Abstract
Purpose
Mutations in rod photoreceptor genes can cause retinitis pigmentosa (RP). Rod gene expression is regulated by the nuclear hormone receptor, Nr2e3. Genetic deletion of Nr2e3 reprograms rods into cells that resemble cone photoreceptors, and might therefore prevent their death from some forms of RP. There are no identified ligands for Nr2e3; however, reverse agonists might mimic the genetic rescue effect and may be therapeutically useful for the treatment of RP.
Methods
We screened for small molecule modulators of Nr2e3 using primary retinal cell cultures and characterized the most potent, which we have named photoregulin1 (PR1), in vitro and in vivo. We also tested the ability of PR1 to slow the progression of photoreceptor degeneration in two common mouse models of autosomal dominant RP, the RhoP23H and the Pde6brd1 mutations.
Results
In developing retina, PR1 causes a decrease in rod gene expression and an increase in S opsin+ cones. Photoregulin1 continues to inhibit rod gene expression in adult mice. When applied to two mouse models of RP, PR1 slows the degeneration of photoreceptors.
Conclusions
Chemical compounds identified as modulators of Nr2e3 activity may be useful for the treatment of RP through their effects on expression of disease-causing mutant genes.
Keywords: retina, retinal degeneration, reprogramming, transcription factors
Death of photoreceptors is a common endpoint of several retinal degenerative diseases and often eventually results in blindness. In many cases of the retinal degenerative disease retinitis pigmentosa (RP), mutations in rod photoreceptor genes result in rod photoreceptor dysfunction and subsequent cell death.1 The majority of mutations in Rhodopsin (Rho) that cause RP are associated with the autosomal dominant form (adRP). These mutations lead to activation of the unfolded protein response (UPR)2 in the rods, due to a mislocalization of the mutant protein. A number of strategies are currently being pursued to reduce the UPR in the rods,3 or to reduce the expression of the mutant allele using siRNA.4 An alternative approach to modulate rod gene expression is suggested by developmental studies. During retinal development, the expression of a few key transcription factors regulates photoreceptor cell fate and further specification into rod and cone photoreceptors.5–7 One critical transcription factor in the specification of cone versus rod fate is Nrl.8–10 Mice with mutations in Nrl have retinas without rods, but an increase in the number of cones, because the rod precursors become cones without the expression of Nrl.11 Conversely, overexpression of Nrl in cone precursors results in decreased cone gene expression and a transformation to rod photoreceptors.12
A recent study showed that partial transdifferentiation of mature rods into cones by conditional knockout of Nrl can prevent retinal degeneration in a mouse model of recessive RP (Rho–/– mice). This reprogramming of rods into cone-like cells prevented their death and therefore any secondary cone cell death as well.13 The orphan nuclear receptor subfamily 2 group E member 3 (Nr2e3, also known as photoreceptor nuclear receptor [PNR]) is a direct target of Nrl and is expressed in postmitotic photoreceptors soon after the onset of Nrl expression.14–17 Nr2e3 has a dual role as a transcriptional suppressor and coactivator during retinal development.17–20 It is required for the suppression of cone gene expression, as evidenced by the findings that mutations in Nr2e3 result in increased expression of cone genes.21–25 Additionally, Nr2e3 coactivates the transcription of rod-specific genes like Rho and Gnat1 with Crx and Nrl.18–20
The finding that partial reprogramming of rods to cones can reduce rod death and decrease secondary cone loss to spare cone-mediated vision in a mouse model of RP provides a novel approach to develop therapies for this disorder and other similar degenerative diseases. This pathway is also potentially amenable to pharmacologic manipulation, since Nr2e3, the downstream target of Nrl, is a nuclear hormone receptor and probably capable of antagonism.26,27 To this end, we identified a small molecule modulator of Nr2e3, photoregulin1 (PR1), using dissociated and intact primary retinal cultures and found that it has large and selective effects on photoreceptor gene expression. We found that PR1 decreased the expression of a subset of rod genes and increased the number of S opsin+ cones. Photoregulin1 also slowed degeneration of photoreceptors in two in vitro models of RP, providing evidence that chemical compounds identified in screens for modulators of Nr2e3 activity may be therapeutically useful for the treatment of retinal degenerative diseases by modulating the expression of disease-causing mutant genes.
Methods
Animals
We used C57Bl/6 (Jackson Stock No: 000664; The Jackson Laboratory, Bar Harbor, ME, USA), RhoP23H (Jackson Stock No: 017628; The Jackson Laboratory),28 C3H/HeJ (harboring the Pde6brd1 mutation; Jackson Stock No: 000659; The Jackson Laboratory), and Nrl-eGFP (Jackson Stock No: 02123229; The Jackson Laboratory) mice at the indicated ages. All mice were housed by the Department of Comparative Medicine at the University of Washington and protocols were approved by the University of Washington Institutional Animal Care and Use Committee. The research was carried out in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Small Molecules
Photoregulin1 and analogs were identified by searching previous small molecule screens with a research application (SciFinder; American Chemical Society, Washington, DC, USA) or PubChem for Nr2e3 interacting molecules. The putative Nr2e3 interacting molecules were then obtained commercially from ChemDiv (San Diego, CA, USA) or synthesized. Ample quantities of PR1 to enable the in vivo assays were synthesized and purified in the lab.
Dissociated Retinal Cultures
Retinas were dissected from postnatal day (P)5 mice and dissociated by treatment with 0.5% trypsin diluted in calcium- and magnesium-free HBSS for 10 minutes at 37°C. Trypsin was inactivated by adding an equal volume of fetal bovine serum (FBS) and cells were pelleted by centrifugation at 4°C and resuspended in media (Neurobasal-A containing 1% FBS, 1% N2, 1% B27, 1% Pen/Strep, and 0.5% L-glutamine). Cells were plated into 96-well black-walled, clear-bottomed tissue culture plates (Greiner Bio One; Kremsmünster, Austria) at a density of one retina per five wells. For screen, small molecules diluted in media to 1 μM were added the day following dissociation and media was changed every other day. After 3 days of treatment, cells were fixed with 4% PFA for 20 minutes at room temperature, blocked with blocking solution (10% normal horse serum and 0.5% Triton X-100 diluted in 1X PBS) for 1 hour at room temperature, and incubated overnight at 4°C with primary antibodies generated against Rhodopsin and Otx2 (Supplementary Table S2) diluted in blocking solution. The following day, wells were washed with 1X PBS and then incubated with species appropriate, fluorescently labeled secondary antibodies diluted in blocking solution for 1 hour at room temperature. Wells were washed three times and the entire plate was imaged using an imager (GE Typhoon FLA 9400; Healthcare Life Sciences, Pittsburgh, PA, USA). Optical density measurements were obtained from the plate scans using ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA) and rhodopsin expression was normalized to Otx2 expression.
Retinal Explant Cultures
Intact retinas without RPE from mice of various strains and ages as indicated were explanted on 0.4-μm pore tissue culture inserts as previously described.30,31 Full media (DMEM/F12 [1:1] containing 1% dialyzed FBS, 1% Pen/Strep, 0.3% D+ glucose, 2% B27, and 1% N2 or Neurobasal-A containing 1% FBS, 1% N2, 1% B27, 1% Pen/Strep, and 0.5% L-Glutamine) changes were performed every other day and small molecules were used at 0.1 μM to 10 μM.
Quantitative Real-Time PCR
We isolated RNA from retinas using a commercial reagent (TRIzol; Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized using a commercial synthesis kit (iScript cDNA; Bio-Rad Laboratories, Hercules, CA, USA) following an intervening DNase treatment with RQ1 RNase-free DNase (Promega Corp., Madison, WI, USA) for 1 hour at 37°C. We used a supermix (SSO Fast; Bio-Rad Laboratories) for quantitative real-time PCR with the primer sequences listed in Supplementary Table S1. For analysis, values were normalized to Gapdh (ΔCt) and ΔΔCt between dimethyl sulfoxide (DMSO) and compound-treated samples was expressed as percent of DMSO-treated controls (100*2^ΔΔCt). Student's t-tests were performed on ΔCt values.
Immunofluorescence
Retinal explants or eyecups were fixed in 4% PFA in 1X PBS for 20 minutes at room temperature and then cryoprotected in 30% sucrose in 1X PBS overnight at 4°C. Samples were embedded in OCT (Sakura Finetek, Torrance, CA, USA), frozen on dry ice, and then sectioned at 14 to 16 μm on a cryostat (Leica Microsystems, Wetzlar, Germany). Slides were blocked with a solution containing 10% normal horse serum and 0.5% Triton X-100 in 1X PBS for 1 hour at room temperature and then stained overnight at 4°C with primary antibodies (Supplementary Table S2) diluted in blocking solution. Slides were washed three times with 1X PBS the following day and then incubated in fluorescently labeled secondary antibodies diluted in blocking solution for 2 hours at room temperature, stained with DAPI, washed, and coverslipped in medium (Fluoromount-G; SouthernBiotech, Birmingham, AL, USA). A confocal laser scanning microscope (FluoView FV1000; Olympus, Tokyo, Japan) was used for confocal microscopy. Cells were counted from single plane confocal images taken at fixed settings for each stain. For analysis of the in vivo experiment, we counted positive cells at a standardized position of each retina (the 200 μm region on the superior side of the optic nerve head). For explant experiments, the central 250 μm of at least three sections spanning the retina were counted to avoid bias in area selection or staining.
Western Blots
Retinal explants or retinas were homogenized in lysis buffer (50 mM Tris, 100 mM NaCl, 5 mM EDTA, 0.1% SDS, 1% Triton X-100, 2.5% glycerol, and 1X protease inhibitor cocktail) and equal amounts of protein samples were loaded and run in a 10% or 4% to 20% SDS gels (Bio-Rad Laboratories). Protein was transferred to a polyvinylidene fluoride membrane (Thermo Fisher Scientific, Waltham, MA, USA), blocked (5% BSA and 0.1% Tween 20 in 1X PBS) for at least 1 hour at room temperature and stained with primary antibodies (Supplementary Table S2) diluted in blocking solution overnight at 4°C. Membranes were washed with 0.1% Tween 20 in 1X PBS and then incubated with HRP-conjugated secondaries (Bio-Rad Laboratories) diluted in blocking solution for 1 hour at room temperature. Signals were visualized on X-ray film with a commercial substrate (SuperSignal West Dura Extended Duration Substrate; Thermo Fisher Scientific) and quantified using ImageJ software. For coimmunoprecipitations (Co-IP), HEK293T cells were transfected in 6-well tissue culture plates with reagent (Lipofectamine 3000; Thermo Fisher Scientific) and 800 ng of each hNRL-pCMVSport6 (Open Biosystems), hCRX-pCMVSport6 (Open Biosystems), and hNR2E3-pcDNA3.1/HisC (provided by Shiming Chen) in media (Opti-MEM; Thermo Fisher Scientific). Transfection reagents were removed after 24 hours and replaced with media containing DMSO or PR1 10 μM for 2 days. Cells were lysed with Co-IP lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% glycerol, and 1X protease inhibitor cocktail). Sheep anti-mouse IgG magnetic beads (Dynabeads; Thermo Fisher Scientific) were incubated with anti-Nr2e3 antibody (5 μg/precipitation) diluted in Co-IP buffer for 2 hours at 4°C. Equal volumes of lysate were then added to the antibody-coated beads and incubated overnight at 4°C. The following day, beads were washed four times with Co-IP buffer and then incubated at 85°C for 15 minutes in 1X sample buffer diluted in Co-IP buffer. We then performed SDS-PAGE and Western blots as described above.
Dual Luciferase Assay
We transfected HEK293T cells with 1 μg of the luciferase reporter BR-225Luc (provided by Shiming Chen), 1 ng of the control pRL-CMV (Promega Corp.), and 100 ng of hNRL-pCMVSport6 (Open Biosystems), hCRX-pCMVSport6 (Open Biosystems), or hNR2E3-pcDNA3.1/HisC (provided by Shiming Chen) in 24-well plates using a commercial reagent (Thermo Fisher Scientific). Transfection reagents were diluted in media (Thermo Fisher Scientific) and removed the following day, and replaced with media containing DMSO or PR1 10 μM. Media was changed every day. After 2 days of treatment with DMSO or PR1, cells were passed into 96-well plates (1 well of a 24-well plate into 6 wells of a 96-well plate) and lysed with 20 μL of 1X passive lysis buffer (Promega Corp.) per well. Firefly and renilla luciferase activity was measured from 10 μL of lysate per sample with a reporter assay (Dual-Luciferase Reporter Assay System; Promega Corp.) using a plate reader (1420 Multilabel Victor3V; Perkin Elmer, Waltham, MA, USA).
Injections
For intravitreal injections, adult mice (>P21) were anesthetized with isoflurane and injected with 1.5 μL of PR1 10 mM using a 32-gauge Hamilton needle. Postnatal pups were injected intraperitoneally (IP) with 20 μL of PR1 50 mM (∼190 mg/kg) or 20 μL of DMSO at P2 or P3 with a 32-gauge Hamilton needle. Timed-pregnant dams were injected IP with 100 μL of PR1 50 mM (∼95 mg/kg) at embryonic day (E)14 and E17 with a BD insulin syringe.
TUNEL Staining
We performed TUNEL staining according to manufacturer's protocol (DeadEnd Fluorometric TUNEL System; Promega Corp.) on frozen retinal sections after a brief fixation with cold 100% methanol. For analysis, the number of TUNEL-positive cells along the entire length of explant sections was counted. The number of TUNEL-positive cells was normalized to the area of the outer nuclear layer (ONL) for each section.
Results
Screen of Putative Chemical Probes of Nr2e3 in Primary Retinal Cell Cultures
Using a research application (American Chemical Society) and NCBI's PubChem Bioassays, we cheminformatically searched the results of high throughput screens that aimed to identify small molecule modulators of Nr2e3 function. A high throughput time-resolved fluorescence energy transfer (TR-FRET) biochemical assay (PubChem IDs 651849, 463256), and a high throughput cell-based luminescence assay (PubChem IDs 602229, 624378) were used to screen 315,100 and 362,351 compounds, and active compounds were then screened on confirmatory assays (363 and 1282, respectively). For the cell-based luminescence assay, the nuclear receptor interaction domain of Ncor was fused to a Gal4 DNA binding domain, and the hinge and ligand-binding domains of Nr2e3 were fused to VP16 activation domain. CHO-S cells were cotransfected with the plasmids and a reporter plasmid containing five GAL4 response elements driving luciferase. Interaction with Nr2e3-Ncor resulted in luciferase expression due to recruitment of the VP16 activator to the promoter, and compounds that reduced luciferase were identified. Based on assay scoring and compound structures, we collected a set of putative Nr2e3 interacting compounds and several structural analogs of this initial set. Because the previous high throughput screens were performed using recombinant Nr2e3 in CHO cells or a cell-free TR-FRET assay, we tested the selected compounds for suppression of rhodopsin expression in a physiologically relevant primary culture assay using dissociated retinal cells from P5 wild-type mice. We chose rhodopsin because it is a well-described target of Nr2e3 in rod photoreceptors. After a 3-day treatment period with media containing DMSO or compounds at 1 μM (Supplementary Fig. S1 for all chemical structures), we fixed the cells and assessed rhodopsin and Otx2 expression by immunohistochemistry. Using Otx2 to normalize for plating density, we found that a particular compound we named PR1 (Fig. 1A), substantially decreased rhodopsin expression compared with DMSO (Fig. 1B, Supplementary Fig. S2). This analysis also provided an initial structure-activity relationship. Specifically, the tricyclic core structure, 5,6,7,8-tetrahydrobenzo[4,5]thieno[2,3-d]pyrimidin-4(3H)-one, is important for repressing rhodopsin expression. The investigation of the side chain pyrimidinone 2-propanamide in PR1 revealed that phenylamide is also essential for the antagonistic activity, as evidenced by the fact that heteroaromatic and aliphatic amides lost activity. Additionally, replacement of the 3,5-dimethyl group with hydrogen and other functional groups in the phenyl amide diminished or abolished the compounds' effect, suggesting the unique antagonistic pattern of PR1.
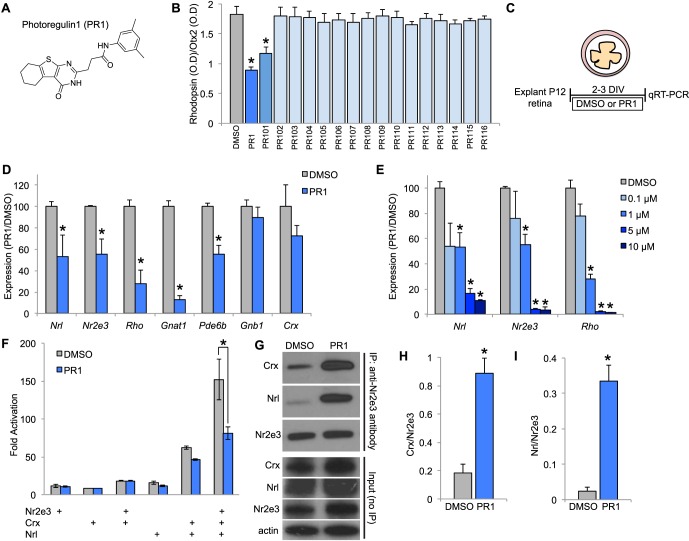
Figure 1.
(A) Chemical structure of PR1. (B) Effect of 1 μM PR1 or PR1 analogs PR101-PR116 on Rhodopsin expression (optical density) normalized to Otx2 expression (optical density) in dissociated retinal cell cultures (n = 4). *P < 0.05, ANOVA and Dunnett's tests. (C) Schematic showing experimental design for testing the effects of Nr2e3 modulators in intact retinal explant cultures for 2 to 3 DIV. (D) Reverse transcription qPCR for rod photoreceptor genes expressed in DMSO-treated controls and 1-μM PR1-treated retinal explants from P12 mice for 3 DIV (n = 4). *P < 0.05, Student's t-test. (E) Dose-response relationship of PR1 on rod-specific genes Nrl, Nr2e3, and Rhodopsin in P12 retinal explants for 2 to 3 DIV (n = 3 to 4). *P < 0.05 from DMSO treatment, Student's t-test. (F) HEK293T cells were transfected with Nr2e3, Crx, or Nrl and BR-225Luc (firefly luciferase driven by the bovine Rhodopsin promoter) and pRL-CMV (renilla luciferase driven by the CMV promoter; internal transfection control) and then treated with DMSO or PR1 10 μM for 2 days. We found PR1 decreased Rho promoter activity after transfection of Nr2e3, Crx, and Nrl (n ≥ 3). *P < 0.05, ANOVA, and Tukey's multiple comparison test. (G) Western blot analysis of DMSO and 10-μM PR1-treated HEK293T cells after transfection with Nr2e3, Crx, and Nrl followed by immunoprecipitation with an antibody generated against Nr2e3. Western blots for input (unprecipitated lysates) are shown below. (H) Quantification of Crx after immunoprecipitation with an anti-Nr2e3 antibody in DMSO and PR1-treated HEK293T cells (n = 3). *P < 0.05, Student's t-test. (I) Quantification of Nrl after immunoprecipitation with an anti-Nr2e3 antibody after treatment with DMSO or PR1 (n = 3). *P < 0.05, Student's t-test.
Loss-of-function mutations of Nr2e3 lead to a reduction in rod gene expression. To determine whether the same was true for PR1, we treated intact explant cultures of P12 retina with DMSO or PR1 1 μM and assayed rod gene expression (Fig. 1C). After 2 days in culture, PR1 decreased the expression of the rod-specific genes Nrl, Nr2e3, Rho, and Gnat1 compared with DMSO treatment by quantitative RT-qPCR analysis (Fig. 1D). However, PR1 did not significantly decrease the expression of Gnb1 or Crx, suggesting that PR1 was not causing a general loss of photoreceptors. To determine the dose-response relationship between PR1 concentration and expression of rod genes, we explanted retinas from P12 mice in media containing DMSO or PR1 at 0.1, 1, 5, or 10 μM for 2 to 3 days and assayed Nrl, Nr2e3, and Rho expression by RT-quantitative (q)PCR. We saw some effect of PR1 at a concentration as low as 0.1 μM, but statistically significant reductions were observed with the higher concentrations (Fig. 1E).
Nr2e3, Crx, and Nrl are known to activate rod gene expression by forming a complex at the promoters of rod genes. To test whether PR1 might interfere with this complex, we used luciferase reporter and co-IP assays. For the luciferase reporter assay, the Rho promoter driving firefly luciferase was cotransfected with Nrl, Nr2e3, or Crx or all of these transcription factors together into HEK293T cells. Consistent with previous reports, we found a large synergy in the activation of the Rho reporter with the combination of these three transcription factors (Fig. 1F).19 However, when PR1 was added to the cells, we observed a large reduction in the activation of the Rho promoter reporter after transfection with all three factors (Fig. 1F). These data support the hypothesis that PR1 interferes with the ability of Nr2e3, Crx, and Nrl to synergistically activate transcription of rod genes.
The previous data from the high throughput screening demonstrated that PR1 interacts with the Nr2e3 through the ligand-binding domain to inhibit its interaction with Ncor. In addition, counter screens for other nuclear hormone receptors, ROR-gamma and PPAR-gamma, did not show activity, providing evidence of specificity. However, we further asked whether PR1 interacts with the transcriptional complex of Nr2e3, Crx, and Nrl using co-IP. We transfected HEK293T cells with Nrl, Nr2e3, and Crx, and then treated the cells with DMSO or PR1. After lysis and immunoprecipitation with an antibody generated against Nr2e3, we found that PR1 significantly altered the binding of Nr2e3 with Crx and Nrl (Figs. 1G–I). The co-IP results show that PR1 causes the proteins in this complex to interact more strongly with one another; although we do not know how the increase in the interaction of these factors leads to a reduction in the ability of the complex to activate Rho transcription, the results provide evidence that PR1 interacts with this complex.
Effects of PR1 on Developing Retina
Loss-of-function mutations in Nr2e3 cause a decrease in rod gene expression and an increase in the number of S opsin+ photoreceptors. To determine whether PR1 has similar effects on developing photoreceptors, we explanted retinas from P0 mice in media containing DMSO or PR1 1 μM and assessed the level of rod gene expression with Western blots and RT-qPCR after 2 to 5 days in vitro (Fig. 2A). When the P0 retina explants were analyzed by Western blot, those treated with PR1 expressed less rhodopsin protein than DMSO controls (Figs. 2B, 2C). We tested expression of additional genes using RT-qPCR. After 2 days in culture, PR1 decreased the expression of the rod-specific genes Nrl, Nr2e3, Rho, and Gnat1 compared with DMSO (Fig. 2D). However, PR1 did not significantly decrease the expression of Gnb1, Crx, or Otx2, suggesting that PR1 was not causing a general loss of photoreceptors. We also carried out immunohistochemical analysis of sections of the explants, and observed an almost complete absence of rhodopsin immunoreactivity in the PR1-treated explants (Fig. 2E). Interestingly, PR1 treatment increased the expression of the cone gene, Thrb (Fig. 2D) by RT-qPCR analysis, and there was an overall increase in the level of S opsin immunoreactivity in sectioned explants treated with PR1 (Fig. 2E, inset) when compared with DMSO-treated control retinal explants. To determine if PR1 affects late progenitor proliferation, we cultured P0 retina explants in media with DMSO or PR1 1 μM for 3 days, and included EdU in the media for the last 24 hours of the 3-day culture period. We found no difference in the number of EdU-positive cells in DMSO and PR1-treated retinas (Supplementary Fig. S3).
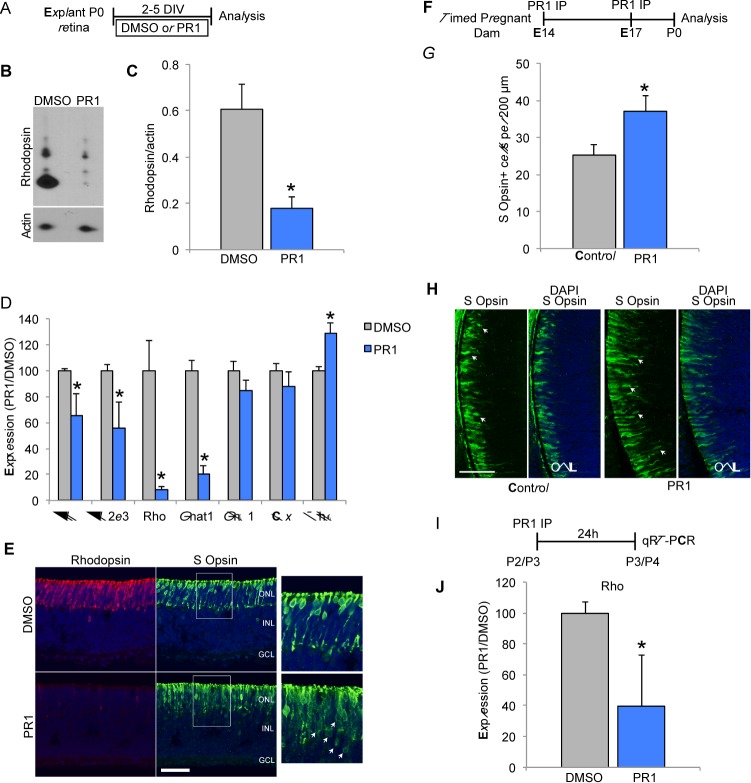
Figure 2.
(A) Schematic showing experimental design for testing the effects of PR1 in intact retinal explant cultures from P0 mice (B–E). (B) Western blot for rhodopsin shows significant reduction in P0 explants treated with PR1 1 μM for 5 DIV. (C). Rhodopsin expression was normalized to β-actin. Explants treated with PR1 had less relative expression than DMSO controls (n = 4). *P < 0.05, Student's t-test. (D) Reverse transcription qPCR for P0 explants treated with DMSO or PR1 1 μM for 2 DIV. We found Nrl, Nr2e3, Rhodopsin, and Gnat1 were significantly reduced with PR1, while Gnb1 and Crx did not change. The expression of the cone gene Thrb was increased in PR1-treated explants (n = 3). *P < 0.05, Student's t-test. (E) Sections from DMSO and 1-μM PR1-treated P0 explants stained for rhodopsin and S-opsin demonstrate a decrease in rhodopsin+ cells and an increase in S-opsin+ cells (arrows). Scale bar: 50 μm. F. Schematic for experimental design for (G–H). Timed-pregnant dams were injected with PR1 at E14 and E17 and pups were euthanized for analysis at P0. (G) Pups treated with PR1 had more S-opsin+ cells per 200 μm of central retina compared with controls (n ≥ 6 retinas). *P < 0.05, Student's t-test. (H) Sections of central retina from control and PR1 pups stained for S-opsin (arrows) and DAPI. Scale bar: 50 μm. (I) Schematic for experimental design for (J). Postnatal pups were IP injected with DMSO or PR1 at P2 or P3 and then euthanized 24 hours later for RT-qPCR analysis. (J) Photoregulin1 decreased Rho expression in P2 and −3 pups (n ≥ 4). *P < 0.05, Student's t-test.
Nr2e3 is expressed in developing photoreceptors prior to birth; therefore, to test whether PR1 could affect prenatal rod development, we injected wild-type, timed-pregnant dams with PR1 (96 mg/kg) at E14 and E17, during peak S-cone photoreceptor genesis and the onset of Nr2e3 expression (Fig. 2F). We euthanized the pups at P0 and collected their retinas to stain and quantify S opsin+ photoreceptors. We found that pups born from the PR1-injected dams had an increase in the number of S opsin+ cells when compared with controls (Figs. 2G, 2H). Next, to determine if PR1 affected rhodopsin expression in developing rods in vivo, we injected pups with PR1 (190 mg/kg) or an equal volume of DMSO at P2 or P3, during the onset of rhodopsin expression in immature rods (Fig. 2I). We euthanized the pups the following day and analyzed Rho expression by RT-qPCR. We found that pups intraperitoneally (IP) injected with PR1 had decreased Rho compared with pups injected with DMSO (Fig. 2J). Together these results show that PR1 affects rhodopsin and S-opsin expression in developing retina like a Nr2e3 loss-of-function mutation.
Effects of PR1 at Later Stages of Retinal Development
Previous loss of function genetic data has shown that Nr2e3 is required for rod photoreceptor development, but it is not known whether there is a continuing requirement for Nr2e3 in mature rods. The results from our studies of retinal explants at P12 described above suggest a continuing role for Nr2e3 in rhodopsin expression (Figs. 1D, 1E). To further examine the effects of PR1 at these late developmental stages, we analyzed the effects of PR1 on P12 retinal explants by immunohistochemical analysis (Fig. 3A). Explants from P12 Nrl-GFP mice were cultured for 5 days in either DMSO or PR1 1 μM containing media, and then fixed and processed for immunofluorescent labeling with anti-rhodopsin and anti-S opsin antibodies. The PR1-treated explants showed a dramatic decrease in rhodopsin staining in Nrl-GFP-positive rods. We also observed an increase in the number of S opsin+ cells in the ONL (Figs. 3B, 3C). Thus, PR1 has effects on rod and cone gene expression, even at late stages of photoreceptor development, suggestive of a continued requirement for Nr2e3 in rod differentiation, after their initial cell fate commitment.
Figure 3.
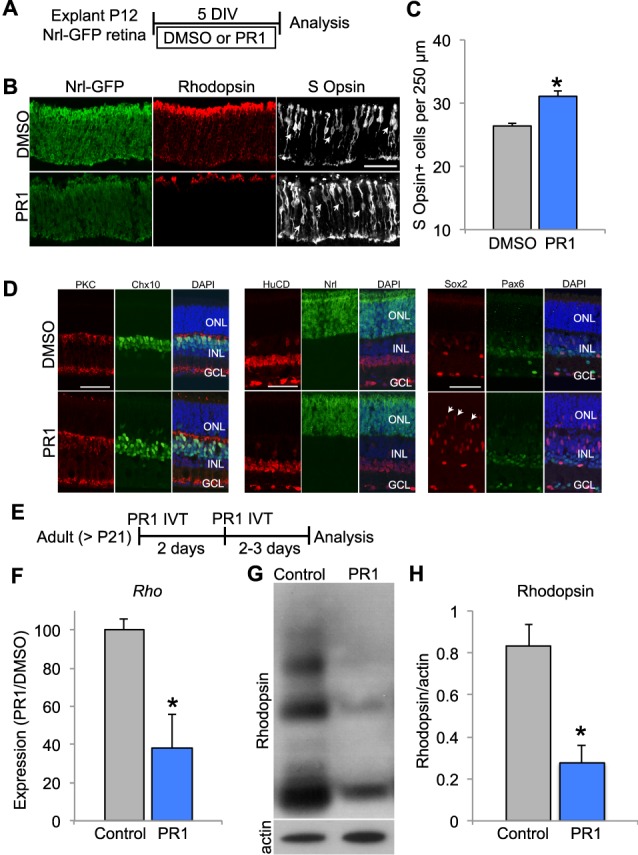
(A) Schematic for experimental design for (B–D). Retinas from P12 Nrl-eGFP mice were explanted in DMSO or PR1 1 μM for 5 DIV. (B) Staining for rhodopsin and S-opsin, and GFP (unstained) in P12 explants from Nrl-GFP mice demonstrate a decrease in rhodopsin expression and an increase in S-opsin+ cells in PR1-treated retinas. Scale bar: 50 μm. (C) Photoregulin1-treated retinas had more S-opsin+ cells (arrows) than DMSO controls per 250 μm of central retina (n = 3). *P < 0.05, Student's t-test. (D) Quantification of bipolar cells (PKCα, Chx10); amacrine cells (HuCD); and Müller glia (Sox2) revealed no difference in the number of these cells (n = 3; P > 0.05, Student's t-test). However, we observed the migration of Sox2+ Müller glia into the ONL of PR1-treated retinas (arrows). (E) Schematic for experimental design for (F–H). Adult mice received two intravitreal (IVT) injections 2 days apart of PR1 10 mM into one eye. (F) Compared with the control retina, PR1 decreased Rho expression in adult retinas (n = 3). *P < 0.05, Student's t-test. (G) Western blot for rhodopsin shows that IVT injection of PR1 decreases expression compared with the uninjected, contralateral retina of adult mice. (H) Rhodopsin expression was normalized to β-actin expression. Photoregulin1 decreased the relative expression of rhodopsin after IVT injection in adult mice (n = 5). *P < 0.05, Student's t-test.
To determine if PR1 affects retinal cells other than photoreceptors, we quantified bipolar cells (PKCα, Chx10); amacrine cells (HuCD); and Müller glia (Sox2) in explants treated with DMSO or PR1 1 μM for 5 days in vitro (DIV). We found no differences in the appearance of these other types of neurons between the DMSO and PR1-treated retinas. However, we consistently observed an increase in the migration of the Müller glial (Sox2+) nuclei into the ONL in the treated retinas (Fig. 3D). To determine if PR1 was causing reactive gliosis, we assessed GFAP expression by staining and Western blots. We saw no difference in GFAP between DMSO and PR1 retinas, suggesting that PR1 was not inducing reactive gliosis (Supplementary Fig. S4).
PR1 Reduces Rhodopsin Expression in the Retinas of Adult Mice
Conditional deletion of Nr2e3 in mature photoreceptors of adult mice has not been reported; however, conditional deletion of Nrl in mature photoreceptors leads to a partial “reprogramming” of the rods: the cells have reduced rod gene expression and an increase in the expression of some cone genes.13 Since PR1 reduces rod gene expression, including Nrl, in rods even at late stages in their development (P12), we reasoned that it might have similar effects in the retinas of adult mice. To determine if PR1 affects adult photoreceptor gene expression in vivo, we made intravitreal injections into one eye of an adult mouse (>P21) and compared expression with the other eye. We found that two intravitreal injections of PR1 made 2 days apart (Fig. 3E) decreased the expression of Rho mRNA assessed by RT-qPCR (Fig. 3F) and protein, by Western blot analysis (Figs. 3G, 3H). Interestingly, PR1 increased the expression of the cone photoreceptor marker TRβ2 in adult retinas (Supplementary Fig. S5), though we did not observe an increase in S-opsin at this age. Additionally, we examined sections from explants of adult mice treated with PR1. Similar to the explants from younger mice, adult explants had reduced Rhodopsin expression and no difference in GFAP immunoreactivity (Supplementary Fig. S6).
PR1 Slows Degeneration of Photoreceptors in RhoP23H and Pde6brd1 Retinas
The effect of PR1 on rhodopsin expression in adult rods might provide a way to slow the degeneration of these cells in dominant forms of retinitis pigmentosa (adRP), like RhoP23H. In this disease, the affected individuals express a mutant form of Rhodopsin that is likely inappropriately processed and ultimately leads to the death of the rods. To test whether reducing rhodopsin expression with PR1 might slow the degeneration of these cells, we explanted retina from RhoP23H transgenic mice at P8 in media containing DMSO or PR1 2 μM and maintained the explants for 3 days. We found that PR1 effectively reduced rhodopsin expression in the mutant rods (Figs. 4A, 4B), similar to what occurs in the wild-type retina. The majority of rod cell death in the RhoP23H transgenic line occurs between P14 and P21.28 Therefore, we made explants of retinas from RhoP23H mice at P12 and treated the explants with PR1 1 μM or DMSO. After 6 days in vitro, the retinas were processed for histology. We counted the number of nuclei (DAPI and Otx2 labeled) in the ONL of each retina in the central region. We found that PR1-treated retinas had on average approximately twice the number of photoreceptors in the ONL than DMSO-treated controls (Figs. 4C, 4D). We also monitored Gnb1 expression as another measure of rod photoreceptor preservation, since this is not directly affected by PR1 and thus serves as a surrogate for rod number. The photoregulin1-treated explants had significantly more Gnb1 expression than the sister cultures treated with DMSO (Fig. 4E).
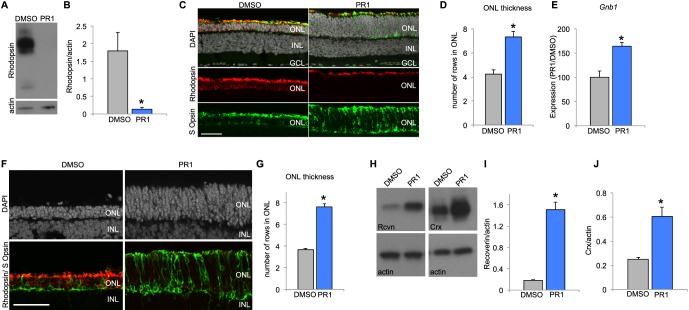
Figure 4.
(A) Retinas from P8 RhoP23H mice were explanted in DMSO or PR1 2 μM for 3 DIV. Western blot analysis shows that PR1-treated RhoP23H retinas have less Rhodopsin expression than DMSO controls. (B) Rhodopsin expression was normalized to β-actin expression. Photoregulin1–RhoP23H retinas had less relative expression of rhodopsin than DMSO-treated RhoP23H controls (n = 3). *P < 0.05, Student's t-test. (C) Retinas from P12-RhoP23H mice were explanted in DMSO or PR1 1 μM for 6 DIV. Staining with DAPI demonstrates that PR1-treated RhoP23H retinas had thicker ONLs in the central retina compared with DMSO-treated RhoP23H retinas. (D) Quantification of DAPI+ cells in the ONL of the central retina of DMSO and PR1-treated RhoP23H retinas (n = 5). *P < 0.05, Student's t-test. (E) Reverse transcription qPCR analysis of Gnb1 expression in DMSO and PR1-RhoP23H retinas suggests greater rod survival with PR1 treatment, since PR1 does not affect expression of this rod-specific transcript (n = 3). *P < 0.05, Student's t-test. (F) Retinas from P7 Pde6brd1 mice were explanted in DMSO or PR1 1 μM for 14 DIV. Retinas treated with PR1 had more DAPI+ cells in the ONL of the central retina than DMSO controls. (G) Quantification of DAPI+ cells in the ONL of DMSO and PR1-treated Pde6brd1 retinas (n = 4). *P < 0.05, Student's t-test. (H) Retinas from P7 Pde6brd1 mice were explanted in DMSO or PR1 1 μM for 12 DIV and analyzed by Western Blot. Photoregulin1-treated retinas expressed more recoverin and Crx than DMSO controls, indicating greater photoreceptor survival. (I) Recoverin expression was normalized to β-actin expression. Photoregulin1-Pde6brd1 retinas expressed more relative recoverin than DMSO-Pde6brd1 retinas (n = 3). *P < 0.05, Student's t-test. (J) Expression of Crx was normalized to β-actin expression. Photoregulin1-treated retinas had greater relative expression of Crx than DMSO controls, indicating greater photoreceptor survival (n = 3). *P < 0.05, Student's t-test.
We tested the ability of PR1 to prevent photoreceptor degeneration in another mouse model of RP, the Pde6brd1 mutation. Because the onset of rod degeneration is earlier in this model, we explanted retinas from Pde6brd1 mice in media containing DMSO or PR1 1 μM at P7. After 14 days in vitro, we collected the retinas for histology. Similar to the effect in RhoP23H mice, we found that PR1-treated retinas had thicker ONL than DMSO controls (Figs. 4F, 4G). Additionally, PR1-treated Pde6brd1 retinas had fewer TUNEL-positive nuclei than DMSO controls, indicating less apoptosis of photoreceptors with PR1 treatment (Supplementary Fig. S7). To confirm our histologic finding, we performed Western blot analysis on whole retina lysates from P7 Pde6brd1 retinas treated with DMSO or PR1 1 μM for 12 days. We found PR1-treated Pde6brd1 retinas had more relative expression of photoreceptor markers recoverin and Crx compared with DMSO controls, confirming greater photoreceptor preservation.
Discussion
In this study, we report the first demonstration of small molecule repression of rod gene expression for the potential treatment of dominant retinitis pigmentosa. The expression of Crx, Otx2, Nrl, and Nr2e3 in mature rods is consistent with their role in the maintenance of proper photoreceptor gene expression and homeostasis.32 We screened compounds that were previously identified as Nr2e3 interacting compounds (using high-throughput FRET and cell-based Nr2e3:Ncor interaction screens) for their ability to modulate rod gene expression in developing retinal cells in vitro. We found that a subset of these compounds inhibited the expression of rhodopsin in the screen and we further characterized one of these, PR1, for its ability to inhibit expression of rod genes in developing and mature rod photoreceptors. The activity of PR1 on rod photoreceptors is similar to loss-of-function mutations in Nr2e3, and significantly reduces expression from the rhodopsin promoter in HEK293T cells transfected with Nr2e3, Crx, and Nrl, and affects the binding of Nr2e3 to Crx and Nrl. Together the data support the conclusion that PR1 interacts with Nr2e3 and acts to antagonize its activity, most likely through its binding with transcriptional cofactors.
The results from the previous screens indicate that PR1 can interact with Nr2e3, most likely through the ligand binding domain, and inhibit the interactions with the corepressor Ncor. Our data also demonstrate that PR1 can inhibit expression from the rhodopsin promoter, driven by Nr2e3-Crx-Nrl cotransfection in HEK cells. Somewhat paradoxically, immunoprecipitation shows that PR1 causes an increase in the binding of Nr2e3 with Crx and Nrl. This may be due to changes in the ligand binding domain of Nr2e3 upon PR1 binding. The protein Crx is thought to interact with Nr2e3 through the DNA binding domain (DBD),19 and so it is unlikely that PR1 would affect this interaction directly. Several Nr2e3 variants associated with enhanced S-cone syndrome reduce Nr2e3 binding with Nrl (p.L336P, p.L353V, and p.R385P); however, two of these impair Nrl/Crx-mediated transactivation of the Rhodopsin promoter (p.L336P,p.L353V ), while another potentiates transactivation.33 Thus, there is a complex relationship between the binding affinity of Nrl and Nr2e3 and the ability of this complex to activate rod gene transcription, and this may relate to interactions with corepressors and coactivators, which are not completely understood for this system. Additionally, Nr2e3 homodimerization is affected by many disease-causing mutations in the DBD of Nr2e3, and these mutations can both potentiate and repress transactivation of the Rhodopsin promoter.34 It is also possible that the effect of PR1 on decreasing transcription factor-mediated activation of the Rho promoter could be in part due to changes in Nr2e3 homodimerization, and we cannot detect changes in homodimer formation from the co-IP results.
The primary effect we observe after treatment of the retina, either in vitro or in vivo, is a reduction in the expression of rod photoreceptor expressed genes, like Rho and Gnat1. However, not all rod genes are reduced to the same extent; we see no changes in Gnb1 expression, for example. Moreover, both Nrl and Nr2e3 are significantly reduced by PR1 treatment at either P0 or P12, and some of the effects we observe on rod gene expression may be due to the reduction in these transcription factors. Similar results are seen after conditional knockout of Nrl in adult mice, in that rod genes are more affected than cone genes.13 Additionally, fewer cone genes are upregulated following knockout in the adult compared to germline knockout of Nrl, possibly due to developmental changes in the methylation status of cone gene promoters.13 Nonetheless, this partial “reprogramming” via conditional knockout of Nrl is sufficient to prevent photoreceptor degeneration in the Rho knockout model of RP,13 similar to our findings with the RhoP23H and Pde6brd1 models in vitro. Together, these studies demonstrate that downregulation of rod gene program in degenerative diseases that primarily affect rods may be an effective strategy for treatment in humans. Suppression of rhodopsin or other commonly mutated rod genes with siRNAs may also be similarly effective.35,36
Interestingly, our effects on rod gene expression are much more pronounced than the effects reported from Nr2e3 loss-of-function models, such as the rd7 mouse and the targeted knockout of Nr2e3.19,21,23,24,32 In these mice, rod genes like Rho and Gnat1 are only modestly reduced, while we observed large changes with PR1 treatment. This difference may be due to our finding that PR1 also decreases the expression of Nrl, while these genetic mutations do not, and Nrl may be sufficient to drive rod gene expression in the absence of Nr2e3. It also remains possible that PR1 inhibits Nrl expression through an Nr2e3-independent mechanism. In addition to changes in the expression of rod genes, rd7 mice show a derepression of some cone genes. Similarly, mutations in Nr2e3 can cause enhanced S-cone syndrome and patients present increased sensitivity to blue light.37 We did not observe very large increases in S-opsin expression that we expected from antagonism of Nr2e3, particularly in the mature retina. It is possible that PR1 selectively affects the ability of Nr2e3 to act as a transcriptional activator (inhibiting its ability to form a complex with Crx and Nrl), but has less of an effect on its repressor functions.38 However, it is also possible that acute loss of Nr2e3 has different effects than developmental deletions. Future studies will be needed to distinguish between these alternatives.
In addition to Nr2e3, there are several other nuclear receptors that regulate photoreceptor development and maintenance (see Ref. 39 for review), and might constitute targets for the treatment of retinal disease. Retinoid X receptors, thyroid hormone receptor-beta 2, and Coup-TF are important in cone photoreceptor development, regulating the expression of S- and M-opsin. In addition to Nr2e3, Nr1d1 (RevErbα), and Nr3b2 (ERRβ) are important in rod photoreceptor development and maintenance.40,41 Retinoic acid receptor–related orphan receptor beta (RORβ) coordinates the development of rods and cones, by activating expression of Nrl.42 Although our evidence suggests that Nr2e3 is a potential target of PR1, it is also possible that one of these other nuclear receptors binds PR1 as well. For example, if PR1 also inhibits the activity of RORβ, this might explain the changes we observe in Nrl expression.
In conclusion, we demonstrate that a small molecule can regulate Rho expression in developing and mature retina, and could provide a novel approach to the treatment of dominant forms of RP. The ability to modulate rod and cone gene expression may also have utility in recessive forms of the disease, since conditional deletion of Nrl in mature mice provided rod protection in the Rho–/– mouse. The possibility for small molecule reprogramming of a cell affected with a disease causing mutation to a related cell that does not express the mutant allele might also have application in other genetic disorders.
Supplementary Material
Acknowledgments
The authors thank members of the Reh lab for their critique, J. Brzezinski, PhD, for comments on the manuscript, and Shiming Chen, PhD, for the Rhodopsin reporter and expression constructs.
Supported by NIH R01 EY021374 (to TAR, SD, and KZ).
Disclosure: P.A. Nakamura, None; S. Tang, None; A.A. Shimchuk, None; S. Ding, Inception Biosciences (C); T.A. Reh, Inception Biosciences (C)
References
- 1. Hartong DT,, Berson EL,, Dryja TP. Retinitis pigmentosa. Lancet. 2006; 368: 1795– 1809. [DOI] [PubMed] [Google Scholar]
- 2. Nguyen AT,, Campbell M,, Kiang AS,, Humphries MM,, Humphries P. Current therapeutic strategies for P23H RHO-linked RP. Adv Exp Med Biol. 2014; 801: 471– 476. [DOI] [PubMed] [Google Scholar]
- 3. Gorbatyuk MS,, Knox T,, LaVail MM,, et al. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proc Natl Acad Sci U S A. 2010; 107: 5961– 5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mao H,, Gorbatyuk MS,, Rossmiller B,, Hauswirth WW,, Lewin AS. Long-term rescue of retinal structure and function by rhodopsin RNA replacement with a single adeno-associated viral vector in P23H RHO transgenic mice. Hum Gene Ther. 2012; 23: 356– 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carter-Dawson LD,, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979; 188 245– 262. [DOI] [PubMed] [Google Scholar]
- 6. Swaroop A,, Kim D,, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010; 11: 563– 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brzezinski JA,, Reh TA. Photoreceptor cell fate specification in vertebrates. Development. 2015; 142: 3263– 3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitton KP,, Swain PK,, Chen S,, Xu S,, Zack DJ,, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000; 275: 29794– 29799. [DOI] [PubMed] [Google Scholar]
- 9. Pittler SJ,, Zhang Y,, Chen S,, et al. Functional analysis of the rod photoreceptor cGMP phosphodiesterase alpha-subunit gene promoter: Nrl and Crx are required for full transcriptional activity. J Biol Chem. 2004; 279: 19800– 19807. [DOI] [PubMed] [Google Scholar]
- 10. Yoshida S,, Mears AJ,, Friedman JS,, et al. Expression profiling of the developing and mature Nrl-/- mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004; 13: 1487– 1503. [DOI] [PubMed] [Google Scholar]
- 11. Mears AJ,, Kondo M,, Swain PK,, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001; 29: 447– 452. [DOI] [PubMed] [Google Scholar]
- 12. Oh EC,, Khan N,, Novelli E,, Khanna H,, Strettoi E,, Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci U S A. 2007; 104: 1679– 1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montana CL,, Kolesnikov AV,, Shen SQ,, Myers CA,, Kefalov VJ,, Corbo JC. Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proc Natl Acad Sci U S A. 2013; 110: 1732– 1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kobayashi M,, Takezawa S-I,, Hara K,, et al. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci U S A. 1999. 96: 4814– 4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oh EC,, Cheng H,, Hao H,, Jia L,, Khan NW,, Swaroop A. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 2008; 1236: 16– 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bumsted O'Brien KM,, Cheng H,, Jiang Y,, Schulte D,, Swaroop A,, Hendrickson AE. Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Invest Ophthalmol Vis Sci. 2004; 45: 2807– 2812. [DOI] [PubMed] [Google Scholar]
- 17. Haider NB1,, Naggert JK,, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001; 10: 1619– 1626. [DOI] [PubMed] [Google Scholar]
- 18. Cheng H,, Khanna H,, Oh EC,, Hicks D,, Mitton KP,, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004; 13: 1563– 1575. [DOI] [PubMed] [Google Scholar]
- 19. Peng G,, Ahmad O,, Ahmad F,, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005; 14: 747– 764. [DOI] [PubMed] [Google Scholar]
- 20. Haider NB,, Mollema N,, Gaule M,, et al. Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res. 2009; 89: 365– 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J1,, Rattner A,, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005; 25: 118– 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng H,, Aleman TS,, Cideciyan AV,, Khanna R,, Jacobson SG,, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet. 2006; 15: 2588– 2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corbo JC,, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005; 1: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng H,, Khan NW,, Roger JE,, Swaroop A. Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3 originate from early-born photoreceptor precursors. Hum Mol Genet. 2011; 20: 4102– 4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milam AH,, Rose L,, Cideciyan AV,, et al. The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc Natl Acad Sci U S A. 2002; 99: 473– 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolkenberg SE,, Zhao Z,, Kapitskaya M,, et al. Identification of potent agonists of photoreceptor-specific nuclear receptor (NR2E3) and preparation of a radioligand. Bioorg Med Chem Lett. 2006; 16: 5001– 5004. [DOI] [PubMed] [Google Scholar]
- 27. Zhao Z,, Wang L,, Wen Z,, et al. Systematic analyses of the cytotoxic effects of compound 11a, a putative synthetic agonist of photoreceptor-specific nuclear receptor (PNR), in cancer cell lines. PLoS One. 2013; 8: e75198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sakami S,, Maeda T,, Bereta G,, et al. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011; 286: 10551– 10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akimoto M,, Cheng H,, Zhu D,, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006; 103: 3890– 3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ueki Y,, Karl MO,, Sudar S,, et al. P53 is required for the developmental restriction in Muller glial proliferation in mouse retina. Glia. 2012; 60: 1579– 1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ueki Y,, Wilken MS,, Cox KE,, Chipman LB,, Bermingham-McDonogh O,, Reh TA. A transient wave of BMP signaling in the retina is necessary for Müller glial differentiation. Development. 2015; 142: 533– 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Webber AL,, Hodor P,, Thut CJ,, et al. Dual role of Nr2e3 in photoreceptor development and maintenance. Exp Eye Res. 2008; 87: 35– 48. [DOI] [PubMed] [Google Scholar]
- 33. von Alpen D,, Tran HV,, Guex N,, et al. Differential dimerization of variants linked to enhanced S-cone sensitivity syndrome (ESCS) located in the NR2E3 ligand-binding domain. Hum Mutat. 2015; 36: 599– 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roduit R,, Escher P,, Schorderet DF. Mutations in the DNA-binding domain of NR2E3 affect in vivo dimerization and interaction with CRX. PLoS One. 2009; 4: e7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernan I,, Gamundi MJ,, Planas E,, Borràs E,, Maseras M,, Carballo M. Cellular expression and siRNA-mediated interference of rhodopsin cis-acting splicing mutants associated with autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2011; 52: 3723– 3729. [DOI] [PubMed] [Google Scholar]
- 36. Gorbatyuk M,, Justilien V,, Liu J,, Hauswirth WW,, Lewin AS. Suppression of mouse rhodopsin expression in vivo by AAV mediated siRNA delivery. Vision Res. 2007; 47: 1202– 1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haider NB,, Jacobson SG,, Cideciyan AV,, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000; 24: 127– 131. [DOI] [PubMed] [Google Scholar]
- 38. Tan MH,, Zhou XE,, Soon FF,, et al. The crystal structure of the orphan nuclear receptor NR2E3/PNR ligand binding domain reveals a dimeric auto-repressed conformation. PLoS One. 2013; 8: e74359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Forrest D,, Swaroop A. Minireview: the role of nuclear receptors in photoreceptor differentiation and disease. Mol Endocrinol. 2012; 26: 905– 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mollema NJ,, Yuan Y,, Jelcick AS,, et al. Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS One. 2011; 6: e17494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Onishi A,, Peng GH,, Poth EM,, et al. The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc Natl Acad Sci U S A. 2010; 107: 11579– 11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia L,, Oh EC,, Ng L,, et al. Retinoid-related orphan nuclear receptor RORbeta is an early-acting factor in rod photoreceptor development. Proc Natl Acad Sci U S A. 2009; 106: 17534– 17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.