Extended Data Figure 7. Conservation of PAM recognition elements.
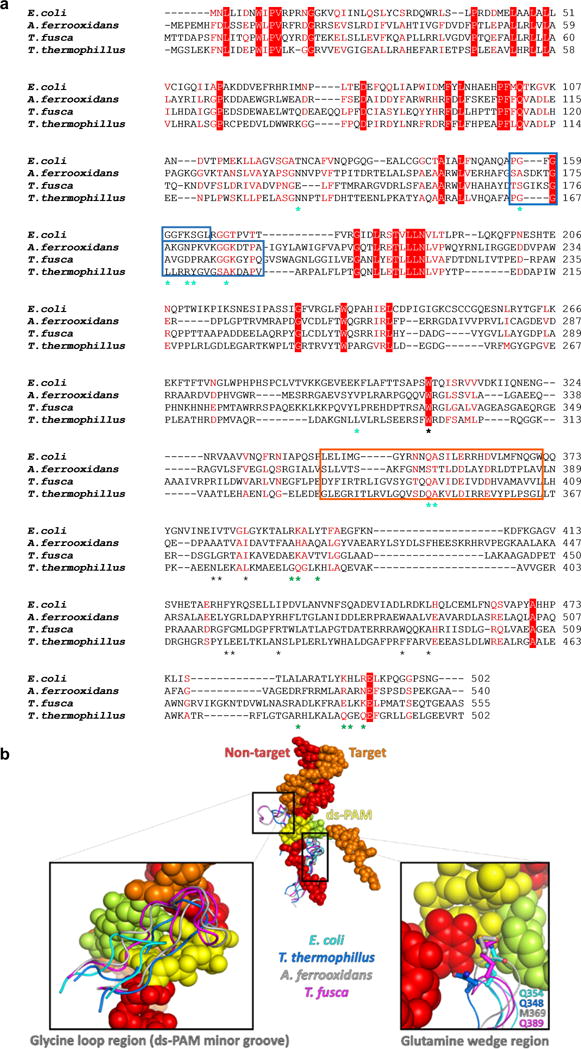
a, Sequence alignment of Cse1 proteins with known structures. Identical residues are highlighted in red, conserved residues in red text. DNA-contacting residues in the Cse1-NTD and CTD are marked by cyan and green asterisks, respectively. The glycine loop and glutamine wedge are in blue and orange boxes, respectively. Residues mediating the W307 ball-and-socket interaction are marked with black asterisks. b, Structural alignment of Cse1 PAM recognition elements. Apo Cse1 from T. thermophillus (PDB: 4AN8), A. ferrooxidans (PDB: 4H3T) and T. fusca (PDB: 3WVO) were superimposed with the Cse1 in our partial R-loop forming E. coli Cascade structure. Despite sequence variation, a glycine-rich loop is present in each Cse1 structure, and likely plays a similar function to recognize PAM from the minor groove (left inset). The glutamine-wedge protrusion is highly conserved in 3-D. Each wedge features a long side chain at the tip (right inset), which likely stacks underneath PAM in a similar fashion.
