Abstract
The IDH1/2 gene mutations, ATRX loss/mutation, 1p/19q status, and MGMT promoter methylation are increasingly used as prognostic or predictive biomarkers of gliomas. However, the effect of their combination on radiation therapy outcome is discussable. Previously, we demonstrated that the IDH1 c.G395A; p.R132H mutation was associated with longer survival in grade II astrocytoma and GBM (Glioblastoma). Here we analyzed the MGMT promoter methylation status in patients with a known mutation status in codon 132 of IDH1, followed by clinical and genetic data analysis based on the two statuses. After a subtotal tumor resection, the patients were treated using IMRT (Intensity-Modulated Radiation Therapy) with 6 MeV photons. The total dose was: 54 Gy for astrocytoma II, 60 Gy for astrocytoma III, 60 Gy for glioblastoma, 2 Gy per day, with 24 h intervals, five days per week. The patients with MGMT promoter methylation and IDH1 somatic mutation (OS = 40 months) had a better prognosis than those with MGMT methylation alone (OS = 18 months). In patients with astrocytoma anaplasticum (n = 7) with the IDH1 p.R132H mutation and hypermethylated MGMT, the prognosis was particularly favorable (median OS = 47 months). In patients with astrocytoma II meeting the above criteria, the prognosis was also better than in those not meeting those criteria. The IDH1 mutation appears more relevant for the prognosis than MGMT methylation. The IDH1 p.R132H mutation combined with MGMT hypermethylation seems to be the most advantageous for treatment success. Patients not meeting those criteria may require more aggressive treatments.
Keywords: glioma, IDH1, MGMT, radiotherapy, survival
1. Introduction
DNA methylation plays a crucial role in many biological processes—this epigenetic modification contributes in the temporal and spatial regulation of gene expression [1], X chromosome inactivation [2], genome imprinting [3], male infertility [4] and carcinogenesis [5], as reviewed in [6]. It has been observed that cancer cells have an excessive methylation of genes involved in cell cycle regulation (p16INK4a, p15INK4a, Rb, p14ARF), DNA damage repair (BRCA1, MGMT), apoptosis (DAPK, TMS1), detoxification, cell differentiation, angiogenesis and metastasis [7]. The CpG islands hypermethylation of the promoter region of tumor suppressor genes is a key mechanism of gene activation in cancer [8,9]. On the other hand, hypomethylation is a player in oncogenesis through the activation of oncogenes, such as cMYC and H-RAS [10], as well as chromosomal instability [11].
Glioblastoma multiforme was the first cancer selected for epigenome analysis in the pilot The Cancer Genome Atlas (TCGA) project. Analysis of the promoter DNA methylation alteration in 272 glioblastoma tumors identified a group of patients with a repetitive methylation pattern called glioma-CpG island methylator phenotype (G-CIMP phenotype). These G-CIMP tumors had distinct molecular features, including a high frequency of mutations in the IDH1 gene and characteristic copy number alterations. Patients with the G-CIMP phenotype were younger at the time of diagnosis in a statistically significant manner and presented significantly improved outcomes [5]. Our previous observation in a Polish group of glioma patients was consistent with those results. The IDH1 c.G395A; p.R132H mutation was associated with longer survival in grade II astrocytoma, glioblastoma, and pooled groups of patients with WHO grade II glioma [12]. Characterization of the molecular landscape of lower-grade gliomas confirmed the presence of a known IDH1 mutation and 1p/19q codeletion (whole-arm loss of the long arm of chromosome 1 and the short arm of chromosome 19) and their role as a predictive markers [13,14]. The latest evaluation of molecular subgroups of gliomas identified ATRX, CIC, and FUBP1 mutations, and allowed distinguishing patients with IDH1/CIC/FUBP1 or IDH1/ATRX mutations [15]. The first group had the longest median overall survival (96 months) in comparison with patients with IDH1 mutations and ATRX loss (51 months), while the shortest median overall survival was in glioma patients who did not harbor any of those signatures (13 months) [15].
In the presented study, we analyzed the MGMT methylation levels in 83 glioma patients with a known IDH1 mutation status and re-evaluated the clinical data with two years extended novel observations.
2. Results
2.1. Patient Characteristics
In this study, based on our previously reported group of 139 glioma patients [12], we randomly selected 90 patients for further evaluation of MGMT promoter methylation. Seven patients were excluded and 83 patients were screened for MGMT methylation. Additional clinical features of the patients with MGMT methylation and IDH1 statuses, including a small number of patients treated with temozolomide, are shown in Table 1. Among the chosen 83 patients, 55 had a previously detected IDH1 c.G395A; p.R132H mutation and 28 were wild-type in codon 132 of the IDH1 gene [12]. Among the histological types, we evaluated MGMT promoter methylation in 57 astrocytomas WHO II, 13 astrocytomas WHO III and 13 glioblastomas (WHO IV). The frequency of MGMT hypermethylation was observed in 53% astrocytoma III and GBM samples, and the highest hypermethylation was observed in astrocytoma II (61.4%) (Table 2).
Table 1.
Clinical features of the study population of 83 glioma patients.
| No. of Patients | All Patients | 83 |
|---|---|---|
| Age | Median | 36 |
| ≤40 | 51 | |
| >40 | 32 | |
| Gender | Male | 48 |
| Female | 35 | |
| Histopathological diagnosis | Astrocytoma II | 57 |
| Astrocytoma III | 13 | |
| Glioblastoma | 13 | |
| Treatment | Radiotherapy | 83 |
| Chemotherapy | 7 | |
| ECOG performance status | I | 56 |
| II | 25 | |
| III | 2 | |
| IV | 0 |
ECOG, Eastern Cooperative Oncology Group.
Table 2.
Distribution of the mutation in IDH1 and MGMT promoter methylation among the patients according to their histopathological diagnosis (WHO II, III, IV). OS, Overall Survival.
| Astrocytoma II (n = 57) (Median OS, Month) | Astrocytoma III (n = 13) (Median OS, Month) | Glioblastoma (n = 13) (Median OS, Month) | ||||
|---|---|---|---|---|---|---|
| MGMT− | MGMT+ | MGMT− | MGMT+ | MGMT− | MGMT+ | |
| IDH1 pR132H | 33 (n = 11) | 47 (n = 24) | 28 (n = 5) | 47 (n = 7) | 22 (n = 2) | 22.5 (n = 6) |
| IDH1 WT | 21 (n = 11) | 19 (n = 11) | 14 (n = 1) | 0 | 7.5 (n = 4) | 2 (n = 1) |
2.2. Molecular Evaluation
We previously demonstrated that the IDH1 c.G395A; p.R132H mutation is more frequently observed in higher-grade astrocytoma cases. In the subsequent studies, we showed that methylation correlates with the presence of the somatic mutation in IDH1:37/55 samples (67.27%), with the somatic mutation having the MGMT promoter methylated. On the other hand, 12/28 (42.85%) samples confirmed as wild-type IDH1 had their MGMT promoter methylated. Moreover, we evaluated the methylation status based on the comparison between the methylated (M) and unmethylated (U) amplicons for each of the 83 randomly selected DNA samples. With the M/U ratio of >1, the MGMT promoter methylation status was evaluated as high. Such a high methylation was observed in 53.6% of IDH1 pR132H samples (30/56 total) and in 18.5% (5/27) of Wild Type IDH1 samples. On the contrary, low methylation status (M/U < 1) was found mainly in samples with WT IDH1 (55.5%; 15/27) and was definitely less frequent (34%; 19/56) in tumor samples with the somatic mutation in the IDH1 codon 132 (Table 3).
Table 3.
Assessment of the methylation status based on the comparison between the methylated (M) and unmethylated (U) amplicons for each of the 83 randomly selected DNA samples.
| Methylation Ratio | M/U < 1 | M/U = 1 | M/U > 1 |
|---|---|---|---|
| IDH1 pR132H (n = 56) | n = 19 | n = 7 | n = 30 |
| IDH1 WT (n = 27) | n = 15 | n = 7 | n = 5 |
2.3. Overall Survival
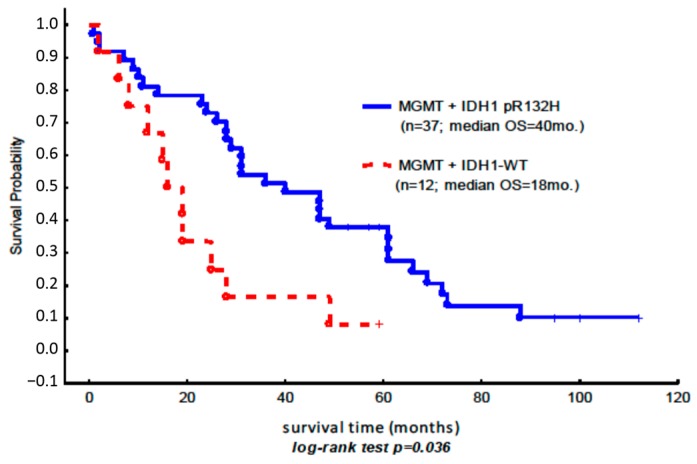
In the studied population, the patients with methylation in the MGMT gene promoter and the concurrent somatic mutation in the IDH1 gene (OS = 40 months) had a better prognosis than the subgroup of patients with the methylated MGMT gene alone (OS = 18 months) (Figure 1).
Figure 1.
Kaplan–Meier overall survival curves for the subgroup of patients with MGMT promoter methylation, in which the assessment of the methylation status based on the comparison between the methylated (M) and unmethylated (U) amplicons was equal to or higher than 1 (Table 3). The blue line shows the Kaplan–Meier overall survival curves for the subgroup of patients with the somatic mutation IDH1 p.R132H and the concurrent hypermethylation of MGMT (n = 37 (7 + 30)); the red line indicates the methylation of MGMT (n = 12 (7 + 5)) and the wild-type IDH1 gene (Table 3).
In a small subgroup of patients with astrocytoma anaplasticum (n = 7) characterized by the presence of the somatic mutation IDH1 p.R132H and the concurrent intermediate degree of MGMT methylation, the prognosis was particularly favorable (median OS = 47 months) compared to the other subgroups of patients (Table 2).
3. Discussion
The IDH1/2 mutations, 1p/19q status and MGMT promoter methylation are commonly recognized biomarkers in patients with gliomas [16,17,18]. Their role as prognostic or predictive biomarkers—that can be used in diagnostics—is growing. However, the question of how the outcome of radiation therapy depends on the combination of these two biomarkers remains open and is still debatable. It was demonstrated that in malignant gliomas of WHO grades III/IV with the IDH1 mutation, MGMT promoter methylation was associated with prolonged Progression-free survival (PFS) with chemotherapy ± radiation therapy (RT) or RT-only groups [19]. Moreover, a recent investigation (in which a molecular registry of 274 glioblastoma patients was used) showed that the poorest OS and PFS were observed in wild-type IDH1 glioblastomas with an unmethylated MGMT promoter [18]. Our results are in concordance with the above results: the best OS was observed in all patients, regardless of their histopathological type, who had hypermethylated MGMT and mutated IDH1, with a median of 33.5 months (n = 30). Furthermore, the status of the IDH1 mutation is more relevant for the prognosis than the methylation of MGMT.
In the subgroup with astrocytoma II, patients meeting the above criteria also had a considerably better prognosis than those not meeting those criteria (Table 2). In every histological subgroup, e.g., that with astrocytoma II, patients with a better prognosis were those with the IDH1 p.R132H mutation, regardless of the MGMT status, compared to those with wild-type IDH1. The most advantageous situation for RT treatment success seems to be the combination of the somatic mutation IDH1 p.R132H with the hypermethylation of MGMT. In an important report by Yang et al., a role of the mutated IDH1 gene in the resistance to temozolomide therapy is suggested. The article confirms the hypothesis in astrocytoma/glioblastoma cell lines, in which exogenous expression of the mutated IDH1 results in a three- to 10-fold increase in temozolomide resistance after long-term passage [18]. Unfortunately, our group of seven patients who received Temozolomide (TMZ) was too small for similar conclusions or any statistical evaluation (only three patients were diagnosed with astrocytoma II and 4 with GBM, and received chemotherapy; all of them, excluding one, had the mutation in IDH1 and the methylated MGMT; data not shown).
As clinical and molecular data were updated for 83 patients in 2015, we can discuss whether adding the MGMT status brings light to future clinical decisions. Previously, we demonstrated that the median overall survival observed in astrocytoma II patients, grouped on the basis of the presence of the IDH1 mutation only, was 24 months longer [12]. Currently, with the additional two years of clinical observation and division of the patients into groups with mutated/WT IDH1 and methylated/nonmethylated MGMT genes, we observed a 28-month-longer overall survival between the IDH1-mutated and MGMT-methylated group of 24 patients versus the IDH1 wild-type and MGMT-methylated group of 11 patients (Table 2). Furthermore, there was a 14-month difference in the median OS in the IDH1-mutated subgroup of patients with methylated MGMT versus the nonmethylated subgroup. We must also point it that a surprising proportion of IDH mutant gliomas are unmethylated—this may reflect the relative insensitivity of MS-PCR (methylation-specific polymerase chain reaction) compared to other methods, for example DNA pyrosequencing, for MGMT promoter methylation [20].
On the other hand, looking at the methylation level in the IDH1-WT subgroup of patients, we did not notice any significant variance between patients with different methylation status. In view of the above results, the somatic mutation IDH1 p.R132H and the concurrent methylation of the MGMT promoter has the most favorable prognosis.
In conclusion, we found a small population of patients with astrocytoma anaplasticum (WHO grade III), characterized by the presence of the somatic mutation IDH1 p.R132H and the concurrent hypermethylation of MGMT, for whom the prognosis was particularly favorable (median OS = 47 months). In the subgroup with astrocytoma II, patients meeting the above criteria also had a considerably better prognosis than those not meeting those criteria. The results indicate that the status of the IDH1 mutation is more relevant for the prognosis than the methylation of MGMT, and this is consistent within every histological subgroup. The most advantageous situation for treatment success seems to be the combination of the somatic mutation IDH1 p.R132H with the hypermethylation of MGMT. In view of the above results, patients not meeting those criteria may have to be subjected to more aggressive oncological treatments [14].
4. Materials and Methods
4.1. Patient Specimens
We randomly selected 83 patients with glioma (WHO II, III or IV) from a previously reviewed cohort of 139 patients with histopathologically confirmed glioma diagnosed between 2000 and 2011 [12]. Additionally, clinicopathological evaluation was performed in 2015 in order to update the data for further statistical analysis. All selected patients had a previously evaluated mutation status in codon 132 of isocitrate dehydrogenase 1 gene, and a novel MGMT promoter methylation analysis was conducted. The patients were divided into groups for the correlation of clinical and genetic data based on the molecular indications: the presence or absence of mutations in the IDH1 gene, as well as methylation status. The degree of tumor resection was assessed using an MRI examination conducted within 12 h after the surgery. In all patients participating in the study, a subtotal tumor resection was conducted. Subsequently, the patients were treated postoperatively using a routine IMRT radiation therapy with 6 MeV photons within four weeks after the procedure. The total dose was: 54 Gy for astrocytoma II, 60 Gy for astrocytoma III, 60 Gy for glioblastoma, divided into fractions of 2 Gy per day, with 24 h intervals between exposures, five days per week. The Planning Target Volume (PTV) and critical organs included all structures in accordance with the ICRU Report 62 [21].
The clinical characteristics of the patients are presented in Table 1.
4.2. Methylation Analysis
For methylation analysis, we used previously isolated DNA with a known status of IDH1 mutation. Then 1 μg of DNA was used as a template for bisulfite DNA conversion according to the manufacturer instructions EpiTect Bisulfite Kit, QIAGEN (Hilden, Germany). Subsequently, MS-PCR was conducted with sets of primers specific for the methylated and unmethylated sequence of the MGMT promoter, according to the previously described methylation-specific polymerase-chain-reaction assay [22,23,24]. The PCR products were evaluated using horizontal electrophoresis (stained with ethidium bromide and visualized under ultraviolet illumination). During methylation analysis, the intensity of the bands present on the gel (93 bp for unmethylated and 81 bp for methylated MGMT promoter sequence) was assessed using a five-point scale (from 1 to 5). As a control, ready-made samples of methylated DNA (EpiTect Control DNA, human, cat. no. 59665) and non-methylated DNA (EpiTect Control DNA, human, cat. no. 59655) were used.
4.3. Statistical Analysis
For statistical analysis, the STATISTICA (version 12.0; lic. no: JGVP502E256520ARCN-4) computer software (StatSoft, Inc., Tulsa, OK, USA). The association between overall survival and the status of the IDH1 (isocitrate dehydrogenase 1) c.G395A; p.R132H mutation and MGMT promoter methylation were estimated using the Kaplan–Meier method and assessed using the log-rank test. The results were considered as statistically significant at p < 0.05.
Author Contributions
Krzysztof Roszkowski, Jacek Furtak and Marzena A. Lewandowska conceived the study, analyzed and interpreted data, and wrote the manuscript; Jacek Furtak and Marzena A. Lewandowska designed the experiments; Marzena A. Lewandowska performed the experiments, Krzysztof Roszkowski, Jacek Furtak designed and performed clinical evaluation, Tadeusz Szylberg contributed biological materials (FFPE), Jacek Furtak contributed reagents, Bogdan Zurawski prepared patients characteristics and helped with interpretation of chemiotherapeutic results. Krzysztof Roszkowski, Jacek Furtak and Marzena A. Lewandowska revised the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lewandowska M.A., Costa F.F., Bischof J.M., Williams S.H., Soares M.B., Harris A. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am. J. Respir. Cell Mol. Biol. 2010;43:334–341. doi: 10.1165/rcmb.2009-0149OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boumil R.M., Ogawa Y., Sun B.K., Huynh K.D., Lee J.T. Differential methylation of Xite and CTCF sites in Tsix mirrors the pattern of X-inactivation choice in mice. Mol. Cell. Biol. 2006;26:2109–2117. doi: 10.1128/MCB.26.6.2109-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 4.Houshdaran S., Cortessis V.K., Siegmund K., Yang A., Laird P.W., Sokol R.Z. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nalejska E., Maczynska E., Lewandowska M.A. Prognostic and predictive biomarkers: Tools in personalized oncology. Mol. Diagn. Ther. 2014;18:273–284. doi: 10.1007/s40291-013-0077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das P.M., Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 8.Steemers F.J., Chang W., Lee G., Barker D.L., Shen R., Gunderson K.L. Whole-genome genotyping with the single-base extension assay. Nat. Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 9.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg A.P., Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem. Biophys. Res. Commun. 1983;111:47–54. doi: 10.1016/S0006-291X(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 11.Tuck-Muller C.M., Narayan A., Tsien F., Smeets D.F., Sawyer J., Fiala E.S., Sohn O.S., Ehrlich M. DNA hypomethylation and unusual chromosome instability in cell lines from ICF syndrome patients. Cytogenet. Cell Genet. 2000;89:121–128. doi: 10.1159/000015590. [DOI] [PubMed] [Google Scholar]
- 12.Lewandowska M.A., Furtak J., Szylberg T., Roszkowski K., Windorbska W., Rytlewska J., Jozwicki W. An analysis of the prognostic value of IDH1 (isocitrate dehydrogenase 1) mutation in Polish glioma patients. Mol. Diagn. Ther. 2014;18:45–53. doi: 10.1007/s40291-013-0050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reifenberger J., Reifenberger G., Liu L., James C.D., Wechsler W., Collins V.P. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am. J. Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 14.Buckner J.C., Shaw E.G., Pugh S.L., Chakravarti A., Gilbert M.R., Barger G.R., Coons S., Ricci P., Bullard D., Brown P.D., et al. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N. Engl. J. Med. 2016;374:1344–1355. doi: 10.1056/NEJMoa1500925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y., Killela P.J., Reitman Z.J., Rasheed A.B., Heaphy C.M., de Wilde R.F., Rodriguez F.J., Rosemberg S., Oba-Shinjo S.M., Nagahashi Marie S.K., et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson B.E., Mazor T., Hong C., Barnes M., Aihara K., McLean C.Y., Fouse S.D., Yamamoto S., Ueda H., Tatsuno K., et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang P., Zhang W., Wang Y., Peng X., Chen B., Qiu X., Li G., Li S., Wu C., Yao K., et al. IDH mutation and MGMT promoter methylation in glioblastoma: Results of a prospective registry. Oncotarget. 2015;6:40896–40906. doi: 10.18632/oncotarget.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wick W., Meisner C., Hentschel B., Platten M., Schilling A., Wiestler B., Sabel M.C., Koeppen S., Ketter R., Weiler M., et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81:1515–1522. doi: 10.1212/WNL.0b013e3182a95680. [DOI] [PubMed] [Google Scholar]
- 20.Li M.Y., Wang Y.Y., Cai J.Q., Zhang C.B., Wang K.Y., Cheng W., Liu Y.W., Zhang W., Jiang T. Isocitrate dehydrogenase 1 Gene Mutation Is Associated with Prognosis in Clinical Low-Grade Gliomas. PLoS ONE. 2015;10:e0130872. doi: 10.1371/journal.pone.0130872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International Commission on Radiation Units and Measurements . Prescribing, Recording, and Reporting Photon Beam Therapy. Volume 62 ICRU; Bethesda, MD, USA: 1999. Supplement to ICRU Report 50. [Google Scholar]
- 22.Herman J.G., Graff J.R., Myohanen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esteller M., Hamilton S.R., Burger P.C., Baylin S.B., Herman J.G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 24.Esteller M., Garcia-Foncillas J., Andion E., Goodman S.N., Hidalgo O.F., Vanaclocha V., Baylin S.B., Herman J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]