Abstract
The past decade brought a revolution in understanding of the structure, topology and disease-inducing lesions of RNA and DNA, fueled by unprecedented progress in next-generation sequencing. This technological revolution has also affected understanding of the epigenome and has provided unique opportunities for the analysis of DNA and histone modifications, as well as the first map of the non–protein-coding genome and three-dimensional (3D) chromosomal interactions. Overall, these advances have facilitated studies that combine genetic, transcriptomics and epigenomics data to address a wide range of issues ranging from understanding the role of the epigenome in development to targeting the transcription of noncoding genes in human cancer. Here we describe recent insights into epigenetic dysregulation characteristic of the malignant differentiation of blood stem cells based on studies of alterations that affect epigenetic complexes, enhancers, chromatin, long noncoding RNAs (lncRNAs), RNA splicing, nuclear topology and the 3D conformation of chromatin.
Large-scale efforts to map mutations and other genomic events in blood malignancies (leukemias and lymphomas) have suggested that these are ‘epigenetic’ diseases—that is, diseases driven by mutations in genes encoding regulators of DNA-cytosine modifications and post-translational modifications of histone, as well as by mutations in noncoding regions of the genome. For example, genetic analysis of acute myeloid leukemia (AML) has consistently revealed common somatic mutations that target genes encoding proteins that regulate DNA methylation (DNMT3A, TET2, IDH1 and IDH2), post-translational modifications of histone (EZH2, KDM6A, ASXL1 and others), RNA splicing and 3D chromosomal interactions (such as the insulator protein CTCF and the cohesin complex). When protein fusions involving epigenetic modifiers and/or transcription factors are also considered, it becomes obvious that the vast majority of patients with AML bear at least one somatic mutational event that affects the epigenome. Such findings are not AML specific, as they can also be found in patients affected by various blood malignancies, including acute and chronic leukemias and numerous subtypes of lymphoma. Although mutations that affect epigenetic modifiers in hematological malignancies have been extensively reviewed1, here we discuss emerging modes of epigenetic regulation in leukemia, including dysregulation of enhancers, noncoding RNAs, DNA looping–3D chromosomal topology and RNA splicing.
Leukemogenic enhancers and ‘super-enhancers’
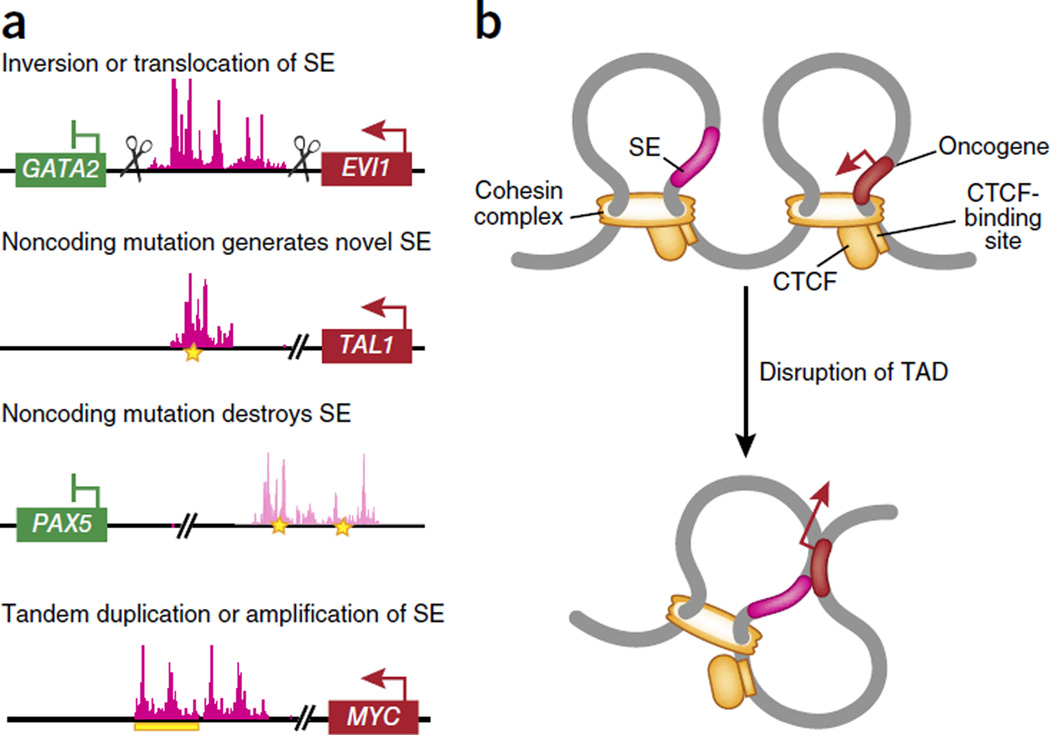
The systematic identification of regions of the genome that encode transcriptional enhancers in normal and malignant blood cell types has revealed enrichment for disease-associated loci in cell-type-specific regulatory regions relative to their abundance in other regions of the genome2. This has included the identification of somatic alterations in gene promoters, as well as in DNA sequences distal from the transcriptional start site (TSS) that are needed to regulate transcription, called ‘enhancers’ and ‘super-enhancers’ (here we define ‘super-enhancers’ as regions of the genome with putative or proven enhancer activity identified by the abundance of MED1 (a core component of the large, multiprotein MEDIATOR complex required for transcription by RNA polymerase II) and acetylation of histone H3 at Lys27 (H3K27ac), as initially noted)2–4. For example, it has been shown that leukemogenic translocations or inversions on chromosome 3 ((inv(3)(q21q26.2) or t(3;3)(q21;q26.2)) link a pre-existing enhancer to the proto-oncogene MECOM (EVI1) while simultaneously creating haploinsufficiency for the gene encoding the tumor suppressor GATA-2 (refs. 5,6) (Fig. 1a). Analysis of the chromatin marks H3K27ac and H3K4me1 (mono-methylation of histone H3 at Lys4) by chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) and chromosome-conformation-capture profiling has revealed substantial interactions between an enhancer on 3q21 and the promoter of EVI1, which encodes the proto-oncogenic transcription factor EVI, whose expression is linked to myeloid leukemogenesis5. Interestingly, the enhancer that promotes EVI1 expression in cells bearing 3q rearrangements is a 9-kilobase region of DNA that normally promotes expression of GATA2 in cells without 3q rearrangements. Thus, 3q rearrangements physically move an enhancer to ectopically promote EVI1 transcription while simultaneously diminishing GATA2 transcription. That finding has been supported by the identification of monoallelic expression of GATA2 from the non-rearranged allele in inv(3)/t(3;3) samples from patients with AML5. Through the use of a similar strategy, acquired mutations that create a novel super-enhancer in the TAL1 locus (which encodes a proto-oncogenic transcription factor) have been identified in a subset of patients with T cell acute lymphoblastic leukemia (T-ALL)7 (Fig. 1a). The authors identified heterozygous insertion or deletion mutations that clustered upstream from the TSS of TAL1. These mutations generate a novel binding motif for the transcription factor c-Myb and a broad domain of H3K27Ac7. This aberrant de novo super-enhancer drives mono-allelic overexpression of TAL1 (ref. 7).
Figure 1.
Genomic alterations that affect gene expression in leukemia. (a) Genetic alterations that link enhancers and super-enhancers (SE) to aberrant upregulation of oncogenes and/or downregulation of genes encoding tumor suppressors in leukemia. These include inversions and/or translocations of regulatory elements that aberrantly drive the expression of an oncogene and/or reduce the expression of genes encoding tumor suppressors (as has been shown for alterations in chromosome 3, whereby the super-enhancers for GATA2 are aberrantly linked to drive EVI1 expression); mutations in the noncoding genome that generate a novel super-enhancer to drive oncogene expression (as has been shown for TAL1) and abolish super-enhancers to reduce the expression of genes encoding tumor suppressors (as has been shown for PAX5); and duplications and/or amplifications of super-enhancers linked to an oncogene (as has been shown for MYC). (b) Chromatin is organized into topologically associated domains (TAD), which are normally restricted from one another through the action of CTCF in association with the cohesin complex. Alterations in the 3D organization of chromatin, through disruption of the binding of CTCF and/or disruption of cohesin expression, might result in aberrant promoter-enhancer interactions to drive cancer.
Following such studies, systematic efforts to integrate epigenomic and mutational data have been most advanced for chronic lymphocytic leukemia (CLL), for which an effort by the International Cancer Genome Consortium has mapped recurrently mutated noncoding regions in genomes of patients with CLL on the basis of whole-genome sequencing of 250 patients with CLL8. While most of the regions repeatedly affected by noncoding mutations in CLL seem to be targets of somatic hypermutation, several regions densely affected by mutations and that are not known targets of somatic hypermutation have been identified. These include mutations in an intergenic region of chromosome 9p13 that, given its abundance of H3K27Ac and H3K4me1, contains an enhancer element important to CLL8 (Fig. 1a). Samples from patients with CLL with mutations at this locus have decreased expression of the gene encoding the transcription factor Pax5, which suggests that disruption of this enhancer element is associated with downregulation of PAX5 expression.
In addition to structural rearrangements and mutations that create or abolish enhancers, several interesting examples of amplifications and duplications of disease-causative enhancers have also been identified. For example, two groups have identified a enhancer in the gene encoding the proto-oncoprotein (MYC) that is regulated by the transmembrane receptor NOTCH1 and is targeted by duplication in T-ALL9,10 (Fig. 1a). Data obtained by array comparative genomic hybridization have identified focal duplications at an enhancer located on 8q24 in 5% of patients with T-ALL9. Further analysis has revealed that this duplicated enhancer interacts with the MYC promoter to drive MYC expression9,10. These data suggest that while individual enhancers or super-enhancers are highly lineage specific, alterations to these elements might be a common mechanism for the upregulation of oncogenes such as MYC in diverse types of cancer.
Alterations in the 3D conformation of chromatin
Alterations in DNA elements that regulate the 3D organization of chromatin itself have been suggested to result in aberrant promoter-enhancer interactions to drive cancer development11. The 3D structure of chromatin is defined in part by the organization of chromatin into conserved loops that are restricted from one another, called ‘topologically associating domains’12,13. These domains are partitioned (or ‘insulated’) from each other through the action of CTCF in association with the cohesin complex14–16 (Fig. 1b). Accumulating evidence suggests that DNA looping is important for the induction and maintenance of leukemia. Indeed, it has been shown that a NOTCH1-orchestrated event brings together the long noncoding RNA (lncRNA) LUNAR1 (‘leukemia-induced noncoding activator RNA’) and the enhancer of IGF1R, which encodes a receptor for insulin-like growth factor that is essential for the survival of T-ALL cells17. Silencing of LUNAR1 leads inhibition of the growth of T-ALL cells due to the loss of IGF1R expression and loss of trophic signals.
Analysis of the enhancer landscape and 3D conformation of chromatin has been particularly useful in generating new insights into the understanding of B cell tumorigenesis. It has been known for quite some time that AID (‘activation-induced cytidine deaminase’), a B cell–specific enzyme necessary for the class-switch recombination and somatic hypermutation of immunoglobulin-encoding genes in mature B cells, promotes mutations and translocations in genes that do not encode immunoglobulins18. Until recently, however, it was not known why loci that do not encode immunoglobulins, some of which are megabases away from the immunoglobulin-encoding locus, are susceptible to AID activity. Several reports have identified AID targets with specific genomic features, such as loci that contain super-enhancers and have a 3D conformation that promotes physical interactions of these genes that do not encode immunoglobulins with the immunoglobulin-encoding locus19,20. It is these physical interactions that promote the occurrence of intra-chromosomal translocations that are recurrently seen in B cell lymphomas and explain the ‘predilection’ of AID for genes (such as PAX5, IRF8 and CD79B) that are expressed mainly in B cells and contain super-enhancers in B cells. Similar observations involving RAG-endonuclease-mediated translocations in early B cells21 have further solidified the concept that frequent 3D interactions between regions of the genome in the nucleus underlie the development of recurrent inter-chromosomal translocations.
Published work has identified recurrent mutations in all genes encoding members of the cohesin complex (STAG2, SMC1A, SMC3 and RAD21) in myeloid leukemia and other cancers22–24. Downregulation of cohesin function in mouse or human cells promotes aberrant self-renewal and myeloid skewing of hematopoiesis25–27. Given that the most well-established function of the cohesin complex is to regulate the alignment of sister chromatids during mitosis28, it was a surprise to find no trace of genomic instability in mouse or human myeloid leukemia. In contrast, cohesin-deficient cells and cells overexpressing mutant cohesin have widespread alterations in chromatin accessibility25–27. It is currently unclear how alterations in the expression or function of cohesin can alter the accessibility or 3D conformation of chromatin. For example, analysis of human hematopoietic cells expressing mutant RAD21 or SMC1A has revealed global reductions in chromatin accessibility and increased accessibility at specific binding sites for transcription factors (including GATA-2 and RUNX1) known to be critical regulators of hematopoietic stem cells25. However, analysis of mouse hematopoietic cells by ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing) has not revealed a genome-wide reduction in chromatin accessibility at promoters. On the other hand, increases in chromatin accessibility at specific loci have been seen in mouse cells deficient in SMC3 (ref. 27) or STAG2 (ref. 26). Thus, additional studies are needed to define the global versus site-specific effects of chromatin accessibility and transcription-factor-binding sites in wild-type versus cohesin-mutant cells.
Spliceosomal gene mutations in leukemia
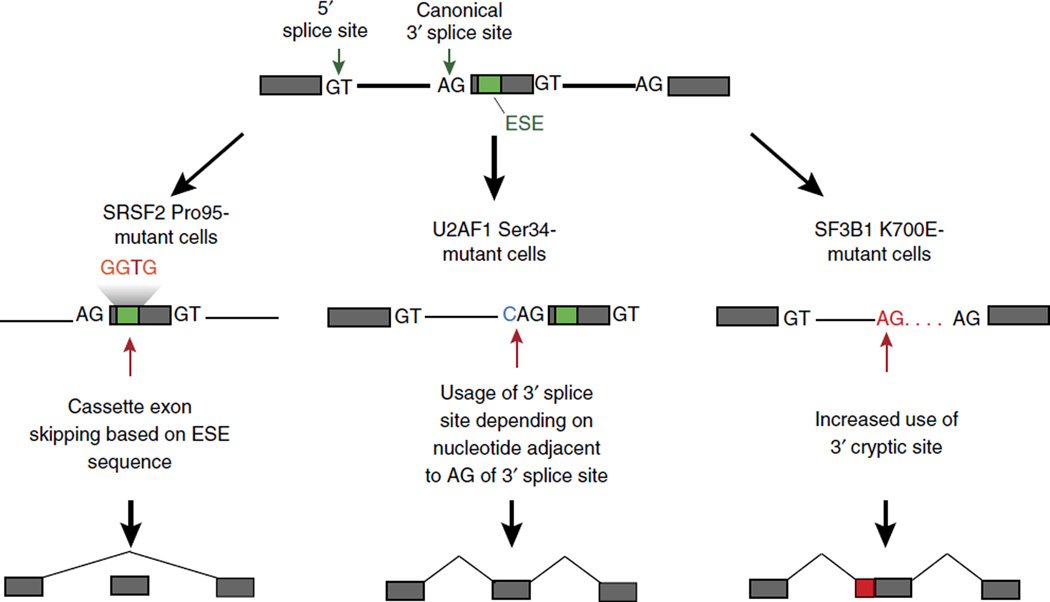
In addition to alterations in the chromatin state that affect gene expression in leukemia, genomic analyses of leukemias have also revealed that patients with myeloid leukemias29–31 and lymphoid leukemias32,33 also show substantial enrichment for mutations in genes encoding proteins essential for mRNA splicing. These include mutations in genes encoding SF3B1 and U2AF1 (core members of the major spliceosome), ZRSR2 (a core member of the minor spliceosome) and SRSF2 (an auxiliary splicing factor essential for constitutive as well as alternative splicing). These genetic data suggest that mutations in genes encoding components of the spliceosome confer an alteration of function. Functional investigation of the effect of these mutations on splicing have revealed that mutations in the genes encoding SRSF2 (refs. 34,35), U2AF1 (ref. 36) and SF3B1 (refs. 37,38) alter splicing in a manner distinct from loss of function39 (Fig. 2). For example, mutations in the gene encoding SRSF2 alter the normal sequence-specific RNA-binding avidity and splicing ‘preference’ of the protein34,35. SRSF2 normally binds to exonic regions called ‘exonic splicing enhancers’ at cytosine- and guanine-rich sequences to promote spliceosome recruitment and splicing40. However, mutant SRSF2 has greatly diminished avidity for guanine-rich exonic splicing enhancers and instead promotes splicing at cytosine-rich exonic splicing enhancers more efficiently than does the wild-type protein. In contrast, loss of SRSF2 is associated with global splicing failure with widespread skipping of constitutive and alternative cassette exons, as well as intron retention34,35. Similarly, mutations in the gene encoding U2AF1 alter splicing ‘preference’ based on the nucleotide sequence immediately 5′ or 3′ of the conserved adenosine-gunanine dinucleotide of the canonical 3′ splice site36 (Fig. 2).
Figure 2.
Targeting the spliceosome in myeloid neoplasms. Mutations in genes encoding spliceosomal proteins alter RNA splicing in a sequence-specific manner to drive aberrant gene expression. Mutations in the gene encoding SRSF2 result in alterations at Pro95 and result in aberrant splice-site ‘preference’ based on the nucleotide sequence of exonic splicing enhancer (ESE). In contrast, U2AF1 and SF3B1 both recognize the 3′ splice site, and mutations that affect Ser34 in U2AF1 result in altered 3′ splice-site usage based on the nucleotide sequence immediately 5′ of the canonical 3′ AG dinucleotide. In contrast, cells bearing mutations that result in the K700E substitution in SF3B1 have increased use of cryptic 3′ splice sites.
Functional analysis of the effects of mutations in genes encoding splicing factors has revealed that these mutations broadly alter gene expression through widespread effects on splicing as well as through altering the expression and function of proteins necessary for epigenetic regulation34,36,41. For example, mutations in the gene encoding SRSF2 promote expression of a specific isoform of the H3K27-methyltransferase EZH2, which subjects the transcript to nonsense-mediated decay. As a result, cells bearing mutant SRSF2 have diminished expression of EZH2 and a lower abundance of H3K27me3 (ref. 34). That discovery provided the molecular basis for the observation of mutually exclusive loss-of-function mutations in EZH2 with alteration-of-function mutations in SRSF2 in patients with myeloid leukemias42,43. Similar links between mutant splicing factors and altered global epigenetic regulation have also been observed for mutant U2AF1. Several studies have confirmed that cells bearing Ser34-mutant U2AF1 ‘preferentially’ express the macroH2A.1 splice variant of the histone variant H2A36,41. Further work will be needed to determine how altered global abundance of H3K27me3 in cells bearing mutant SRSF2, as well as altered histone acetylation in cells expressing Ser34-mutant U2AF1, might be linked to the aberrant phenotype of spliceosome-mutant hematopoietic cells.
There is also emerging evidence that spliceosomal proteins might influence the chromatin state independently of their roles in splicing. For example, heterozygous Sf3b1+/− mice develop skeletal transformations due to inappropriate activation of expression genes encoding the Hox family of transcription factors44. That work identified direct physical interactions between SF3B1 and the polycomb repressive complex 1 (PRC1) protein RNF2 (RING1B), an E3 ubiquitin-protein ligase that monoubiquitinates H2A at Lys119 (ref. 44). Consistent with that, the authors noted coincident co-localization of SF3B1 and RNF2 at specific Hox-encoding loci. Subsequently, SF3B1 was suggested to directly bind nucleosomes in a manner that does not depend on the presence of RNA or on interactions with large protein complexes45. Given that the overall effects of mutations of the gene encoding SF3B1 on splicing are modest and that the implications of such mutations on the structure of SF3B1 are not well understood, it might be important to determine whether cancer-associated mutations in the gene encoding SF3B1 alter the genome-wide binding of this protein.
lncRNAs in hematologic malignancies
Interest in epigenetic regulation and RNA processing in hematological malignancies has resulted in further efforts to understand how lncRNAs, which are RNAs with a length of at least 200 nucleotides and no protein-coding potential, might be involved in disease pathogenesis. Alterations in the expression of lncRNAs are widely suspected to be prevalent in leukemias and lymphomas. For example, one of the most common copy-number alterations in CLL, deletion of an intergenic region at 13q14, targets the lncRNAs DLEU1 and DLEU2 as well as the microRNAs miR-15 and mi-R16 (refs. 46,47). Indeed, deletion of this syntenic region in mice results in the development of B cell malignancies48 (although the contribution of the deletion of DLEU1 and DLEU2 to this phenotype versus that of miR-15 and mi-R16 is not clear). Beyond this example in CLL and the aforementioned NOTCH-regulated lncRNAs in T-ALL, there are currently only a few additional examples of functional involvement of lncRNAs in the pathogenesis of leukemias. One experimental example whereby altering expression of a lncRNA directly results in a fully penetrant hematopoietic disorder is deletion of the 17-kilobase lncRNA Xist (‘X-inactive specific transcript’)49. Xist targets polycomb complexes to the X-chromosome in cis to promote X-chromosome inactivation. It was initially believed that Xist is dispensable once X-chromosome inactivation, which occurs at embryonic days 4.5–5.5 (E4.5–E5.5) in mice, is established. However, deletion of Xist in a hematopoietic-cell-specific manner at E10.5 results in a fully penetrant myeloproliferative-myelodysplastic disorder in vivo49. This is coincident with a failure to maintain X-chromosome inactivation and the generation of defects in DNA replication and chromosome segregation. Further efforts to systematically annotate lncRNAs that have dysregulated expression in disease and to link these lncRNAs to somatic mutations and chromatin state will hopefully promote similar efforts to reveal causal roles of lncRNAs in hematologic malignancy pathogenesis.
Histone modifications and the role of the histone code
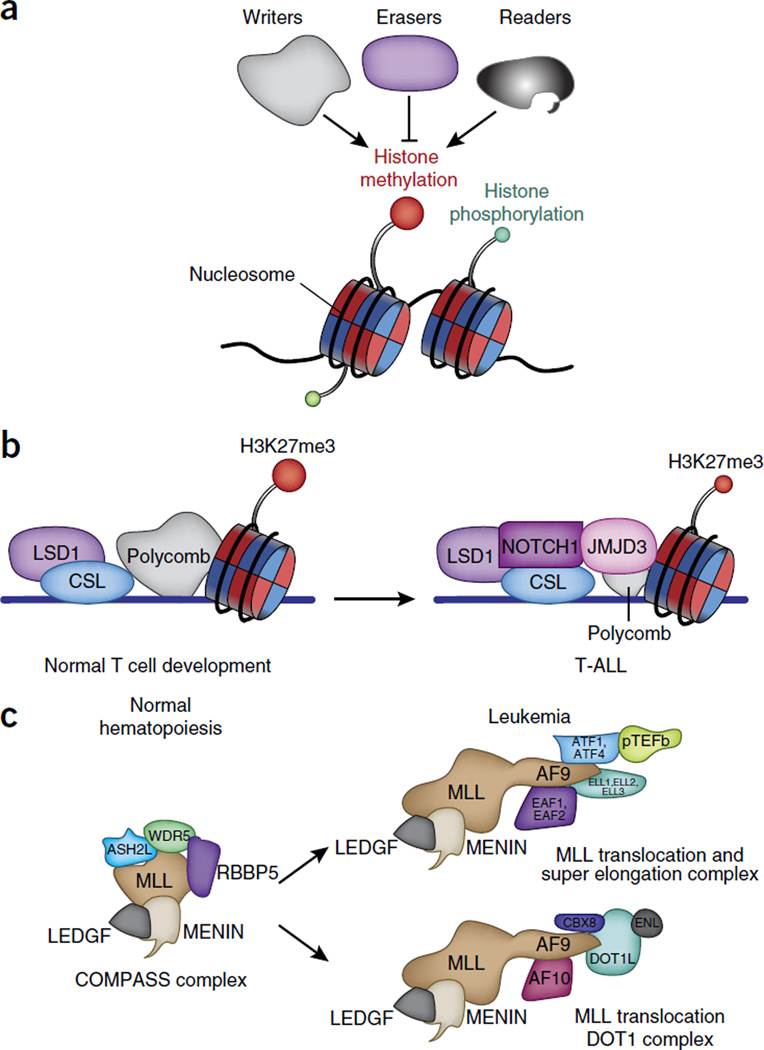
As noted above, post-translational modifications of amino-terminal histone tails50,51 are established mechanisms of regulating gene expression in development and disease (Fig. 3a). 67 different types of modifications of lysine or arginine residues have been described so far52. The coexistence of activating and repressive histone marks can be simultaneously found on bivalent promoters, which contain both H3K4me3 and H3K27me3 (refs. 53,54). Expression of genes at the Hox-encoding locus is a paradigmatic model of epigenetic regulation of gene expression, as Hox-encoding genes are maintained in an active state or an inactive state through activity of the trithorax complex (also known as the MLL (‘mixed-lineage leukemia’) complex), which catalyzes the trimethylation of H3K4, or the polycomb complex (PRC2), which regulates H3K27me3, respectively. Below we discuss the effect of mutations in genes encoding members of the MLL and PRC2 complexes in leukemias and lymphomas, as well as that of mutations in genes encoding components of the histone-demethylase complexes potentially intertwined with the function of the MLL and PRC2 complexes.
Figure 3.
Physiological and leukemia-associated functions of epigenetic complexes. (a) Three main categories of epigenetic modulators: ‘writers’ add the histone marks; ‘readers’ have domains that recognize the modifications; and ‘erasers’ remove the modifications. (b) NOTCH1 is a typical example of oncogenic transcription factor in leukemia. LSD1 acts as both transcriptional corepressor (when associated with the CSL repressor complex) and a coactivator of NOTCH1 (when NOTCH is activated via demethylation of histone H3 at Lys4 and Lys9). (c) In a physiological context, MLL1 is the catalytic subunit of the COMPASS complexes and methylates histone H3 at Lys4. The DNA-binding amino terminus of MLL is a frequent partner in translocations. AF9, a member of the super elongation complex and the DOT1L elongation complex, is a frequent translocation partner of MLL (associated with MENIN and LEDGF); this lead to aberrant activation of genes that are targets of MLL. LEDGF, lens epithelium-derived growth factor.
PRC2 in leukemia and lymphoma
PRC2 can serve as an oncoprotein or a tumor suppressor depending on the tissue lineage55–57. Recurrent heterozygous mutations that alter Tyr641 in the catalytic SET domain of EZH2 occur in ~30% of germinal-center diffuse large B cell lymphomas and 10% of follicular lymphomas58–61. Tyr641-mutant EZH2, while impaired in its ability to mono- or dimethylate H3K27, acts together with wild-type EZH2 to enhance the trimethylation of H3K27. This cooperation between the wild-type EZH complexes and mutant EZH2 complexes fails to recognize unmodified H3K27 but is hyperactive in converting mono- or dimethylated H3K27 to the trimethylated state in diffuse large B cell lymphoma and follicular lymphoma. In contrast, in T-ALL, loss-of-function mutations affecting members of the PRC2 complex result in more aggressive disease than that of tumors with wild-type PRC2 (refs. 62–64), suggestive of a tumor-suppressor role for PRC2. Mutations in the genes encoding components of PCR2 exist in 25% of patients with T-ALL and are particularly prevalent in the aggressive early T cell progenitor ALL, in which they are associated with poor disease outcome64. Alterations in PRC2 in T-ALL consist mainly of nonsense mutations that affect protein sequences upstream of the catalytic domain of EZH2 and larger deletions or truncations of the locus, suggestive of a true loss-of-function outcome (Fig. 3b). A study modeling early T cell progenitor ALL in mice through the use of a mutant oncogenic form of the small GTPase NRAS (NRAS-Q61K) has shown that genetic inactivation of EZH2 or EED (another core subunit of the PRC2 complex) act cooperatively with mutant NRAS in leukemogenesis through enhancement of a stem-cell-related transcriptional program and increased phosphorylation and activation of the transcription factor STAT3 (ref. 65).
The biology of PRC2 seems to be more complex in myeloid malignancies, in which the complex can have either oncogenic roles or tumor-suppressive roles in different disease subtypes. For example, loss-of-function mutations in EZH2 are associated with poor survival in myelodysplastic syndrome (MDS) and myeloproliferative neoplasms (MPNs)55, whereas in mouse models of AML in which the gene encoding MLL is rearranged, PRC2 is required for efficient transformation, suggestive of a role for the complex in contributing to the aberrant self-renewal of leukemia-initiating cells66,67. Such results suggest that proper maintenance of H3K27me3 is critical for the homeostasis of normal cells. We speculate that the oncogenic activity of PRC2 might be mediated through epigenetic silencing of genes encoding key tumor suppressors, such as the cell-cycle regulators CDKN1A and CDKN1B68 or CDKN2A-CDKN2B69. In contrast, PRC2 might act antagonistically to oncogenic transcription-factor networks in cases in which the disease is driven by strong transcription factors, such as T-ALL (Fig. 3b). One open question is what determines the binding of PRC2 to different genomic positions. In contrast to organisms such as Drosophila, in which the binding of PRC2 is determined by the presence of polycomb response elements, vertebrates have other factors, such as lncRNAs, nucleosomal density, previous H3K27 methylation (mediated via the EED chromodomain), DNA methylation in CpG islands, the polycomb protein JARID2 and transcription factors that can control binding of the Polycomb complex70,71. HOTAIR is a characteristic example of a lncRNA that interacts with the polycomb complex (and the dual (Lys4 and Lys9) demethylase LSD1) and can go awry in cancer72. Interestingly, the homeobox protein HHEX is required for the PRC2-mediated repression of the Cdkn2a locus in AML in which the gene encoding MLL is rearranged73, and it will be important to determine if such mechanisms are present in different tumor types. Mutations that affect different parts of the complex can lead to different outcomes associated with altered binding or activity of the complex.
MLL rearrangements in cancer
Rearrangements in the gene encoding MLL (KMT2A) can be seen in any form of acute leukemia, in which they represent ~10% of all translocations. KMT2A is the human homolog of the Drosophila gene trithorax. MLL is a key component of mammalian COMPASS-like complexes, which have critical roles in both embryonic development and hematopoiesis. COMPASS complexes can contain hSET1A and hSET1B, MLL1, MLL2, MLL3 or MLL4 as catalytic subunits and have a critical role in activating transcription by catalyzing the mono-, di- and trimethylation of H3K4. Other important core subunits include WDR5, RBBP5 and ASH2L, which can all stimulate the activity of the methyltransferase74,75. Genetic deletion of those core subunits causes global loss of H3K4me3, but such deletion of the methyltransferase subunits does not74,75, which indicates possible redundancy among members of the MLL family.
MLL (encoded at 11q23) can be rearranged to more than 60 different partner loci via 100 different translocation events76. The resulting MLL fusion proteins contain the amino-terminal (DNA-binding) domain of MLL and the carboxy-terminal domain of each translocation partner. As the fusion proteins no longer contain the SET domain of MLL, the oncogenic action of such chimeric proteins is independent of the methylation of H3K4. The majority of MLL fusion partners are nuclear proteins, including members of the super elongation complex (SEC), SEC-L1 and SEC-L2 (the SEC-like complex), and the Dot1-methyltransferase-containing elongation complex DotCom (which catalyzes di- or trimethylation of H3K79). Members of the complexes include CDK9, cyclin-T1 and cyclin-T2 (collectively known as the elongation factor p-TEFb), ELL1–ELL3 (‘eleven-nineteen lysine-rich leukemia proteins’), and AFF1–AFF4 (which are part of the SEC complex), as well as ENL proteins and AF9 (shared by SEC and DotCom) (Fig. 3c). Methylation of H3K79 is associated with actively transcribed genes and is essential for transformation by the fusion protein MLL-AF9 (refs. 77,78). In addition, the physical interaction of MLL-AF9 with the PRC1 protein CBX8, combined with the essential role of CBX8 in myeloid leukemia, demonstrate the intricate biological mechanisms between MLL fusion proteins and polycomb complexes79. Indeed, CBX8 interacts directly with MLL-AF9 through the carboxy-terminal domain of AF9 and is necessary for Hoxa9 expression, potentially through involvement of the acetyltransferase Tip60. This activating role of CBX8 in the context of its interaction with MLL-AF9 is independent of PRC1 and repression of the Cdkn2a locus.
Histone demethylases in leukemia
The amine oxidases LSD1 and LSD2 (refs. 80,81) demethylate mono- and di-methylated lysine residues exclusively. Two groups have demonstrated a role for LSD1 in AML82,83. Through the use of inhibitors of LSD1, such as tranylcypromine and its analogs, they have shown that LSD1 is crucial for MLL-AF9–dependent leukemogenesis and that downregulation of LSD1 leads to de-repression of polycomb targets. It is likely that LSD1 has roles beyond AML in which the gene encoding MLL is rearranged. For example, although acute promyelocytic leukemia (APL) associated with fusion of the phosphoprotein transcription factor PML and the nuclear retinoic acid receptor RARA (PML-RARA) can be treated with all-trans-retinoic acid (ATRA) as a means of differentiation therapy, ATRA-based treatment has not been effective for patients with non-APL AML. Inhibition of LSD1 can sensitize non-APL AML to an ATRA-driven therapeutic response82,83. Such studies highlight a role for LSD1 as an essential regulator of leukemia stem cells in AML. They also suggest inhibition of LSD1 via tranylcypromine analogs, alone or in combination with ATRA, as a proof-of-principle model for the treatment of acute leukemia.
LSD1 has been shown to interact directly with the oncoprotein NOTCH1 and TAL1 in T-ALL. In this cancer type, LSD1 is a component of a NOTCH-containing multifunctional complex and is involved in the transcriptional programs of NOTCH1’s target genes84 (Fig. 3b). In the absence of NOTCH1, LSD1 binds to the transcription repressor CSL and effects gene repression by removing H3K4me2 at NOTCH1’s targets, while in the presence of NOTCH1 it demethylates H3K9me2, which leads to the activation of target genes. Genomic knockdown of LSD1 and chemical inhibition of LSD1 (with tranylcypromine) in T-ALL lead to results similar to those achieved via inhibition of NOTCH1, including cell-cycle arrest. LSD1 has also been found to associate with the hematopoietic-cell-specific transcription factor TAL1-SCL, and this interaction is controlled by phosphorylation of TAL1 at Ser172 by the kinase PKA. Disruption of this interaction leads to hypermethylation of H3K4 and gene activation85.
Beyond LSD1-LSD2, the largest family of histone demethylases are the Jumonji C (JmjC)-domain-containing dioxygenases86. Two of the best characterized demethylases are those that affect H3K27me3: UTX (KDM6A) and JMJD3 (KDM6B)87–92 (Fig. 3b). JMJD3 and UTX are examples of enzymes with similar biochemical activities that can have physiologically contrasting roles in the same disease context. UTX, encoded by a gene (KDM6A) located on the X chromosome93, is a tumor suppressor in NOTCH1-associated T-ALL94,95, and the gene encoding UTX is affected by mutations in 5% of patients with T-ALL. Mutations in the gene encoding UTX occur together with aberrant expression of the oncogene TLX3, as well as with NOTCH1 mutations and mutations or deletions that target the putative chromatin-reading factor PHF6 (ref. 94). Deletion of the gene encoding UTX leads to accelerated development of leukemia in NOTCH1-driven mouse models, potentially because of accumulation of H3K27me3 at the TSSs of genes encoding putative tumor suppressors61, such as Rbbp6 and Fbxw7 (ref. 96). Loss of UTX enhances the sensitivity of cancer cells to the EZH2 inhibitor 3-DZNep, which further emphasizes the clinical importance of H3K27me94. ChIP studies have shown that JMJD3 and UTX have a repertoire of unique targets89,96. UTX controls genes encoding tumor suppressors, at least in NOTCH1-associated T-ALL94,95, whereas JMJD3 is part of the NOTCH1 transcriptional complex and controls oncogenic targets such as HES1 in T-ALL96.
DNA methylation and leukemia initiation and progression
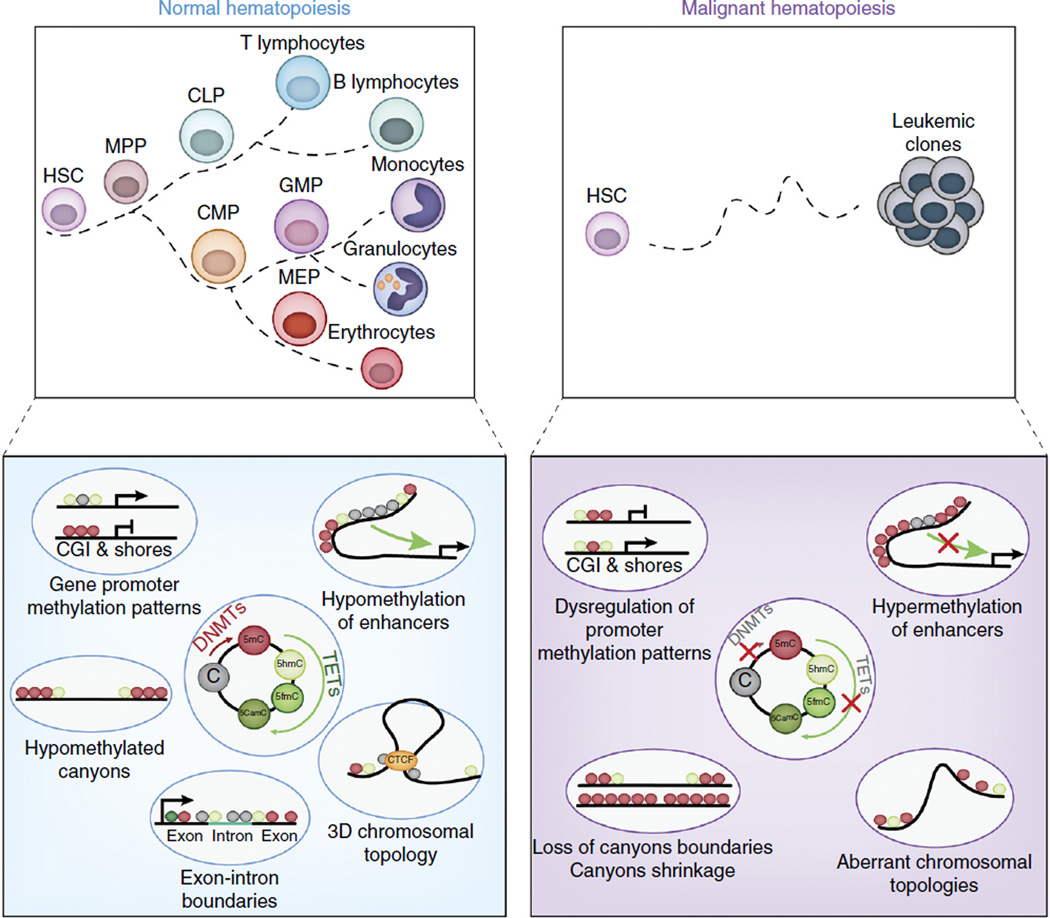
Methylation of CpG dinucleotides is one of the most frequent forms of epigenetic modification and is controlled by the maintenance methyltransferase DNMT1 and the de novo methyltransferases DNMT3A and DNMT3B. On the flip side, DNA demethylation is also dynamically regulated. An accumulating amount of evidence has revealed an additional layer of epigenetic regulation in which methyl groups at the 5′ position of cytosine bases (5mC) can be modified to 5-hydroxymethyl cytosine (5hmC) by the enzymes TET1, TET2 and TET3. The presence of 5hmC has been proposed to contribute to both passive DNA demethylation and active DNA demethylation. Maintenance methylation follows replication and is mediated by DNMT1, which might be unable to recognize 5hmC, causing passive loss of 5mC during cell division. The TET proteins might also actively prevent hypermethylation and promote demethylation by a sequential process involving AID-APOBEC-mediated deamination of 5hmC to 5hmU, followed by base-excision repair97 (Fig. 4).
Figure 4.
Roles of the cytosine methylation and hydroxymethylation of DNA in normal and malignant hematopoiesis. The balance of the function of DNA methyltransferases (DNMTs) and TET enzymes is critical to the regulation of genome-wide localization and abundance of DNA cytosine modifications that in turn regulate gene-expression patterns required for normal hematopoiesis (top left). DNA methyltransferases catalyze the conversion of cytosine (C) into 5-methylcytosine (5mc), which serve as a substrate for the conversion of 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fmC) and 5-carboxymethylcytosine (5cmC) by TET enzymes (bottom left). Loss or mutation of genes encoding DNA methyltransferases or TET enzymes, which commonly occur in a variety of leukemias, due to somatic mutations, perturbs the genome-wide distribution of 5mC and its oxidized derivatives (bottom right) and promotes leukemogenesis (top right). For example, hypermethylation might affect promoters or enhancers and result in gene silencing, or it might disrupt CTCF-binding sites to alter chromosomal structure and allow aberrant promoter-enhancer interactions to occur. Moreover, extended regions of low methylation that span domains containing transcription-factor-binding sites (so-called ‘canyons’) might shrink or widen with alterations in TET or DNMT function. MPP, multipotent progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocytic-erythroid progenitor; CGI, CpG island.
DNA methylation is important for both normal hematopoiesis and malignant hematopoiesis. Array-based methylation analysis and bisulfite sequencing has created a comprehensive ‘methylome’ (pattern of methylated DNA in the genome) map of hematopoiesis98–100. Interestingly, DNA-methylation mapping in AML has produced similar findings. Methylation signatures can identify distinct subtypes of AML, reflective of different hematopoietic lineages and genetic lesions101, and can be used to predict clinical outcome. In parallel to such mapping studies, accumulating evidence of the role of DNA methylation in hematopoiesis has been provided by the mutational landscapes of blood tumors. Indeed, the genes encoding DNMT3A, TET2 and IDH1-IDH2 are frequently targeted by mutations in leukemia and lymphoma. DNMT3A is somatically mutated in approximately 20% of AML and 10% of MDS and MPNs102. TET2 is frequently mutated in AML, chronic myelomonocytic leukemia, MPNs and MDS, in which it is the most prevalent genetic abnormality (affecting 25–35% of patients)103. Finally, focal mutations in IDH1 (which affecting Arg132 in the protein encoded) and IDH2 (which affect Arg140 and Arg172 in the protein encoded) have been found in AML104. These genetic events are also present (albeit less frequently) in T-ALL105, but they are almost totally absent from B-ALL, which suggests intriguing correlations between lineage commitment and DNA methylation. Interestingly, both direct data and indirect data suggest that mutations in TET2 and DNMT3A are early events that can take place in pre-leukemic hematopoietic stem cells (HSCs)22,106.
Various laboratories have generated mouse models for studying the effect of loss of TET-encoding genes (either individually or in combination) on disease progression107–109 and DNA methylation. Loss of Tet2 (as in Tet2−/− mice) leads to both myeloid bias and much greater HSC self-renewal, followed by overt hematological disease characterized by splenomegaly, extra-medullary hematopoiesis and an MPN-like disease phenotype in most mice, while a small number of mice develop T cell lymphoma107,110. The disease progression of Tet2−/− mice is almost indistinguishable from that of Tet2−/− mice, which suggests that TET2 has a haploinsufficient effect as a gatekeeper of HSC self-renewal and differentiation. Moreover, loss of TET2 in animal models acts together with additional genetic lesions (in the genes encoding the oncoprotein FLT3-ITD or genomic loss of the tumor suppressor EZH2), which leads to aggressive AML, while a combination of deficiencies in TET2 and ASXL1 leads to MDS110,111. Finally, TET2 and TET3 might have redundant roles in early hematopoiesis, as their combined deletion in mice leads to rapid and aggressive AML. Tet1−/− mice develop B cell non-Hodgkins lymphoma that is probably initiated by pro-B cells with aberrant 5hmC, increased DNA damage and acquisition of mutations in genes frequently mutated in human B cell lymphoma112.
It is currently unclear how deletion of either DNMT3a or TET2, two proteins with opposing enzymatic functions, results paradoxically in a somewhat similar spectrum of hematologic malignancies. Genomic analysis of the distribution of 5mC and 5hmC throughout the genome suggests that CpG islands at promoters normally show enrichment for 5mC, while active enhancers tend to show enrichment for 5hmC113,114. Loss of TET2 in embryonic stem cells results in the loss of 5hmC at promoters and a resultant increase in 5mC at the same sites and gene silencing. These data clearly suggest dynamic interplay of TET2 and DNMT3a in regulating gene expression and indicate a scenario in which an imbalance in the function of either enzyme might alter gene expression in a manner that promotes transformation (Fig. 4). That concept has been reinforced by the discovery of extended regions of the genome that coincide with the loci of genes encoding important products linked to leukemogenesis and marked by hypomethylation (so-called ‘methylation canyons’) that are regulated by DNMT3a as well as TET2.
Neomorphic mutations in the genes encoding IDH1 and IDH2 drive synthesis of the ‘oncometabolite’ R-2-hydroxyglutarate. Mice carrying the recurrent R132H mutation knocked into the Idh1 locus have increased numbers of stem and progenitor cells and develop splenomegaly, anemia and extramedulary hematopoiesis with age115. Combining the R140Q and R172K mutations of Idh2 with the oncogenic allele encoding FLT3-ITD and the NRAS G12D mutation in mice results in the development of AML116. Interestingly, deficiency in DNMT3a in mice leads to a wide array of blood malignancies, including, most frequently, MDS and AML, but also T-ALL and chronic myelomonocytic leukemia. Mutational analysis of spontaneous genetic events that occur with loss of DNMT3a in vivo has shown cooperation of genetic events that fits with previous knowledge of the mutational and transcriptional regulation of distinct disease entities. For example, Notch1 mutations have been found in mice developing T-ALL, while NPM1 mutations have been seen in AML tumors117. Subsequently, deletion of a single Dnmt3a allele has been shown to efficiently act together with duplications of the gene FLT3-ITD to induce AML in vivo, which has revealed the oncogenic potential of DNMT3a haploinsuffiency118.
Conclusions
A large number of open questions remain about the effects of mutant epigenetic modifiers on leukemogenesis. For example, while it has been largely assumed that the leukemogenic effects of mutations in the genes encoding TET2, DNMT3A, and IDH1-IDH2 drive disease through effects on DNA methylation (Fig. 4), the possibility remains that proteins that regulate DNA methylation might also drive disease development through direct roles in other biological processes. Is it possible that TET2 mutations alter differentiation or transcriptional programs due to the ability of TET2 to bind to regulators of transcription or to influence the DNA-damage-and-repair machinery. This is definitely plausible, given the ability of TET2 to interact with histone regulators such as OGT1 and SIN3A119,120. Moreover, it is also possible that mutations in genes encoding DNA-methylation-regulatory enzymes also affect the 3D chromatin confirmation in a manner that promotes leukemogenesis. Thus, mutations in IDH-encoding genes lead to defective binding of CTCF and altered 3D chromosomal topology in brain tumors121. It will thus be intriguing to connect changes in DNA methylation caused by such genetic events to higher-order regulation of gene expression in the nucleus of leukemic cells. Finally, complexes such as PRC1–PRC2 and MLL (COMPASS) or chromatin-remodeling complexes (such as SWI–SNF) consist of several subunits, and the exact pattern with which of each of the subunits is recruited onto target regions and their biological interactions are not known. The exploitation of ‘synthetic lethality’ (in which two perturbations that are normally tolerated well individually result in cell death when combined) in ARID2-mutant tumors through targeting of EZH2 (refs. 122,123) is a classic example of how such knowledge can lead to targeted therapies. Indeed, therapeutic targeting of various epigenetic regulatory proteins, including inhibitors of EZH2 (refs. 124,125), LSD1 (refs. 82,83), DOT1L126,127, mutant IDH1-IDH2 (ref. 128) and BET-bromodomain-containing proteins129–131, is now in clinical trials of leukemias, lymphomas and myelomas (several reviews focusing on epigenetic targeted therapies have also been published). For example, there are currently more than five separate ongoing clinical trials of patients with leukemia, lymphoma or myeloma testing the BET inhibitors BMS-986158, CPI-0610, FT-1101, GOTX015, I-BET762 and TEN-010.
Clearly more efforts are now needed for systematic mapping of genetic alterations throughout the coding and noncoding genome to the expression and function of coding genes, noncoding RNAs and chromatin conformation. At the same time, a robust effort to generate better and more reliable biochemical tools, including antibodies to the wild-type and mutant gene products, needs to be implemented. Moreover, increased appreciation of the role of RNA splicing in providing further means for regulating gene expression will hopefully promote efforts to understand the intersection of chromatin state and splicing in normal and malignant hematopoiesis further. Given that each of these transcriptional regulatory processes involves a large number of enzymes and will probably identify numerous novel mechanisms of oncogenesis, understanding of the diverse means by which the epigenome is altered in leukemogenesis will also hopefully lead to new therapeutic approaches.
Acknowledgments
We thank the members of all laboratories, and M. Guillamot and L. Cimmino for comments and illustrations. Supported by the US National Institutes of Health (1K08CA160647-01 and R01 HL128239 for the Abdel-Wahab laboratory; and RO1CA133379, RO1CA105129, RO1CA149655, 5RO1CA173636, 1RO1CA194923 and R01 CA190509 for the Aifantis laboratory); the National Cancer Institute (R00CA188293-02), the American Society of Hematology, the Zell Foundation and the Chicago Region Physical Science-Oncology Center (all for the Ntziachristos laboratory); the Department of Defense (BM150092 and W81XWH-12-1-0041), the Damon Runyon Foundation, the Edward P. Evans Foundation, the V Foundation, the Starr Foundation, the Josie Robertson Investigator Program and the Pershing Square Sohn Cancer Research Alliance (all for the Abdel-Wahab laboratory); and the Leukemia and Lymphoma Society, the NYSTEM program of the New York State Health Department, The William Lawrence and Blanche Hughes Foundation and the Chemotherapy Foundation (all for the Aifantis laboratory).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016;17:284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 5.Gröschel S, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki H, et al. A remote GATA2 hematopoietic enhancer drives leukemogenesis in inv(3)(q21;q26) by activating EVI1 expression. Cancer Cell. 2014;25:415–427. doi: 10.1016/j.ccr.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 9.Herranz D, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat. Med. 2014;20:1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc. Natl. Acad. Sci. USA. 2014;111:705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 15.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 16.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimarchi T, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keim C, Kazadi D, Rothschild G, Basu U. Regulation of AID, the B-cell genome mutator. Genes. Dev. 2013;27:1–17. doi: 10.1101/gad.200014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng FL, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan M, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kon A, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 24.Thota S, et al. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124:1790–1798. doi: 10.1182/blood-2014-04-567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazumdar C, et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullenders J, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viny AD, et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watrin E, Kaiser FJ, Wendt KS. Gene regulation and chromatin organization: relevance of cohesin mutations to human disease. Curr. Opin. Genet. Dev. 2016;37:59–66. doi: 10.1016/j.gde.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 30.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 34.Kim E, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. USA. 2015;112:E4726–E4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilagan JO, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome. Res. 2014;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darman RB, et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell. Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 38.Alsafadi S, et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue D, Bradley RK, Abdel-Wahab Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes. Dev. 2016;30:989–1001. doi: 10.1101/gad.278424.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daubner GM, Cléry A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirai CL, et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27:631–643. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isono K, Mizutani-Koseki Y, Komori T, Schmidt-Zachmann MS, Koseki H. Mammalian polycomb-mediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes. Dev. 2005;19:536–541. doi: 10.1101/gad.1284605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kfir N, et al. SF3B1 association with chromatin determines splicing outcomes. Cell. Rep. 2015;11:618–629. doi: 10.1016/j.celrep.2015.03.048. [DOI] [PubMed] [Google Scholar]
- 46.Garding A, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the in cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stilgenbauer S, et al. Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Oncogene. 1998;16:1891–1897. doi: 10.1038/sj.onc.1201764. [DOI] [PubMed] [Google Scholar]
- 48.Klein U, et al. The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 51.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 52.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes. Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 55.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 56.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Béguelin W, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc. Natl. Acad. Sci. USA. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sneeringer CJ, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon C, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes. Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danis E, et al. Ezh2 controls an early hematopoietic program and growth and survival signaling in early T cell precursor acute lymphoblastic leukemia. Cell. Rep. 2016;14:1953–1965. doi: 10.1016/j.celrep.2016.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi J, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013;32:930–938. doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neff T, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. USA. 2012;109:5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Velichutina I, et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes. Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Kruijsbergen I, Hontelez S, Veenstra GJ. Recruiting polycomb to chromatin. Int. J. Biochem. Cell Biol. 2015;67:177–187. doi: 10.1016/j.biocel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shields BJ, et al. Acute myeloid leukemia requires Hhex to enable PRC2-mediated epigenetic repression of Cdkn2a. Genes. Dev. 2016;30:78–91. doi: 10.1101/gad.268425.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol. Biol. Cell. 2007;18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol. Cell. Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 77.Bernt KM, Armstrong SA. A role for DOT1L in MLL-rearranged leukemias. Epigenomics. 2011;3:667–670. doi: 10.2217/epi.11.98. [DOI] [PubMed] [Google Scholar]
- 78.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan J, et al. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 81.Allis CD, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 82.Harris WJ, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 83.Schenk T, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yatim A, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol. Cell. 2012;48:445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li Y, et al. Dynamic interaction between TAL1 oncoprotein and LSD1 regulates TAL1 function in hematopoiesis and leukemogenesis. Oncogene. 2012;31:5007–5018. doi: 10.1038/onc.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 87.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 88.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 89.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 90.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 91.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 92.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greenfield A, et al. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 94.Van der Meulen J, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benyoucef A, et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes. Dev. 2016;30:508–521. doi: 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hogart A, et al. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome. Res. 2012;22:1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bock C, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith AE, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 104.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grossmann V, et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosom. Cancer. 2013;52:410–422. doi: 10.1002/gcc.22039. [DOI] [PubMed] [Google Scholar]
- 106.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.An J, et al. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 2015;6:10071. doi: 10.1038/ncomms10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lobry C, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J. Exp. Med. 2013;210:301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shih AH, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27:502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 114.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen C, et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes. Dev. 2013;27:1974–1985. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mayle A, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meyer SE, et al. DNMT3A haploinsufficiency transforms FLT3ITD myeloproliferative disease into a rapid, spontaneous, and fully penetrant acute myeloid leukemia. Cancer Discov. 2016;6:501–515. doi: 10.1158/2159-8290.CD-16-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 121.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat. Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim KH, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 125.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 126.Chen CW, et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 2015;21:335–343. doi: 10.1038/nm.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 129.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]