 A two-component hybrid system consisting of organic dye nanoparticles (ash) and a protein (blue) at different ratios allows for the design of narrow range tunable nanoprobes for sensing and imaging of pH variations in live cells through ratiometric signalling.
A two-component hybrid system consisting of organic dye nanoparticles (ash) and a protein (blue) at different ratios allows for the design of narrow range tunable nanoprobes for sensing and imaging of pH variations in live cells through ratiometric signalling.
Abstract
Accurate monitoring of pH variations inside cells is important for the early diagnosis of diseases such as cancer. Even though a variety of different pH sensors are available, construction of a custom-made sensor array for measuring minute variations in a narrow biological pH window, using easily available constituents, is a challenge. Here we report two-component hybrid sensors derived from a protein and organic dye nanoparticles whose sensitivity range can be tuned by choosing different ratios of the components, to monitor the minute pH variations in a given system. The dye interacts noncovalently with the protein at lower pH and covalently at higher pH, triggering two distinguishable fluorescent signals at 700 and 480 nm, respectively. The pH sensitivity region of the probe can be tuned for every unit of the pH window resulting in custom-made pH sensors. These narrow range tunable pH sensors have been used to monitor pH variations in HeLa cells using the fluorescence imaging technique.
Introduction
pH plays a vital role in regulating many cellular events, including proliferation, apoptosis, enzyme activity and protein degradation.1,2 Monitoring of pH variations inside living cells is thus of great importance due to its direct link with many common diseases such as cancer, Alzheimer's disease, and others.3,4 While several types of probes are available, protein modified fluorescent sensors are desirable to map the pH distribution and fluctuation with high spatio-temporal resolution in living cells.5–9 The different types of pH sensors currently available are based on fluorescent organic molecules,10,11 fluorescent proteins,12 polymer nanoparticles,13 metal nanoparticles,14 quantum dots,15 dendrimers,16 hydrogels17 and DNA nanomachines.18,19 In order to achieve a highly sensitive probe that detects pH in different ranges, two or more fluorophores having different pK a values have to be covalently linked20 or embedded into different matrices such as a polymer matrix,21 nanoparticles22 or carbon nanodots.23 While most of these probes respond to broad pH variations, the use of a composition dependent two-component system to accurately monitor minute pH variations in a narrow pH range remains elusive.
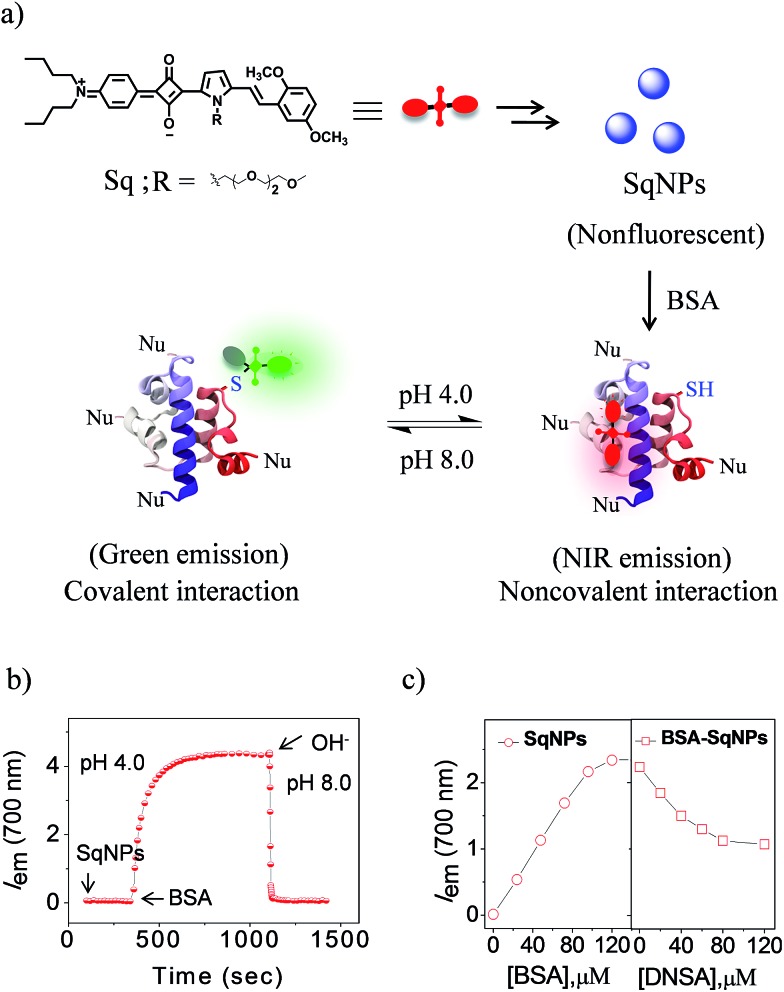
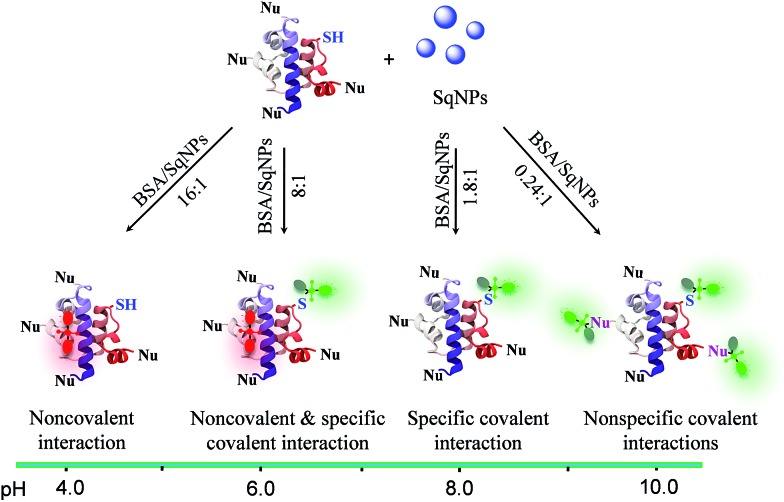
In order to achieve the above objective, we explore the pH dependent interaction of squaraine dyes (Sq) with thiols resulting in changes in the absorption and emission properties.24 Recently, we have shown that Sq nanoparticles (SqNPs) specifically interact with serum albumin proteins (SAP), resulting in BSA and HSA specific sensors.25 In the present work, we report a unique two-component hybrid pH sensor whose sensitivity range can be tuned for narrow pH windows by choosing different ratios of the individual components, for monitoring minute pH variations in live cells. These nanosensors have a fast and variable fluorescence response that is ideal for measuring minor pH variations with a linear response. The mode of interaction between the dye and the protein is depicted in Fig. 1a. The dye nanoparticles undergo pH dependent reversible noncovalent and covalent interactions with BSA as evident by the fluorescence changes of SqNPs at 700 nm (λ ex@640 nm, phosphate buffer, pH 4.0) and the formation of a new band at 480 nm (λ ex@380 nm, phosphate buffer, pH 8.0), respectively.
Fig. 1. (a) The mode of pH controlled covalent and noncovalent interaction of SqNPs with BSA protein. (b) Time dependent fluorescence response of 16 : 1 BSA–SqNPs at 700 nm (λ ex@640 nm). (c) Fluorescence response of SqNPs towards BSA protein (0–96 μM) followed by the addition of DNSA (0–120 μM). Experiments were performed using a 6 μM SqNP solution in a 25 mM phosphate buffer at pH 4.0.
Results and discussion
In order to establish the mode of interaction of the SqNPs with BSA, first we examined the stability of the dye and the protein under different pH conditions and found that they are stable (Fig. S1 and S2†). Then we added SqNPs to BSA and the fluorescence changes were monitored. The fluorescence intensity of BSA–SqNPs (16 : 1) at 700 nm with respect to time under an acidic pH condition (pH 4.0) indicates a gradual enhancement which became saturated and remained at the saturation point for a long time (Fig. 1b). To this solution, addition of NaOH led to a sudden decrease in the fluorescence intensity to the initial state. Thus, it is evident that, although the dye under basic condition interacts covalently, under an acidic condition the interaction becomes noncovalent as depicted in Fig. 1a. This switching of dye interaction with protein was observed for several cycles revealing the reversibility of the interaction between Sq and BSA (Fig. S3†).
The noncovalent interaction of the dye with protein under an acidic condition was further verified from the electronic absorption spectral measurements. Addition of BSA protein (96 μM) to a solution of SqNPs (6 μM) in phosphate buffer at pH 4.0 resulted in the narrowing of the broad absorption band from 550–850 nm to 650–700 nm (Fig. S4a†). The spectral feature of this absorption band is similar to that of the Sq dye in acetonitrile in which the molecule exists in the monomeric state. The corresponding fluorescence changes (λ ex@640 nm) upon addition of BSA protein (0–96 μM) exhibited a gradual increase in the fluorescence intensity at 700 nm (Fig. S4b† and 1c). These data indicate the disassembly of SqNPs in the presence of BSA which was further confirmed by DLS analysis (Fig. S5†). Addition of BSA (96 μM) into SqNPs (6 μM, phosphate buffer) at pH 4.0 showed the disappearance of the peak at 250 nm corresponding to the SqNPs with an increase in the intensity of the band at 10 nm, corresponding to the size of the globular protein. For a deeper understanding of the noncovalent binding of Sq with the protein, ligand displacement studies were performed. Addition of dansylamide (DNSA), a site specific ligand for proteins, with the Sq-bound BSA showed a quenching of fluorescence intensity at 700 nm due to the displacement of the bound dye by DNSA and its subsequent aggregation (Fig. 1c). DLS analysis of the BSA–SqNPs complex after the addition of DNSA exhibited a peak at 250 nm corresponding to the size of the SqNPs, which confirms the above observation (Fig. S6†).
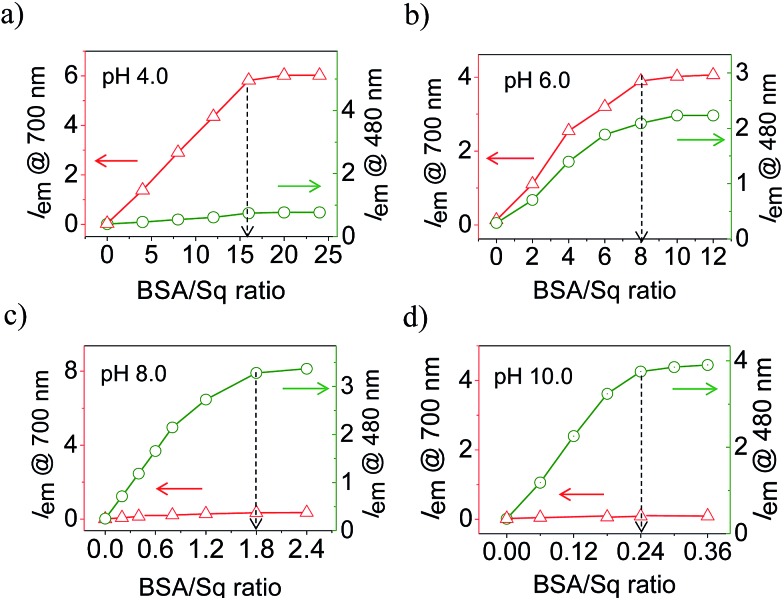
Fig. 2a–d shows the ratio dependent fluorescence intensity variation of BSA–SqNPs at the 700 (λ ex@640 nm) and 480 nm (λ ex@380 nm) region under different pH conditions. These data reveal the strong influence of the ratio between BSA and SqNPs on the emission properties. For example, at pH 4.0, a large amount of protein is required to turn-on the original emission of the dye at 700 nm which got saturated with the addition of 16 equiv. of BSA (Fig. 2a). In this case, no signal was observed at 480 nm, indicating the absence of a covalent interaction of Sq with BSA. At pH 6.0, 8 equiv. of protein was required to get the maximum fluorescence response (Fig. 2b). Noticeably, in this case, both noncovalent and covalent interactions are active as evident from the 700 and 480 nm fluorescence responses, respectively. Surprisingly, at pH 8.0, the maximum response of fluorescence occurred with 1.8 equiv. of BSA, with a reversal of the fluorescence response from 700 nm to 480 nm, indicating the specific covalent interaction of the dye with the protein (Fig. 2c). At pH 10.0, the dye nanoparticles showed a maximum fluorescence response at 480 nm with an extremely small amount of protein (0.24 equiv.) (Fig. 2d). These data clearly suggest that the ratio of the fluorescence intensities at 700 and 480 nm are strongly dependent upon the ratio between the SqNPs and BSA.
Fig. 2. (a)–(d) Emission intensity variation of SqNPs at 700 (λ ex@640 nm, -△-) and 480 nm (λ ex@380 nm, -○-) with different BSA concentrations under different pH conditions. Experiments were performed using a 6 μM SqNP solution in 25 mM phosphate buffer.
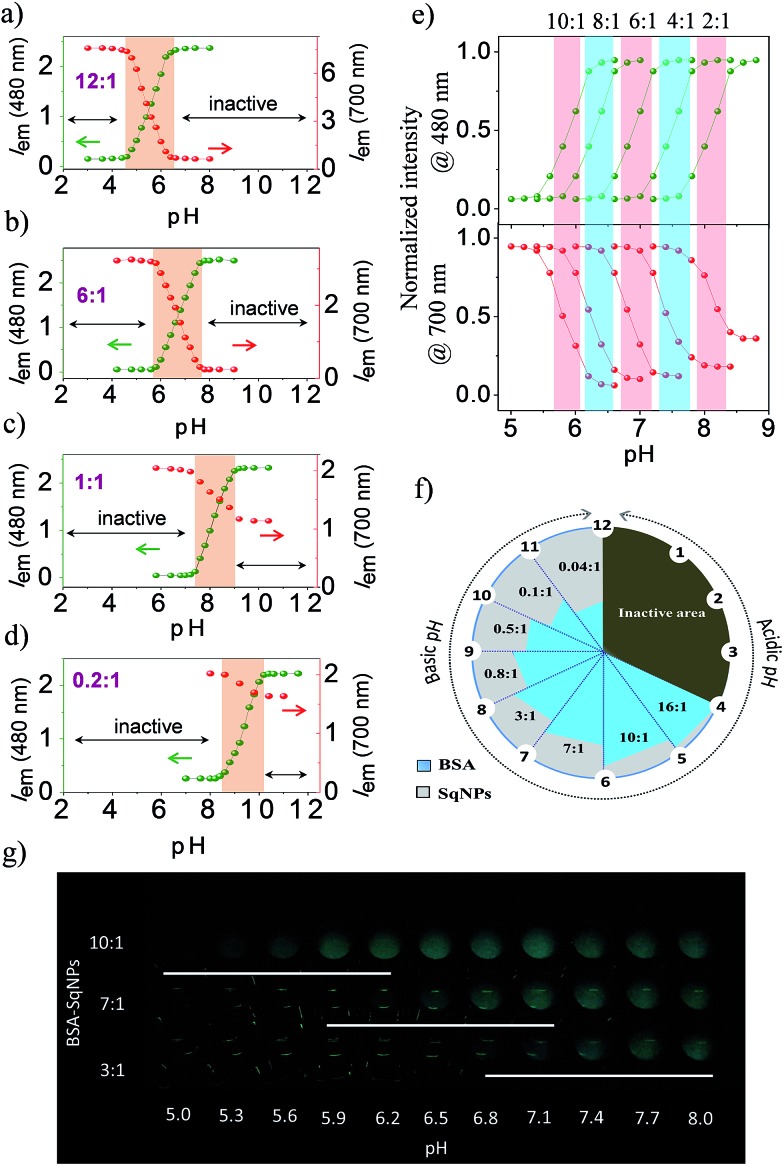
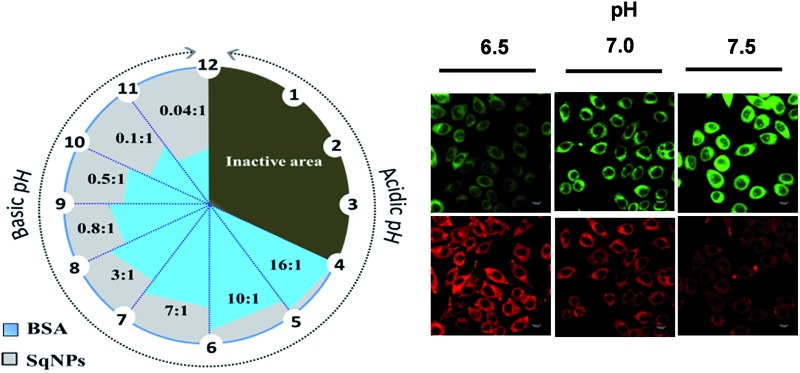
Since the interaction of the dye with protein could be controlled by changing the ratio between SqNPs and the protein at different pH conditions, we hypothesized that an array of hybrid probes that respond to different pH values can be designed. In order to establish this point, we prepared a series of nanosensors with BSA–SqNP ratios of 12 : 1, 6 : 1, 1 : 1 and 0.2 : 1. Each of these combinations was specific to a narrow pH compartment and allowed for continuous pH sensing over a broad pH window of 4.0 to 10.0, each with a detection range of 1.5–2.0 units as shown in the respective ratiometric emission plots (Fig. 3a–d). At a pH range of 2–4.6, a 12 : 1 BSA–SqNP combination exhibited a maximum fluorescence response at 700 nm and a minimum response at 480 nm due to the preferential noncovalent interaction of the dye with protein (Fig. 3a). As the pH of the solution is increased from 4.6, the intensity of the green fluorescence at 480 nm is increased with the simultaneous quenching of the NIR emission at 700 nm due to the covalent modification of the dye with protein. This noncovalent to covalent transformation of the dye with the protein allows ratiometric monitoring of pH. Therefore, the 12 : 1 BSA–SqNP complex exhibits optical signaling in a ratiometric fashion in the pH range of 4.6–6.4 and becomes inactive beyond pH 6.4. For sensing pH above 6.4, the amount of protein has to be decreased to a ratio of 6 : 1. The effect of pH on this combination is shown in Fig. 3b. At this combination, the sensor becomes active above pH 5.8 and inactive at pH 7.6. Therefore, a 6 : 1 combination becomes a pH sensor in the range of 5.8–7.6, however above 7.6, this combination turns inactive. Hence, we further reduced the amount of protein and a 1 : 1 combination was prepared. This combination exhibited activity in the pH range of 7.4–9.0 (Fig. 3c). For pH > 9.0, we tried a ratio of 0.2 : 1 BSA–SqNPs. This combination exhibited a pH response between 8.6 and 10.2 and the emission became saturated above this pH (Fig. 3d). Thus, by varying the ratio of BSA and SqNPs, we could design a series of sensors for monitoring pH over a narrow range in a broad window. From the above data we found that a 6 : 1 ratio provides good response in the biological window. In order to further fine tune the ratio required for minute pH variations in the biological window, we tried other combinations of BSA and SqNPs between 10 : 1 and 2 : 1. The results of these studies are shown in Fig. 3e. These studies revealed that the best combinations to monitor the pH variations in the biological pH window are between 10 : 1 and 4 : 1. For example, the maximum response of the 10 : 1 ratio is at pH 5.8 whereas the response of the 8 : 1 ratio is at 6.3. On the other hand, 6 : 1 and 4 : 1 ratios showed responses at 6.7 and 7.4 respectively. Thus, we could design probes whose maximum responses are narrowed down to 0.4–0.6 pH units. In this way, by swiftly adjusting the ratio between BSA and the dye nanoparticles, we could custom-make a series of hybrid nanosensors for monitoring pH variations in narrow pH segments over a broad pH scale as shown in Fig. 3f. From this figure, it is easy to decide upon the optimum ratio of the protein and the dye nanoparticles required for accurate pH monitoring of a given sample, particularly of a biological specimen. In addition, we have demonstrated the pH dependent fluorescence changes of the hybrids in the in vitro solution condition. UV illumination (365 nm) of hybrids of ratios 10 : 1, 7 : 1 and 3 : 1 at different pH in a microwell plate displaying sensitivity in different ranges, enables naked eye detection (Fig. 3g).
Fig. 3. (a)–(e) Fluorescence intensity of BSA–SqNP hybrid nanosensors in the 480 (λ ex@380 nm) and 700 nm (λ ex@640 nm) region at different pH conditions. The active pH range for different ratios is indicated by the shadow color. (f) A pictorial representation of the construction of the narrow range tunable pH sensors with different ratios of BSA and SqNPs and the corresponding pH sensitivity range within a unit of one pH scale. (g) Photograph of BSA–SqNP hybrids with ratios of 10 : 1, 7 : 1 and 3 : 1 filled in wells of pH 5.0–8.0 in a microwell plate under UV illumination (365 nm).
After having a clear idea of the effect of pH on various combinations of BSA–SqNPs, their potential for fluorescence imaging of pH variations in the intracellular environment of living cells was investigated. Prior to this study, the effect of the intracellular environment on the probe was examined by monitoring the fluorescence spectra of a protein–dye hybrid with a 6 : 1 ratio in the presence of physiologically important metal ions and chemicals (such as Na+, K+, Ca2+, Zn2+, Mg2+, Mn2+, Cu2+, Fe2+, Fe3+, glutathione (GSH), cysteine (Cys) and homocysteine (Hcy)) and H2O2 at pH 5.8 (I em@700 nm, λ ex@640 nm) and 7.6 (I em@480 nm, λ ex@380 nm) (Fig. S7†). No appreciable spectroscopic changes were observed under these conditions, which imply that the probe could be used to measure the intracellular pH changes without interference from other biomolecules at their physiological concentrations. To further confirm this point, HeLa cells were incubated with SqNPs in the absence of BSA and imaged using confocal fluorescence microscopy (Fig. S8†). No appreciable fluorescence signal was observed from the cells confirming that the SqNPs could not interact with any of the biologically relevant analytes.
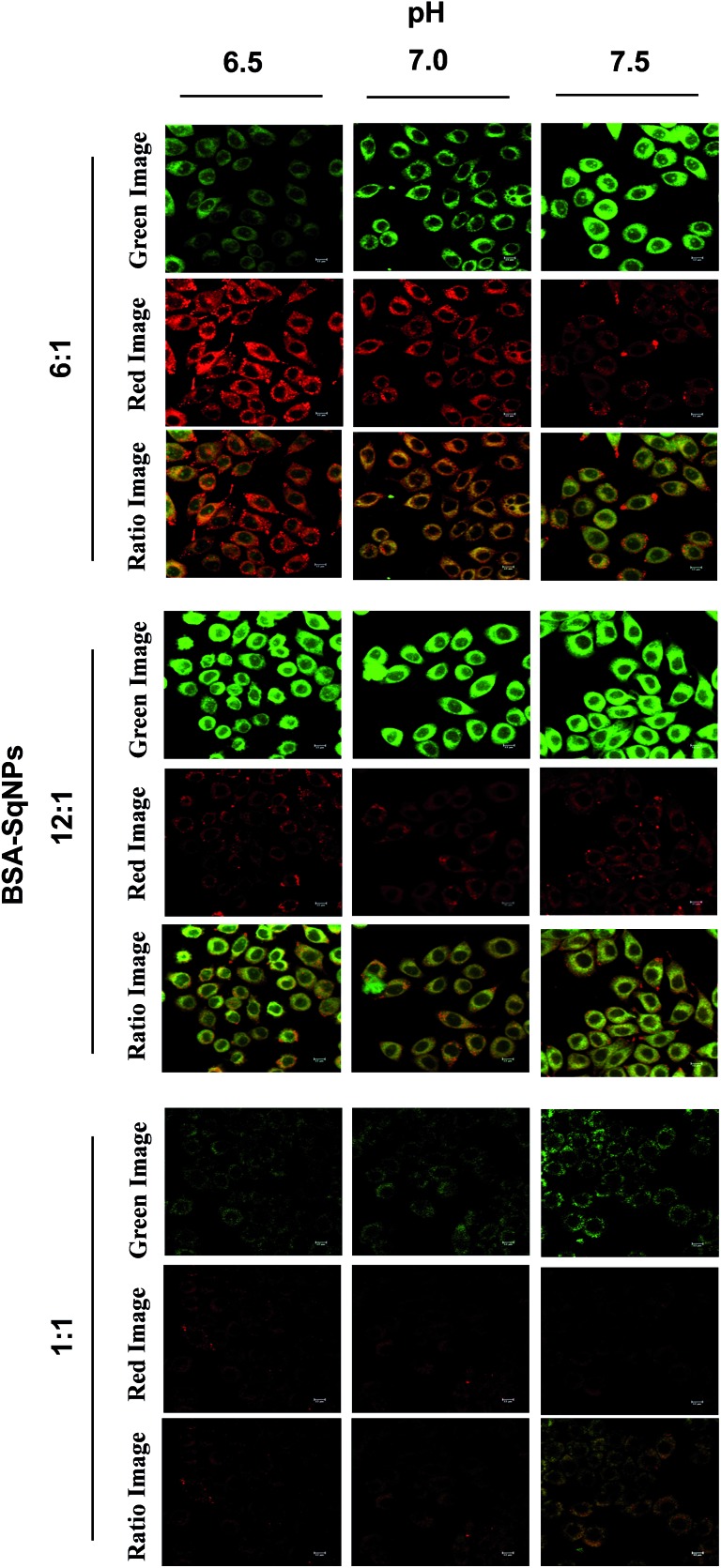
The pH variations inside cells were imaged using BSA–SqNP hybrid sensors of different ratios. Before the pH monitoring, the cytotoxicity of the probe was evaluated using the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. S9†). The result revealed that SqNPs and BSA–SqNPs exhibit minimal toxicity to the cells under the concentrations measured, demonstrating their suitability for biological applications. Furthermore, we investigated the cellular uptake and intracellular distribution of BSA–SqNPs. After incubation of HeLa cells with a 6 : 1 BSA–SqNP combination (in phosphate buffer, pH 7.0) for 30 min, significant red and green fluorescence was detected in the cytoplasm, however not in the nucleus or extracellular environment (Fig. S10†); indicative of the excellent membrane permeability of the probe. Hence, we used a 6 : 1 combination of the BSA–SqNP hybrid to monitor minor pH fluctuations inside live cells. For this purpose, the protein–dye hybrid (6 : 1) was imported into HeLa cells grown in wells of different pH values (6.5–7.5). The intracellular pH was equilibrated to the surrounding medium using the H+/K+ ionophore, nigericin. After the uptake of the BSA–SqNP conjugate for 30 min, confocal fluorescence images of the cells were recorded (Fig. 4). At acidic pH (6.5), the probe gave a strong red emission and a weak green emission due to the noncovalent interaction of the dye with BSA. As the pH of the cells is increased to the basic region (7.5), the probe exhibited a strong green emission and a weak red emission as a result of the covalent interaction of the dye with BSA. The corresponding fluorescence intensity (Fig. S11a and S12b†) obtained from the ratio image exhibits good linearity, which shows the ability of the probe to measure the intracellular pH variation over a small pH range. Cells of pH 6.5–7.5 were incubated with BSA–SqNP hybrids with a 12 : 1 and 1 : 1 ratio (Fig. 4). The cells treated with the 12 : 1 hybrid gave an “always on” signal in the green window and an “always off” signal in the red window irrespective of pH as shown in Fig. 4, S12a and b,† which implies that at the given experimental pH window, this combination is not sensitive. As expected, the 1 : 1 combination gives an “always off” signal in both green and red windows, since it works above the pH range that we chose.
Fig. 4. Confocal fluorescence images of HeLa cells incubated with BSA–SqNP hybrids in the ratio of 6 : 1, 12 : 1 and 1 : 1 at pH 6.5, 7.0 and 7.5 for 30 min at 37 °C. Green and red images were obtained by exciting at 405 and 640 nm respectively. The corresponding ratio images are also shown.
The role of the ratio between BSA and SqNPs on the pH response is explained based on the pH dependent reactivity of various functional groups in the protein (Scheme 1). At lower pH, the thiol group of BSA is less reactive towards the SqNPs and the interaction is exclusively noncovalent as indicated by the turn-on emission at 700 nm. In this case, a large amount of BSA is required for the emission response to become saturated. As the pH of the medium is increased, the thiol group of BSA becomes increasingly reactive towards the SqNPs, triggering the emission at 480 nm with a relatively lower amount of the protein. At pH > 8.0, in addition to the thiol group, the other nucleophilic groups of BSA also become reactive towards the Sq dyes and the interaction becomes exclusively covalent. Thus, at higher pH, a single protein can be covalently labeled with more than one dye molecule.26 Thus, the maximum fluorescence response at 480 nm occurs even with a lower amount of the protein. In all these cases, the protein facilitates the disassembly of SqNPs. The optimum amount of protein required for the complete disassembly of SqNPs at different pH values was determined by DLS analysis (Fig. S13†).
Scheme 1. The mode of interaction of SqNPs with BSA at different pH conditions: BSA protein contains several amino acids having different nucleophilic functional groups. SqNPs interact with BSA differently with respect to pH variation at different BSA–SqNP compositions.
In conclusion, we have demonstrated a two-component protein–dye hybrid sensor system for monitoring narrow range pH variations in live cells. The ratio of the protein and the dye nanoparticles controls the subtle balance between the noncovalent and the covalent interactions of the protein and the dye nanoparticle which is the key for the observed properties. The “turn-on” fluorescence at distinct emission wavelengths allows ratiometric monitoring of the pH variations. By systematic investigation, we were able to design sensor combinations that are suitable for the imaging of pH variations in live cells. These custom-made nanoprobes can be used in the spatio-temporal imaging of narrow range pH fluctuations, which can provide information on the physiological conditions and the general health of live cells.
Experimental
Materials and reagents
Bovine serum albumin (BSA), dansylamide (DNSA), nigericin and other amino acids were purchased from commercial suppliers. All other reagents were of analytical grade and were used without further purification. Reactions were performed under an inert atmosphere of nitrogen unless specified otherwise.
Preparation of the BSA–SqNP hybrid nanosensor array for compartmental pH sensing
Sq was prepared and characterized as reported earlier.25 A stock solution of Sq with a concentration of 5 × 10–3 M was prepared in acetonitrile. 25 μL of this solution was injected into 5 mL of phosphate buffer (25 mM NaH2PO4, 10 mM NaCl) having a pH of 7.0 at room temperature (25 °C). The green color of the stock solution turned blue indicating aggregation of the dye. Stock solutions of BSA protein at four different concentrations (7.5 × 10–3 M, 3.75 × 10–3 M, 6.25 × 10–4 M and 1.25 × 10–4 M respectively for preparing 12 : 1, 6 : 1, 1 : 1 and 0.2 : 1 complex) were prepared in phosphate buffer (25 mM NaH2PO4, 10 mM NaCl) maintained at pH 7.0. Concentrations of these stock solutions were calculated from the absorbance at a particular wavelength and molar extinction coefficient values. 200 μL of BSA from the respective stock was slowly added to the stirring solution of SqNPs (2.5 × 10–5 M). The resulting solution was vigorously stirred for 30 min and then equilibrated at 25 °C for a period of 15 min. A series of standard pH buffers of different pH with a difference of 0.2 were prepared in 5 mL volumetric flasks. The pH value was measured by a Delta 320 pH-meter. 200 μL of the BSA–SqNP complex was added to each of these solutions and shaken well. After keeping the solution for 30 min, each solution was taken into a 3 mL quartz cuvette with a path length of 1 cm at room temperature (25 °C). The emission spectra at 480 nm (λ ex@380 nm) as well as 700 nm (λ ex@640 nm) were recorded using a spectrofluorimeter. The pH cycling experiments of the SqNP–BSA hybrid were performed by the alternate addition of 4 μL of 0.5 N HCl and 4 μL of 0.5 N KOH respectively.
Site-selective binding of BSA by the Sq dye
The BSA–SqNP hybrid at a ratio of 16 : 1 was prepared by mixing 50 μL of BSA (5.7 × 10–3 M) to a stirring solution of SqNPs (6 × 10–6 M, 25 mM NaH2PO4, 10 mM NaCl, pH 4.0) in a 3 mL quartz cuvette. The solution then continued to stir for another 30 min. 0–150 μL of DNSA, a known site-I specific binding reagent, was added from a stock solution (2.4 × 10–3 M) and emission spectra were recorded each time after 30 min.
Cell culture
HeLa cells were maintained in DMEM containing 10% fetal bovine serum. It was incubated in a humidified CO2 incubator (5% CO2) at 37 °C for 2 days so that the cells attained about 70% confluency after cell revival. Then the medium was removed and cells were given a wash with phosphate buffered saline (PBS), the cells were trypsinized using 0.05% trypsin–EDTA by incubating at 37 °C for 5 min. These cells were counted and were seeded into optibottom 96 well glass culture plates with approximately 5000 cells per well in DMEM containing 10% FBS. The cells were used for the experiment 24–30 h after this.
In vitro cytotoxicity assay
The MTT assay test was carried out by following standard procedures. HeLa cells were seeded into a 96-well plate (1 × 104 cells per well) in DMEM (Dulbecco's Modified Eagle's Medium) containing 10% fetal bovine serum (FBS) and grown under a humidified atmosphere with 5% CO2 at 37 °C. After a 12 h incubation, the media in the wells were replaced with fresh DMEM (100 μL per well) containing Sq and BSA–SqNPs complexes with different concentrations, and the cells were further incubated for 12 h. Then, the medium was changed with DMEM (100 μL per well) containing MTT (0.5 mg mL–1), followed by incubation for another 4 h. The culture medium was removed and the frozen crystals were dissolved in freshly prepared DMSO (100 μL). Before the cytotoxicity measurement, the plate was agitated gently for 15 min, and then the absorbance intensity at 560 nm was recorded using a micro plate reader. The relative cell viability (%) for each sample related to the control well was finally calculated.
Fluorescence imaging and pH monitoring of HeLa cells using the SqNP–protein hybrid nanosensor
pH monitoring of HeLa cells was performed by the confocal laser scanning fluorescence microscopy technique. For fluorescence imaging, the culture medium was removed and the cells were washed with PBS (pH 7.4) twice. The cells were incubated with high K+ buffer (30 mM NaCl, 120 mM KCl, 1 mM CaCl2, 0.5 mM MgSO4, 1 mM NaH2PO4, 5 mM glucose, 20 mM HEPES, and 20 mM NaOAc) at various pH values (6.5–7.5) in the presence of 10.0 μM nigericin. After 30 min, 10 μL of the nanosensor (2.0 × 10–4 M, phosphate buffer (25 mM NaH2PO4, 10 mM NaCl) at pH 7.0) at different ratios (6 : 1, 12 : 1, 1 : 1) was added to each well and incubated for 30 min at 37 °C. The adherent cells were washed three times with PBS (pH 7.4) to remove excess probes from the medium. Fluorescence imaging experiments were performed with a Nikon A1Rsi confocal microscope with a 405 nm diode laser and 640 nm far red lasers for excitation using a 40× (1.4) NA objective. Optical sections were acquired at 0.8 μm. The fluorescence was collected in the ranges of 420–650 nm (green) and 650–800 nm (red), respectively. The background fluorescence for both the wavelengths was nullified by adjusting the voltage of the photomultiplier tube and adjusting the threshold. The region of interest (ROI) was drawn around every cell in a field for the purpose of quantitation. Image processing and analysis were performed on NIS elements version 4.0 software, and the ratio of intensities was calculated. All data were expressed as mean ± standard deviation.
Acknowledgments
A. A. is grateful to CSIR, Govt. of India, New Delhi for financial support under CSIR 12 FYP network programme M2D-CSC-0134 and DST-SERB, Govt. of India for a J. C. Bose Fellowship. P. A. and K. V. S. thank CSIR for research fellowship. A. R. C. and R. V. O. were financially supported by Rajiv Gandhi Centre for Biotechnology.
Footnotes
References
- (a) Gottlieb R. A., Nordberg J., Skowronski E., Babior B. M. Proc. Natl. Acad. Sci. U. S. A. 1996;93:654–658. doi: 10.1073/pnas.93.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ma-tsuyama S., Llopis J., Deveraux Q. L., Tsien R. Y., Reed J. C. Nat. Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- (a) Nishi T., Forgac M. Nat. Rev. Mol. Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]; (b) Casey J. R., Grinstein S., Orlowski J. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- (a) Izumi H., Torigoe T., Ishiguchi H., Uramoto H., Yoshida Y., Tanabe M., Ise T., Murakami T., Yoshida T., Nomoto M., Kohno K. Cancer Treat. Rev. 2003;29:541–549. doi: 10.1016/s0305-7372(03)00106-3. [DOI] [PubMed] [Google Scholar]; (b) Webb B. A., Chimenti M., Jacobson M. P., Barber D. L. Nat. Rev. Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhou K., Huang G., Hensley C., Huang X., Ma X., Zhao T., Sumer B. D., DeBerardinis R. J., Gao J. A. Nat. Methods. 2014;13:204–212. doi: 10.1038/nmat3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Mizakami S., Hori Y., Kikuchi K. Acc. Chem. Res. 2014;47:247–256. doi: 10.1021/ar400135f. [DOI] [PubMed] [Google Scholar]; (b) Grover A., Schmidt B. F., Salter R. D., Watkins S. C., Waggoner A. S., Bruchez M. P. Angew. Chem., Int. Ed. 2012;51:4838–4842. doi: 10.1002/anie.201108107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chen Q., Liu X., Chen J., Zeng J., Cheng Z., Liu Z. Adv. Mater. 2012;27:6820–6827. doi: 10.1002/adma.201503194. [DOI] [PubMed] [Google Scholar]
- (a) Wang H., Nakata E., Hamachi I. ChemBioChem. 2009;10:2560–2577. doi: 10.1002/cbic.200900249. [DOI] [PubMed] [Google Scholar]; (b) Takaoka Y., Ojida A., Hamachi I. Angew. Chem., Int. Ed. 2013;52:4088–4106. doi: 10.1002/anie.201207089. [DOI] [PubMed] [Google Scholar]
- (a) Mizuhara T., Saha K., Moyano D. F., Kim C. S., Yan B., Kim Y. K., Rotello V. M. Angew. Chem., Int. Ed. 2015;54:6567–6570. doi: 10.1002/anie.201411615. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rana S., Le N. D. B., Mout R., Saha K., Tonga G. Y., Bain R. E. S., Miranda O. R., Rotello C. M., Rotello V. M. Nat. Nanotechnol. 2015;8:65–69. doi: 10.1038/nnano.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Gao F. P., Lin Y. X., Li L. L., Liu Y., Mayerhöffer U., Spenst P., Su J. G., Li J. Y., Würthner F., Wang H. Biomaterials. 2014;35:1004–1014. doi: 10.1016/j.biomaterials.2013.10.039. [DOI] [PubMed] [Google Scholar]; (b) Zhang D., Zhao Y. X., Qiao Z. Y., Mayerhöffer U., Spenst P., Li X. J., Würthner F., Wang H. Bioconjugate Chem. 2014;25:2021–2029. doi: 10.1021/bc5003983. [DOI] [PubMed] [Google Scholar]
- (a) Ghimire A., Kasi R. M., Kumar C. V. J. Phys. Chem. B. 2014;118:5026–5033. doi: 10.1021/jp500310w. [DOI] [PubMed] [Google Scholar]; (b) Deshapriya I. K., Stromer B. S., Pattammattel A., Kim C. S., Iglesias-Bartolome R., Gonzalez-Fajardo L., Patel V., Gutkind S., Lu X., Kumar C. V. Bioconjugate Chem. 2015;11:605–618. doi: 10.1021/bc500621h. [DOI] [PubMed] [Google Scholar]
- (a) Urano Y., Asanuma D., Hama Y., Koyama Y., Barrett T., Kamiya M., Nagano T., Watanabe T., Hasegawa A., Choyke P. L., Kobayashi H. Nat. Med. 2009;15:104–109. doi: 10.1038/nm.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Myochin T., Kiyose K., Hanaoka K., Kojima H., Terai T., Nagano T. J. Am. Chem. Soc. 2011;133:3401–3409. doi: 10.1021/ja1063058. [DOI] [PubMed] [Google Scholar]; (c) Lee M. H., Park N., Yi C., Han J. H., Hong J. H., Kim K. P., Kang D. H., Sessler J. L., Kang C., Kim J. S. J. Am. Chem. Soc. 2014;136:14136–14142. doi: 10.1021/ja506301n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Li P., Xiao H., Cheng Y., Zhang W., Huang F., Zhang W., Wang H., Tang B. Chem. Commun. 2014;50:7184–7187. doi: 10.1039/c4cc01390e. [DOI] [PubMed] [Google Scholar]; (e) Sarkar A. R., Heo C. H., Xu L., Lee H. W., Si H. Y., Byun J. W., Kim H. M. Chem. Sci. 2016;7:766–773. doi: 10.1039/c5sc03708e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Han J., Burgess K. Chem. Rev. 2010;110:2709–2728. doi: 10.1021/cr900249z. [DOI] [PubMed] [Google Scholar]; (b) Schremla S., Meierb R. J., Wolfbeis O. S., Landthalera M., Szeimiesa R. M., Babilasa P. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2432–2437. doi: 10.1073/pnas.1006945108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chen S., Hong Y., Liu Y., Liu J., Leung C. W. T., Li M., Kwok R. T. K., Zhao E., Lam J. W. Y., Yu Y., Tang B. Z. J. Am. Chem. Soc. 2013;135:4926–4929. doi: 10.1021/ja400337p. [DOI] [PubMed] [Google Scholar]; (d) Shundo A., Ishihara S., Labuta J., Onuma Y., Sakai H., Abe M., Ariga K., Hill J. P. Chem. Commun. 2013;49:6870–6872. doi: 10.1039/c3cc42859a. [DOI] [PubMed] [Google Scholar]
- (a) Llopis J., McCaffery J. M., Miyawaki A., Far-quhar M. G., Tsien R. Y. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Miesenbock G., De Angelis D. A., Rothman J. E. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]; (c) Tantama M., Hung Y. P., Yellen G. J. Am. Chem. Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Sun G., Cui H., Lin L. Y., Lee N. S., Yang C., Neumann W. L., Freskos J. N., Shieh J. J., Dorshow R. B., Wooley K. L. J. Am. Chem. Soc. 2011;133:8534–8543. doi: 10.1021/ja200182t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ling D., Park W., Park S. J., Lu Y., Kim K. S., Hackett M. J., Kim B. H., Yim H., Jeon Y. S., Na K., Hyeon T. J. Am. Chem. Soc. 2014;136:5647–5655. doi: 10.1021/ja4108287. [DOI] [PubMed] [Google Scholar]; (c) Ma X., Wang Y., Zhao T., Li Y., Su L. C., Wang Z., Huang G., Sumer B. D., Gao J. J. Am. Chem. Soc. 2014;136:11085–11092. doi: 10.1021/ja5053158. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wang X.-D., Stolwijk J. A., Lang T., Sperber M., Meier R. J., Wegener J., Wolfbeis O. S. J. Am. Chem. Soc. 2012;134:17011–17014. doi: 10.1021/ja308830e. [DOI] [PubMed] [Google Scholar]; (e) Yin L. Y., He C. S., Huang C. S., Zhu W. P., Wang X., Xu Y. F., Qian X. H. Chem. Commun. 2012;48:4486–4488. doi: 10.1039/c2cc30404j. [DOI] [PubMed] [Google Scholar]; (f) Paek K., Yang H., Lee J., Park J., Kim B. J. ACS Nano. 2014;8:2848–2856. doi: 10.1021/nn406657b. [DOI] [PubMed] [Google Scholar]
- (a) Kim S., Pudavar H. E., Prasad P. N. Chem. Commun. 2011:2071–2073. doi: 10.1039/b600926c. [DOI] [PubMed] [Google Scholar]; (b) Benjaminsen R. V., Sun H., Hen-riksen J. R., Christensen N. M., Almdal K., Andresen T. L. ACS Nano. 2011;5:5864–5873. doi: 10.1021/nn201643f. [DOI] [PubMed] [Google Scholar]; (c) Ariga K., Ji K. Q., McShane M. J., Lvov Y. M., Hill J. P. Chem. Mater. 2012;24:728–737. [Google Scholar]
- (a) Medintz I. L., Uyeda H. T., Goldman E. R., Mattoussi H. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]; (b) Snee P. T., Somers R. C., Nair G., Zimmer J. P., Bawendi M. G., Nocera D. G. J. Am. Chem. Soc. 2006;128:13320–13321. doi: 10.1021/ja0618999. [DOI] [PubMed] [Google Scholar]; (c) Jin T., Sasaki A., Kinjo M., Miyazaki J. A. Chem. Commun. 2010;46:2408–2410. doi: 10.1039/b921602b. [DOI] [PubMed] [Google Scholar]; (d) Medintz I. L., Stewart M. H., Trammell S. A., Susumu K., Delehanty J. B., Mei B. C., Melinger J. S., Blanco-Canosa J. B., Dawson P. E., Mattoussi H. Nat. Mater. 2010;9:676–684. doi: 10.1038/nmat2811. [DOI] [PubMed] [Google Scholar]; (e) Orte A., Alvarez-Pez J. M., Ruedas-Rama M. J. ACS Nano. 2013;7:6387–6395. doi: 10.1021/nn402581q. [DOI] [PubMed] [Google Scholar]; (f) Paek K., Chung S., Cho C.-H., Kim B. J. Chem. Commun. 2011;47:10272–10274. doi: 10.1039/c1cc13848k. [DOI] [PubMed] [Google Scholar]; (g) Wu Y., Chakrabortty S., Gropeanu R. A., Wilhelmi J., Xu Y., Er K. S., Kuan S. L., Koynov K., Chan Y., Weil T. ACS Nano. 2012;6:2917–2924. [Google Scholar]; (h) Ji X., Palui G., Avellini T., Na H. B., Yi C., Knappenberger Jr K. L., Mattoussi H. J. Am. Chem. Soc. 2012;134:6006–6017. doi: 10.1021/ja300724x. [DOI] [PubMed] [Google Scholar]
- (a) Almutairi A., Guillaudeu S. J., Berezin M. Y., Achilefu S., Frechet J. M. J. Am. Chem. Soc. 2008;130:444–445. doi: 10.1021/ja078147e. [DOI] [PubMed] [Google Scholar]; (b) Albertazzi L., Storti B., Marchetti L., Beltram F. J. Am. Chem. Soc. 2010;132:18158–18167. doi: 10.1021/ja105689u. [DOI] [PubMed] [Google Scholar]; (c) Srikun D., Albersa A. E., Chang C. J. Chem. Sci. 2011;2:1156–1165. [Google Scholar]
- Peng H. S., Stolwijk J. A., Sun L. N., Wegener J., Wolfbeis O. S. Angew. Chem., Int. Ed. 2010;49:4246–4249. doi: 10.1002/anie.200906926. [DOI] [PubMed] [Google Scholar]
- (a) Modi S., Swetha M. G., Goswami D., Gupta G. D., Mayor S., Krishnan Y. Nat. Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]; (b) Modi S., Nizak C., Surana S., Halder S., Krishnan Y. Nat. Nanotechnol. 2013;8:459–467. doi: 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- (a) Idili A., Vallee-Belisle A., Ricci F. J. Am. Chem. Soc. 2014;136:5836–5839. doi: 10.1021/ja500619w. [DOI] [PubMed] [Google Scholar]; (b) Chen G., Liu D., He C., Gannett T. R., Lin W., Weizmann Y. J. Am. Chem. Soc. 2015;137:3844–3851. doi: 10.1021/ja512665z. [DOI] [PubMed] [Google Scholar]
- (a) Lee M. H., Han J. H., Lee J. H., Park N., Kumar R., Kang C., Kim J. S. Angew. Chem., Int. Ed. 2013;52:6206–6209. doi: 10.1002/anie.201301894. [DOI] [PubMed] [Google Scholar]; (b) Chen S., Hong Y., Liu Y., Liu J., Leung C. W. T., Li M., Kwok R. T. K., Zhao E., Lam J. W. Y., Yu Y., Tang B. Z. J. Am. Chem. Soc. 2013;135:4926–4929. doi: 10.1021/ja400337p. [DOI] [PubMed] [Google Scholar]
- Sun H., Almdal K., Andresen T. L. Chem. Commun. 2011;47:5268–5270. doi: 10.1039/c1cc10439j. [DOI] [PubMed] [Google Scholar]
- (a) Zhou K., Wang Y., Huang X., Luby-Phelps K., Sumer B. D., Gao J. Angew. Chem., Int. Ed. 2011;50:6109–6114. doi: 10.1002/anie.201100884. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhou K., Liu H., Zhang S., Huang X., Wang Y., Huang G., Sumer B. D., Gao J. J. Am. Chem. Soc. 2012;134:7803–7811. doi: 10.1021/ja300176w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Shi W., Li X., Ma H. Angew. Chem., Int. Ed. 2012;51:6432–6435. doi: 10.1002/anie.201202533. [DOI] [PubMed] [Google Scholar]; (b) Nie H., Li M. J., Li Q. S., Liang S. J., Tan Y. Y., Sheng Z., Shi W., Zhang S. X.-A. Chem. Mater. 2014;26:3104–3112. [Google Scholar]
- (a) Ros-Lis J. V., Garcia B., Jimenez D., Mar-tinez-Manez R., Sancenon F., Soto J., Gonzalvo F., Valldecabres M. C. J. Am. Chem. Soc. 2004;126:4064–4065. doi: 10.1021/ja031987i. [DOI] [PubMed] [Google Scholar]; (b) Sreejith S., Divya K. P., Ajayaghosh A. Angew. Chem., Int. Ed. 2008;47:7883–7887. doi: 10.1002/anie.200803194. [DOI] [PubMed] [Google Scholar]; (c) Hewage H. S., Anslyn E. V. J. Am. Chem. Soc. 2009;131:13099–13106. doi: 10.1021/ja904045n. [DOI] [PubMed] [Google Scholar]
- Anees P., Sreejith S., Ajayaghosh A. J. Am. Chem. Soc. 2014;136:13233–13239. doi: 10.1021/ja503850b. [DOI] [PubMed] [Google Scholar]
- The reactivity of various functional groups of BSA with Sq dye at various pH conditions, were examined with various amino acids such as serine, tyrosine and threonine in 30% ACN/phosphate buffer at different pH (Fig. S13)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.