ABSTRACT
Gene expression patterns change dramatically during neuronal development. Proliferating cells, including neural stem cells (NSCs), express telomere repeat-binding factor 2 (TRF2), a nuclear protein that associates with telomeric proteins, DNA, and RNA telomeres. In NSCs TRF2 also binds to the transcription regulator REST to facilitate repression of numerous neuron-specific genes, thereby keeping the NSCs in a self-renewing state. Upon neuronal differentiation, TRF2 levels decline, REST-regulated neuronal genes are derepressed, and a short isoform of TRF2 arises (TRF2-S) which localizes in the cytoplasm, associates with different subsets of proteins and transcripts, and mobilizes axonal G-rich mRNAs. We recently identified two RNA-binding proteins, HNRNPH1 and H2 (referred to jointly as HNRNPH due to their high homology), which mediate the alternative splicing of an exon required for the expression of full-length TRF2. As HNRNPH levels decline during neurogenesis, TRF2 abundance decreases and TRF2-S accumulates. Here, we discuss the shared and unique functions of TRF2 and TRF2-S, the distinct subcellular compartment in which each isoform resides, the subsets of proteins and nucleic acids with which each interacts, and the functional consequences of these ribonucleoprotein interactions. This paradigm illustrates the dynamic mechanisms through which splicing regulation by factors like HNRNPH enable distinct protein functions as cells adapt to developmental programs such as neurogenesis.
KEYWORDS: Neuronal development, RNA-binding proteins, splicing, ribonucleoprotein complex, REST, HNRNPH
Introduction
In mammalian cells, more than 100,000 proteins are predicted to be synthesized from 20,000–25,000 protein-coding genes1,2 at least in part by rearrangement of protein-coding exons in a tissue- and developmental stage-specific manner. This process, known as alternative splicing, is essential for maintaining proteomic complexity.1 Indeed, more than 90% of human genes that contain more than one exon undergo alternative splicing with variable expression among tissues.3-5 The inclusion or exclusion of particular exons from the final, processed mRNA leads to the formation of distinct protein isoforms that harbor specific sets of domains or signaling peptides, which in turn alters the functionality and/or subcellular localization of final protein products.2
Distinct from dividing cells, postmitotic neurons exhibit extreme polarity that allows signal propagation from dendrites to axons. Dendrites are short protrusions around the cell body and receive input from afferent neuron axons, whereas axons are long neurites that propagate action potentials to trigger neurotransmitter release from presynaptic terminals at synapses. The establishment and maintenance of neuronal polarity in part requires the precise localization of mRNAs in dendrites and axons so that protein synthesis can be regulated locally by synaptic activation and signals such as neurotrophic factors.6,7 In addition, neurons exhibit an unusually high number of alternative splicing events, whereby neuron-specific protein isoforms are produced to control neuronal properties such as excitability, neurite outgrowth and synaptogenesis.8
Regulation of TRF2 splicing and function during neurogenesis
In mammalian nuclei, the ends of chromosomes or telomeres, comprise stretches of TTAGGG repeats. Telomeres are protected by a protein complex termed shelterin that protects chromosome termini against nucleolytic processing events and prevents the triggering of a DNA damage response.9-13 Yet, during S-phase, this telomere-capping structure is disassembled in order to proceed with DNA replication and is reassembled after replication.14,15 Among the shelterin proteins, telomere repeat-binding factor 2 (TRF2, also known as TERF2 and TRBF2) has emerged as a key scaffolding molecule for remodeling telomeres throughout the replicative stages of a cell.10,15,16 TRF2 was originally discovered as a telomeric DNA-binding protein via its C-terminal SAND/Myb-like DNA-binding domain.13 TRF2 also functions as a protein hub to recruit numerous shelterin and non-shelterin proteins through its N-terminal GAR domain and its TRFH dimerization domain (Table 1).
Table 1.
TRF2- and TRF2-S-associated proteins and nucleic acids. Top, molecules (proteins, DNA, RNA) that associate with TRF2 (column 1), domain of TRF2 implicated (column 2), influence of these interactions on TRF2 function (column 3), and bibliography describing these functions (column 4). Bottom, molecules (proteins and RNAs) that associate with TRF2-S (column 1), domain of TRF2-S implicated (column 2), influence of these interactions on TRF2-S function (column 3), and bibliography describing these functions (column 4).
| TRF2 binding partners | TRF2 domain | TRF2 function | References |
|---|---|---|---|
| Duplex telomeric DNA | Myb | TRF2 protects telomeres from end-to-end fusions | 64 |
| DNA replication fork | GAR | TRF2 promotes telomere replication | 65 |
| TERRA RNA | GAR | TRF2 recruits ORC and TERRA to telomeres | 32 |
| PARP1, poly(ADP-ribose) polymerase | GAR | TRF2 recruits PARP1 to damaged telomeres | 66 |
| PRMT1, protein methyltransferase | GAR | Arginines in TRF2 GAR domain are methylated | 36 |
| OPC1 (origin recognition complex) | GAR | TRF2 is recruited to telomeres at S-phase | 32 |
| REST (neuron-restrictive transcription repressor) | TRFH | TRF2 prevents REST degradation | 46 |
| hREST4 (REST splicing isoform) | TRFH-F120 | TRF2 prevents hREST4 degradation | 49 |
| RAP1 (repressor/activator1) | TRFH-F120 | TRF2 forms shelterin complex | 67 |
| SLX4 (endonuclease) | TRFH-F120 | TRF2 assembles with multiple endonucleases | 68 |
| Apollo (nuclease) | TRFH-F120 | TRF2 protects telomeres in S phase | 38 |
| RTEL1 (helicase) | TRFH | TRF2 unwinds telomere t-loops in S phase | 11 |
| WRN (RecQ helicase) | N/A | TRF2 lacking GAR domain forms telomeric circles, telomere recombination, and senescence | 69 |
| ATM (sensor of double-stranded DNA damage) | N/A | TRF2 inhibits ATM activation at telomeres | 14 |
| MRE11 (nuclease) |
N/A |
TRF2 prevents assembly of TERRA-LSD1-MRE11 at damaged telomeres |
31 |
| TRF2-S binding partners |
TRF2-S domain |
TRF2-S function |
References |
| G-rich mRNAs | GAR | TRF2-S binds to and facilitates neuron mRNA trafficking | 50 |
| FMRP | GAR | TRF2-S binding to mRNA is suppressed by FMRP | 50 |
| REST | TRFH | TRF2-S sequesters REST in the cytoplasm | 17 |
Although the telomeric role of TRF2 in dividing cells has been extensively investigated for nearly 2 decades, it is poorly understood whether TRF2 and other shelterin proteins expressed in postmitotic neurons function to safeguard telomeres. While studying this question, we discovered a C-terminal truncated TRF2 splicing isoform (TRF2-S) that was expressed as neurons differentiated and grew their axons and dendrites,17 and 5 years later we identified the splicing factor HNRNPH as being important for regulating neuronal TRF2 alternative splicing.18 We also reported recently that instead of telomeric DNA, TRF2-S displayed a preference for binding RNA, and that TRF2-S is present in axons where it regulated mRNA trafficking.19 In the present Extra Views, we briefly review the distinct functions of the nuclear, telomere-protective TRF2, and the cytoplasmic RNA-regulatory TRF2-S. We discuss the generation of the two TRF2 splicing isoforms and the role of HNRNPH in regulating the production of TRF2 in neural stem cells (NSCs) and TRF2-S in mature neurons.
Nuclear role of TRF2
TRF2 was originally identified as part of the shelterin complex that protects telomeres.10,11 TRF2 was found to bind selectively to double-stranded telomeric DNA aided by other components of the shelterin complex, TRF1, POT1, TPP1, TINs and RAP1.20 Progressive telomere attrition or deficiency of the shelterin complex is recognized as a break in double-stranded DNA and elicits genotoxic response which can trigger apoptosis.21,22 To prevent such genotoxic response, TRF2 protects telomeres and thereby prevents activation of the ATM (ataxia telangiectasia mutated)-mediated DNA damage response23,24; TRF2 further inhibits ATM by interacting with an upstream regulator of ATM, the checkpoint kinase 2 (CHEK2)25 and suppressing CHEK2 phosphorylation. Loss of TRF2 induces chromosome end-to-end fusions mediated by the non-homologous end-joining (NHEJ) pathway.26 TRF2 was proposed to achieve this function by folding and maintaining the telomeric DNA in a lasso-like structure called a ‘t-loop’, a configuration that sequesters the telomere end and blocks a DNA damage response.24 This structure likely limits the loading of the Ku70/80 heterodimer onto the telomeres and prevents NHEJ.21,26 TRF2 also contributes to the synthesis of functional 3′ G-overhangs, which are required for folding telomeres into t-loop structures,27,28 to the unwinding of t-loops,10 and to the maintenance of telomere integrity following the topological stress encountered during DNA replication in S phase.29,30 In addition, TRF2 prevents excess accumulation of long non-coding telomeric repeat-containing RNAs, TERRA,31 which are transcribed from subtelomeric regions and function as structural components of telomeres.32-34
TRF2 acts as a protein ‘hub’ to recruit shelterin and non-shelterin proteins through its N-terminal GAR domain and its central TRFH dimerization domain (Table 1). The lack of a GAR domain can induce DNA recombination at telomeres and telomere deletion,35 whereas amino acid substitutions can promote telomere doublet formation.36 These findings further support a key role of TRF2 in maintaining telomere integrity. In the C-terminus, TRF2 contains a DNA-binding Myb/SANT domain (Telobox) through which it interacts with sequence-specific telomeric DNA.37 The homodimerization domain of TRF2 in the middle of the protein forms a horseshoe structure in its dimeric form (TRFH for TRF homology domain).38 This domain is responsible for preventing ATM activation24 and for the transcriptional control of TERRA expression.31,33 It was recently discovered that this domain interacts with DNA through lysines and arginines, and that a 90-bp long DNA segment wraps around the homodimer.16
Given its function in the DNA damage response, TRF2 is implicated in development and in diseases such as cancer.39 TRF2-deficient mice die during embryonic development and TRF2 is essential for adult skin homeostasis.40,41 TRF2 is a potent oncogene in telomerase-deficient mice42; conversely, since TRF2 inhibits the recruitment of natural killer (NK) cells to tumors, cells with reduced TRF2 are more easily eliminated by NK cells.43 In addition, TRF2 overexpression promotes telomere shortening, an effect not caused by inhibition of telomerase44 but instead by replication stalling in duplex telomeric regions and the formation of ultrafine anaphase bridges (UFBs), triggering chromosome fusions similar to those seen in human cancers.45
Besides its function on telomeres, extra-telomeric functions of TRF2 have recently emerged. TRF2 is upregulated during human embryonic stem cell differentiation and promotes differentiation by binding to and preventing the proteasomal degradation of REST (Repressor Element‐1 Silencing Transcription Factor).46 Since REST binds to the promoters of many genes that are critical for neuronal differentiation and inhibits their transcription,47 REST stabilization represses neuronal gene transcription in NSCs, thereby inhibiting neuronal differentiation and enabling continued proliferation of the NSCs.46 An isoform of REST, hREST4, counters the gene-silencing effect of REST in a dominant negative fashion. Depending on the cell context, TRF2 could either stabilize REST to maintain self-renewal of NSCs,46,48 or prevent hREST4 from ubiquitin-mediated proteasomal degradation to promote neuronal differentiation.46,49 A dominant negative form of TRF2 triggers a DNA damage response and senescence in mitotic neural cells and TRF2 inhibition enhances the differentiation of hippocampal neurons.50
Molecular switch of TRF2 to TRF2-S by alternative splicing
Alternative splicing of the TRF2 mRNA (Trf2 mRNA in rodents) leads to the production of a short protein isoform named TRF2-S.18 Alternative splicing is a regulated process because recognition and recruitment of the spliceosomal machinery is suboptimal on alternative splice sites.51 Trf2 mRNA contains 10 exons. In rodents, TRF2-S arises when an alternative 5′ splice site on exon 7 is utilized; this creates a variant mRNA that contains only part of exon 7. As a consequence, a shift occurs in the open reading frame that gives rise to a premature termination codon on exon 8 and a smaller protein isoform is synthesized. TRF2-S lacks the DNA-binding domain and the nuclear localization signal (NLS) and instead possesses a short sequence that retains the protein in the cytoplasm17 and enables distinct functions (Fig. 1A and B).
Figure 1.
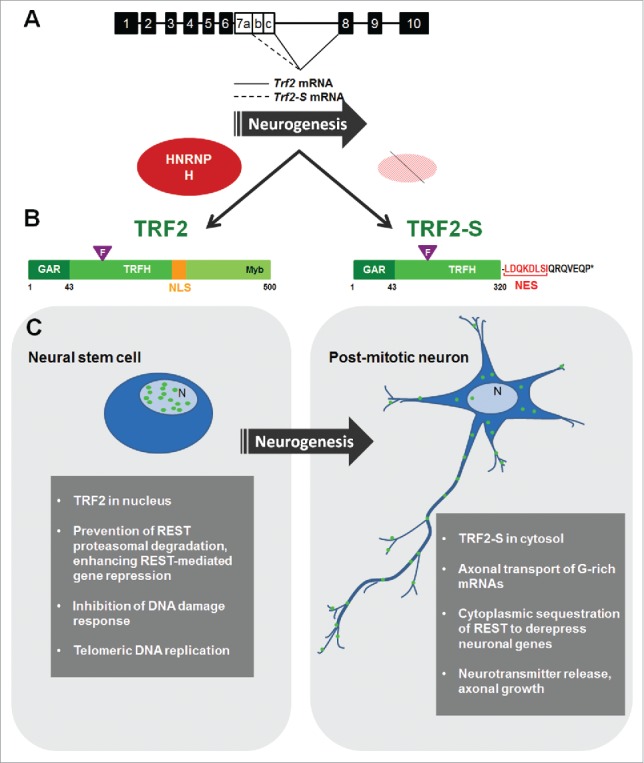
Regulation and impact of alternatively spliced TRF2 and TRF2-S on neurogenesis. (A) Schematic of the alternatively spliced exon 7 giving rise to full length TRF2 (left) and a truncated, short form, TRF2-S (right). In dividing neural cells, the splicing factor HNRNPH1/H2 (HNRNPH) enables a splicing pattern leading to high levels of TRF2. During neurogenesis, HNRNPH levels decline, favoring the formation of the short isoform TRF2-S. (B) The two isoforms share the GAR and TRFH domains, but TRF2 has a nuclear localization signal (NLS), whereas TRF2-S has a nuclear export signal (NES) that mobilizes TRF2-S to the cytoplasm. ‘F’ indicates the TRF2-Phe120 site that interacts with the [F/Y]xL signature motif on hREST4. (C) Schematic representation of the distinct subcellular localizations TRF2 (green ovals) and TRF2-S (green circles) in dividing (left) and post-mitotic (right) neural cells, respectively. The gray shaded boxes summarize the major functions of TRF2 (left) and TRF2-S (right) in different differentiation states.
To regulate alternative splicing events, pre-mRNA sequences within the regulated exon or near the splice sites (<300 nucleotides apart) act as cis-elements recognized by RNA-binding proteins (RBPs) to bind and block or promote the recruitment of the spliceosome to the site, in turn driving the skipping or inclusion of alternatively spliced exons in the mature transcript.52 This regulatory process was observed for TRF2 and TRF2-S. By conducting a proteomic screen, we identified several RBPs that bind to rat Trf2 pre-mRNA using as bait exon 7 along with flanking intronic sequences, where we expected to find regulatory cis-elements. Among the RBPs that were pulled down from rodent brain lysate, HNRNPH1 and HNRNPH2 (2 highly conserved proteins, >95%) positively regulated exon 7 inclusion and inhibited TRF2-S production, although HNRNPH1 appeared to suppress splicing and TRF2-S production more robustly.18 Crosslinking and immunoprecipitation revealed that human HNRNPH1 also bound TRF2 pre-mRNA, and that the binding sequence was a G-rich tract containing A residues.53,54 HNRNPH binding sites on and near exon 7 were further identified using in vitro binding assays. Interestingly, 3 binding sites were identified as potentially regulating splicing and all 3 appeared to be partially redundant.18 Both paralogs were implicated in regulating alternative splicing. The significance of HNRNPH in controlling alternative splicing of neural genes was previously shown in oligodendrocytes.55,56 Of note, HNRNPH1 regulates alternative splicing of the neuron-specific exon N of REST.57
The role of HNRNPH on TRF2 splicing in neuronal differentiation was tested in two model systems. In one, treatment of rat pheochromocytoma PC12 cells with nerve growth factor for 7 days to induce neuritogenesis caused a gradual decrease in HNRNPH1/2 levels, suggesting an inhibitory role of the protein during differentiation. In another, primary cortical neurons from rats at embryonic day 18 differentiated over the course of 15 days; during this time, a similar decrease in HNRNPH1/2 protein was observed, coinciding with a rise in TRF2-S expression. Interestingly, Hnrnph1 and Hnrnph2 mRNAs did not decline, suggesting that HNRNPH proteins were regulated via changes in protein translation or stability. To test directly the hypothesis that HNRNPH was involved in neuronal differentiation, HNRNPH2 was suppressed in PC12 cells using CRIPSR-Cas9 or shRNA. Inhibition of HNRNPH2 increased the number of differentiated cells earlier in the process, implicating it in suppressing neuronal differentiation through inhibition of TRF2-S.18
Cytoplasmic TRF2-S shifts its binding preference to other RNAs and proteins
Mammalian telomeric DNA and RNAs (TERRA) contain tandem repeats of the G-rich sequences (TTAGGG)n and (UUAGGG)n, respectively.58 Both sets of sequences can form a 4-stranded helical structure named G-quadruplex or G-quartet with multiple stacking of G·G·G·G tetrads, although their topologies may differ if they consist of single or double strands of oligonucleotide.59 G-quadruplexes are commonly present in RNAs as a distinct motif recognized by a subset of RBPs and participate in many aspects of mRNA metabolism, including localization, translation, and turnover. RBPs can bind G-quartet-containing RNAs via the GAR domain, also known as an ‘RGG box’ because it harbors arrays of glycine-arginine repeats (Fig. 1B).19 For example, the RGG box in the fragile X mental retardation protein (FMRP) binds intramolecular G-quartets in mRNAs encoding proteins important for neuronal function, and regulates local protein synthesis in dendrites.60,61 Nonetheless, this general model was challenged by the recent discovery that a single FMRP target, DgkK mRNA, encoding a key regulator of signaling lipid production (diacylglycerol kinase κ), is translationally activated by FMRP and was proposed to elicit the major FMRP actions.62
TRF2 was reported to bind TERRA RNAs via its N-terminal GAR domain,32 and DNA via its C-terminal Myb-DNA binding domain. Distinct from its telomere-binding counterpart TRF2, TRF2-S retains the GAR and TRFH domains, but its nuclear localization signal is replaced by a nuclear export signal, and it lacks a Myb DNA-binding domain.17,19 Using E18 rat embryonic cortical neuron cultures, we performed RNP immunoprecipitation (RIP) analysis to identify TRF2-S-associated neuronal mRNAs and identified 140 mRNAs that were highly enriched in the TRF2-S RIP. Using approaches such as in vitro crosslinking and IP (CLIP) and biotin RNA pulldown analyses, we were able to map the TRF2-S–mRNA-binding sequences in Trf2-S, Aplp1, and Rab3a mRNAs. Tagging RNA with MS2 hairpins, further revealed that G-rich, TRF2-S target mRNAs localized in axons.19 These studies suggest that the TRF2-S GAR domain is involved in assembling and transporting G-rich mRNA-protein complexes in the cytoplasm of axons in neurons. We therefore hypothesize that when TRF2 is bound to the DNA-shelterin complex, the GAR domain binds TERRA RNAs; however, since TRF2-S is cytoplasmic and thus not bound to shelterin, the GAR domain binds other RNAs (e.g. neuronal G-rich mRNAs) and participates in the post-transcriptional regulation of neuronal protein production.
Cytoplasmic TRF2-S interacts with protein subsets
TRF2 and TRF2-S can bind to a broad spectrum of non-shelterin proteins (Table 1). In particular, we and others showed that in the cytoplasm of neurons, TRF2-S binds and stabilizes REST, in turn maintaining the self-renewal of NSCs,17,48 or binds the truncated, dominant negative isoform hREST4, thus promoting neuronal differentiation.46,49 In addition, TRF2-S binds and sequesters REST in the cytoplasm of neurons to antagonize REST nuclear activity and to maintain neuronal traits.17 It is worth noting that hREST4 harbors a [F/Y]xL signature motif that can be recognized by a TRF2-Phe120 binding site in TRFH domain.49 Since the TRF2-Phe120 is conserved in both TRF2 and TRF2-S (Fig. 1B and C), it is reasonable to predict that TRF2-S may interact dynamically with nuclear or/and cytoplasmic protein networks during neurogenesis.63,64
Besides the TRFH domain, the GAR domain in TRF2-S is also capable of binding non-shelterin proteins [e.g., PARP1, ORC, and FMRP (Table 1)]. Interestingly, we recently reported that FMRP binds to the TRF2-S GAR domain in the absence of RNA, thereby blocking the interaction of TRF2-S with its target mRNAs. On the other hand, the TRF2 GAR domain binds to TERRA RNAs, facilitating heterochromatin formation and the recruitment of origin recognition complex at telomeres.32 Therefore, there are rich and dynamic interactions of the GAR domain in TRF2 and TRF2-S with RNAs and proteins. These interactions provide molecular insight into the regulation of gene expression patterns by TRF2/TRF2-S in dividing NSCs and postmitotic neurons.
Concluding remarks
In neuronal cells, alternative splicing gives rise to TRF2 and TRF2-S, two proteins with distinct functions, subcellular localization, and interacting molecules. Proliferating neural cells, such as NSCs and glial cells, express the long isoform, TRF2, which resides in the nucleus, associates with telomeres and the transcription repressor REST, which together preserve telomere integrity and suppress neuronal differentiation. As shown in dividing rodent neural precursors, the splicing factor HNRNPH enhances the inclusion of exons giving rise to TRF2. Upon differentiation, HNRNPH levels decline and exon-skipping leads to the rise of the short isoform, TRF2-S. TRF2-S resides in the cytoplasm, and associates with a distinct subset of proteins and RNAs (including G-rich RNAs) and via such molecular interactions may regulate mRNAs transport and protein translation.
These new findings raise interesting questions aimed at elucidating the underlying molecular mechanisms. For example, how does HNRNPH regulate the TRF2/TRF2-S splicing switch? Does HNRNPH inhibit spliceosome recruitment to the 5′ alternative splice site? Are there binding partners in this regulation? New questions concerning the functions of the two isoforms at the molecular and physiologic level are also emerging. For instance, does TRF2 have the ability to bind TERRA RNA or other RNAs through the GAR domain? Is TRF2-S involved in the post-transcriptional metabolism of target mRNAs? What other cellular roles does TRF2-S have in neuronal differentiation?
In sum, this is an interesting paradigm in which a single alternative splicing event retools a protein with one function and one set of interacting factors during a proliferative state (TRF2), into another protein that has a different function and a different set of interacting factors during a differentiated state (TRF2-S). This mechanism sets the stage for the identification of other alternatively spliced protein isoforms which are responsible for different developmental tasks dictated by their molecular structure and interacting factors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded in its entirety by the NIA-IRP, NIH, project Z01-AG000518-11.
References
- [1].Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 2010; 11:345-55; PMID:20376054; http://dx.doi.org/ 10.1038/nrg2776 [DOI] [PubMed] [Google Scholar]
- [2].Moore AD, Bjorklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem Sci 2008; 33:444-51; PMID:18656364; http://dx.doi.org/ 10.1016/j.tibs.2008.05.008 [DOI] [PubMed] [Google Scholar]
- [3].Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413-5; PMID:18978789; http://dx.doi.org/ 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- [4].Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456:470-6; PMID:18978772; http://dx.doi.org/ 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010; 463:457-63; PMID:20110989; http://dx.doi.org/ 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 2006; 127:49-58; PMID:17018276; http://dx.doi.org/ 10.1016/j.cell.2006.09.014 [DOI] [PubMed] [Google Scholar]
- [7].Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci 2010; 33:173-82; PMID:20303187; http://dx.doi.org/ 10.1016/j.tins.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: Recent insights into mechanisms and functional roles. Neuron 2015; 87:14-27; PMID:26139367; http://dx.doi.org/ 10.1016/j.neuron.2015.05.004 [DOI] [PubMed] [Google Scholar]
- [9].de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005; 19:2100-10; PMID:16166375; http://dx.doi.org/ 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- [10].Sarek G, Vannier JB, Panier S, Petrini JH, Boulton SJ. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol Cell 2015; 57:622-35; PMID:25620558; http://dx.doi.org/ 10.1016/j.molcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rai R, Chen Y, Lei M, Chang S. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun 2016; 7:10881; PMID:26941064; http://dx.doi.org/ 10.1038/ncomms10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 1997; 17:231-5; PMID:9326950; http://dx.doi.org/ 10.1038/ng1097-231 [DOI] [PubMed] [Google Scholar]
- [13].Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol 2004; 2:E240; PMID:15314656; http://dx.doi.org/ 10.1371/journal.pbio.0020240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012; 149:795-806; PMID:22579284; http://dx.doi.org/ 10.1016/j.cell.2012.03.030 [DOI] [PubMed] [Google Scholar]
- [15].Feuerhahn S, Chen LY, Luke B, Porro A. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem Sci 2015; 40:275-85; PMID:25845889; http://dx.doi.org/ 10.1016/j.tibs.2015.03.003 [DOI] [PubMed] [Google Scholar]
- [16].Benarroch-Popivker D, Pisano S, Mendez-Bermudez A, Lototska L, Kaur P, Bauwens S, Djerbi N, Latrick CM, Fraisier V, Pei B, et al.. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Mol Cell 2016; 61:274-86; PMID:26774283; http://dx.doi.org/ 10.1016/j.molcel.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang P, Casaday-Potts R, Precht P, Jiang H, Liu Y, Pazin MJ, Mattson MP. Nontelomeric splice variant of telomere repeat-binding factor 2 maintains neuronal traits by sequestering repressor element 1-silencing transcription factor. Proc Natl Acad Sci U S A 2011; 108:16434-9; PMID:21903926; http://dx.doi.org/ 10.1073/pnas.1106906108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grammatikakis I, Zhang P, Panda AC, Kim J, Maudsley S, Abdelmohsen K, Yang X, Martindale JL, Motino O, Hutchison ER, et al.. Alternative splicing of neuronal differentiation Factor TRF2 regulated by HNRNPH1/H2. Cell Rep 2016; 15:926-34; PMID:27117401; http://dx.doi.org/ 10.1016/j.celrep.2016.03.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang P, Abdelmohsen K, Liu Y, Tominaga-Yamanaka K, Yoon JH, Ioannis G, Martindale JL, Zhang Y, Becker KG, Yang IH, et al.. Novel RNA- and FMRP-binding protein TRF2-S regulates axonal mRNA transport and presynaptic plasticity. Nat Commun 2015; 6:8888; PMID:26586091; http://dx.doi.org/ 10.1038/ncomms9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Janouskova E, Necasova I, Pavlouskova J, Zimmermann M, Hluchy M, Marini V, Novakova M, Hofr C. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res 2015; 43:2691-700; PMID:25675958; http://dx.doi.org/ 10.1093/nar/gkv097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 2010; 29:2230-41; PMID:20551906; http://dx.doi.org/ 10.1038/emboj.2010.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 2013; 155:345-56; PMID:24120135; http://dx.doi.org/ 10.1016/j.cell.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 2013; 494:502-5; PMID:23389450; http://dx.doi.org/ 10.1038/nature11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007; 448:1068-71; PMID:17687332; http://dx.doi.org/ 10.1038/nature06065 [DOI] [PubMed] [Google Scholar]
- [25].Buscemi G, Zannini L, Fontanella E, Lecis D, Lisanti S, Delia D. The shelterin protein TRF2 inhibits Chk2 activity at telomeres in the absence of DNA damage. Curr Biol 2009; 19:874-9; PMID:19375317; http://dx.doi.org/ 10.1016/j.cub.2009.03.064 [DOI] [PubMed] [Google Scholar]
- [26].Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell 2013; 51:141-55; PMID:23850488; http://dx.doi.org/ 10.1016/j.molcel.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell 1999; 97:503-14; PMID:10338214; http://dx.doi.org/ 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- [28].Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J 2001; 20:5532-40; PMID:11574485; http://dx.doi.org/ 10.1093/emboj/20.19.5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Muraki K, Nabetani A, Nishiyama A, Ishikawa F. Essential roles of Xenopus TRF2 in telomere end protection and replication. Genes Cells 2011; 16:728-39; PMID:21554499; http://dx.doi.org/ 10.1111/j.1365-2443.2011.01520.x [DOI] [PubMed] [Google Scholar]
- [30].Saint-Leger A, Koelblen M, Civitelli L, Bah A, Djerbi N, Giraud-Panis MJ, Londono-Vallejo A, Ascenzioni F, Gilson E. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle 2014; 13:2469-74; PMID:25483196; http://dx.doi.org/ 10.4161/cc.29422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun 2014; 5:5379; PMID:25359189; http://dx.doi.org/ 10.1038/ncomms6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 2009; 35:403-13; PMID:19716786; http://dx.doi.org/ 10.1016/j.molcel.2009.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol 2010; 30:4808-17; PMID:20713443; http://dx.doi.org/ 10.1128/MCB.00460-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J 2009; 28:2503-10; PMID:19629047; http://dx.doi.org/ 10.1038/emboj.2009.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, et al.. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J 2009; 28:641-51; PMID:19197240; http://dx.doi.org/ 10.1038/emboj.2009.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mitchell TR, Glenfield K, Jeyanthan K, Zhu XD. Arginine methylation regulates telomere length and stability. Mol Cell Biol 2009; 29:4918-34; PMID:19596784; http://dx.doi.org/ 10.1128/MCB.00009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep 2005; 6:39-45; PMID:15608617; http://dx.doi.org/ 10.1038/sj.embor.7400314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 2008; 319:1092-6; PMID:18202258; http://dx.doi.org/ 10.1126/science.1151804 [DOI] [PubMed] [Google Scholar]
- [39].Munoz P, Blanco R, Blasco MA. Role of the TRF2 telomeric protein in cancer and ageing. Cell Cycle 2006; 5:718-21; PMID:16582635; http://dx.doi.org/ 10.4161/cc.5.7.2636 [DOI] [PubMed] [Google Scholar]
- [40].Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 2005; 7:712-8; PMID:15968270; http://dx.doi.org/ 10.1038/ncb1275 [DOI] [PubMed] [Google Scholar]
- [41].Martinez P, Ferrara-Romeo I, Flores JM, Blasco MA. Essential role for the TRF2 telomere protein in adult skin homeostasis. Aging Cell 2014; 13:656-68; PMID:24725274; http://dx.doi.org/ 10.1111/acel.12221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Blanco R, Munoz P, Flores JM, Klatt P, Blasco MA. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev 2007; 21:206-20; PMID:17234886; http://dx.doi.org/ 10.1101/gad.406207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Biroccio A, Cherfils-Vicini J, Augereau A, Pinte S, Bauwens S, Ye J, Simonet T, Horard B, Jamet K, Cervera L, et al.. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat Cell Biol 2013; 15:818-28; PMID:23792691; http://dx.doi.org/ 10.1038/ncb2774 [DOI] [PubMed] [Google Scholar]
- [44].Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol 2002; 22:3474-87; PMID:11971978; http://dx.doi.org/ 10.1128/MCB.22.10.3474-3487.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nera B, Huang HS, Lai T, Xu L. Elevated levels of TRF2 induce telomeric ultrafine anaphase bridges and rapid telomere deletions. Nat Commun 2015; 6:10132; PMID:26640040; http://dx.doi.org/ 10.1038/ncomms10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang P, Pazin MJ, Schwartz CM, Becker KG, Wersto RP, Dilley CM, Mattson MP. Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Curr Biol 2008; 18:1489-94; PMID:18818083; http://dx.doi.org/ 10.1016/j.cub.2008.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 2005; 121:645-57; PMID:15907476; http://dx.doi.org/ 10.1016/j.cell.2005.03.013 [DOI] [PubMed] [Google Scholar]
- [48].Bai Y, Lathia JD, Zhang P, Flavahan W, Rich JN, Mattson MP. Molecular targeting of TRF2 suppresses the growth and tumorigenesis of glioblastoma stem cells. Glia 2014; 62:1687-98; PMID:24909307; http://dx.doi.org/ 10.1002/glia.22708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ovando-Roche P, Yu JS, Testori S, Ho C, Cui W. TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells 2014; 32:2111-22; PMID:24740933; http://dx.doi.org/ 10.1002/stem.1725 [DOI] [PubMed] [Google Scholar]
- [50].Zhang P, Furukawa K, Opresko PL, Xu X, Bohr VA, Mattson MP. TRF2 dysfunction elicits DNA damage responses associated with senescence in proliferating neural cells and differentiation of neurons. J Neurochem 2006; 97:567-81; PMID:16539655; http://dx.doi.org/ 10.1111/j.1471-4159.2006.03779.x [DOI] [PubMed] [Google Scholar]
- [51].Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods 2005; 37:331-40; PMID:16314262; http://dx.doi.org/ 10.1016/j.ymeth.2005.07.015 [DOI] [PubMed] [Google Scholar]
- [52].Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature 2010; 465:53-9; PMID:20445623; http://dx.doi.org/ 10.1038/nature09000 [DOI] [PubMed] [Google Scholar]
- [53].Uren PJ, Bahrami-Samani E, de Araujo PR, Vogel C, Qiao M, Burns SC, Smith AD, Penalva LO. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol 2016; 13:400-11; PMID:26760575; http://dx.doi.org/ 10.1080/15476286.2015.1138030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al.. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 2012; 1:167-78; PMID:22574288; http://dx.doi.org/ 10.1016/j.celrep.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mandler MD, Ku L, Feng Y. A cytoplasmic quaking I isoform regulates the hnRNP F/H-dependent alternative splicing pathway in myelinating glia. Nucleic Acids Res 2014; 42:7319-29; PMID:24792162; http://dx.doi.org/ 10.1093/nar/gku353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang E, Aslanzadeh V, Papa F, Zhu H, de la Grange P, Cambi F. Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS ONE 2012; 7:e51266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ortuno-Pineda C, Galindo-Rosales JM, Calderon-Salinas JV, Villegas-Sepulveda N, Saucedo-Cardenas O, De Nova-Ocampo M, Valdes J. Binding of hnRNP H and U2AF65 to respective G-codes and a poly-uridine tract collaborate in the N50-5′ss selection of the REST N exon in H69 cells. PLoS ONE 2012; 7:e40315; PMID:22792276; http://dx.doi.org/ 10.1371/journal.pone.0040315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 2008; 10:228-36; PMID:18157120; http://dx.doi.org/ 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- [59].Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J 2010; 277:1107-17; PMID:19951353; http://dx.doi.org/ 10.1111/j.1742-4658.2009.07464.x [DOI] [PubMed] [Google Scholar]
- [60].Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001; 107:489-99; PMID:11719189; http://dx.doi.org/ 10.1016/S0092-8674(01)00566-9 [DOI] [PubMed] [Google Scholar]
- [61].Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al.. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011; 146:247-61; PMID:21784246; http://dx.doi.org/ 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tabet R, Moutin E, Becker JA, Heintz D, Fouillen L, Flatter E, Krężel W, Alunni V, Koebel P, Dembélé D, et al.. Fragile X Mental Retardation Protein (FMRP) controls diacylglycerol kinase activity in neurons. Proc Natl Acad Sci U S A 2016;113:E3619-28; PMID:27233938; http://dx.doi.org/ 10.1073/pnas.1522631113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Giannone RJ, McDonald HW, Hurst GB, Shen RF, Wang Y, Liu Y. The protein network surrounding the human telomere repeat binding factors TRF1, TRF2, and POT1. PLoS ONE 2010; 5:e12407; PMID:20811636; http://dx.doi.org/ 10.1371/journal.pone.0012407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol 2009; 16:372-9; PMID:19287395; http://dx.doi.org/ 10.1038/nsmb.1575 [DOI] [PubMed] [Google Scholar]


