Abstract
In this work, we present a novel microfluidic biosensor for sensitive fluorescence detection of DNA based on 3D architectural MoS2/multi-walled carbon nanotube (MWCNT) nanocomposites. The proposed platform exhibits a high sensitivity, selectivity, and stability with a visible manner and operation simplicity. The excellent fluorescence quenching stability of a MoS2/MWCNT aqueous solution coupled with microfluidics will greatly simplify experimental steps and reduce time for large-scale DNA detection.
Keywords: MoS2/MWCNT nanocomposites, DNA fluorometric detection, microfluidic biosensing
1. Introduction
Over the last two decades, various nanomaterials including graphene oxide [1,2,3,4], Au nanoparticles [5,6,7], metal-organic framework (MOF) [8,9,10], carbon nanotubes [11,12], and MoS2 nanosheets [13,14,15,16,17,18,19] have been synthesized to develop nanomaterial-based biosensors for a variety of applications. In particular, there has been increasing interest in developing rapid, cost-effective, and sensitive methods for DNA detection because of its vital importance in molecular diagnostics, environmental monitoring, and anti-bioterrorism [3]. Among these nanomaterials for DNA detection, 2D MoS2, evolved as a fluorescence nanoquencher, has recently attracted extensive research efforts due to its unique properties [20,21,22]. Despite the advantages of MoS2, such as high quenching efficiency, easy synthesis at a large scale, and good dispersion in aqueous solutions [23], its quenching ability is not very stable and would be strongly affected by moisture and oxygen ubiquitous in the environment [24]. Furthermore, similar to the 2D graphene, the freshly synthesized MoS2 nanosheets have a tendency to aggregate in practical applications, which can greatly reduce the electrochemical active sites [25]. In our previous work [15], we found that the quenching ability of MoS2 aqueous solutions can substantially degrade within one day due to oxidization. It is vital to perform experiments immediately after preparation of the MoS2 aqueous solution, which is not convenient for a routine DNA test. The combination of MoS2 and other materials may overcome this deficiency. A MoS2/multi-walled carbon nanotube (MWCNT) composite as an electrode in a Li battery has shown a higher capacity and better cycling stability compared with pure MoS2 [26,27]. It was also reported that the thermal stability of the MoS2/MWCNT composites was better than MWCNT and MoS2 [28]. Huang et al. applied MoS2/MWCNT as an electrochemical biosensor that used graphene oxide as a redox marker while a thiol-tagged DNA probe was assembled on a MoS2/MWCNT and AuNP-modified electrode [28]. However, little attention has been paid to its application as a nanoquencher for DNA sensing. Here, to solve the degradation problem of pure MoS2 nanosheets for DNA detection, a novel 3D nanomaterial with MoS2 nanosheets grown on MWCNT is proposed in this work as a new fluorescence nanoquencher with high stability and sensitivity to detect single-stranded DNA (ssDNA) that can be adsorbed by van der Waals forces between MoS2 and nucleobases [13] and even helical wrapping to the MWCNT surface through п-stacking [29]. The novel MoS2/MWCNT composite is in a three dimensional (3D) hierarchical structure with 1D MWCNTs as back bones and the 2D MoS2 grown on the outer surface of MWCNTs with partially standing branch features [26], which provides a much larger material surface area for DNA sensing. The MoS2/MWCNT-based DNA sensing is performed in a microfluidic channel device that helps maintain uniform fluorescence intensity and thus ensures consistent fluorescence measurements in different locations. Using microfluidic channels can substantially reduce the sample volume needed for fluorescence measurement, typically less than 0.2 μL. This microfluidic chip has five identical channels, which offers a simple, rapid, portable, and high-throughput analysis method for screening of DNA.
2. Experimental Section
2.1. Reagents and Apparatus
Synthesized and purified DNA oligonucleotides were directly purchased from Integrated DNA Technologies Pte. Ltd. in Singapore. The sequences of all DNA oligonucleotides are shown in Table 1.
Table 1.
Sequences of DNA oligonucleotides used in this study.
| DNA Name | Sequence (5′–3′) |
|---|---|
| Probe ssDNA (P1) | 5′-TAMRA-TGCGAACCAGGAATT-3′ |
| Perfect complementary ssDNA (T1) | 5′-AATTCCTGGTTCGCA-3′ |
| Single-base mismatched ssDNA (M1) | 5′-AATTCCTTGTTCGCA-3′ |
| Non-complementary ssDNA (N1) | 5′-CTGCAAGACCGGATT-3′ |
MWCNTs (L-MWCNTs-60100) (SEM image is shown as the supplementary information in Figure S1) were purchased from Shenzhen Nanotech Port Co., Ltd., Shenzhen, China. Other chemicals were purchased from Sigma-Aldrich Pte. Ltd., St. Louis, MO, USA. The deionized water used in all the experiments was purified using a Millipore filtration system. Fluorescence images were taken with a CCD camera (Leica, Model MC120HD, Wetzlar, Germany) on an inverted fluorescence microscope (Leica, Model DMIL LED). Fluorescence spectra were measured by Raman spectroscopy (WITec alpha300 R, WITec Pte. Ltd., Ulm, Germany) at the excitation wavelength of 532 nm. Laser spot was focused into a circle with a diameter of ~1.3 µm, and the fluid volume in the optical lightpath was 5.309 × 10−8 µL. All measurements were performed at room temperature in 1× phosphate buffered saline (PBS, pH 7.4), which contains 11.9 mM phosphatase, 2.7 mM potassium chloride, and 137 mM sodium chloride.
2.2. Synthesis of MoS2/MWCNT
MoS2/MWCNT nanocomposites were obtained using solvent thermal method [26], in which N,N-dimethylformamide (DMF) and (NH4)2MoS4 were used as the solvent and single reactant, respectively. In a typical experiment, briefly explained, 220 mg (NH4)2MoS4 powder and 100 mg MWCNTs were mixed first with the 30 mL solvent and sonicated to achieve uniform dispersion. Thereafter, an autoclave at 200 °C for 10 h was applied to form MoS2/MWCNT composites. After separation of the product by centrifugation (9391 g, 5 min), the purified MoS2/MWCNT composites were further washed up by DI water at least 5 times. Three different ratios of MoS2/MWCNT were synthesized by varying the Mo/Carbon ratio of 1:20, 1:10, and 1:5.
2.3. Optimization of Detection Conditions
The loading effect of MoS2 and MWCNT on the DNA detection was investigated by using three different MoS2/MWCNT ratios: 1:20, 1:10, and 1:5. A portion of 100 nM dye-labeled probe DNA was incubated for 10 min with different ratios of MoS2/MWCNT at room temperature. The concentrations of the different MoS2/MWCNT ratios are all 150 µg/mL. The fluorescence spectra were measured using Raman spectroscopy at the same location near the end of the microchannel. Similar procedures for the best performance ratio were also performed to investigate the effect of different concentrations of MoS2/MWCNT nanocomposites on the fluorescence quenching ability. Fluorescence spectra of pure PBS solution tested in the microchannel were used as the baseline, and results presented in this work were spectra minus this baseline. Error bars are obtained from at least six groups of data.
2.4. Fluorescent DNA Assays
Zigzag-shaped polydimethylsiloxane (PDMS) microchannels (Figure S2) were used as a platform for fluorescence measurement and imaging. The mixed samples were added into each channel in the device by a micro pipet in a volume of 0.2 µL and Raman spectroscopy was used to measure the fluorescence spectra at the end of the microchannels. In a typical DNA assay, the fluorescent probe P1 was hybridized with the targets in 1 × PBS (pH 7.4) for 10 min, after which the resulting solution was uniformly mixed with MoS2/MWCNT aqueous solution using a vortex mixer. All the concentrations mentioned in the following measurements are final concentrations in the mixture.
3. Results and Discussion
3.1. Working Mechanism
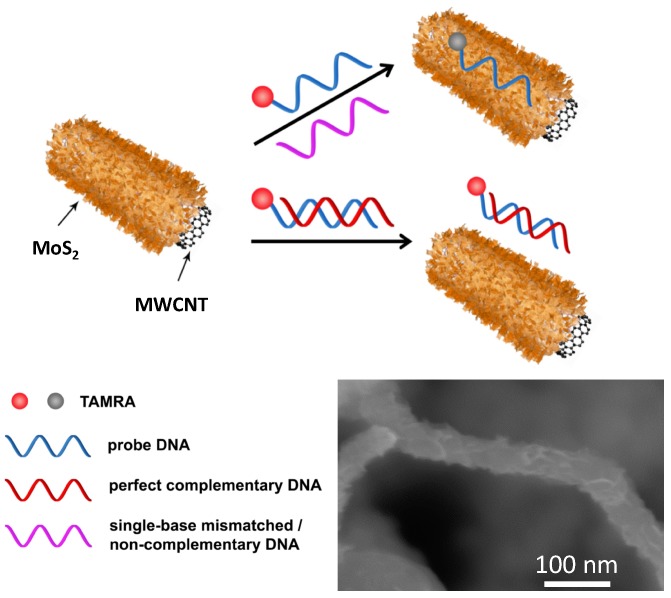
As depicted in Figure 1, a probe DNA (P1) labeled with TAMRA is utilized to detect its perfect complementary DNA (T1) where TAMRA was excited at 532 nm and the emission wavelength was 580 nm. MoS2/MWCNT (SEM image is shown in Figure 1, inset) can adsorb a TAMRA-labeled single-stranded probe DNA (P1) via the van der Waals force between nucleobases and the basal plane of MoS2 nanosheets, as well as helical wrapping to the MWCNT surface through п-stacking, and the fluorescence of the probe DNA is thus quenched. A double-stranded DNA (dsDNA) is formed when the perfect complementary target DNA (T1) is hybridized with P1. The fluorescence of P1 is retained well even with the presence of MoS2/MWCNT due to the weak binding of MoS2/MWCNT to dsDNA. As a result, P1 fluorescence intensity can offer a quantitative assessment of T1. However, when single-base mismatched (M1) and non-complementary DNA (N1) are used for hybridization, the strong fluorescence of P1 will be largely quenched in the presence of MoS2/MWCNT because of the formation of non-perfect dsDNA. Furthermore, the novel MoS2/MWCNT composite has a 3D hierarchical structure providing a large surface area for interacting with the DNA strand.
Figure 1.
Schematic illustration of the MoS2/MWCNT nanocomposites-based fluorescence DNA sensing assay. Inset shows an SEM image of the MoS2/MWCNT nanocomposites.
3.2. Fluorescence Quenching Ability of MoS2/MWCNT Nanocomposites
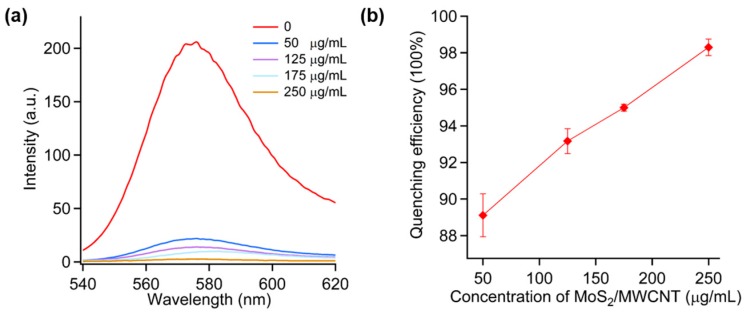
Figure S3 shows that a 1:5 ratio of MoS2/MWCNT has the highest quenching ability. This could be attributed to the formation of the hierarchical structure of MoS2 and MoS2 layers extruding from the sidewall of MWCNTs with the enhancement of the Mo/carbon ratio [26], which provides a larger interacting surface area for the DNA strand. The effect of the concentration of MoS2/MWCNT nanocomposites in the aqueous solution on the fluorescence quenching ability was investigated by measuring the fluorescence intensity in mixtures of 100 nM probe DNA and different concentrations of MoS2/MWCNT with the 1:5 ratio. Figure 2a depicts the fluorescence spectra of the 100 nM probe DNA quenched by different concentrations of MoS2/MWCNT. It is shown that the fluorescence intensity decreased with the increase of MoS2/MWCNT concentration. Figure 2b displays the quenching efficiency of MoS2/MWCNT that is defined as (F0 − F)/F0 × 100%, where F and F0 are the fluorescence intensities of the solutions at the wavelength of λ = 580 nm in the presence and absence of MoS2/MWCNT, respectively. It was found that the quenching efficiency is higher than 88%, even at a low concentration (50 μg/mL) of MoS2/MWCNT. Since the quenching efficiency of MoS2/MWCNT is very important for the proposed DNA sensor, we chose a 1:5 ratio and 250 μg/mL concentration reaching 98.3% quenching efficiency for the other assays in this work.
Figure 2.
(a) Fluorescence spectra of P1 (100 nM) in the presence of various concentrations of MoS2/MWCNT (50, 125, 175, and 250 μg/mL); (b) Quenching efficiency of corresponding concentration of MoS2/MWCNT.
3.3. DNA Detection Sensitivity
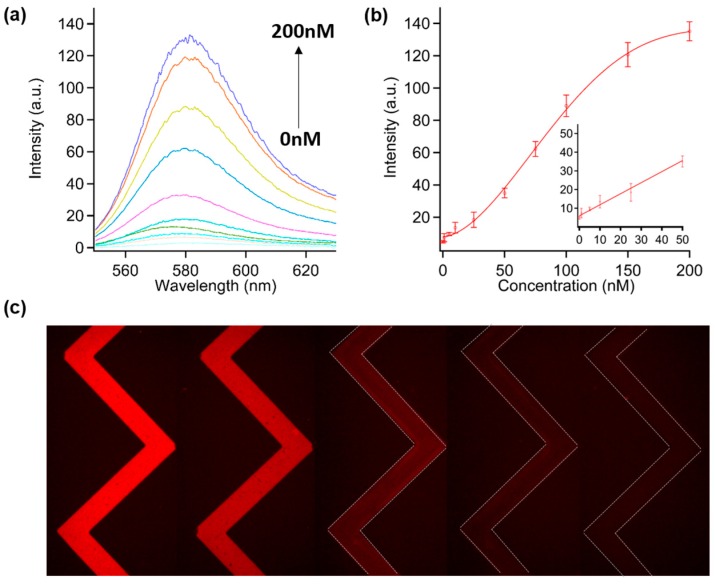
The sensitivity of the proposed biosensor was investigated by changing the target DNA concentration. In a typical experiment, after 10 min incubation of P1 mixed with T1 at different concentrations (1–200 nM) at ambient temperature, MoS2/MWCNT nanocomposites were added to this hybridized P1T1 solution and mixed by using a vortex mixer and then added into microchannels. The detection time was less than 5 min and the DNA hybridization time was about 10 min. The fluorescence spectra of the above experiments with its corresponding derived calibration can be seen in Figure 3. The measurement of the spectra was performed at the end of the microchannels (Figure 3a). With the increase in T1 concentration, a larger amount of P1 was hybridized with T1 to form a dsDNA and resulted in higher fluorescence of P1 remained. It should be mentioned that, when the T1 concentration (150 and 200 nM) exceeded P1 (100 nM), there was still an increase in the fluorescence intensity. This might have arisen as a result of the fact that the redundant T1 has a stronger interaction with MoS2/MWCNT in comparison to hybridized P1T1 and could replace the P1T1 duplex adsorbed on MoS2/MWCNT; hence, a higher amount of P1T1 duplex remained in the solution. According to the derived calibration curve (Figure 3b), the detection limit of 1 nM with good linearity from 0 to 50 nM was achieved by this DNA biosensor. The linear range is wider than previous studies on MoS2-based fluorescence assays [13,30]. Notably, because the effective volume of DNA solution was less than 0.2 µL inside the microchannel, the presented microfluidic biosensor can detect the target DNA as low as 1 fmol, which is comparable to our previous work with pure MoS2 nanosheets [15] and much lower than other nanoprobe-based fluorescence methods in bulk solutions [13,30]. In addition, a table (Table S1) with a comprehensive comparison of various MoS2-based biosensors is provided in the supplementary information. Figure 3c demonstrates that the P1T1 fluorescence decreased along with the reduction of the T1 concentration, and ∼fmol of T1 produced a visible red color in the presence of MoS2/MWCNT.
Figure 3.
(a) Fluorescence spectra of P1 with a variety of T1 (0, 1, 5, 10, 25, 50, 75, 100, 150, and 200 nM) in the presence of MoS2/MWCNT (250 μg/mL); (b) Calibration curve for DNA detection; (c) Fluorescence images of P1T1 in the presence of various T1 concentrations (left to right; 150, 100, 75, 50 and 10 nM). The concentration of P1 is 100 nM.
3.4. DNA Detection Selectivity
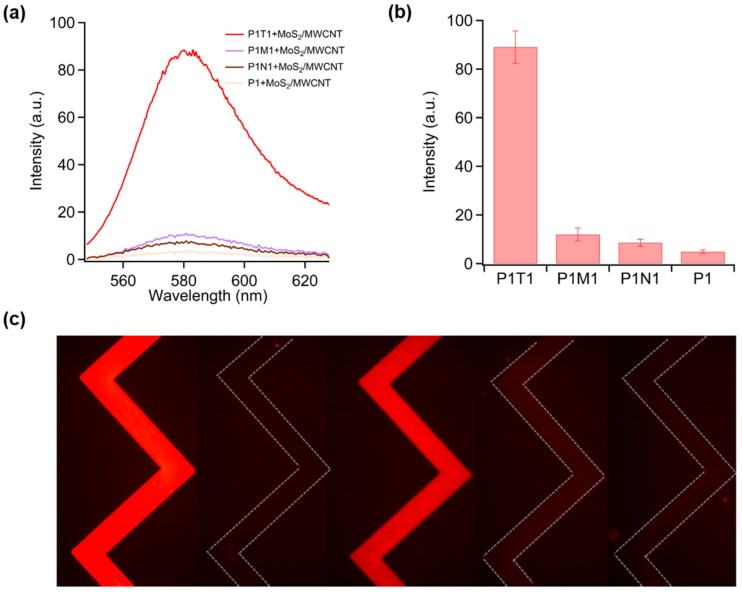
The selectivity of the proposed methods was also examined by introducing the control experiments into the sensing system, including single-base mismatched ssDNA (M1) and non-complementary ssDNA (N1). As shown in Figure 4a,b, even at a very high concentration (100 nM), neither M1 nor N1 could produce a significant fluorescence increase. P1T1 showed a much stronger fluorescence than P1, P1M1, and P1N1 in the MoS2/MWCNT solution. It was also found that the fluorescence intensity of P1 was slightly lower than P1M1 and P1N1. This might be attributed to the redundant M1 or N1 that would compete with P1 and replace a small amount of the adsorbed P1 on MoS2/MWCNT; as a result, more P1 was maintained to produce slightly higher fluorescence intensity. Figure 4c illustrates that T1 induced a much stronger red color than M1 and N1 at the same concentration. The color of P1 appeared a bit weaker than that of P1 mixed with 100 nM M1 and N1 in the presence of MoS2/MWCNT, which is in good agreement with our fluorescence measurement (shown in Figure 4a,b).
Figure 4.
(a) Fluorescence spectra of 100 nM P1, P1T1, P1M1, and P1N1 in the presence of MoS2/MWCNT; (b) Selectivity of the MoS2/MWCNT-based target DNA (T1) sensor over single-base mismatched (M1) and non-complementary (N1) sequences; (c) Fluorescence images of P1, P1/(MoS2/MWCNT), P1T1/(MoS2/MWCNT), P1M1/(MoS2/MWCNT), and P1N1/(MoS2/MWCNT) from left to right. P1, T1, M1, and N1 have the same concentration of 100 nM.
3.5. Stability of MoS2/MWCNT Aqueous Solution
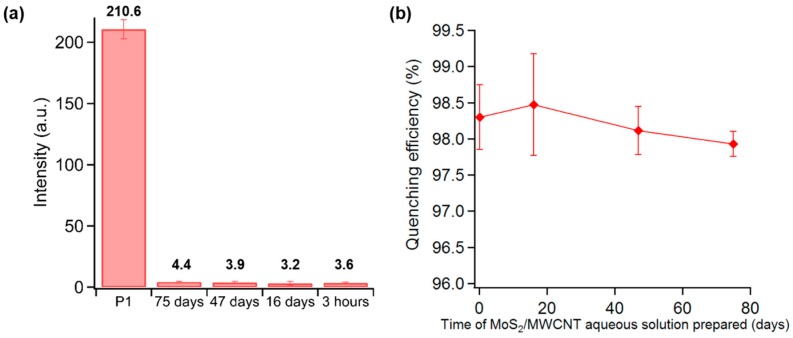
A significant advantage of the proposed biosensor stems from the high quenching stability of the MoS2/MWCNT aqueous solution compared with the pure MoS2 nanosheet aqueous solution. In a typical experiment, MoS2/MWCNT aqueous solutions prepared at different days (75, 47, and 16 days prior, as well as 3 h prior) were sonicated again for 10 min, mixed with 100 nM P1 by vortex mixer and then added into microchannels for fluorescence measurement. Figure 5a,b show that MoS2/MWCNT aqueous solution has a high stability of quenching ability, which can reach 98% even for MoS2/MWCNT aqueous solutions prepared 75 days prior. Therefore, prolonged storage of MoS2/MWCNT solutions for more than two months does not affect its quenching efficiency and stability. It is worth mentioning that these prepared MoS2/MWCNT solutions were kept in ambient temperature without any special ventilation or prevention from the average humidity of 80% in Singapore. This quenching stability will greatly reduce the time for sample preparation. Moreover, this simple, high-throughput, and homogeneous assay can be completed within a few minutes. Hence, the proposed method will greatly reduce time for large-scale DNA sensing studies.
Figure 5.
(a) Fluorescence intensity of 100 nM P1 at the emission wavelength of λ = 580 nm without and with MoS2/MWCNT prepared 75, 47, and 16 days prior, as well as 3 h prior; (b) Quenching efficiency of MoS2/MWCNT prepared 75, 47, and 16 days prior, as well as 3 h prior.
4. Conclusions
We developed a novel 3D architectural MoS2/MWCNT nanocomposite-based microfluidic biosensor for the sensitive detection of DNA molecules. Several advantages are summarized as follows: (1) It can detect 1 nM DNA with 0–50 nM linear range; (2) Compared with other nanomaterials such as MoS2, the MoS2/MWCNT aqueous solution has an excellent quenching stability for storage more than 75 days without further processing, which is a great convenience for routine DNA tests; (3) The sample volume can be largely reduced with the use of microfluidic channels; hence, ~fmol DNA detection could be achieved with a visible color of fluorescence within a few minutes through the microfluidic assay. It offers a simple and rapid approach for high-throughput DNA analysis. The excellent quenching stability of the MoS2/MWCNT aqueous solution coupled with microfluidics can greatly simplify steps of DNA testing experiments and reduce time for large-scale DNA sensing studies, demonstrating that MoS2/MWCNTs can be a promising nanocomposite for versatile fluorometric biosensing.
Acknowledgments
We gratefully acknowledge the support received from SUTD Startup Research Grant (SREP13053) and CGH-SUTD HealthTech Innovation Research Fund (T1MOECGH1501) awarded to Y.A.
Supplementary Materials
The following are available online at http://www.mdpi.com/1424-8220/16/11/1911/s1. Figure S1: Scanning electron microscopy (SEM) image of pristine multi-walled nanotubes (MWNTs). Figure S2: Channel geometry of the polydimethylsiloxane (PDMS) microfluidic device. The microchannel height is 40 µm and width is 100 µm. Figure S3: Fluorescence spectra of P1 (100 nM) in the presence of different ratios of MoS2/MWCNT (1:5, 1:10, and 1:20). The concentrations of the different MoS2/MWCNT ratios are all 150 µg/mL. Table S1: DNA detection performance of existing MoS2-based biosensors.
Author Contributions
D.Y., Y.H., H.Y.Y. and Y.A. developed the idea. D.Y., M.T. and Y.A. designed the experiments. H.Y.Y. synthesized MoS2/MWCNT nanocomposite. D.Y. and M.T. performed all the rest of experiments. D.Y., M.T. and Y.A. analyzed the results and wrote the paper. All authors read the paper and confirmed all results.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zhang Q., Kong D.-M. A general fluorescent sensor design strategy for “turn-on” activity detection of exonucleases and restriction endonucleases based on graphene oxide. Analyst. 2013;138:6437–6444. doi: 10.1039/c3an01447a. [DOI] [PubMed] [Google Scholar]
- 2.Tao Y., Lin Y., Huang Z., Ren J., Qu X. DNA-templated silver nanoclusters-graphene oxide nanohybrid materials: A platform for label-free and sensitive fluorescence turn-on detection of multiple nucleic acid targets. Analyst. 2012;137:2588–2592. doi: 10.1039/c2an35373c. [DOI] [PubMed] [Google Scholar]
- 3.Lu C.H., Li J., Liu J.J., Yang H.H., Chen X., Chen G.N. Increasing the sensitivity and single-base mismatch selectivity of the molecular beacon using graphene oxide as the “nanoquencher”. Chemistry. 2010;16:4889–4894. doi: 10.1002/chem.200903071. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y., Yang H.Y., Ai Y. DNA single-base mismatch study using graphene oxide nanosheets-based fluorometric biosensors. Anal. Chem. 2015;87:9132–9136. doi: 10.1021/acs.analchem.5b03037. [DOI] [PubMed] [Google Scholar]
- 5.Ma J.L., Yin B.C., Le H.N., Ye B.C. Label-free detection of sequence-specific DNA based on fluorescent silver nanoclusters-assisted surface plasmon-enhanced energy transfer. ACS Appl. Mater. Interfaces. 2015;7:12856–12863. doi: 10.1021/acsami.5b03837. [DOI] [PubMed] [Google Scholar]
- 6.Lee H., Lee K., Kim I.-K., Park T.G. Fluorescent gold nanoprobe sensitive to intracellular reactive oxygen species. Adv. Funct. Mater. 2009;19:1884–1890. doi: 10.1002/adfm.200801838. [DOI] [Google Scholar]
- 7.Deng D., Gu Y., Li Y., Xue J., Wang J., Ai G., Li X. Gold nanoparticle-based beacon to detect STAT5B mRNA expression in living cells: A case optimized by bioinformatics screen. Int. J. Nanomed. 2015;10:3231–3244. doi: 10.2147/IJN.S81754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye T., Liu Y., Luo M., Xiang X., Ji X., Zhou G., He Z. Metal-organic framework-based molecular beacons for multiplexed DNA detection by synchronous fluorescence analysis. Analyst. 2014;139:1721–1725. doi: 10.1039/c3an02077k. [DOI] [PubMed] [Google Scholar]
- 9.Jung S., Kim Y., Kim S.-J., Kwon T.-H., Huh S., Park S. Bio-functionalization of metal-organic frameworks by covalent protein conjugation. Chem. Commun. 2011;47:2904–2906. doi: 10.1039/c0cc03288c. [DOI] [PubMed] [Google Scholar]
- 10.Liu W.-L., Lo S.-H., Singco B., Yang C.-C., Huang H.-Y., Lin C.-H. Novel trypsin–fitc@ mof bioreactor efficiently catalyzes protein digestion. J. Mater. Chem. B. 2013;1:928–932. doi: 10.1039/c3tb00257h. [DOI] [PubMed] [Google Scholar]
- 11.Chen H., Wang J., Liang G., Zhang P., Kong J. A novel exonuclease III aided amplification method for sensitive nucleic acid detection based on single walled carbon nanotube induced quenching. Chem. Commun. 2012;48:269–271. doi: 10.1039/C1CC16127J. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z., Yang R., You M., Zhang X., Wu Y., Tan W. Single-walled carbon nanotube as an effective quencher. Anal. Bioanal. Chem. 2010;396:73–83. doi: 10.1007/s00216-009-3192-z. [DOI] [PubMed] [Google Scholar]
- 13.Zhu C., Zeng Z., Li H., Li F., Fan C., Zhang H. Single-layer MoS2-based nanoprobes for homogeneous detection of biomolecules. J. Am. Chem. Soc. 2013;135:5998–6001. doi: 10.1021/ja4019572. [DOI] [PubMed] [Google Scholar]
- 14.Balcioglu M., Rana M., Robertson N., Yigit M.V. DNA-length-dependent quenching of fluorescently labeled iron oxide nanoparticles with gold, graphene oxide and MoS2 nanostructures. ACS Appl. Mater. Interfaces. 2014;6:12100–12110. doi: 10.1021/am503553h. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y., Shi Y., Yang H.Y., Ai Y. A novel single-layered MoS2 nanosheet based microfluidic biosensor for ultrasensitive detection of DNA. Nanoscale. 2015;7:2245–2249. doi: 10.1039/C4NR07162J. [DOI] [PubMed] [Google Scholar]
- 16.Huang J., Ye L., Gao X., Li H., Xu J., Li Z. Molybdenum disulfide-based amplified fluorescence DNA detection using hybridization chain reactions. J. Mater. Chem. B. 2015;3:2395–2401. doi: 10.1039/C4TB01986E. [DOI] [PubMed] [Google Scholar]
- 17.Jin K., Xie L., Tian Y., Liu D. Au-modified monolayer MoS2 sensor for DNA detection. J. Phys. Chem. C. 2016;120:11204–11209. doi: 10.1021/acs.jpcc.6b01193. [DOI] [Google Scholar]
- 18.Kalantar-Zadeh K., Ou J.Z. Biosensors based on two-dimensional MoS2. ACS Sens. 2015;1:5–16. doi: 10.1021/acssensors.5b00142. [DOI] [Google Scholar]
- 19.Kalantar-zadeh K., Ou J.Z., Daeneke T., Strano M.S., Pumera M., Gras S.L. Two-dimensional transition metal dichalcogenides in biosystems. Adv. Funct. Mater. 2015;25:5086–5099. doi: 10.1002/adfm.201500891. [DOI] [Google Scholar]
- 20.Radisavljevic B., Radenovic A., Brivio J., Giacometti I.V., Kis A. Single-layer MoS2 transistors. Nat. Nanotechnol. 2011;6:147–150. doi: 10.1038/nnano.2010.279. [DOI] [PubMed] [Google Scholar]
- 21.Mak K.F., Lee C., Hone J., Shan J., Heinz T.F. Atomically thin MoS2: A new direct-gap semiconductor. Phys. Rev. Lett. 2010;105:136805. doi: 10.1103/PhysRevLett.105.136805. [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Zeng Z., Cao X., Lu G., Wang L.H., Fan Q.L., Huang W., Zhang H. Preparation of MoS2-polyvinylpyrrolidone nanocomposites for flexible nonvolatile rewritable memory devices with reduced graphene oxide electrodes. Small. 2012;8:3517–3522. doi: 10.1002/smll.201200999. [DOI] [PubMed] [Google Scholar]
- 23.Xiang X., Shi J., Huang F., Zheng M., Deng Q., Xu J. MoS2 nanosheet-based fluorescent biosensor for protein detection via terminal protection of small-molecule-linked DNA and exonuclease III-aided DNA recycling amplification. Biosens. Bioelectron. 2015;74:227–232. doi: 10.1016/j.bios.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 24.Loan P.T., Zhang W., Lin C.T., Wei K.H., Li L.J., Chen C.H. Graphene/MoS2 heterostructures for ultrasensitive detection of DNA hybridisation. Adv. Mater. 2014;26:4838–4844. doi: 10.1002/adma.201401084. [DOI] [PubMed] [Google Scholar]
- 25.Ren D., Jiang H., Hu Y., Zhang L., Li C. Self-assembling few-layer MoS2 nanosheets on a Cnt backbone for high-rate and long-life lithium-ion batteries. RSC Adv. 2014;4:40368–40372. doi: 10.1039/C4RA08604J. [DOI] [Google Scholar]
- 26.Shi Y., Wang Y., Wong J.I., Tan A.Y.S., Hsu C.-L., Li L.-J., Lu Y.-C., Yang H.Y. Self-assembly of hierarchical MoSx/Cnt nanocomposites (2 < x < 3): Towards high performance anode materials for lithium ion batteries. Sci. Rep. 2013;3:2169. doi: 10.1038/srep02169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y.M., Yu X.Y., Li Z., Paik U., Lou X.W.D. Hierarchical MoS2 tubular structures internally wired by carbon nanotubes as a highly stable anode material for lithium-ion batteries. Sci. Adv. 2016;2:e1600021. doi: 10.1126/sciadv.1600021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang K.J., Liu Y.J., Wang H.B., Wang Y.Y., Liu Y.M. Sub-femtomolar DNA detection based on layered molybdenum disulfide/multi-walled carbon nanotube composites, au nanoparticle and enzyme multiple signal amplification. Biosens. Bioelectron. 2014;55:195–202. doi: 10.1016/j.bios.2013.11.061. [DOI] [PubMed] [Google Scholar]
- 29.Aravind S.S.J., Ramaprabhu S. Noble metal dispersed multiwalled carbon nanotubes immobilized ss-DNA for selective detection of dopamine. Sens. Actuators B Chem. 2011;155:679–686. doi: 10.1016/j.snb.2011.01.029. [DOI] [Google Scholar]
- 30.Li J., Huang Y., Wang D., Song B., Li Z., Song S., Wang L., Jiang B., Zhao X., Yan J., et al. A power-free microfluidic chip for SNP genotyping using graphene oxide and a DNA intercalating dye. Chem. Commun. 2013;49:3125–3127. doi: 10.1039/c3cc40680f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.