Abstract
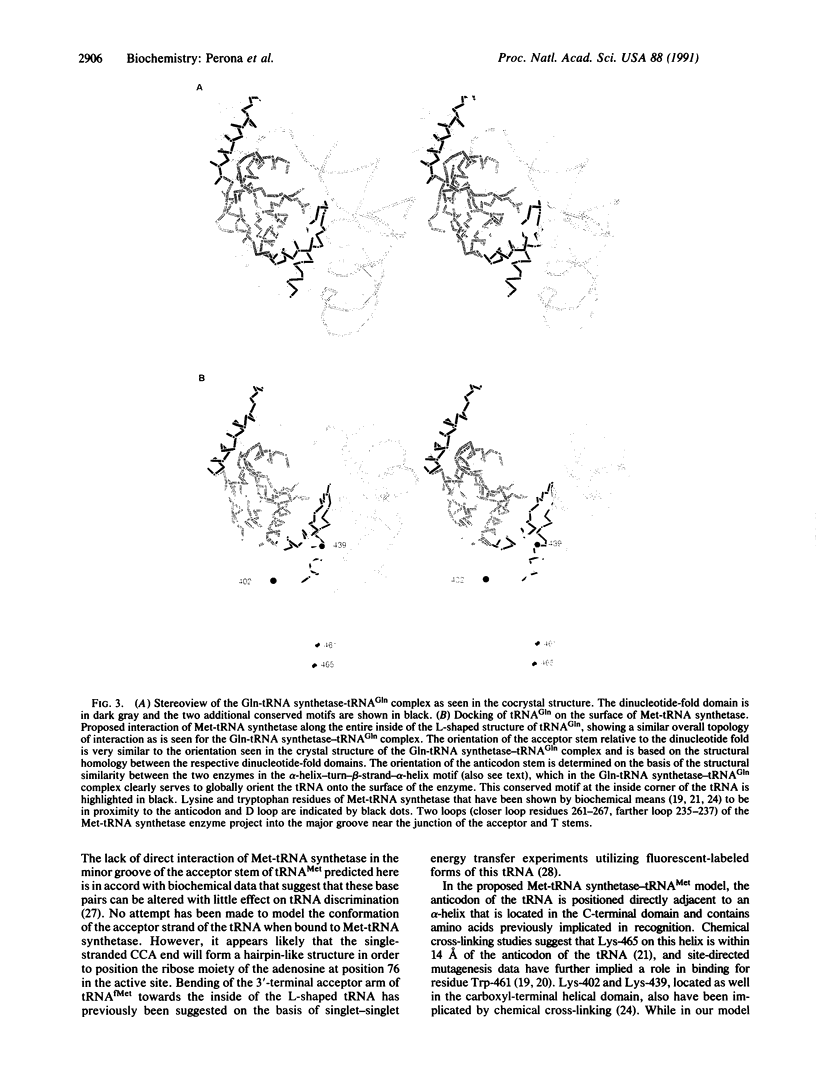
Detailed comparisons between the structures of the tRNA-bound Escherichia coli glutaminyl-tRNA (Gln-tRNA) synthetase [L-glutamine:tRNA(Gln) ligase (AMP-forming), EC 6.1.1.18] and recently refined E. coli methionyl-tRNA (Met-tRNA) synthetase [L-methionine:tRNA(Met) ligase (AMP-forming), EC 6.1.1.10] reveal significant similarities beyond the anticipated correspondence of their respective dinucleotide-fold domains. One similarity comprises a 23-amino acid alpha-helix-turn-beta-strand motif found in each enzyme within a domain that is inserted between the two halves of the dinucleotide binding fold. A second correspondence, which consists of two alpha-helices connected by a large loop and beta-strand, is located in the Gln-tRNA synthetase within a region that binds the inside corner of the "L"-shaped tRNA molecule. This structural motif contains a long alpha-helix, which extends along the entire length of the D and anticodon stems of the complexed tRNA. We suggest that the positioning of this helix relative to the dinucleotide fold plays a critical role in ensuring the proper global orientation of tRNA(Gln) on the surface of the enzyme. The structural correspondences suggest a similar overall orientation of binding of tRNA(Met) and tRNA(Gln) to their respective synthetases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Dardel F., Fayat G., Blanquet S. Molecular cloning and primary structure of the Escherichia coli methionyl-tRNA synthetase gene. J Bacteriol. 1984 Dec;160(3):1115–1122. doi: 10.1128/jb.160.3.1115-1122.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen P., Zaccaï G., Blanquet S. Neutron scattering studies of escherichia coli tyrosyl-trna synthetase and of its interaction with trna tyr. J Mol Biol. 1982 Aug 25;159(4):651–664. doi: 10.1016/0022-2836(82)90106-1. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Isolation and characterization of the gene coding for Escherichia coli arginyl-tRNA synthetase. Nucleic Acids Res. 1989 Jul 25;17(14):5725–5736. doi: 10.1093/nar/17.14.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. Q., Yang D. C. Topographic modeling of free and methionyl-tRNA synthetase bound tRNAfMet by singlet-singlet energy transfer: bending of the 3'-terminal arm in tRNAfMet. Biochemistry. 1986 Oct 21;25(21):6572–6578. doi: 10.1021/bi00369a035. [DOI] [PubMed] [Google Scholar]
- Ghosh G., Pelka H., Schulman L. H. Identification of the tRNA anticodon recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1990 Mar 6;29(9):2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- Hall C. V., vanCleemput M., Muench K. H., Yanofsky C. The nucleotide sequence of the structural gene for Escherichia coli tryptophanyl-tRNA synthetase. J Biol Chem. 1982 Jun 10;257(11):6132–6136. [PubMed] [Google Scholar]
- Heck J. D., Hatfield G. W. Valyl-tRNA synthetase gene of Escherichia coli K12. Primary structure and homology within a family of aminoacyl-TRNA synthetases. J Biol Chem. 1988 Jan 15;263(2):868–877. [PubMed] [Google Scholar]
- Hountondji C., Blanquet S., Lederer F. Methionyl-tRNA synthetase from Escherichia coli: primary structure at the binding site for the 3'-end of tRNAfMet. Biochemistry. 1985 Feb 26;24(5):1175–1180. doi: 10.1021/bi00326a018. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Härtlein M., Frank R., Madern D. Nucleotide sequence of Escherichia coli valyl-tRNA synthetase gene valS. Nucleic Acids Res. 1987 Nov 11;15(21):9081–9082. doi: 10.1093/nar/15.21.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlein M., Madern D. Molecular cloning and nucleotide sequence of the gene for Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Res. 1987 Dec 23;15(24):10199–10210. doi: 10.1093/nar/15.24.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouze E., Bedouelle H. Structural and kinetic bases for the recognition of tRNATyr by tyrosyl-tRNA synthetase. J Mol Biol. 1989 Feb 20;205(4):729–735. doi: 10.1016/0022-2836(89)90317-3. [DOI] [PubMed] [Google Scholar]
- Leon O., Schulman L. H. tRNA recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1987 Aug 25;26(17):5416–5422. doi: 10.1021/bi00391a030. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Le Corre D., Panvert M., Blanquet S., Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Argos P. Exploring structural homology of proteins. J Mol Biol. 1976 Jul 25;105(1):75–95. doi: 10.1016/0022-2836(76)90195-9. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Burbaum J. J., Schimmel P. Insertion of new sequences into the catalytic domain of an enzyme. Biochemistry. 1989 Oct 17;28(21):8479–8484. doi: 10.1021/bi00447a031. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Vambutas A., Akai A. Characterization of MSM1, the structural gene for yeast mitochondrial methionyl-tRNA synthetase. Eur J Biochem. 1989 Feb 1;179(2):365–371. doi: 10.1111/j.1432-1033.1989.tb14562.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela D., Schulman L. H. Identification of peptide sequences at the tRNA binding site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1986 Aug 12;25(16):4555–4561. doi: 10.1021/bi00364a015. [DOI] [PubMed] [Google Scholar]
- Walter P., Gangloff J., Bonnet J., Boulanger Y., Ebel J. P., Fasiolo F. Primary structure of the Saccharomyces cerevisiae gene for methionyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1983 May;80(9):2437–2441. doi: 10.1073/pnas.80.9.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward W. H., Fersht A. R. Tyrosyl-tRNA synthetase acts as an asymmetric dimer in charging tRNA. A rationale for half-of-the-sites activity. Biochemistry. 1988 Jul 26;27(15):5525–5530. doi: 10.1021/bi00415a021. [DOI] [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- Winter G., Koch G. L., Hartley B. S., Barker D. G. The amino acid sequence of the tyrosyl-tRNA synthetase from Bacillus stearothermophilus. Eur J Biochem. 1983 May 2;132(2):383–387. doi: 10.1111/j.1432-1033.1983.tb07374.x. [DOI] [PubMed] [Google Scholar]
- Yamao F., Inokuchi H., Cheung A., Ozeki H., Söll D. Escherichia coli glutaminyl-tRNA synthetase. I. Isolation and DNA sequence of the glnS gene. J Biol Chem. 1982 Oct 10;257(19):11639–11643. [PubMed] [Google Scholar]