Abstract
SWI/SNF-type chromatin remodelers, such as BRAHMA (BRM), and H3K27 demethylases both have active roles in regulating gene expression at the chromatin level1–5, but how they are recruited to specific genomic sites remains largely unknown. Here we show that RELATIVE OF EARLY FLOWERING 6 (REF6), a plant-unique H3K27 demethylase6, targets genomic loci containing a CTCTGYTY motif via its zinc-finger (ZnF) domains and facilitates the recruitment of BRM. Genome-wide analyses showed that REF6 colocalizes with BRM at many genomic sites with the CTCTGYTY motif. Loss of REF6 results in decreased BRM occupancy at BRM–REF6 co-targets. Furthermore, REF6 directly binds to the CTCTGYTY motif in vitro, and deletion of the motif from a target gene renders it inaccessible to REF6 in vivo. Finally, we show that, when its ZnF domains are deleted, REF6 loses its genomic targeting ability. Thus, our work identifies a new genomic targeting mechanism for an H3K27 demethylase and demonstrates its key role in recruiting the BRM chromatin remodeler.
Chromatin-mediated control of gene expression is achieved mainly by chromatin remodelers and enzymes covalently modifying histones7–10. In Arabidopsis thaliana, the SWI/SNF-type chromatin-remodeling ATPase BRM and the H3K27 demethylase REF6 have critical roles in many developmental processes11–16. Both proteins have been shown to antagonize Polycomb group proteins at target loci6,17–20, but how they are recruited to specific genomic sites and whether their activities are coordinated remain largely unknown. To address these questions, we mapped the genome-wide occupancy of BRM and REF6 by chromatin immunoprecipitation followed by sequencing (ChIP-seq). ChIP-seq analyses using transgenic plants expressing a BRM-GFP fusion protein from the native BRM promoter in the brm-1 mutant background (pBRM::BRM-GFP brm-1; refs. 19,21) identified 5,278 genes occupied by BRM (Supplementary Data 1), including previously reported BRM targets19,22–25 (Supplementary Fig. 1). To map the genomic occupancy of REF6, we generated a transgenic line expressing a REF6-GFP fusion protein from the native REF6 promoter (pREF6::REF6-GFP). The transgene was functional in vivo, as its expression fully rescued the morphological defects of REF6-1 plants (Supplementary Fig. 2). ChIP-seq analyses identified 3,164 REF6 target genes (Supplementary Data 2), including previously identified direct REF6 targets6 (Supplementary Fig. 3).
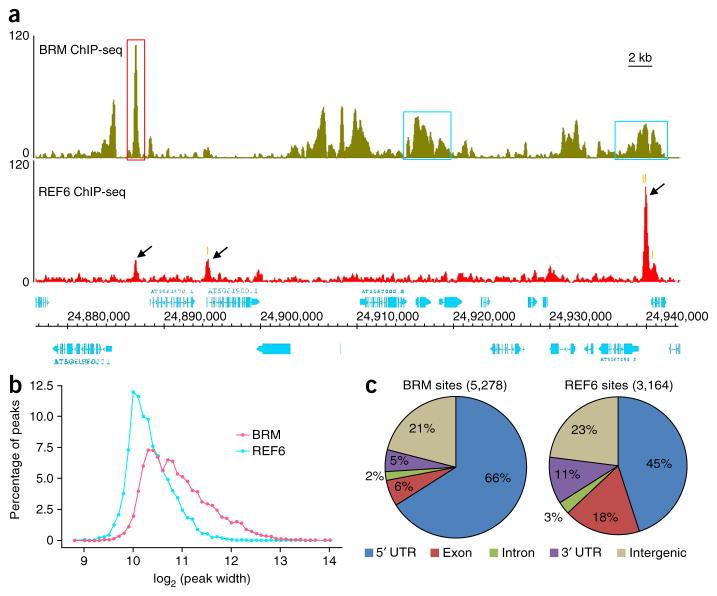
Occupancy profiles of individual genomic regions showed that both BRM and REF6 proteins occupied defined locations within the genome. We found two major types of BRM sites characterized by either sharp, narrow peaks or broad peaks (Fig. 1a). The average size of a BRM site was 2,155 bp (Fig. 1b). In contrast, only one major type of REF6 site was observed, characterized by single, defined narrow peaks (Fig. 1a), with an average size of 1,355 bp (Fig. 1b). Examination of the distribution of both BRM- and REF6-associated sites showed that ~80% of these sites were located in promoters and gene bodies, whereas ~20% of the sites were in intergenic regions (Fig. 1c). When the occupancy profiles were compared with published genome-wide histone modification data26, both BRM and REF6 were found to colocalize with active histone marks but not with repressive ones (Supplementary Fig. 4). Notably, genes involved in responses to different types of stimuli were highly enriched in BRM or REF6 target genes (Supplementary Fig. 5).
Figure 1.
Genome-wide occupancy of BRM and REF6. (a) ChIP-seq genome browser views of occupancy of BRM (top) and REF6 (bottom) at the same genomic coordinates on chromosome 5. The red box highlights a single, defined BRM peak, and the blue boxes highlight broad BRM peaks. Black arrows mark REF6 peaks. The positions of the CTCTGYTY motifs underlying the REF6 peaks (Fig. 4) are indicated by orange vertical bars. Gene structures are shown underneath the panel. The y-axis scales represent shifted merged MACS tag counts for every 10-bp window. (b) The average peak widths of BRM and REF6 sites. The x axis shows log2-transformed values for peak width. The y axis shows the percentage of peaks with a specific width. (c) Pie charts showing the distribution of BRM and REF6 at annotated genic and intergenic regions in the genome.
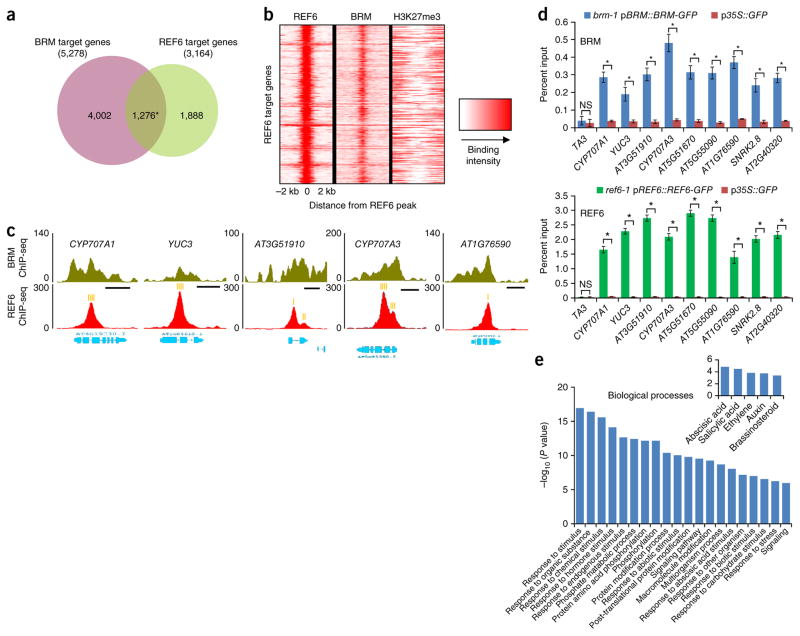
Comparing their genomic distribution patterns, we found that BRM and REF6 co-occupied a total of 1,276 genes (Fig. 2a and Supplementary Data 3), a number much larger than expected by chance alone (hypergeometric test, P < 7 × 10−162). Consistently, BRM was strongly enriched right at the summits of REF6-occupied sites but not at loci marked by trimethylation of histone H3 at lysine 27 (H3K27me3) (Fig. 2b). ChIP-seq data showing the colocalization of BRM and REF6 at a set of selected loci are presented in Figure 2c, and the colocalization was further validated by ChIP–qPCR (Fig. 2d). Again, genes involved in responses to various stimuli were highly enriched among BRM–REF6 co-targets (Fig. 2e).
Figure 2.
BRM and REF6 co-occupy a large number of genomic regions. (a) A Venn diagram displaying a statistically significant overlap between genes occupied by BRM and those occupied by REF6 (1,276 genes; *P < 7 × 10−162, hypergeometric test). (b) Heat map representation of the co-occupancy of BRM and REF6 in the genome. Each horizontal line represents a REF6-bound region, and signal intensity is shown for REF6 binding (left), BRM binding (middle), and H3K27me3 (right). Columns show the genomic region surrounding each REF6 peak summit. Signal intensity is indicated by the shade of red. (c) ChIP-seq genome browser views of BRM and REF6 co-occupancy at selected genes. Gene structures are shown underneath each panel. The positions of the CTCTGYTY motifs underlying the REF6 peaks (Fig. 4) are indicated by orange vertical bars. Scale bars (black), 1 kb. (d) ChIP–qPCR validation of BRM (top) and REF6 (bottom) occupancy at shared targets using ChIP DNA samples independent from those used for ChIP-seq. Data are shown as percentage of input. p35S::GFP plants were used as the negative-control sample, and the TA3 locus was used as the negative-control locus. Error bars, s.d. from three biological replicates. *P < 0.01; NS, not significant. (e) Gene Ontology (GO) analysis of the BRM–REF6 co-target genes showing that BRM and REF6 co-regulate a large number of genes involved in responses to stress. Inset, genes involved in plant responses to hormones are highly enriched.
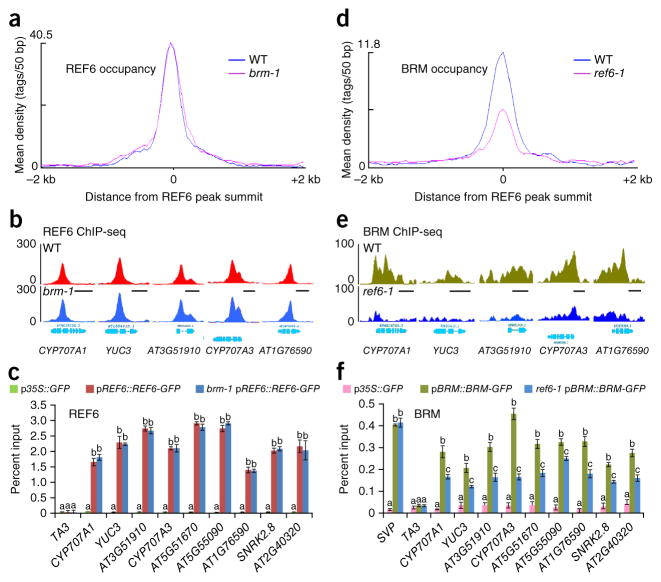
We wondered whether the co-occupancy of chromatin by BRM and REF6 reflects mutually dependent occupancy by these proteins. To test whether the association of REF6 with chromatin is dependent on BRM, we introduced the brm-1 mutation into the pREF6::REF6-GFP transgenic line by genetic crossing (brm-1 pREF6::REF6-GFP) and then performed ChIP-seq analyses to compare the genome-wide occupancy profiles of REF6 in brm-1 mutant plants with those in plants with wild-type BRM. We could not detect a significant decrease in REF6 occupancy in brm-1 plants (Fig. 3a–c). In control analyses, we observed that loss of BRM activity had no effect on REF6 RNA (Supplementary Fig. 6a) and protein (Supplementary Fig. 6b) levels, as well as nuclear localization of the REF6 protein (Supplementary Fig. 6c). These data suggest that BRM is likely not required for the occupancy of REF6 on chromatin. To examine whether the occupancy of BRM on chromatin depends on REF6, we carried out ChIP-seq analyses comparing the BRM occupancy profiles in REF6-1 plants with those in plants with wild-type REF6 (REF6-1 pBRM::BRM-GFP versus pBRM::BRM-GFP). We found that BRM occupancy on target chromatin was significantly reduced in REF6-1 plants (see Fig. 3d for a global view and Fig. 3e,f for individual loci), indicating that the association of BRM with its target genes requires REF6. A total of 443 genes showed a marked reduction or elimination of BRM binding (false discovery rate (FDR) < 0.001) in the absence of REF6 (Supplementary Data 4). Notably, loss of REF6 did not affect BRM RNA and protein levels or the nuclear localization of BRM protein (Supplementary Fig. 7a–c), excluding the possibility of reduced BRM occupancy being a result of decreased BRM abundance in nuclei. These results indicate that REF6 mediates the recruitment of BRM to its target loci, but its own genomic targeting is independent of BRM.
Figure 3.
REF6-dependent recruitment of BRM to genomic loci. (a) Mean density of REF6 occupancy at all REF6-associated sites in brm-1 plants as compared to plants with wild-type BRM (WT). The average REF6 binding signal within 2-kb genomic regions flanking the center of the REF6 peaks is shown. (b) ChIP-seq genome browser views of REF6 occupancy at selected loci in brm-1 plants and those with wild-type BRM. Gene structures are shown underneath each panel. (c) REF6 occupancy at selected genes as determined by ChIP–qPCR in brm-1 pREF6::REF6-GFP and pREF6::REF6-GFP plants. ChIP signals are shown as percentage of input. TA3 was used as a negative-control locus. Error bars, s.d. from three biological replicates. Lowercase letters indicate significant differences between genetic backgrounds, as determined by the post hoc Tukey’s HSD test. (d) Mean density of BRM occupancy at all REF6-associated sites in REF6-1 plants as compared to those with wild-type REF6 (WT). The average BRM binding signal within 2-kb genomic regions flanking the center of the REF6 peaks is shown. (e) ChIP-seq genome browser views of BRM occupancy in REF6-1 plants and those with wild-type REF6. (f) Decreased BRM occupancy at selected genes in REF6-1 pBRM::BRM-GFP plants as compared to pBRM::BRM-GFP plants as determined by ChIP–qPCR. SVP, a BRM target gene19 showing no difference in BRM occupancy between the two backgrounds in ChIP-seq analysis, was also included as a control. Error bars, s.d. from three biological replicates. Lowercase letters indicate significant differences between genetic backgrounds, as determined by the post hoc Tukey’s HSD test.
To examine whether REF6 physically interacts with BRM, we performed immunoprecipitation for GFP followed by mass spectrometry (IP–MS) with the transgenic line encoding BRM-GFP. We observed coimmunoprecipitation of several known and predicted subunits of SWI/SNF complexes, including BRM, SWP73A, SWP73B, SWI3A, SWI3B, SWI3C, SWI3D, and SYD (Supplementary Fig. 8a), indicating that we had successfully isolated the BRM-containing SWI/SNF complexes in Arabidopsis. Notably, the IP–MS analysis also identified REF6 peptides (Supplementary Fig. 8a), indicating that REF6 physically associates with BRM-containing SWI/SNF complexes. We validated the physical interaction between REF6 and BRM by bimolecular fluorescence complementation (BiFC) assay (Supplementary Fig. 8b). An unrelated nucleus-localized protein27 was used as a negative control and did not interact with REF6 or BRM (Supplementary Fig. 8b,c).
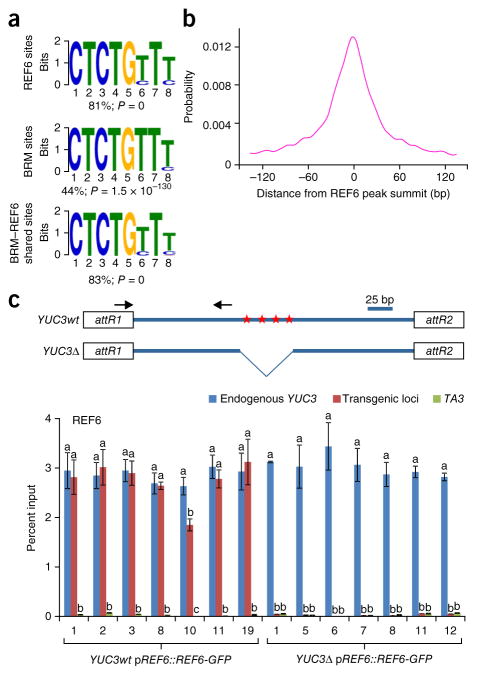
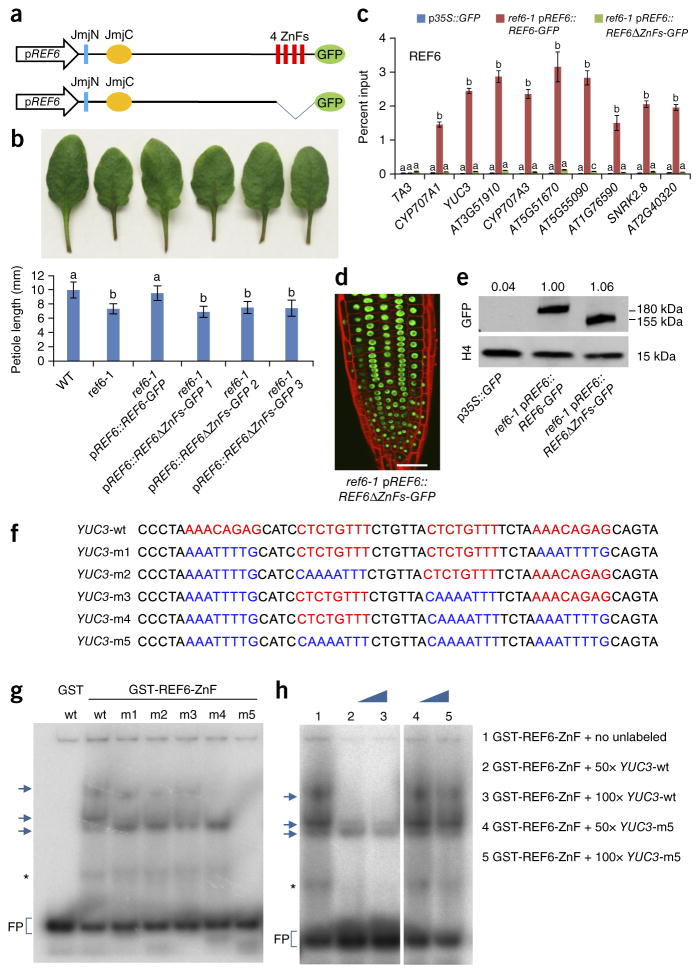
REF6 contains four repeats of a Cys2His2 ZnF domain28, which could potentially bind DNA directly29,30. We therefore attempted to define the consensus sequence(s) targeted by REF6 through a motif discovery analysis using MEME-ChIP31. This analysis showed that 81% of REF6-occupied sites contained a CTCTGYTY motif, where Y represents T or C (Fig. 4a), which was mostly located in the center of REF6 binding peaks (Fig. 4b). Consistent with the genome-wide colocalization of BRM and REF6 (Fig. 2a–c), this motif was also highly over-represented among BRM–REF6 co-targets (83%), as well as BRM targets (44%) (Fig. 4a). We further investigated whether the CTCTGYTY motif is required for the genomic targeting of REF6. We cloned a genomic fragment from a BRM–REF6 co-target, YUC3, that harbors four repeats of the CTCTGTTT motif (YUC3wt) and generated a version with the motifs deleted (YUC3Δ) (Fig. 4c and Supplementary Fig. 9). These constructs were then transformed into pREF6::REF6-GFP plants, and REF6 occupancy levels at the transgenes—YUC3wt and YUC3Δ—were then analyzed by ChIP–qPCR. As a positive control, REF6 occupancy at the endogenous YUC3 locus was also measured simultaneously. REF6 occupancy was enriched at the transgene containing the CTCTGTTT motifs, and this enrichment was completely abolished when the motifs were deleted (Fig. 4c). These data suggest that the CTCTGYTY motif is necessary for REF6 recruitment in vivo. Consistent with the observation that REF6 is required to recruit BRM (Fig. 3d–f), we found that BRM occupancy was significantly lower at YUC3Δ than at YUC3wt (Supplementary Fig. 10).
Figure 4.
A DNA motif required for REF6 genomic targeting. (a) The CTCTGYTY motif is present in REF6 and BRM targets and in BRM–REF6 co-targets. MEME-ChIP was used for de novo motif discovery. The percentage of peaks containing the motif is shown. P values were determined by MEME-ChIP. (b) Distribution of the CTCTGYTY motif across REF6 peaks. (c) The CTCTGYTY motif is necessary for the recruitment of REF6. Shown at the top is a schematic of the transgene constructs derived from the YUC3 gene. Red stars indicate the positions of the CTCTGTTT sequences; attR1 and attR2 are recombination sites in the Gateway-compatible vector. For the full sequence of the transgene, see Supplementary Figure 9. At the bottom are ChIP–qPCR results showing that REF6 binds the transgene containing the wild-type motifs (YUC3wt) but not the transgene without the motifs (YUC3Δ). Seven independent transgenic lines were analyzed for each construct. ChIP signals are shown as percentage of input. The endogenous YUC3 locus and the TA3 locus were used as positive and negative controls, respectively. Error bars, s.d. from three biological replicates. Lowercase letters indicate significant differences between genetic backgrounds, as determined by the post hoc Tukey’s HSD test.
We examined how REF6 might be recruited to target chromatin by testing whether the ZnF domains of REF6 mediate its genomic targeting. We stably expressed a truncated version of REF6 lacking the ZnF domains from the native REF6 promoter in REF6-1 plants (REF6-1 pREF6::REF6ΔZnFs) (Fig. 5a). We found that the ZnF-deleted version of REF6 (REF6ΔZnFs) failed to rescue the short-petiole and late-flowering phenotypes of the REF6-1 mutant (Fig. 5b and Supplementary Fig. 11), suggesting that the ZnF domains are essential for the biological function of REF6. We then examined the chromatin occupancy of REF6ΔZnFs by ChIP–qPCR. No enrichment of REF6ΔZnFs at the selected loci was detected, demonstrating that the ZnF domains are essential for the association of REF6 with chromatin (Fig. 5c). As neither the nuclear localization (Fig. 5d) nor the abundance (Fig. 5e) of REF6 was affected by deletion of the ZnF domains, the most likely explanation for the loss of chromatin occupancy by REF6ΔZnFs is that the ZnF domains of REF6 mediate the recruitment of REF6 to chromatin.
Figure 5.
The REF6 zinc-finger domains are essential for the binding of REF6 to chromatin. (a) A schematic of the proteins encoded by the transgene constructs. The conserved domains of REF6 are shown. (b) Image of petioles (top) and quantification of the petiole length (bottom) in plants with the different genotypes. Error bars, s.d. from 17 plants. Lowercase letters indicate significant differences between genetic backgrounds, as determined by the post hoc Tukey’s HSD test. (c) ChIP–qPCR results showing genomic occupancy by the wild-type and ZnF-deleted REF6-GFP fusion proteins. p35S::GFP was used as the negative-control transgene, and the TA3 locus was used as the negative-control locus. Error bars, s.d. from three biological replicates. (d) Confocal image of root tips showing nuclear localization of the REF6ΔZnFs-GFP fusion protein. Red fluorescent signal is from propidium iodide staining. Scale bar, 20 μm. (e) Immunoblot analyses showing the relative protein levels of REF6-GFP and REF6ΔZnFs-GFP (numbers at the top represent amounts normalized to the loading control, histone H4). (f) Sequences of the DNA probes used in the EMSAs. Wild-type and mutated sequences are shown in red and blue, respectively. (g) EMSA showing that GST-REF6-ZnF but not GST by itself specifically binds the YUC3-wt, YUC3-m1, YUC3-m2, YUC3-m3, and YUC3-m4 probes but not the YUC3-m5 probe. Arrows indicate the shifted bands. FP, free probe. The asterisk indicates a band likely corresponding to degraded GST-REF6-ZnF. (h) The addition of excess unlabeled wild-type probe (lanes 2 and 3) but not YUC3-m5 mutant probe (lanes 4 and 5) outcompetes the strong interactions visible in lane 1. The uncropped scan is shown in Supplementary Data 6.
To test whether the ZnF domains of REF6 could directly bind the CTCTGYTY motif, we performed electrophoretic mobility shift assays (EMSAs) using a recombinant GST-tagged C-terminal part of REF6 containing all four ZnF domains (GST-REF6-ZnF; residues 1175–1360) and a DNA fragment of the YUC3 gene that contains four copies of the CTCTGTTT motif (YUC3-wt) (Fig. 5f). GST-REF6-ZnF but not GST alone bound the YUC3-wt DNA probe (Fig. 5g). Moreover, GST-REF6-ZnF recognized probes containing one or two CTCTGTTT motifs (YUC3-m1, YUC3-m2, YUC3-m3, and YUC3-m4) (Fig. 5g). In contrast, GST-REF6-ZnF failed to bind the DNA probe in which all four motifs were mutated (YUC3-m5) (Fig. 5g). The addition of excess unlabeled wild-type probe (YUC3-wt) but not mutant probe YUC3-m5 was sufficient to outcompete the specific interactions, as evidenced by reduced intensity for the shifted bands (Fig. 5h). Together, these data strongly suggest that REF6 uses its ZnF domains to bind to genomic sites containing CTCTGYTY motif(s).
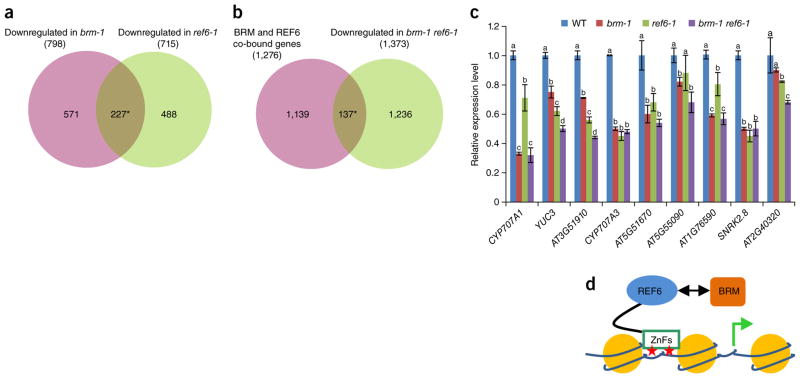
To examine whether BRM and REF6 regulate a shared set of genes, we performed RNA-seq analyses in the brm-1, REF6-1, and brm-1 REF6-1 backgrounds (see Supplementary Fig. 12 for the morphologies of the single and double mutants and Supplementary Data 5 for lists of differentially expressed genes). We found that 227 genes were downregulated (Fig. 6a) and 31 genes were upregulated (Supplementary Fig. 13a) in both the brm-1 and REF6-1 mutants. In contrast, the overlap between the downregulated genes in brm-1 and the upregulated genes in REF6-1, and vice versa, did not seem significant (Supplementary Fig. 13a). These data suggest that BRM and REF6 preferentially co-activate a common set of genes. To define genes regulated by both BRM and REF6, we compared our ChIP-seq data with the RNA-seq data. For genes associated with both BRM and REF6, there was a statistically significant overlap with genes downregulated in the brm-1, REF6-1, and brm-1 REF6-1 mutants (Fig. 6b and Supplementary Fig. 13b). In contrast, the overlap of BRM and REF6 co-targets with upregulated genes in the brm-1, REF6-1, and brm-1 REF6-1 mutants was not significant (Supplementary Fig. 13c), indicating that BRM and REF6 preferentially co-occupy expressed genes. The RNA-seq data were further validated by qRT–PCR at selected genes (Fig. 6c). In the brm-1 REF6-1 double mutant, there was no additive effect on expression at most of the genes examined relative to the single mutants (Fig. 6c), suggesting that BRM and REF6 act in the same pathway to activate transcription of these loci.
Figure 6.
Expression of BRM–REF6 co-target genes in the brm-1, REF6-1, and brm-1 REF6-1 backgrounds. (a,b) Venn diagrams showing statistically significant overlaps between genes downregulated in both brm-1 and REF6-1 (*P = 5.5 × 10−185) (a) and between BRM–REF6 co-bound genes and genes downregulated in brm-1 REF6-1 (*P = 5.8 × 10−21) (b). (c) qRT–PCR analyses showing decreased expression of selected genes in brm-1, REF6-1, and brm-1 REF6-1 plants as compared with wild-type plants. The expression level of each gene was normalized to that of GAPDH. Error bars, s.d. from three biological replicates. Lowercase letters indicate significant differences between genetic backgrounds, as determined by the post hoc Tukey’s HSD test. (d) A proposed model showing that REF6 directly binds to chromatin DNA containing CTCTGYTY motifs (red stars) via its ZnF domains and subsequently facilitates the recruitment of BRM, predominantly resulting in activation of gene expression.
To examine whether BRM could facilitate the function of REF6 as an H3K27 demethylase, we performed H3K27me3 ChIP-seq analyses in wild-type, brm-1, REF6-1, and brm-1 REF6-1 plants. In comparison with wild-type plants, an increase in H3K27me3 levels at REF6 target genes was observed in REF6-1 mutant plants (Supplementary Fig. 14), confirming that REF6 is an H3K27 demethylase. In contrast, no increase in H3K27me3 levels at REF6 target genes was detected in brm-1 mutants, suggesting that BRM might not be required for REF6 to remove methyl groups from H3K27me3-modified sites. These data are consistent with our results showing that BRM is not required for the genomic targeting of REF6. Further supporting this notion, we found that the H3K27me3 levels in the brm-1 REF6-1 double mutant were similar to those in the REF6-1 single mutant (Supplementary Fig. 14).
In summary, our findings have identified a mechanism by which REF6 and BRM are recruited to specific genomic sites (Fig. 6d). First, we demonstrate that REF6 directly binds to a specific DNA motif (CTCTGYTY) through its ZnF domains. This conclusion is supported by the observations that deletion of the CTCTGYTY motifs from a REF6 target gene abolishes the binding of REF6 in vivo (Fig. 4), that the ZnF-deleted version of REF6 fails to bind to its target genes (Fig. 5c), and that the ZnF domains of REF6 directly bind to DNA in vitro (Fig. 5f–h). Second, we demonstrate that BRM occupies many REF6 target genes that contain the CTCTGYTY DNA motif (Figs. 2a–d and 4a). Finally, we show that loss of REF6 leads to an impairment in BRM occupancy at many genes (Fig. 3d–f). Thus, this work highlights a paradigm for genomic targeting of H3K27 demethylases, as well as BRM-containing SWI/SNF complexes. It is worth noting that the H3K27me3 mark has been shown to have a feedback role in recruiting its ‘writer’, Polycomb complex PRC2 (refs. 32–34). It will be interesting to investigate whether it also facilitates the recruitment of its ‘erasers’ such as REF6. Finally, our BRM and REF6 ChIP-seq data are also expected to be important community resources for future dissection of the roles of these two global chromatin regulators in controlling specific genes and pathways.
URLs
BINGO, http://www.psb.ugent.be/cbd/papers/BiNGO/Home.html; GeneProf, http://www.geneprof.org/GeneProf/tools/hypergeometric.jsp.
ONLINE METHODS
Plant materials and growth conditions
Arabidopsis seeds were stratified for 4 d at 4 °C in darkness. The seeds were then sown on soil or on agar plates containing 4.3 g/L Murashige and Skoog (MS) nutrient mix (Sigma-Aldrich), 1.5% sucrose (pH 5.8), and 0.8% agar. Plants were grown in growth rooms with 16-h light/8-h dark cycles at 22 °C. Transfer DNA (T-DNA) insertion mutants were obtained from the Arabidopsis Biological Resource Center (ABRC), unless otherwise indicated. The brm-1 (SALK_030046) and REF6-1 (SALK_001018) mutants are both in the Col background and have been described previously13,35. Homozygous T-DNA insertion mutants were identified by PCR-based genotyping. The brm-1 pBRM::BRM-GFP and p35S::GFP transgenic plants have been described19,21,36.
Generation of transgenic plants
Genomic regions corresponding to full-length REF6 and REF6ΔZnFs including a 2-kb promoter and the coding region without the stop codon were amplified and subcloned into the pDONR221 vector (Invitrogen) by BP reaction. The resulting entry vectors were sequenced to ensure that no mutation was introduced during PCR amplification. The inserts were then transferred into the pMDC107 vector37 by LR reaction (to generate pREF6::REF6-GFP and pREF6::REF6-GFP-ΔZnFs). The constructs were introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform REF6-1 mutant plants using the floral dip method38.
A segment of the YUC3 genomic DNA sequence located 701–977 bp downstream of the ATG start codon was amplified and subcloned into the pDONR221 vector by BP reaction. The insert was then transferred into the pEarleyGate 201 vector by LR reaction. To delete CTCTGTTT motifs, the Phusion Site-Direct Mutagenesis kit (Finnzymes) was used according to the manufacturer’s instructions. The constructs were introduced into A. tumefaciens strain GV3101, which was then used to transform REF6-1 pREF6::REF6-GFP plants using the floral dip method38. Sequences for the primers used are listed in Supplementary Table 1.
ChIP assays
ChIP was carried out as described39,40, with minor modifications. Briefly, 2 g of 14-d-old seedlings grown on MS agar were collected and cross-linked with 1% formaldehyde for 20 min under vacuum and then ground into fine powder in liquid nitrogen. Chromatin was isolated and sheared into 200- to 800-bp fragments by sonication. The sonicated chromatin was incubated with 10 μl of antibody to GFP (Abcam, ab290) or H3K27me3 (Millipore, 07-449) overnight at 4 °C. Precipitated DNA was then recovered with the MinElute PCR Purification kit (Qiagen) according to the manufacturer’s instructions. ChIP–qPCR was performed with three technical replicates, and results were calculated as percentage of input DNA according to the Champion ChIP–qPCR user manual (SABioscience). ChIP experiments were performed at least three times. Sequences for the primers used for ChIP–qPCR are listed in Supplementary Table 1.
ChIP-seq and data analyses
Ten nanograms of DNA from at least ten ChIPs was pooled to ensure enough starting DNA for library construction. Two biological replicates were prepared and sequenced for each ChIP-seq experiment. The ChIP DNA was first tested by qRT–PCR and then used to prepare ChIPseq libraries. End repair, adaptor ligation, and amplification were carried out using the Illumina Genomic DNA Sample Prep kit according to the manufacturer’s protocol. An Illumina HiSeq 2500 instrument was used for high-throughput sequencing of the ChIP-seq libraries. The raw sequence data were processed using the Illumina sequence data analysis pipeline GAPipeline 1.3.2. Bowtie41 was then employed to map the reads to the Arabidopsis genome (TAIR10)42. Only perfectly and uniquely mapped reads were retained for further analysis. A summary of the number of reads for each replicate is given in Supplementary Table 2. To determine the correlation between biological repeats, Pearson correlation was computed using R statistical software on normalized signal intensity for ChIP binding peaks. Correlation (R2) was 0.94, 0.90, 0.87, and 0.88 for pREF6::REF6-GFP, pBRM::BRM-GFP, REF6-1 pBRM::BRM-GFP, and brm-1 pREF6::REF6-GFP, respectively (Supplementary Fig. 15), indicating that our ChIP-seq experiments are reliable. The alignments were first converted to Wiggle (WIG) files using MACS43. The data were then imported into the Integrated Genome Browser (IGB)44 for visualization. Second, the program SICER45 was used to identify ChIP-enriched domains (peaks) and for qualitative comparisons of BRM binding levels in wild-type and REF6-1 plants. Third, the program seqMINER46 was used to generate the heat map in Figure 2b and to compare the global changes in BRM and REF6 binding levels, as shown in Figure 3a,d and Supplementary Figure 14. Published ChIP-seq data for H3K27me3 were used19. To assign the peaks to proximal genes, the distance between each peak summit and the nearby transcriptional start site (TSS) of a gene was calculated. A peak summit that was positioned within 2 kb upstream or 2 kb downstream of a TSS was assigned to the corresponding gene. If multiple genes could be assigned to a peak, the one with the closest TSS was selected. If no TSS was found in this window, the peak was left unassigned. To identify DNA motifs enriched at REF6- and BRM-associated sites, 300-bp sequences encompassing each peak summit (150 bp upstream and 150 bp downstream) were extracted and searched for enriched DNA motifs with an oligomer length of 6–8 bp using MEME-ChIP31. Searches were performed using default parameters.
Gene ontology term and gene list overlap analyses
The BINGO 2.44 plugin for Cytoscape47 was used to determine which GO categories were statistically enriched. To test whether the overlap between two groups of genes was statistically significant, a hypergeometric probability test was performed using GeneProf.
Electrophoretic mobility shift assays
GST and GST-REF6-ZnF recombinant fusion protein were expressed in Escherichia coli (BL21-CodonPlus, Stratagene) and purified using Glutathione Sepharose 4B beads (GE Healthcare). Complementary oligonucleotides (Supplementary Table 1) were labeled with [α-32P]dATP using T4 polynucleotide kinase (New England BioLabs, M0201) and annealed. Approximately 100 ng of GST or GST-REF6-ZnF protein and 0.3 pM of 32P-labeled probe were incubated in a 10-μl reaction mixture (containing 25 mM Tris-HCl, 100 mM NaCl, 2.5 mM MgCl2, 0.1% CA-630, 10% glycerol, 1 μM ZnSO4, and 1 mM DTT) for 1 h on ice, and the reaction mixture was then separated on a 6% polyacrylamide gel in Tris-glycine buffer (50 mM Tris-HCl, 380 mM glycine, and 2 mM EDTA, pH 8.5) for 1 h at 80 V. For the competition assays, 50- or 100-fold more non-labeled competitor DNA than labeled probe was added to the reaction 10 min before addition of the labeled probe. The uncropped scan is shown in Supplementary Data 6.
Immunoblotting
Two grams of 14-d-old seedlings were collected, and nuclei were isolated according to the ChIP protocol but without the tissue fixation step. Nuclear proteins were released by incubating the nuclei preparation in 120 μl of lysis buffer (50 mM Tris-HCl, 10 mM EDTA, 1% SDS, and 1× protease inhibitors) for 3 h at 4 °C. The extract was then diluted with 1 volume of ChIP dilution buffer (16.7 mM Tris-HCl, 167 mM NaCl, and 1.1% Triton X-100, pH 8.0) and centrifuged at 15,000g for 10 min at 4 °C to remove debris. Proteins were resolved on a 4–20% Mini-PROTEAN TGX Precast Protein Gel (Bio-Rad) by electrophoresis and detected by antibody to GFP (Invitrogen, A11122; 1:5,000 dilution), HA (Sigma, H6908; 1:5,000 dilution), FLAG (Sigma, F7425; 1:5,000 dilution), or histone H4 (Millipore, 07-108; 1:20,000 dilution). Histone H4 was used as the loading control. Quantification of protein signal was performed using ImageJ software. Uncropped scans of immunoblotting results are shown in Supplementary Data 6.
Immunoprecipitation–mass spectrometry
Seedlings of brm-1 pBRM::BRM-GFP and p35S::YFP transgenic Arabidopsis lines were collected and frozen in liquid nitrogen. They were then ground into fine powder with a mortar. Ten grams of tissue powder was mixed with 20 ml of extraction buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 1 mM EDTA, 1% Triton X-100, and 1× protease and phosphatase inhibitors (Thermo Fisher)). After filtering and centrifugation, the protein extract was mixed with 50 μl of protein A–conjugated magnetic beads preincubated with 20 μg of polyclonal antibody to GFP (custom made) for 2 h at 4 °C. After incubation, the beads were washed three times with wash buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 1 mM EDTA, and 0.1% Triton X-100), and the proteins were eluted by incubating beads twice with SDS loading buffer for 10 min at 95 °C.
The immunoprecipitated proteins were separated on a NuPAGE Novex 4–12% Bis-Tris gel, and the gel was stained using the Colloidal Blue Staining kit (Invitrogen). Each gel lane was cut into 15 bands. After in-gel digestion with trypsin, the samples were analyzed on a Thermo Scientific Q Exactive mass spectrometer using the data-dependent mode. The spectrum data were searched against the TAIR10 database using Protein Prospector.
BiFC assays
Full-length BRM and REF6 coding sequences were amplified and transformed into the pDONR221 vector by BP reaction. The resulting entry vectors were confirmed by sequencing to ensure that no errors were introduced by PCR amplification. The inserts were then transferred into the modified pEarleyGate 201-nYFP or pEarleyGate 202-cYFP vector48 by LR reaction. The constructs were introduced into A. tumefaciens strain GV3101 individually, and the resulting bacteria were used to infiltrate the lower epidermis of tobacco (Nicotiana benthamiana) leaves. After 48 h, the fluorescence signals were visualized using a confocal microscope (Leica Microsystems). Sequences for the primer used are listed in Supplementary Table 1.
Gene expression analyses
Total RNA was isolated from ~50 mg of plant tissue using the Plant/Fungi Total RNA Purification kit (Norgen). All RNA samples were treated with RNase-free DNase (Qiagen). One microgram of RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Random primers from the kit were used as primers. Real-time qPCR was conducted using the SsoFast EvaGreen Supermix kit with the Bio-Rad CFX96 real-time PCR detection system. Results are repeated for two additional independent RNA samples (biological replicates). GAPDH was used as the internal reference. Sequences for the PCR primers used in real-time PCR are listed in Supplementary Table 1.
RNA-seq analyses
For genome-wide expression analysis, RNA from 14-d-old seedlings of wild-type, brm-1, REF6-1, and brm-1 REF6-1 plants was isolated using the Plant/Fungi Total RNA Purification kit and treated with RNase-free DNase. RNA from three biological replicates was sequenced separately. Sequencing libraries were built using the Illumina TruSeq RNA library preparation protocol. The libraries were sequenced on the Illumina HiSeq 2500 platform using a paired-end scheme (2 × 100 bp) with TruSeq v3 chemistry. Reads were mapped to the TAIR10 Arabidopsis genome using TopHat v2.0.4 (ref. 49) with default settings, except that a minimum intron length of 20 bp and a maximum intron length of 4,000 bp were required. Reads that mapped to multiple regions were discarded. Calculations to identify differentially expressed genes were performed as described50. Genes with at least a twofold change in expression (FDR = 5%, P < 0.01) were considered to be differentially expressed.
Assessment of flowering time and petiole length
Wild-type and mutant plants were grown side by side in soil at 22 °C with 16-h light/8-h dark cycles. The number of rosette leaves was determined when the first flower opened. The petiole length of the fifth true leaf was measured for each plant 40 d after germination. For each genotype, at least 17 plants were analyzed, and the analysis was repeated three times independently.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (ABRC) for seeds for T-DNA insertion lines; A. Molnar for help with figure preparation; X. Shi of the Clinical Genomics Centre at Mount Sinai Hospital for overseeing the next-generation sequencing; and S. Rothstein for critical reading of the manuscript. C.C. is supported by a graduate fellowship from the Chinese Scholarship Council. This work was supported by funding from the Agriculture and Agri-Food Canada A-base and the National Science and Engineering Research Council of Canada (R4019A01) to Y.C., the Natural Science Foundation of China (31128001 to K.W. and Y.C. and 31210103901 to X. Cao and X. Chen), the State Key Laboratory of Plant Genomics (2015B0129-01 to X. Cao), and the US National Institutes of Health to X. Chen (GM061146) and Z.-Y.W. (R01GM066258).
Footnotes
Accession codes. The ChIP-seq and RNA-seq data sets have been deposited in the Gene Expression Omnibus (GEO) under accession GSE72736. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD003583.
Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
C.L. and Y.C. conceived the project. C.L. performed most of the experiments. C.-Q.W., L.-F.A., C.-W.C., M.P.S., L.J., A.L.B., and Z.-Y.W. performed BRM-GFP IP–MS assays. L. Gu, L. Gao, C.L., and C.C. conducted bioinformatics analyses. C.L., Q.Q., S.W., Y.Q., S.Y., C.-Y.C., V.N., S.E.K., S.H., X. Cao., and K.W. analyzed data. C.L., Y.C., and X. Chen wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–668. doi: 10.4161/epi.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 5.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 6.Lu F, Cui X, Zhang S, Jenuwein T, Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet. 2011;43:715–719. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 11.Reyes JC. The many faces of plant SWI/SNF complex. Mol Plant. 2014;7:454–458. doi: 10.1093/mp/sst147. [DOI] [PubMed] [Google Scholar]
- 12.Jerzmanowski A. SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta. 2007;1769:330–345. doi: 10.1016/j.bbaexp.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Noh B, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu X, et al. Modulation of brassinosteroid-regulated gene expression by Jumonji domain–containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:7618–7623. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008;147:1143–1157. doi: 10.1104/pp.108.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao M, et al. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 2015;11:e1005125. doi: 10.1371/journal.pgen.1005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu MF, et al. SWI2/SNF2 chromatin remodeling ATPases overcome Polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA. 2012;109:3576–3581. doi: 10.1073/pnas.1113409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho L, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating Polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, et al. The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates Polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet. 2015;11:e1004944. doi: 10.1371/journal.pgen.1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S, et al. The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell. 2015;27:1670–1680. doi: 10.1105/tpc.15.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smaczniak C, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vercruyssen L, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014;26:210–229. doi: 10.1105/tpc.113.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han SK, et al. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell. 2012;24:4892–4906. doi: 10.1105/tpc.112.105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efroni I, et al. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell. 2013;24:438–445. doi: 10.1016/j.devcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archacki R, et al. BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in Arabidopsis. PLoS One. 2013;8:e58588. doi: 10.1371/journal.pone.0058588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C, et al. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J. 2013;73:77–90. doi: 10.1111/tpj.12017. [DOI] [PubMed] [Google Scholar]
- 27.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu F, et al. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol. 2008;50:886–896. doi: 10.1111/j.1744-7909.2008.00692.x. [DOI] [PubMed] [Google Scholar]
- 29.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 30.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margueron R, et al. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 34.Yuan W, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 35.Hurtado L, Farrona S, Reyes JC. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 2006;62:291–304. doi: 10.1007/s11103-006-9021-2. [DOI] [PubMed] [Google Scholar]
- 36.Masiero S, et al. INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development. 2004;131:5981–5990. doi: 10.1242/dev.01517. [DOI] [PubMed] [Google Scholar]
- 37.Curtis MD, Grossniklaus U. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 39.Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 40.Li C, et al. Regulation of oleosin expression in developing peanut (Arachis hypogaea L.) embryos through nucleosome loss and histone modifications. J Exp Bot. 2009;60:4371–4382. doi: 10.1093/jxb/erp275. [DOI] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye T, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 48.Lu Q, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]