Abstract
Background
Plasmid expression is a popular method in studies of MVA pathway for isoprenoid production in Escherichia coli. However, heterologous gene expression with plasmid is often not stable and might burden growth of host cells, decreases cell mass and product yield. In this study, MVA pathway was divided into three modules, and two heterologous modules were integrated into the E. coli chromosome. These modules were individually modulated with regulatory parts to optimize efficiency of the pathway in terms of downstream isoprenoid production.
Results
MVA pathway modules Hmg1-erg12 operon and mvaS-mvaA-mavD1 operon were integrated into E. coli chromosome followed by modulation with promoters with varied strength. Along with activation of atoB, a 26% increase of β-carotene production with no effect on cell growth was obtained. With a combinatory modulation of two key enzymes mvas and Hmg1 with degenerate RBS library, β-carotene showed a further increase of 51%.
Conclusions
Our study provides a novel strategy for improving production of a target compound through integration and modulation of heterologous pathways in both transcription and translation level. In addition, a genetically hard-coded chassis with both efficient MEP and MVA pathways for isoprenoid precursor supply was constructed in this work.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0607-3) contains supplementary material, which is available to authorized users.
Keywords: Escherichia coli, Isoprenoid, β-carotene, MVA pathway, RBS
Background
Isoprenoids, also referred to as terpenes or terpenoids, are the most diverse class of natural products consisting of over 55,000 structurally different compounds, which has lots of applications in pharmaceuticals, nutraceuticals, cosmetics and food [1–4]. These valuable compounds are commonly isolated from plant, microbes and marine organisms. But the supply of these compounds has been limited by scarce resources from which they were originally extracted. Production by chemical synthesis is uneconomical due to the complex structure of these products [1]. For these reasons, microbial metabolic engineering has been explored in the past decade for isoprenoid production, including artemisinin, limonene paclitaxel (Taxol), astaxanthin, β-carotene and lycopene etc. [5–8].
Isoprenoids are all derived from two five-carbon building blocks called isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), which are synthesized either by the mevalonate (MVA) pathway in eukaryotes, archaea, and some bacteria or 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway in other prokaryotes and plastids in plants [2, 3, 9]. In the past 15 years, to increase the yield of terpene production, considerable effort has been focused on improving precursor supply by overexpression or deletion of upstream pathway genes, and altering global metabolic network by rational strategies or random mutagenesis [10–12]. MEP pathway has been engineered to increase IPP and DMAPP in E. coli for increased synthesis of carotenoids. Yuan et al. showed that four enzymes in the MEP pathway were rate limiting [13]. Similarly, when intrinsic dxs and idi were modulated by artificial modulation parts, the resultant strains had increased β-carotene production [14]. On the other hand, to address precursor IPP/DMAPP limitations in E. coli, heterologous MVA pathway genes were overexpressed using plasmid to improve isoprenoid production [15–19]. Higher isoprenoid production were achieved by strains equiped with the bottom portion of MVA pathway of Streptococcus pneumoniae, and cultured in media with MVA supplementation [18, 19]. Lycopene production of E. coli harboring the whole MVA pathway from Streptomyces sp. CL190 was two-fold higher than strain with only native MEP pathway [16]. However, high-level expression of mevalonate pathway enzymes might inhibit cell growth. Pitera et. al found that accumulation of MVA pathway intermediate 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) inhibited cell growth with overexpressed atoB, mvaS and hmg1 [20]. Mevalonate kinase (MK), encoded by erg12, was identified as another rate-limiting enzyme when the MVA pathway was used to increase in amorphadiene production [21].
To balance MVA pathway flux, it is necessary to express the HMG-CoA reductase and MK at a higher level to decrease accumulation of HMG-CoA, and to eliminate the rate-limiting step. In our previous work, β-carotene synthetic gene operon (crtEXYIB) from P. agglomerans CGMCC No. 1.2244 controlled by trc promoter and rrnB transcriptional terminator was integrated into wild type E. coli ATCC 8739 at ldhA site, resulting in strain QL002. The inducible promoter of crtEXYIB in QL002 was replaced with strong constitutive promoter M1-12 to obtain strain QL105. Activation of dxs, idi genes and crt operon in QL105 led to increase of β-carotene production, and the resulting stain was named CAR001 [14]. In this study, the MVA pathway genes were divided into three modules, (i) hmg1 and erg12, which need to be expressed at high level, (ii) atoB, which is an endogenous gene of E. coli, and (iii) mvaS, mvaA and mvaD1, which are the other genes of the MVA pathway (Fig. 1a). The objective of the study was to increase β-carotene production by integrating heterologous genes of MVA pathway into E. coli chromosome as two operons, modulate involved heterologous and endogenous genes individually, as well as illustrate relationship between gene expression level and β-carotene production in hyper producer strain.
Fig. 1.

Genes used for β-carotene synthesis in engineered E. coli strains, vector constructs and the two artificial MVA operons. a Genes involved in β-carotene Production via both MEP and MVA pathways. The abbreviations for enzymes and pathway intermediates are as follows: atoB, gene of acetoacetyl-CoA thiolase; mvaS, gene of HMG-CoA synthase; Hmg1, truncated HMG-CoA reductase; erg12, gene of mevalonate kinase; mvaA, gene of HMG-CoA reductase; mvaD1, gene of mevalonate pyrophosphate decarboxylase; idi, gene of IPP isomerase; ispA, FPP synthase; crtEYIB, β-carotene synthesis operon from Pantoea agglomerans; b Plasmid maps of vectors with heterologous MVA genes; c Integrated artificial MVA pathway operons in E. coli chromosome at pflB and frdB sites
Methods
Strains, medium and growth conditions
Strains used in this study are listed in Additional file 1: Table S6. During strain construction, cultures were grown aerobically at 30, 37, or 39 °C in Luria broth (per liter: 10 g Difco tryptone, 5 g Difco yeast extract and 5 g NaCl). For β-carotene production, single colonies were picked from the plate and inoculated into 15 × 100 mm tubes containing 4 ml LB with or without 34 mg/l chloramphenicol and 100 mg/l ampicillin, and grown at 30 °C and 250 rpm overnight. Seed culture was then inoculated into 100 ml flask containing 10 ml LB, with or without 34 mg/l chloramphenicol and 100 mg/l ampicillin (with an initial OD600 of 0.05), and grown at 30 °C and 250 rpm. After 24 h growth, cells were collected for measurement of β-carotene production. For strains using trc promoter for induction of MVA pathway genes, 1 mM IPTG was added for induction 3 h after inoculation, followed by 21 h growth [14].
Construction of plasmids for expressing MVA pathway genes
All plasmids used in this study are listed in Additional file 1: Table S5. The Hmg1 and erg12 genes, which need be expressed at high level, were placed in one operon; while mvas, mvaA and mavD1 genes were put in another operon (Fig. 1a). Hmg1 and erg12 were isolated by PCR with Pfu DNA polymerase (NEB) from chromosomal DNA of Saccharomyces cerevisiae. Individual genes were spliced together (sequence named as He) using overlapping extensions from primers HMG1-XmaI-f, HMG1-r, ERG12-f, ERG12-SalI-r (Additional file 1: Table S1). mvas, mvaA and mavD1 genes were isolated and spliced together (sequence named as Mmm) by overlapping extensions from primers ERG13-BamHI-f, ERG13-r, ERG8-f, ERG8-r, MVD1-f and MVD1-SalI-r (Additional file 1: Table S1). Plasmid pTrc99A-M were digested by XmaI and SalI ligated by T4 DNA ligase, and transformed into Trans T1 competent cells (Transgen, Beijing, CN). Plasmid carrying Hmg1 and erg12 genes was screened, selected and designated as pTrc99A-M-He (Fig. 1b). mvas, mvaA and mavD1 genes were inserted into pACYC184-M at BamHI and SalI site using the same method, and the plasmid was designated as pACYC184-M-Mmm.
Integration of MVA genes into E. coli chromosome
A two-step homologous recombination method [22, 23] was used to integrate Hmg1-erg12 operon into E. coli CAR001 [14] at pflB site, and the mvaS-mvaA-mavD1 operon at frdB site (Fig. 1c). pflB gene was amplified from genomic DNA of E. coli ATCC 8739 using primer set pflB-up/pflB-down (Additional file 1: Table S1), and cloned into pEASY-Blunt (Transgen, Beijing, CN) to produce plasmid pXZ014 (Additional file 1: Table S5). A 1000-fold dilution of this plasmid DNA served as template for inside-out amplification using the pflB-1/pflB-2 primer set (Additional file 1: Table S1). The resulting 4735 bp fragment containing replicon was ligated with cat-sacB cassette from pXZ-CS [24] to produce pXZ015C (Additional file 1: Table S5). PCR fragment amplified from pXZ014 was ligated with the Hmg1-erg12 operon, which was amplified from pTrc99A-M-He with prime set HMG1-XmaI-f/99A-r (Additional file 1: Table S1), to produce plasmid pQL003-He (Additional file 1: Table S5). A two-step recombination method was developed for markerless recombination of Hmg1-erg12 operon expression in CAR001 at pflB site (Additional file 1: Figure S1). In the first recombination, cat-sacB cassette was amplified from pXZ015C with primer set pflB-up/pflB-down (Additional file 1: Table S1), treated with DpnI, and electroporated into competent CAR001 with pKD46. After overnight growth on LB plate with 34 mg/l chloramphenicol and 100 mg/l ampicillin at 30 °C, several colonies were picked for PCR verification using primer set cat-up/pflB-down (Additional file 1: Table S1). In the second recombination, Hmg1-erg12 operon with rrnB terminator were amplified from pQL003-He with a same primer set pflB-up/pflB-down (Additional file 1: Table S1), and used to replace cat-sacB cassette by selection for resistance to sucrose. Cells containing sacB gene accumulated levan during incubation with sucrose and were eliminated. With this mechanism, survived recombinants were highly enriched for colonies without cat-sacB cassette [22, 23]. The resulting strain was designated as CAR006. Plasmid pQL006-Mmm was constructed using the same method as pQL003-He. mvaS-mvaA-mavD1 operon was inserted into frdB site using the same method as integration of Hmg1-erg12 operon. The primers used are listed in Additional file 1: Table S1. Plasmids are listed in Additional file 1: Table S5.
Two-step recombination method for markerless modulation of gene expression
A two-step recombination method was used for markerless modulation of gene expression, which was beneficial for multiple rounds of genome editing [25]. Hmg1-erg12 operon was first modulated in CAR006 using this method (Additional file 1: Figure S2). In the first recombination, cat-sacB cassette was amplified with primer set pflB-up-cat/Hmg1-sacB-down (Additional file 1: Table S1) for insertion at upstream of Hmg1-erg12 operon. In the second recombination, different artificial regulatory parts (M1-46 and M1-93, whose strengths were 2.5 and 5 times of induced E. coli lacZ when cultivated in LB medium [26]) were amplified with a same primer set pflB-up-P/Hmg1-RBS-down (Additional file 1: Table S1) and used to replace cat-sacB cassette by selection for resistance to sucrose. Markerless modulation of other genes was the same as Hmg1-erg12 operon, and primers used are listed in Additional file 1: Table S1. Resulting strains are listed in Additional file 1: Table S6.
One-step recombination method for modulating mvas and Hmg1 expression with RBS library
mvas of CAR012 was modulated with RBS library using a one-step recombination method as described previously [26, 27]. Regulatory parts M1-93 was PCR template for RBS library. The artificial regulatory parts with different resistance gene were constructed to be used as templates for RBS library construction, so that different genes can be modulated in a one strain with resistance genes intact. For construction of M1-cam-93, a chloramphenicol resistance gene fragment followed by 50 nucleotides homologous sequence of FRT in M1-93 was amplified from plasmid pXZ-CS using primer set FRT-cam-up/FRT-cam-down (Additional file 1: Table S1), and electroporated into competent M1-93 with pKD46. After overnight growth on LB plate with 34 mg/l chloramphenicol, several colonies were picked for PCR verification using primer set Cat-g-up/LacZ-373 (Additional file 1: Table S1). The obtained strain was designated M1-cam-93, as listed in Additional file 1: Table S6.
Apramycin resistance regulatory parts construction was the same as M1-cam-93, primers are listed in Additional file 1: Table S1, and the resultant strain was designated as M1-Apr-93 (Additional file 1: Table S6).
A one-step homologous recombination method was used to further modulate mvas and Hmg1 gene with RBS Library. For modulating mvas, DNA fragments were amplified from genomic DNA of M1-cam-93 with primer set frdB-up-FRT/Mvas-RBSL-down (Additional file 1: Table S2). Primer Mvas-RBSL-down was degenerated with six bp random sequence at RBS site (NNNNNNYC) [28]. The resulting PCR product was a degenerated RBS library, and electroporated into competent E. coli CAR012 with pKD46. After overnight growth on LB plate with 34 mg/l chloramphenicol, fifteen colonies were picked for PCR verification using primer set Cat-g-up/MvaS-350-r (Additional file 1: Table S1). Fifteen correct colonies were randomly selected and designated from mvaS-RBSL-1 to mvaS-RBSL-15 (Additional file 1: Table S6), β-carotene production of which were measured (Additional file 1: Table S6). Hmg1 was modulated with RBS Library in the same way of the mvas, except that the RBS library was PCR amplified from M1-Apr-93 with apramycin resistance gene. Primers are listed in Additional file 1: Table S1 and strains are listed in Additional file 1: Table S6.
To further increase the β-carotene production, Hmg1 gene of the Hmg1-erg12 operon in the best strain mvaS-RBSL-13 was also modulated with the RBS Library. In addition, a ten-colony-strain mixture randomly selected from mvaS-RBS Library was also used as parent strain pool for Hmg1 modulation. Fifteen colonies from each resulted library were randomly selected for measuring β-carotene production (Additional file 1: Table S6).
Measurement of β-carotene production
Production of β-carotene was quantified by measuring absorption of acetone-extracted β-carotene at 453 nm as previously reported [14]. One millililitre of cells cultured for β-carotene production were harvested by centrifugation at 4000 rpm for 10 min, suspended in acetone (1 ml), and incubated at 55 °C for 15 min in dark. Samples were then centrifuged at 14,000 rpm for 10 min to obtain supernatant containing β-carotene, whose absorbance was measured at 453 nm using a Shimadzu UV-2550 spectrophotometer (Shimadzu, Kyoto, Japan). The β-carotene production was normalized to the cell density. The results represented the mean ± SD of three independent experiments.
Calculation of mvaS and Hmg1 RBS strength of strains from Re-modulation libraries
RBS sequences along with their context region of mvaS and Hmg1 in representative strains were sequenced. Their theoretical RBS strength characterized by the value of translation initiation rate was calculated with the RBS library calculator [29, 30].
Results
β-carotene production by E coli with heterologous expression of MVA pathway
To eliminate growth inhibition caused by imbalanced overexpression of MVA pathway gene, the heterologous MVA pathway genes from S. cerevisiae were separated into two portions and cloned into plasmids pTrac99A-M and pACYC184-M, respectively. pACYC184-M-Mmm containing mvaS-mvaA-mavD1 operon and pTrac99A-M-He containing Hmg1-erg12 operon were first transformed into QL105 and CAR001 [14]. Optimization of MEP pathway in QL105 led to increased β-carotene production, and the resulting strain was CAR001 (Fig. 2). Both strains were used as parent strains to compare effects of addition of MVA pathway in present of native and engineered MEP pathway. After culturing with IPTG induction, β-carotene yields of the resulting strain QL105 (pACYC184-M-Mmm, pTrac99A-M-He) and CAR001 (pACYC184-M-Mmm, pTrac99A-M-He) were 1.45-, 1.47-fold of the initial strain (Fig. 2b). Meanwhile, the expression of MVA pathway genes on plasmids caused a 12 and 27% decrease in cell mass for QL105 and CAR001, respectively (Fig. 2a). The result showed that the β-carotene production of the native MEP pathway did not affect introduction of heterologous MVA pathway, in terms of β-carotene production. Growth defect was observed in engineered strains, which were probably due to plasmid burden or imbalanced MVA pathway. In addition, 90% of the plasmid bearing strains lost resistance after 24 h of culturing, indicating loss of their plasmids.
Fig. 2.
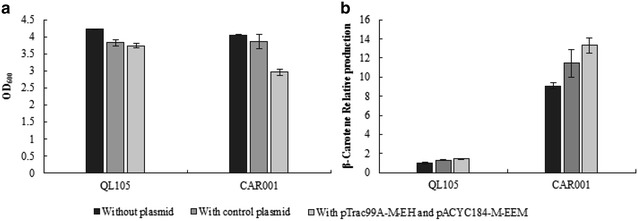
Cell mass and relative β-carotene production by E. coli strains with or without MVA pathway. a Cell mass. b Relative β-carotene production. β-carotene yield was compared to the parent strain QL105 and CAR001. Three repeats were performed for each strain, and the error bars represented standard deviation
Improved β-carotene production by Integration MVA pathway genes into E. coli and expression modulation
In order to eliminate possible growth burden caused by plasmids and obtain genetically stable strains, the Hmg1-erg12 operon without promoter was integrated into chromosome of CAR001 at pflB site, resulting in strain CAR006. Then, this operon was modulated with two regulatory parts (M1-46 and M1-93), resulting in strains CAR007 and CAR008 (Additional file 1: Table S6). Plasmid pACYC184-M-Mmm was transformed into CAR007 and CAR008 to complement MVA pathway. In the resulting strains, the cell mass decreased by nearly 20%, while the β-carotene yield was 1.26- and 1.17-fold that of strain CAR001 respectively (Table 1). To integrate the whole MVA pathway into E. coli chromosome, operon of mvaS-mvaA-mavD1 genes without promoter was integrated into CAR007 at frdB site, resulting in strain CAR009. This operon was then modulated with two regulatory parts (M1-46 and M1-93), resulting strains CAR010 and CAR011 (Additional file 1: Table S6). In the resulting strains, the cell mass decreased by nearly 20%, and the β-carotene yield was only 1.03- and 1.02-fold that of CAR001 respectively (Table 1), suggesting imbalanced expression of the MVA pathway in these two strains.
Table 1.
Integration and modulation of MVA pathway genes for improving β-carotene production
| Strainsa | OD600 | ODb453 | OD453/OD600 | Increase of β-carotene yield- CAR001 |
|---|---|---|---|---|
| QL105 | 4.22 ± 0.15 | 0.27 ± 0.01 | 0.06 ± 0.01 | |
| CAR001 | 4.05 ± 0.10 | 2.36 ± 0.06 | 0.58 ± 0.00 | 1.00 ± 0.00 |
| CAR007 (184-EEM) | 3.31 ± 0.05 | 2.43 ± 0.13 | 0.73 ± 0.04 | 1.26 ± 0.06 |
| CAR008 (184-EEM) | 3.32 ± 0.07 | 2.26 ± 0.09 | 0.68 ± 0.04 | 1.17 ± 0.07 |
| CAR010 | 4.05 ± 0.05 | 2.45 ± 0.07 | 0.60 ± 0.01 | 1.03 ± 0.02 |
| CAR011 | 4.11 ± 0.07 | 2.43 ± 0.05 | 0.59 ± 0.02 | 1.02 ± 0.03 |
| CAR012 | 4.01 ± 0.07 | 2.93 ± 0.20 | 0.73 ± 0.05 | 1.26 ± 0.09 |
| CAR013 | 3.91 ± 0.11 | 2.41 ± 0.12 | 0.62 ± 0.03 | 1.07 ± 0.05 |
aThree repeats were performed for each strain, and the error bars represented standard deviation
bAcetone-extracted β-carotene solution was concentrated 2 times for measuring the absorption at 453 nm
Improved β-carotene production by modulation of atoB gene expression
Escherichia coli is known to contain only low levels of acetoacetyl-CoA [20, 31], which may be the reason for the low β-carotene production of CAR010 and CAR011. To improve β-carotene production, atoB of CAR010 was modulated with three regulatory parts (M1-46, M1-37 and M1-93), which were characterized constitutive promoters with different transcription efficiency [26]. The best strain CAR012 with atoB expressed by M1-37 had a β-carotene production 1.26-fold that of CAR001 (Table 1).
Further improvement by re-modulation of MVA pathway genes
Although modulation of all the MVA pathway genes in strain CAR012 with regulatory parts led to an increase in β-carotene production, the increase was only 26%, and the regulatory parts of two operons of the MVA pathway were the same (M1-46). Strengths of these regulatory parts might not be optimal for β-carotene production, suggesting a possibility of obtaining a higher β-carotene production by modulating the expression of these important genes with different artificial regulatory parts.
With this strategy, mvas gene of the mvas-mvaA-mavD1 operon was modulated firstly with an RBS library. Fifteen colonies were randomly selected from the library for measuring β-carotene production (Additional file 1: Table S6), the β-carotene yield ranged from 1.07 to 1.36 times and cell mass ranged from 0.94 to 1.15 times of CAR001 (Fig. 3). The best strain mvaS-RBSL-13, produced 8% higher than CAR012 and 36% higher than CAR001.
Fig. 3.
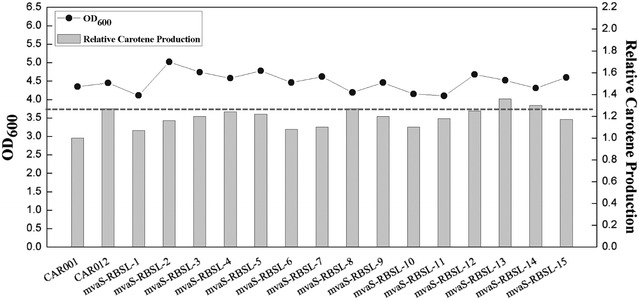
Cell mass and relative β-carotene production by E. coli strains after modulating mvas expression with RBS library in CAR012
To further increase the β-carotene production, Hmg1 gene of the Hmg1-erg12 operon in the best strain mvaS-RBSL-13 was also modulated with the RBS library. In addition, a ten-colony-strain mixture randomly selected from mvaS-RBS Library was used as a parent strain pool for Hmg1 modulation. Two new libraries were obtained, and fifteen colonies from each library were randomly selected for measuring β-carotene production (Additional file 1: Table S6). The resulting β-carotene yield ranged from 0.93 to 1.51 times and cell mass ranged from 0.90 to 0.98 times of CAR001 (Fig. 4).
Fig. 4.

Cell mass and relative production by E. coli strains with modulation of Hmg1 expression with RBS library from mvaS-RBSL-13 or mvaS-RBSL-mix. Gray Square recombinant E. coli strains derived from mvaS-RBSL-13; Black square recombinant E. coli strains derived from the mvaS-RBSL-mix pool
Gray columns in Fig. 4 represent E. coli strains from RBS modulated library of mvaS-RBSL-13, which had β-carotene yield ranged from 0.93 to 1.34 times of CAR001. Most strains had a titer between 0.93 and 1.09 times of CAR001. Black columns represent strains from the RBS modulated library of mvaS-RBSL-mix. Their β-carotene yield ranged from 1.04 to 1.51 times of CAR001, and were mostly between 1.09 and 1.51 times of CAR001. This result suggested that a RBS modulated library from a mixture of strains might be more ideal than those from a single regulatory part, probably due to having more possible regulation patterns. The cell mass of best strains Hmg1-RBSL-M10 was 0.99 times of that of CAR001, which demonstrated that balanced MVA pathway restored growth of the strain (Additional file 1: Table S3). In addition, β-carotene production of best strain Hmg1-RBSL-M10 increased 51% compared with CAR001 (Figs. 4, 5).
Fig. 5.
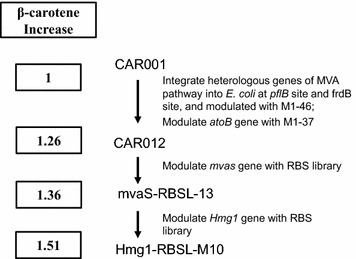
Diagram summarizing steps in the metabolic engineering of E. coli for β-carotene production in this work
Hmg1-RBSL-M10 culture after 48 h fermentation was spread on LB agar plates for measurement of genetic stability. It was found that 100% colonies showing orange color which was the color of β-carotene. This demonstrated the stability of strains with chromosomal integrated heterologous genes. In comparison, when plasmids were used as expression vector we found that 90% strains lost resistance after 24 h of culturing.
Analysis of mvaS and Hmg1 RBS strength of strains from re-modulation libraries
RBS strength was analyzed with the RBS calculator [29, 30] to find the relationship between mvaS and Hmg1 expression status and β-carotene production. Calculated RBS strength was represented by the translation initiation rate and listed in Table 2 and Additional file 1: Table S7. The RBS strength is by no means a very accurate measurement of the expression status of mvaS and Hmg1, however, could give a good estimate of general trend of and optimized regulation pattern of MVA pathway. According to the data, a medium to low expression of both mvaS and Hmg1 improved the efficiency of MVA pathway. The best strain carries the third weakest mvaS RBS and the middle level of hmg1 RBS (Additional file 1: Table S7). Since mvaS and Hmg1 are closest to the promoter and had higher transcription than other genes, the weaker RBSs might help to balance their expression with other genes in the same operon, which suggested an overall balanced expression of MVA pathway genes was beneficial with its efficiency.
Table 2.
Calculated strength of mvaS and Hmg1 RBS from strains from re-modulation libraries
| Strains | Sequence of mvaS RBS | Strengtha of mvaS RBS | Sequence of Hmg1 RBS | Strengtha of Hmg1 RBS | INCREASE of β-carotene yield- CAR001 |
|---|---|---|---|---|---|
| Hmg1-RBSL-2 | AGGAGAGAGAGG | 408 | AGGAGAAACAAC | 3877 | 0.93 |
| Hmg1-RBSL-3 | AGGAGAGAGAGG | 408 | AGGAGGGAAAAA | 9975 | 0.98 |
| Hmg1-RBSL-6 | AGGAGAGAGAGG | 408 | AGGAGAACAGCT | 3238 | 1.00 |
| Hmg1-RBSL-M3 | AGGAGGGTATCG | 2159 | AGGAGGATAAAG | 7965 | 1.09 |
| Hmg1-RBSL-11 | AGGAGAGAGAGG | 408 | AGGAGGAAAAAC | 12,720 | 1.11 |
| Hmg1-RBSL-M1 | AGGAGACAAAAG | 1099 | AGGAGACAAAAG | 2656 | 1.26 |
| Hmg1-RBSL-15 | AGGAGAGAGAGG | 408 | AGGAGGAAAAGG | 4855 | 1.31 |
| Hmg1-RBSL-M10 | AGGAGAGGACTG | 612 | AGGAGAACAGCT | 3238 | 1.52 |
Discussion
In MVA pathway, accumulation of the intermediate 3-hydroxy-3-glutaryl-CoA (HMG-CoA) was found to cause inhibition of cell growth [20]. It is necessary to reduce expression of mvaS and increase Hmg1, which encodes HMG-CoA reductase, for reduction of HMG-CoA accumulation. The mevalonate should be rapidly converted into mevalonate-5 phosphate by mevalonate kinase (MK) or it would cross cell membrane and diffuse into medium [20]. MK is also one of the rate-limiting enzymes [21]. To facilitate modulation of these genes and balance their expression to keep pathway intermediates at a proper level in host cells, MVA pathway was divided and engineered as three separately expressed operons in this work. This strategy was proved to be effective with increased β-carotene production.
Plasmid overexpression of the MVA pathway genes was previously utilized to increase isoprenoid production in E. coli, but caused metabolic burden in host and led to reduced cell growth [32]. In this study, the five heterologous genes of MVA pathway were first cloned into two compatible plasmids, and expressed in QL105 and CAR001 strains. The growth of strains with plasmid had one quarter decrease compared to the initial strains (Fig. 2). In our work, the growth burden was eliminated by integration and of these genes into E. coli chromosome followed by modulation. This result indicated that overexpression of heterologous genes with chromosome integrated form was a better strategy than plasmid expression.
Strong promoters have been generally used to overexpress genes for improved carotenoid production [13, 33, 34]. However, they might not be optimal to obtain maximum metabolic flux towards desired products [35]. Previous studies indicated that better production could be obtained by modulating genes with regulatory parts of varied strengths. dxs gene of MEP pathway was modulated by several artificial promoters, and an optimal strength for improved lycopene production was identified [36]. Similarly, glyceraldehyde-3-phosphate dehydrogenase gene (gapA) was modulated by three artificial promoters to achieve an optimal strength for glycerol production [37]. In both cases, the optimal expression strength was not the strongest one. A similar result was obtained in this work. In the strain with highest β-carotene production, two MVA operons (mvas-mvaD-mvaA and Hmg1-erg12) were expressed with a medium strength promoters M1-46, and atoB was expressed with a weak promoter M1-37. In addition, key enzymes mvas and Hmg1 were under control of medium to weak RBSs. Thus, previous reports and our results suggested an overall balanced expression of MVA pathway genes was relatively efficient, which might also be true for most heterologous pathways.
One of the most important research subjects of metabolic engineering is pursuing a balanced metabolic pathway. In recent years, several combinatorial pathway engineering strategies and methods were established [38, 39]. In this work we dedicated to develop and apply a relatively simple method for pathway balancing and production improvement. Firstly, operons were modulated with limited number of promoters, which were previously defined; then genes within an operon were combinatorially modulated with RBS libraries for multiple rounds. To reduce library size and simplify screening, limited number of strains from previous round were picked as parent strains for next round of modulation with RBS library. By compromising with the goal of finding the most balanced pathways, this method requires minimal lab work for finding a reasonably efficient pathway. In this work, modulating mvaS with RBS libraries led to 36% improvement of β-carotene yield versus CAR001, and 8% improvement of β-carotene yield versus parent strain (Figs. 3, 5; Additional file 1: Table S2). This result illustrated that modulating gene with a library of regulatory parts led to a wider variation in expression strength than using limited regulatory parts with fixed strengths, provided more possibilities to obtain optimal efficiency for pathways. Furthermore, a comparison was made between modulating gene expression from a single strain or from a pool of mixed strains in modulating Hmg1 RBSs. β-carotene production of strains derived from a pool was generally higher than that from a single strain, probably due to similar reasons as above, that more combinations were achieved with parent strains in a mixture pool. This result inspired us to develop methods to build metabolic engineering libraries with more random regulatory parts in higher mathematic dimensions to achieve better possibilities for optimal expression combinations.
In this study, a genetically stable E. coli strain with high β-carotene production was obtained by combined engineering of MEP and MVA (Fig. 5). A genetically stable chassis with activated MEP and MVA pathways for IPP and DMAPP precursor supply to produce various terpenes was obtained. Our study provided a novel strategy for improving production of a target compound through integration and subsequent modulation of heterologous pathways by changes in both promoters and RBSs.
Conclusions
In this study, MVA pathway was divided into three modules, integrated into the E. coli chromosome, and individually modulated with regulatory parts to optimize efficiency of the pathway in terms of downstream isoprenoid production. Hmg1-erg12 operon and mvaS-mvaA-mavD1 operon were integrated into E. coli chromosome followed by modulation with promoters with varied strength, resulting in a 26% increase of β-carotene production with no effect on cell growth. With a combinatory modulation of two key enzymes mvas and Hmg1 with degenerate RBS library, β-carotene showed a further increase of 51%.
Our study provides a novel strategy for improving production of a target compound through integration and modulation of heterologous pathways in both transcription and translation level. In addition, a genetically hard-coded chassis with both efficient MEP and MVA pathways for isoprenoid precursor supply was constructed in this work.
Authors’ contributions
YL, ZC and LQ planned and performed experiments, analyzed and interpreted the data. ZX, ZC and YL supervised the study, designed experiments and analyzed and interpreted the results. YL, LQ and BC wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All supporting data is present in the article and the supplemental material documents. Specifically, plasmid maps and DNA sequence data are repent in Additional file 2.
Funding
This research was supported by grants from National Natural Science Foundation of China (31522002), Natural Science Foundation of Tianjin (15JCYBJC49400), Tianjin Key Technology RD program of Tianjin Municipal Science and Technology Commission (11ZCZDSY08600).
Abbreviations
- LB
lysogeny broth
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- RBS
ribosome-binding site
Additional files
Additional file 1: Table S1. Primers used in this work. Table S2. Modulating genes of mvaS-mvaA-mavD1 operon for improving β-carotene production. Table S3. Modulating genes of Hmg1-erg12 operon for improving β-carotene production. Table S4. Sequences of representative artificial regulatory parts. Table S5. Plasmids used in this work. Table S6. Escherichia coli strains used in this work. Table S7. Calculated strength of mvaS and Hmg1 RBS, RBS sequence and relative β-carotene yield of strains from Re-modulation libraries. Figure S1. Two-step recombination method for inserting Hmg1-erg12 operon in E. coli chromosome. Figure S2. Two-step recombination method for modulating gene expression in E. coli chromosome by different artificial regulatory parts.
Additional file 2. Additional plasmid profiles and gene sequences.
Footnotes
Lijun Ye and Chunzhi Zhang contributed equally to this work
Contributor Information
Lijun Ye, Email: 705458110@qq.com.
Chunzhi Zhang, Email: zhangcz@dlpu.edu.cn.
Changhao Bi, Email: bi_ch@tib.cas.cn.
Qingyan Li, Email: li_qy@tib.cas.cn.
Xueli Zhang, Email: zhang_xl@tib.cas.cn.
References
- 1.Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm. 2008;5:167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- 2.Das A, Yoon SH, Lee SH, Kim JY, Oh DK, Kim SW. An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl Microbiol Biotechnol. 2007;77:505–512. doi: 10.1007/s00253-007-1206-3. [DOI] [PubMed] [Google Scholar]
- 3.Lee PC, Schmidt-Dannert C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl Microbiol Biotechnol. 2002;60:1–11. doi: 10.1007/s00253-002-1101-x. [DOI] [PubMed] [Google Scholar]
- 4.Roberts SC. Production and engineering of terpenoids in plant cell culture. Nat Chem Biol. 2007;3:387–395. doi: 10.1038/nchembio.2007.8. [DOI] [PubMed] [Google Scholar]
- 5.Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Lemuth K, Steuer K, Albermann C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb Cell Fact. 2011;10:29. doi: 10.1186/1475-2859-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang CW, Oh MK, Liao JC. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol Bioeng. 1999;62:235–241. doi: 10.1002/(SICI)1097-0290(19990120)62:2<235::AID-BIT14>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Willrodt C, David C, Cornelissen S, Buhler B, Julsing MK, Schmid A. Engineering the productivity of recombinant Escherichia coli for limonene formation from glycerol in minimal media. Biotechnol J. 2014;9:1000–1012. doi: 10.1002/biot.201400023. [DOI] [PubMed] [Google Scholar]
- 9.Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14:233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer WR, Liao JC. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol Prog. 2001;17:57–61. doi: 10.1021/bp000137t. [DOI] [PubMed] [Google Scholar]
- 11.Kajiwara S, Fraser PD, Kondo K, Misawa N. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem J. 1997;324(Pt 2):421–426. doi: 10.1042/bj3240421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SW, Keasling JD. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng. 2001;72:408–415. doi: 10.1002/1097-0290(20000220)72:4<408::AID-BIT1003>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 13.Yuan LZ, Rouviere PE, Larossa RA, Suh W. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng. 2006;8:79–90. doi: 10.1016/j.ymben.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, Zhang X, Ma Y. Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab Eng. 2013;17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 16.Vadali RV, Fu Y, Bennett GN, San KY. Enhanced lycopene productivity by manipulation of carbon flow to isopentenyl diphosphate in Escherichia coli. Biotechnol Prog. 2005;21:1558–1561. doi: 10.1021/bp050124l. [DOI] [PubMed] [Google Scholar]
- 17.Yoon SH, Lee SH, Das A, Ryu HK, Jang HJ, Kim JY, Oh DK, Keasling JD, Kim SW. Combinatorial expression of bacterial whole mevalonate pathway for the production of beta-carotene in E. coli. J Biotechnol. 2009;140:218–226. doi: 10.1016/j.jbiotec.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Yoon SH, Lee YM, Kim JE, Lee SH, Lee JH, Kim JY, Jung KH, Shin YC, Keasling JD, Kim SW. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng. 2006;94:1025–1032. doi: 10.1002/bit.20912. [DOI] [PubMed] [Google Scholar]
- 19.Yoon SH, Park HM, Kim JE, Lee SH, Choi MS, Kim JY, Oh DK, Keasling JD, Kim SW. Increased beta-carotene production in recombinant Escherichia coli harboring an engineered isoprenoid precursor pathway with mevalonate addition. Biotechnol Prog. 2007;23:599–605. doi: 10.1021/bp070012p. [DOI] [PubMed] [Google Scholar]
- 20.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng. 2007;9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Anthony JR, Anthony LC, Nowroozi F, Kwon G, Newman JD, Keasling JD. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2009;11:13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101:881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Jantama K, Moore JC, Shanmugam KT, Ingram LO. Production of l-alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2007;77:355–366. doi: 10.1007/s00253-007-1170-y. [DOI] [PubMed] [Google Scholar]
- 24.Tan Z, Zhu X, Chen J, Li Q, Zhang X. Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improvement of succinate production. Appl Environ Microbiol. 2013;79:4838–4844. doi: 10.1128/AEM.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi A, Zhu X, Lu J, Zhang X, Ma Y. Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng. 2013;16:1–10. doi: 10.1016/j.ymben.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Tang J, Liu Y, Zhu X, Zhang T, Zhang X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl Microbiol Biotechnol. 2012;93:2455–2462. doi: 10.1007/s00253-011-3752-y. [DOI] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Zhu X, Tan Z, Xu H, Tang J, Xiao D, Zhang X. Activating C4-dicarboxylate transporters DcuB and DcuC for improving succinate production. Appl Microbiol Biotechnol. 2014;98:2197–2205. doi: 10.1007/s00253-013-5387-7. [DOI] [PubMed] [Google Scholar]
- 29.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauli G, Overath P. ato Operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur J Biochem. 1972;29:553–562. doi: 10.1111/j.1432-1033.1972.tb02021.x. [DOI] [PubMed] [Google Scholar]
- 32.Park YC, Kim SJ, Choi JH, Lee WH, Park KM, Kawamukai M, Ryu YW, Seo JH. Batch and fed-batch production of coenzyme Q10 in recombinant Escherichia coli containing the decaprenyl diphosphate synthase gene from Gluconobacter suboxydans. Appl Microbiol Biotechnol. 2005;67:192–196. doi: 10.1007/s00253-004-1743-y. [DOI] [PubMed] [Google Scholar]
- 33.Albermann C, Trachtmann N, Sprenger GA. A simple and reliable method to conduct and monitor expression cassette integration into the Escherichia coli chromosome. Biotechnol J. 2010;5:32–38. doi: 10.1002/biot.200900193. [DOI] [PubMed] [Google Scholar]
- 34.Chiang CJ, Chen PT, Chao YP. Replicon-free and markerless methods for genomic insertion of DNAs in phage attachment sites and controlled expression of chromosomal genes in Escherichia coli. Biotechnol Bioeng. 2008;101:985–995. doi: 10.1002/bit.21976. [DOI] [PubMed] [Google Scholar]
- 35.Pfleger BF, Pitera DJ, Newman JD, Martin VJ, Keasling JD. Microbial sensors for small molecules: development of a mevalonate biosensor. Metab Eng. 2007;9:30–38. doi: 10.1016/j.ymben.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meynial-Salles I, Cervin MA, Soucaille P. New tool for metabolic pathway engineering in Escherichia coli: one-step method to modulate expression of chromosomal genes. Appl Environ Microbiol. 2005;71:2140–2144. doi: 10.1128/AEM.71.4.2140-2144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE. Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. Nucleic Acids Res. 2013;41:10668–10678. doi: 10.1093/nar/gkt809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu P, Rizzoni EA, Sul SY, Stephanopoulos G. Improving metabolic pathway efficiency by statistical model-based multivariate regulatory metabolic engineering. ACS Synth Biol. 2016;11:460–469. doi: 10.1021/acssynbio.6b00187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supporting data is present in the article and the supplemental material documents. Specifically, plasmid maps and DNA sequence data are repent in Additional file 2.


