Track-Etched Membranes, Whatman International Ltd, London, UK) to completely remove bacterial cells. Finally, virus-sized particles were filtered onto 0.03-μm carbonate membrane filters (Nuclepore Track-Etched Membranes, Whatman International Ltd, London, UK). The filters were placed in sterilized 2 mL centrifuge tubes and kept at −20 °C.
DNA extraction and PCR amplification
The 0.03-μm filters were treated with DNase and RNase (40 μg mL−1 each) in 10 mM Tris-HCl buffer (pH 7.5) to decompose free DNA and RNA. Next, viral DNA extraction was performed according to the protocol reported previously12. The extracted DNA was dissolved in 30 μL TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and stored at −20 °C for further analysis.
The phoH sequences were amplified with the degenerate primers vPhoHf and vPhoHr23. PCR reactions were performed in a 50 μL mixture containing 10 μL EasyTaq buffer (TransGen Biotech, Beijing, China), 5 μL dNTPs (2.5 mM each; TransGen Biotech, Beijing, China), 0.5 μL forward and reverse primers (50 pmol each), 1.5 μL DNA template and 2 μL of Easy Taq DNA polymerase (TransGen Biotech, Beijing, China). The reactions were filled to the required volume with sterile Milli-Q water. The negative control contained all reagents and sterile Milli-Q water without the template. The thermal program used for PCR amplification was the same as a paper reported previously23.
Cloning and sequencing
PCR products of approximately 420 bp in length were cut out from a 2% agarose gel and purified using the QIAExII Gel Extraction Kit (QIAGEN, Shanghai, China, Cat. No. 20021). The purified DNA was cloned into the pMD18-T vector (TaKaRa, Dalian, China) and transformed into competent cells of Escherichia coli DH5α. White clones were picked out for PCR amplification using the same primers and PCR program described above. After being harvested by overnight culture, the plasmid DNA from positive clones was sequenced by a commercial company (BGI, Shenzhen, China). The phoH nucleotide sequences obtained in this study were deposited in GenBank under accession numbers KX189635-KX190058.
Phylogenetic analysis
The phoH sequences were translated to deduced amino acid sequences by the EMBOSS Transeq program on the European Bioinformatics Institute website (http://www.ebi.ac.uk/Tools/st/emboss_transeq/). The closest relatives of phoH clones were examined at the amino acid level using the BLASTp search program on the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST). Reference phoH sequences from cultured viruses and bacteria, as well as environmental viral clones were retrieved from GenBank. After the amino acid sequences were aligned with CLUSTALX 1.8132, neighbour-joining phylogenetic trees were constructed using software (MEGA 4.0)33 with 1,000 bootstrap replicates.
Clone library and diversity analyses
Operational taxonomic units (OTUs) were generated based on sequence similarity of 97% at the nucleotide level. Non-metric multidimensional scaling (NMDS) analysis was used to visually display the differences of phage phoH sequence assemblages based on the distances between clone libraries. The NMDS analysis was performed in R-2.15.134 with the “vegan” package35. Accession numbers for the reference sequences used in this analysis are shown in Table S2.
Results
Closest relatives of phoH clones
Out of 486 positive clones submitted for sequencing, 424 clones were identified as phoH sequences. Among these sequences, 166, 97, 36 and 125 clones were from water samples of Daan, Suihua, Mudanjiang, and Yanjiagang, respectively. The phoH fragments had lengths of 381~405 bp, encoding 127~135 amino acid residues (Table S3). The BLASTp search for the closest relatives at the amino acid level showed that 108, 114, 40 and 4 clones had the highest identity to viral phoH clones from the Sargasso Sea, the Mediterranean Sea, the Gulf of Mexico and British Columbia coastal waters, respectively23. Additionally, 75 clones had the highest identity to Synechococcus phage S-SSM7 from the Sargasso Sea36, 31 clones had the highest identity to an uncultured phage uvMED from the Mediterranean Sea, Spain37, 20 clones had the highest identity to Synechococcus phage S-RSM4 from the Red Sea38, and 32 clones had the highest identity to Acinetobacter phage YMC13/03/R2096 from South Korea (Table S3; Table S4). Noticeably, approximately 70% (295/424) of the clones had identity ≤70% with known phoH sequences, which indicated that the majority of phoH clones obtained in this study are novel (Table S3; Table S4).
Phylogeny of phoH sequences
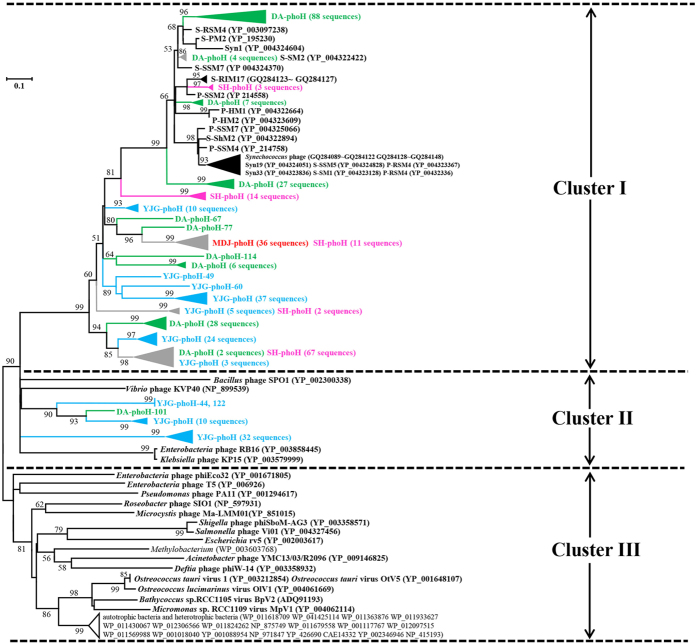
All the phoH clones obtained in this study were used to build a phylogenetic tree with phoH sequences from cultured phages of autotrophs and heterotrophs, and phoH sequences from cultured autotrophic and heterotrophic bacterial hosts (Fig. 1). Overall, the phylogenetic tree could be mainly divided into three clusters (Clusters I, II and III). Cluster I contained phages infecting autotrophic bacteria, Cluster II contained phages infecting heterotrophic bacteria, and Cluster III contained viruses infecting eukaryotes, phages infecting several heterotrophic bacteria, one phage of an autotrophic bacterium (Microcystis phage Ma-LMM01), and numerous host phoH genes from heterotrophic and autotrophic bacteria. It should be noted that approximately 90% (379/424) of phoH clones observed in this study were grouped into Cluster I (phages of autotrophic bacteria), while the remaining 45 clones were grouped into Cluster II (phages of heterotrophic bacteria). No clones from this study were grouped into Cluster III.
Figure 1. Neighbour-joining phylogenetic tree of phoH amino acid sequences obtained in this study with reference phoH sequences of cultured phages of autotrophs and heterotrophs, viruses of eukaryotic autotrophs, and cultured autotrophic and heterotrophic bacterial hosts.
Clones in this study were named to reflect the sampling site, biomarker gene name and clone number. In detail, DA, SH, MDJ and YJG indicated samples from Daan (green words in bold), Suihua (pink words in bold), Mudanjiang (red words in bold) and Yanjiagang (blue words in bold), respectively. The green, pink and blue triangles indicate phoH clusters obtained from Daan, Suihua and Yanjiagang, respectively, and the grey triangles represent phoH clusters obtained from more than one paddy sample. The phoH sequences of isolated virus and phage origins are in bold black font, and their clusters are indicated with black triangles. The phoH sequences of isolated host origins are in normal black font, and their clusters are indicated with white triangles. Bootstrap values <50 are not shown. The scale bar represents the abundance of amino acid substitutions per residue.
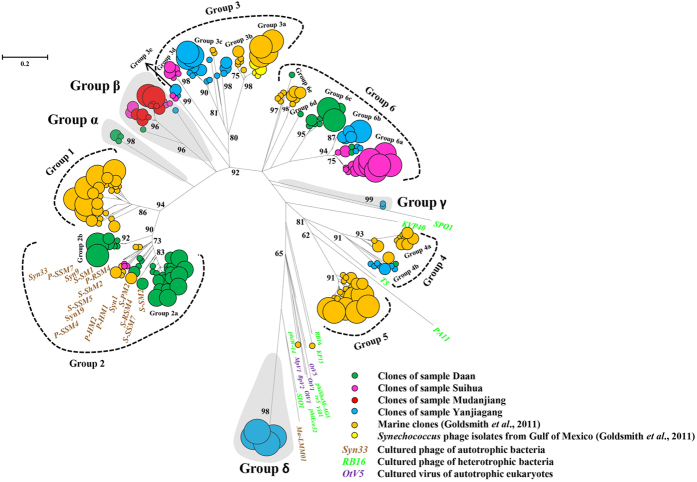
To determine whether novel phage phoH groups exist in paddy waters, the phylogenetic relationships of all sequences observed in this study with phoH sequences coming from marine water clones, Synechococcus phage isolates, cultured phage of autotrophic and heterotrophic bacteria, and cultured viruses of autotrophic eukaryotes at the amino acid level are shown in Fig. 2. According the grouping standard first designed by Goldsmith et al.23, the viral phoH sequences from six worldwide oceans were phylogenetically distributed into Groups 1, 2, 3, 4, 5 and 6, of which Group 3 was further divided into Group 3a, 3b and 3c. In this study, we found that 30.42%, 16.27%, 2.60% and 29.72% of clones across all samples were grouped into the previously designed Groups 2, 3, 4 and 6, respectively. No clones were grouped into Groups 1 and 5. In addition, 6, 49, 2 and 32 clones from this study formed four new groups, named as Group α, Group β, Group γ, and Group δ, respectively. No clones from marine waters were grouped into the four newly designed groups. Furthermore, within formerly named groups, seven new subgroups, i.e., Group 2b, Group 3d, Group 3e, Group 6a, Group 6b, Group 6c and Group 6d, were formed exclusively with the clones obtained in this study.
Figure 2. Unrooted phylogenetic tree comparing phoH amino acid sequences of environmental clones obtained from this study and marine waters, cultured phages of autotrophic and heterotrophic bacteria, as well as cultured eukaryotic viruses.
The bootstrap values <50 are not shown. The size of circles at the end of branches is proportional to the number of clones/phages, and the small, medium and large circles represent one, four and eight clones/phages, respectively. The scale bar represents the abundance of amino acid substitutions per residue.
Biogeography of phage phoH sequences
The distribution proportions of phoH clones into different groups not only differed between marine waters and paddy waters but also differed among paddy water samples (Table 1; Fig. S1). For example, over 60% (99/166) of clones from Daan and 3% of clones from Suihua (but no clones from other two water samples) were grouped into Group 2a. Approximately 69% of clones from Suihua were grouped into Group 6a, while less than 3% of clones from other water samples were grouped into that group. Approximately 26%, 38% and 19% of clones from Yanjiagang were grouped into Group δ, Group 3c, and Group 6b, respectively, and no clones from other samples were grouped into those groups. All clones from Mudanjiang were grouped into Group β, while only about 11% of clones from Suihua and less than 1% of clones from both Daan and Yanjiagang were grouped into Group β.
Table 1. Number and distribution proportion of the phage phoH clones in phylogenetic groups obtained from marine waters and paddy waters.
| Phylogenetic groups | Marinea (275)d | Paddyb (424)d | Daanc (166)d | Suihuac (97)d | Mudanjiangc (36)d | Yanjiagangc (125)d | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of clones | Proportion (%) | Number of clones | Proportion (%) | Number of clones | Proportion (%) | Number of clones | Proportion (%) | Number of clones | Proportion (%) | Number of clones | Proportion (%) | |
| Group α | 6 | 1.42 | 6 | 3.62 | ||||||||
| Group β | 49 | 11.56 | 1 | 0.60 | 11 | 11.34 | 36 | 100 | 1 | 0.80 | ||
| Group γ | 2 | 0.47 | 2 | 1.60 | ||||||||
| Group δ | 32 | 7.55 | 32 | 25.60 | ||||||||
| Group 1 | 91 | 33.09 | ||||||||||
| Group 2a | 13 | 4.73 | 102 | 24.05 | 99 | 59.64 | 3 | 3.09 | ||||
| Group 2b | 27 | 6.37 | 27 | 16.27 | ||||||||
| Group 3a | 33 | 12 | ||||||||||
| Group 3b | 8 | 2.91 | ||||||||||
| Group 3c | 2 | 0.73 | 48 | 11.32 | 48 | 38.40 | ||||||
| Group 3d | 14 | 3.30 | 14 | 14.44 | ||||||||
| Group 3e | 7 | 1.65 | 2 | 2.06 | 5 | 4 | ||||||
| Group 4a | 4 | 1.45 | ||||||||||
| Group 4b | 24 | 8.72 | 11 | 2.59 | 1 | 0.60 | 10 | 8 | ||||
| Group 5 | 77 | 28 | ||||||||||
| Group 6a | 72 | 16.98 | 2 | 1.20 | 67 | 69.07 | 3 | 2.40 | ||||
| Group 6b | 24 | 5.66 | 24 | 19.20 | ||||||||
| Group 6c | 28 | 6.60 | 28 | 16.87 | ||||||||
| Group 6d | 1 | 0.24 | 1 | 0.60 | ||||||||
| Group 6e | 21 | 7.64 | 1 | 0.24 | 1 | 0.60 | ||||||
| Undesigned group | 2 | 0.73 | ||||||||||
aMarine sources including the Sargasso Sea and worldwide oceans (Goldsmith et al.23).
bPaddy sources including the four paddy sampling sites (Daan, Suihua, Mudanjiang and Yanjiagang) in this study.
cPaddy sample in this study.
dNumber in parenthesis is the total number of clones obtained from each source/sample.
To show clearly the distribution patterns of phage communities in different environments, the phage phoH sequence assemblages from paddy waters of this study and from marine environments23 including the Sargasso Sea, the Mediterranean Sea, the Gulf of Mexico, British Columbia coastal waters, Raunefjorden and Kongsfjorden were subjected to NMDS analysis (Table S2). The plot clearly showed that all samples were separated into two groups (Fig. 3). One group contained samples from marine waters, and another group included samples from paddy waters. This finding indicated that phage phoH assemblages in paddy waters were distinct from those in marine waters.
Figure 3. Non-metric multidimensional scaling analysis of phage phoH communities.
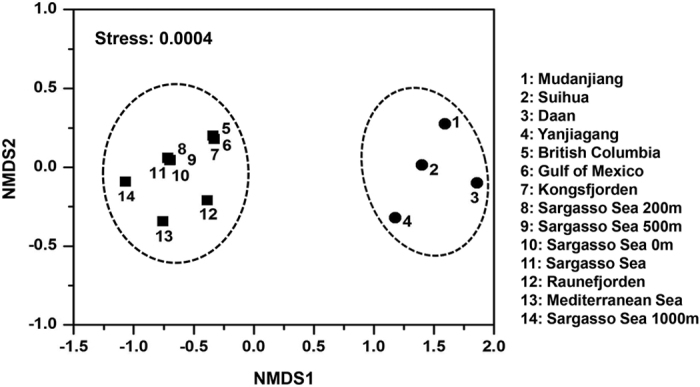
The plot shows the distribution pattern of the phage phoH sequence assemblages obtained from different environments. Samples located in close proximity on the NMDS plot are grouped by dashed circles.
Discussion
PCR amplification of the phoH genes in paddy waters
The primer set vPhoHf/vPhoHr was originally designed by Goldsmith et al.23 to amplify the phage phoH genes from marine environments. They found that the phoH gene is commonly carried in phages that infect heterotrophic and autotrophic bacteria, as well as in viruses infecting autotrophic eukaryotes. Unlike other biomarker genes, such as g23, g20, psbA and DNA pol, which only target a specific family of phages, the phoH gene is not restricted to a certain morphological type of phage, which suggests that it could be a powerful biomarker gene for studying phage diversity. Moreover, by comparing the fully sequenced phage genomes in GenBank, they found that nearly 40% of marine phages contained the phoH gene, while only 4% of nonmarine phages contained this gene. This finding seemed to restrict the application of this gene to study phage diversity in terrestrial environments. However, only a small fraction of bacteria can be cultured39,40, which may hamper our ability to isolate cultured phages. Thus, many viruses in natural environments that might contain this gene may not have been identified due to current culture conditions. In this study, we pioneered the use of this gene to survey the diversity of phages in paddy waters. We collected the viral particles, ultimately excluded contamination from hosts or host’s DNA, and obtained 424 phoH clones from four paddy water samples. We found all clones were phylogenetically grouped into Cluster I and Cluster II (Fig. 1), and Clusters I and II contained several phoH references coming from phages of autotrophic and heterotrophic bacteria, respectively. None of the phoH sequences from viruses of eukaryotes and hosts were grouped into these two clusters (Fig. 1). This finding strongly demonstrated that the phoH gene was contained in phage genomes of terrestrial environments and that this biomarker gene was useful for studying phage ecology in paddy ecosystems.
Phylogenetic position of phoH genes in paddy waters
The pioneering work of Goldsmith et al.23 showed that phage-originated phoH sequences could be separated from host phoH sequences through constructing a phylogenetic tree, and they also stated that autotrophic phages and heterotrophic phages tended to cluster separately. In this study, we found that nearly 90% of phoH clones formed a well-supported (99%) cluster in Cluster I with several cyanophages infecting Synechococcus and Prochlorococcus. No phoH sequences from cultured phages of heterotrophic bacteria, viruses of eukaryotes, or hosts were grouped into Cluster I (Fig. 1). This suggested that Cluster I clones might originate from cyanophages from paddy water. Many clades in Cluster I of Fig. 1 have no reference sequences, which might be due to great diversity of cyanobacteria in the paddy field. Consistent with that speculation, our previous study has already revealed that many unknown groups of picocyanobacteria exist in paddy fields22. In contrast, less than 10% of clones in this study (mainly obtained from Yanjiagang) formed two clades in Cluster II closely related to phages infecting heterotrophic bacteria, suggesting that those clones might be from noncyanophages (Fig. 1). This heavily disproportionate split of phoH clones, between those that group with cyanophages and those that group with noncyanophages, may imply that in paddy waters, this auxiliary metabolic gene is mainly contained in genomes of cyanophages. Future studies using culture-dependent methods are needed to address this implication.
Goldsmith et al.23 obtained 289 phage phoH sequences from the Sargasso Sea, British Columbia coastal waters, the Gulf of Mexico, Raunefjorden, Kongsfjorden and the Mediterranean Sea. They found that the majority of those sequences were grouped into six groups, and they labeled those groups as Groups 1–6. Further sequencing by a high throughput method revealed no new phylogenetic groups in the Sargasso Sea26. In this study, we obtained 424 phage phoH clones from paddy waters. Among them, approximately 80% of clones were grouped into Groups 2, 3, 4 and 6, while 20% of clones formed four new groups (Group α, Group β, Group γ and Group δ). In addition, several new subgroups within individual groups were formed exclusively with the clones obtained from paddy waters (Fig. 2). These findings suggest that the distribution pattern of phage phoH genes in paddy waters is somewhat distinct from that found in marine waters.
Comparing the distribution patterns between Figs 1 and 2, we found that all clones obtained in this study in Cluster I of Fig. 1 were located in Groups 2, 3, 6, α and β, while the clones in Cluster II of Fig. 1 were distributed into Group 4, γ and δ (Fig. 2). As elaborated above, we speculated the sources of phoH sequences obtained in paddy waters in Groups 2, 3, 6, α and β may be cyanophages. This speculation was bolstered by the phylogenetic tree of Fig. 2, in which Groups 1, 2, 3, α and β formed a large well-supported cluster (92%) containing several reference phoH sequences of cyanophages23,26 (Fig. 2). Although no known phage phoH sequences were grouped into Group 6, all the marine phoH sequences in Group 6 were obtained from surface and upper water samples, and no clones from 1000 m depth of the Sargasso Sea (where cyanophages were absent) were grouped into this group23. In addition, all the paddy phoH clones that fell into Group 6 (Fig. 2) were located in Cluster I of Fig. 1. Together, these findings strongly implied that the phoH clones in Group 6 might also originated from cyanophages. Future research is needed to address this possibility.
Biogeographical distribution of phage phoH sequences in paddy waters
Previous studies showed that the composition of viral phoH sequences not only varied throughout the water column but also changed throughout the year in the Sargasso Sea23,26,27. In addition, the composition of viral phoH sequences was also different among six worldwide oceans23. Those findings indicated that the composition of viral phoH sequences in oceans was spatially and temporally distributed. In this study, although the paddy water samples were collected only one time for surveying the diversity of viral phoH genes, we found that the distribution proportions of phoH clones into different groups varied among paddy water samples (Table 1; Fig. S1), which suggested that the phage phoH genes in paddy fields were biogeographically distributed. Furthermore, comparison of the assemblages of viral phoH sequences showed that the viral communities were distinctly different between paddy waters and ocean waters (Fig. 3). Similar findings have also been observed by analysing other biomarker genes in paddy fields, such as g2341, g2015 and DNA pol13.
Although different phoH compositions were observed among paddy waters, and between paddy waters and marine waters, we still found that certain phoH groups were commonly detected in multiple samples, which suggested that some phages are not restricted by geographical separation. Sano et al.42 reported that the phages obtained from soil, freshwater and sediment can propagate on hosts from the marine environment. Moreover, they also showed that marine phages from one location can infect hosts from a different marine location. Others studies on phage ecology targeting different biomarker genes, such as g23 of T4-type phages43, g20 of cyanomyophages44 and DNA pol gene of podophages45, have also detected identical sequences across widely separated geographical locations and different habitats. Those findings suggested that some phages are not host-specific. It is a universal phenomenon in natural conditions that one host can be infected by different phages or a phage can infect different hosts46. This phenomenon promotes horizontal gene transfer between phages and hosts, and even across different hosts, which promotes evolution.
Conclusions
In conclusion, 424 phoH clones were obtained from paddy waters. Among them, approximately 90% of the phoH clones were grouped with cyanophages, while 10% of the clones were grouped with phages of heterotrophic bacteria. This division implied that this auxiliary metabolic gene was carried mainly in the genomes of cyanophages in paddy waters. Four new groups and seven new subgroups were formed exclusively with the clones from paddy waters, suggesting that the distribution pattern of the phage phoH gene in paddy waters was distinct from that in marine waters. In addition, the phage community compositions represented by phoH sequences varied among paddy water samples and were also remarkably different from those observed in marine waters. As far as we know, this is the first study revealing that phages in paddy fields also contain the phoH gene, suggesting this biomarker gene is an effective signature gene for investigating phage diversity both in marine and terrestrial environments.
Additional Information
How to cite this article: Wang, X. et al. Novel groups and unique distribution of phage phoH genes in paddy waters in northeast China. Sci. Rep. 6, 38428; doi: 10.1038/srep38428 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was financially supported by National Nature Science Foundation of China (41271262, 41571246) and Strategic Priority Research Program of Chinese Academy of Sciences (XDB15010103).
Footnotes
Author Contributions G.H.W. conceived and designed the experiments; X.Z.W. performed the experiments; X.Z.W. analysed the data; X.Z.W. and G.H.W. wrote the paper; J.J.L., Z.H.Y., J.J. and X.B.L. revised the manuscript.
References
- Song T., Mårtensson L., Eriksson T., Zheng W. & Rasmussen U. Biodiversity and seasonal variation of the cyanobacterial assemblage in a rice paddy field in Fujian, China. FEMS Microbiol Ecol 54, 131–140 (2005). [DOI] [PubMed] [Google Scholar]
- Ishii S., Ikeda S., Minamisawa K. & Senoo K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ 26, 282–292 (2011). [DOI] [PubMed] [Google Scholar]
- Tun C. C., Ikenaga M., Asakawa S. & Kimura M. Community structure of bacteria and fungi responsible for rice straw decomposition in a paddy field estimated by PCR-RFLP analysis. Soil Sci Plant Nutr 48, 805–813 (2002). [Google Scholar]
- Watanabe T., Kimura M. & Asakawa S. Diversity of methanogenic archaeal communities in Japanese paddy field ecosystem, estimated by denaturing gradient gel electrophoresis. Biolo Fert Soils 46, 343–353 (2010). [Google Scholar]
- Shrestha M., Abraham W. R., Shrestha P. M., Noll M. & Conrad R. Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ Microbiol 10, 400–412 (2008). [DOI] [PubMed] [Google Scholar]
- Chu H. Y., Morimoto S., Fujii T., Yagi K. & Nishimura S. Soil ammonia-oxidizing bacterial communities in paddy rice fields as affected by upland conversion history. Soil Sci Soc AM J 73, 2026–2031 (2009). [Google Scholar]
- Jia Z. J., Ishihara R., Nakajima Y., Asakawa S. & Kimura M. Molecular characterization of T4-type bacteriophages in a rice field. Environ Microbiol 9, 1091–1096 (2007). [DOI] [PubMed] [Google Scholar]
- Nakayama N., Okumura M., Inoue K., Asakawa S. & Kimura M. Morphological analysis of viral communities in the floodwater of a Japanese paddy field. Soil Biol Biochem 39, 3187–3190 (2007). [Google Scholar]
- Nakayama N., Okumura M., Inoue K., Asakawa S. & Kimura M. Seasonal variations in the abundance of virus-like particles and bacteria in the floodwater of a Japanese paddy field. Soil Sci Plant Nutr 53, 420–429 (2007). [Google Scholar]
- Nakayama N., Tsuge T., Asakawa S. & Kimura M. Morphology, host range and phylogenetic diversity of Sphingomonas phages in the floodwater of a Japanese paddy field. Soil Sci Plant Nutr 55, 53–64 (2009). [Google Scholar]
- Kimura M., Jia Z. J., Nakayama N. & Asakawa S. Ecology of viruses in soils: past, present and future perspectives. Soil Sci Plant Nutr 54, 1–32 (2008). [Google Scholar]
- Wang G. H., Murase J., Asakawa S. & Kimura M. Novel cyanophage photosynthetic gene psbA in the floodwater of a Japanese rice field. FEMS Microbiol Ecol 70, 79–86 (2009). [DOI] [PubMed] [Google Scholar]
- Wang G. H., Liu J. J., Yu Z. H., Jin J. & Liu X. B. Unique distribution of cyanobacterial podoviruses and their potential hosts in a paddy field of northeast China. FEMS Microbiol Ecol 90, 331–334 (2014). [DOI] [PubMed] [Google Scholar]
- Liu J. J., Wang G. H., Wang Q., Liu J. D., Jin J. & Liu X. B. Phylogenetic diversity and assemblage of major capsid genes (g23) of T4-type bacteriophages in paddy field soils during rice growth season in Northeast China. Soil Sci Plant Nutr 58, 435–444 (2012). [Google Scholar]
- Jing R. Y., Liu J. J., Yu Z. H., Liu X. B. & Wang G. H. Phylogenetic distribution of the capsid assembly protein gene (g20) of cyanophages in paddy floodwaters in Northeast China. PloS One 9, e88634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack K. E. & Colwell R. R. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol R 64, 69–114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M. G. Ecology of prokaryotic viruses. FEMS Microbiol Rev 28, 127–181 (2004). [DOI] [PubMed] [Google Scholar]
- Chae J. C., Hung N. B., Yu S. M., Lee H. K. & Lee Y. H. Diversity of bacteriophages infecting Xanthomonas oryzae pv. oryzae in paddy fields and its potential to control bacterial leaf blight of ice. J Microbiol Biotechnol 24(6), 740–747 (2014). [DOI] [PubMed] [Google Scholar]
- Wang G. H., Jin J., Asakawa S. & Kimura M. Survey of major capsid genes (g23) of T4-type bacteriophages in rice fields in Northeast China. Soil Biol Biochem 41, 423–427 (2009). [Google Scholar]
- Wang G. H., Hayashi M., Saito M., Tsuchiya K., Asakawa S. & Kimura M. Survey of major capsid genes (g23) of T4-type bacteriophages in Japanese paddy field soils. Soil Biol Biochem 41, 13–20 (2009). [Google Scholar]
- Wang G. H., Murase J., Asakawa S. & Kimura M. Unique viral capsid assembly protein gene (g20) of cyanophages in the floodwater of a Japanese paddy field. Biol Fert Soils 46, 93–102 (2010). [Google Scholar]
- Wang G. H., Asakawa S. & Kimura M. Spatial and temporal changes of cyanophage communities in paddy field soils as revealed by the capsid assembly protein gene g20. FEMS Microbiol Ecol 76, 352–359 (2011). [DOI] [PubMed] [Google Scholar]
- Goldsmith D. B. et al. Development of phoH as a novel signature gene for assessing marine phage diversity. Appl Environ Microbiol 77, 7730–7739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Phosphorus assimilation and control of the phosphate regulon, p. 1357–1381. In Neidhardt F. C. & Curtiss R. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC. (1996). [Google Scholar]
- Hsieh Y. J. & Wanner B. L. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13, 198–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith D. B., Parsons R. J., Beyene D., Salamon P. & Breitbart M. Deep sequencing of the viral phoH gene reveals temporal variation, depth-specific composition, and persistent dominance of the same viral phoH genes in the Sargasso Sea. Peer J 3, e997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith D. B., Brum J. R., Hopkins M., Carlson C. A. & Breitbart M. Water column stratification structures viral community composition in the Sargasso Sea. Aquat Microb Ecol 76, 85–94 (2015). [Google Scholar]
- Filée J., Tétart F., Suttle C. A. & Krisch H. M. Marine T4 type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci 102, 12471–12476 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Chen F., Wilhelm S. W., Poorvin L. & Hodson R. E. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl Environ Microbiol 68, 1576–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidner G. et al. Molecular diversity among marine picophytoplankton as revealed by psbA analyses. Environ Microbiol 5, 212–216 (2003). [DOI] [PubMed] [Google Scholar]
- Chen F. et al. Diversity and dynamic populations of cyanobacterial podoviruses in the Chesapeake Bay unveiled through DNA polymerase gene sequences. Environ Microbiol 11, 2884–2892 (2009). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F. & Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M. & Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24, 1596–1599 (2007). [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R 21. Foundation for Statistical Computing, Vienna, Austria (2006).
- Oksanen J. et al. Vegan: Community Ecology Package. R package version 2.0-7. Available: http://CRAN.R project org/package=vegan (2013).
- Sullivan M. B. et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol 12, 3035–3056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno C. M., Rodriguez-Valera F., Kimes N. E. & Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet 9, e1003987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard A. D., Zwirglmaier K., Downey M. J., Mann N. H. & Scanlan D. J. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ Microbiol 11, 2370–2387 (2009). [DOI] [PubMed] [Google Scholar]
- Ward D. M., Weller R. & Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a nature community. Nature 344, 63–65 (1990). [DOI] [PubMed] [Google Scholar]
- Pace N. R. A molecular view of microbial diversity and the biosphere. Science 276, 734–740 (1997). [DOI] [PubMed] [Google Scholar]
- Liu J. J., Wang G. H., Zheng C. Y., Yuan X. H., Jin J. & Liu X. B. Specific assemblages of major capsid genes (g23) of T4-type bacteriophages isolated from upland black soils in Northeast China. Soil Biol Biochem 43, 1980–1984 (2011). [Google Scholar]
- Sano E., Carlson S., Wegley L. & Rohwer F. Movement of viruses between biomes. Appl Environ Microbiol 70, 5842–5846 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. H. et al. Changes in major capsid genes (g23) of T4-type bacteriophages with soil depth in two Japanese rice fields. Biol Fertil Soils 5, 521–529 (2009). [Google Scholar]
- Short C. M. & Suttle C. A. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl Environ Microbiol 71, 480–486 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M. et al. Diversity and population structure of a near-shore marine-sediment viral community. Proceedings of the Royal Society of London Series B: Biological Sciences 271, 565–574 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M. & Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol 13, 278–284 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.