Abstract
Background:
The molecular mechanisms behind diabetic neuropathy remains to be investigated.
Methods:
This is a secondary study on microarray dataset (GSE24290) downloaded from Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI), which included 18 nerve tissue samples of progressing diabetic neuropathy (fibers loss ≥500 fibers/mm2) and 17 nerve tissue samples of nonprogressing diabetic neuropathy (fibers loss ≤100 fibers/mm2). Differentially expressed genes (DEGs) were screened between progressing and nonprogressing diabetic neuropathy. With the DEGs obtained, a weighted gene coexpression network analysis was conducted to identify gene clusters associated with diabetic neuropathy. Diabetes-related microRNAs (miRNAs) and their target genes were predicted and mapped to the genes in the gene clusters identified. Consequently, a miRNA–gene network was constructed, for which gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed. Potential drugs for treatment of diabetic neuropathy were also predicted.
Results:
Total 370 upregulated and 379 downregulated DEGs were screened between nonprogressing and progressing diabetic neuropathy. Has-miR-377, has-miR-216a, and has-miR-217 were associated with diabetes. Inflammation was the most significant GO term. The peroxisome proliferator-activated receptor (PPAR) pathway and the adenosine monophosphate (AMP)-activated protein kinase (AMPK) signaling pathway were significantly KEGG pathways significantly enriched with PPAR gamma (PPARG), stearoyl-CoA desaturase (SCD), cluster of differentiation 36 (CD36), and phosphoenolpyruvate carboxykinase 1 (PCK1).
Conclusion:
The study suggests that PPARG, SCD, CD36, PCK1, AMPK pathway, and PPAR pathway may be involved in progression of diabetic neuropathy.
Keywords: differentially expressed genes, gene ontology, Kyoto Encyclopedia of Genes and Genomes, microarray analysis, pathway
1. Introduction
Diabetic neuropathy is a leading complication of diabetes characterized by progressive loss of peripheral nerve axons, leading to decreased sensation and pain.[1,2] It tortures approximately 30% of patients with diabetes, and will finally occur in up to half of diabetic patients during the disease course.[3] Nerve damage usually occurs before symptoms and signs are clinically obvious. When the diabetic neuropathy is detected, irreversible damage often has already been caused.[4] It is a challenging task to prevent, slow, or reverse the progression of diabetic neuropathy. Therefore, it is of considerable significance to investigate the molecular mechanisms of diabetic neuropathy for prevention and treatment of the disease.
Omete et al[5] have suggested that inhibition of tumour necrosis factor (TNF) pathway might inhibit the progression of diabetic neuropathy in mice model. A latest study suggests that alterations in ion-channel function and energy metabolism depending on axon–glia crosstalk might also contribute to the development of diabetic neuropathy.[6] Moreover, Chen et al[7] have reported that nucleotide-binding oligomerization domain (NOD)-like receptor 3 (NLRP3) inflammasome activation mediated by ATP–P2X4 signaling might be implicated in diabetic neuropathy-related inflammation. There is in vivo evidence that microRNA (miR)-29c knockdown or miR-27a knockdown compromises progression of diabetic nephropathy.[8,9] Furthermore, Chien et al[10] have found that miR-21, miR-29a/b/c, and miR-192 are involved in pathogenesis of diabetic neuropathy and might be suggested as biomarkers of progression of diabetic neuropathy. Despite these achievements, the molecular mechanisms behind the diabetic neuropathy have not been fully elucidated.
Hur et al[11] have characterized the differentially expressed genes (DEGs) between patient samples with progressing diabetic neuropathy (fibers loss ≥500 fibers/mm2) or nonprogressing diabetic neuropathy (fibers loss ≤100 fibers/mm2), and unraveled that these genes are linked to inflammatory responses and lipid metabolism-related pathways. However, miRNAs are not targeted in their study. Using gene expression dataset (GSE24290) uploaded to NCBI GEO database, this study conducted a series of microarray analysis to shed light on development of promising drugs for prevention of diabetic neuropathy, and biological function of DEGs involved in progression of diabetic neuropathy. A weighted gene coexpression network analysis (WGCNA)[12] was used to search for gene clusters associated with diabetic neuropathy following identification of DEGs between progressing and nonprogressing diabetic neuropathy. Moreover, diabetes-related miRNAs and their target genes were predicted and mapped to the gene coexpression network for construction of a miRNA–gene network. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed for the genes in the miRNAs–target genes network. Additionally, potential drugs for prevention of the disease were also investigated, based on the identified DEGs.
2. Methods
2.1. Data preprocessing
It is a secondary study of gene expression dataset (GSE24290) downloaded from NCBI GEO database[13] (http://www.ncbi.nlm.nih.gov/geo/). The GSE24290 dataset included 18 nerve tissue samples from patients with progressing diabetic neuropathy (fibers loss ≥500 fibers/mm2) and 17 nerve tissue samples from patients with nonprogressing diabetic neuropathy (fibers loss ≤100 fibers/mm2). Preprocessing of the probe-level data in cell intensity (CEL) files was conducted using oligo package (Bioconductor, Seattle, Washington State, USA) of R 3.1.3 language, including conversion into expression measures, background correction, and quartile data normalization.[14] The platform of the data is GPL10526 (Affymetrix GeneChip Human Genome HG-U133 Plus 2 Array, Affymetrix, Santa Clara, CA, USA). No additional subjects were included, thus ethical approval and patient consent are not required.
2.2. Identification of DEGs and two-way hierarchical clustering analysis
Significant DEGs between progressing diabetic neuropathy samples and nonprogressing diabetic neuropathy samples were screened using Linear Models for Microarray Analysis package in R language.[15] The strict threshold was set at fold change (|logFC|) ≥1.2 and P value < 0.05.
2.3. Weighed gene coexpression network analysis
The WGCNA package[12] in R software (GNU Project, Free Software Foundation, Boston, MA, USA) was applied to construct a weighted gene coexpression network. Briefly, the correlation efficient (Smn) between gene m and gene n was calculated as follow: Smn = |cor(m,n)|. Second, the adjacency (amn) was calculated using power adjacency ,function:
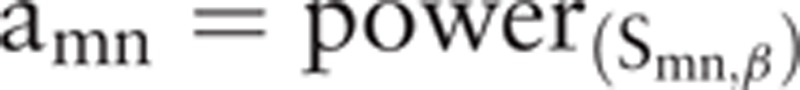 |
. Based on the scale-free behavior of the coexpression network, the weighted value (β value) was determined by linear correlation between the number of edges log (k), and the probability of finding a node with k edges P (k). The correlation coefficient between log (k) and log P (k) was set at ≥0.9. Subsequently, the adjacency matrix was converted into topological matrix.
Hierarchical clustering tree was built using hybrid dynamic shear tree method, with different branches corresponding to different gene modules. At least 30 genes were required for each module. The cut weight was set at 0.9. The module significance (MS) of each module was calculated as the average gene significance (GS) values of all genes in the module, with higher MS value indicating a closer connection of the module with the disease. GS value of the difference in the mRNA expression of each gene between progressing diabetic neuropathy and nonprogressing diabetic neuropathy was calculated using Student t-test.
Based on the GS values of all network modules, the top 3 modules were selected for construction of a gene coexpression network associated with diabetic neuropathy using heatmap.2 function of gplots package (Bioconductor) in R3.1.3.[16]
2.4. Identification of diabetes-related miRNAs and prediction of target genes of miRNAs
The study searched for diabetes-related miRNAs using miR2Diease database.[17] It is a monthly updated database, providing rich information concerning deregulated miRNAs in various human diseases. Its current version included 3273 curated associations between 349 human microRNAs and 163 human diseases by reviewing published papers. Subsequently, target genes of the diabetic-related miRNAs obtained were predicted using miRanda software[18] Computational Biology Center of Memorial Sloan, New York, USA) and TargetScan software [19] (www.targetscan.org, Whitehead Institute for Biomedical Research, Cambridge, MA, USA), which were commonly used tool for scanning target sites of miRNAs. These target genes were mapped to the gene coexpression network, and a miRNA–gene network was then constructed.
2.5. Identification of potential therapeutic small molecules
Connectivity map is a useful approach to mine connections among bioactive small molecules, which share an action mechanism, chemicals and physiological processes, and diseases and drugs.[20] With the help of Connectivity map (cmap), the DEGs were compared with the predefined signatures of drugs and drug-like small molecules, and ranked according to a connectivity score which corresponded to the relative similarity to the imported DEGs. The small molecules with P value <0.05 and |connectivity score| >0.8 were determined to be highly associated with diabetic neuropathy. The connectivity score ranged from −1 to 1. The score closer to −1 indicated that the drug had a stronger effect against the disease.
2.6. GO and KEGG pathway enrichment analysis
In order to unravel the biological function of the genes in the miRNA–gene network, GO function and KEGG pathway enrichment analysis was performed using Gostat software[21] (Bioconductor) and KOBAS software (KOBAS, Surrey, UK),[22] respectively.
3. Results
3.1. Identification of DEGs
Total 749 DEGs were screened between nonprogressing diabetic neuropathy and progressing diabetic neuropathy samples, including 370 upregulated DEGs and 379 downregulated DEGs.
3.2. WGCNA analysis and module selection
In order to guarantee the scale-free topology of the gene coexpression network, power = 6 was selected when the correlation coefficient between log (k) and log P (k) reached 0.9 for the first time (Fig. 1). The constructed gene clustering tree (cut height = 0.9) was shown in Fig. 2. Different modules were marked in different colors. All modules were with MS value >0.8 and MS P value <0.05. According to the MS value, top 3 modules ranked in descending order were selected, consisting of the blue module (156 DEGs), the brown module (55 DEGs), and the green module (47 DEGs) for further analysis (Fig. 3).
Figure 1.
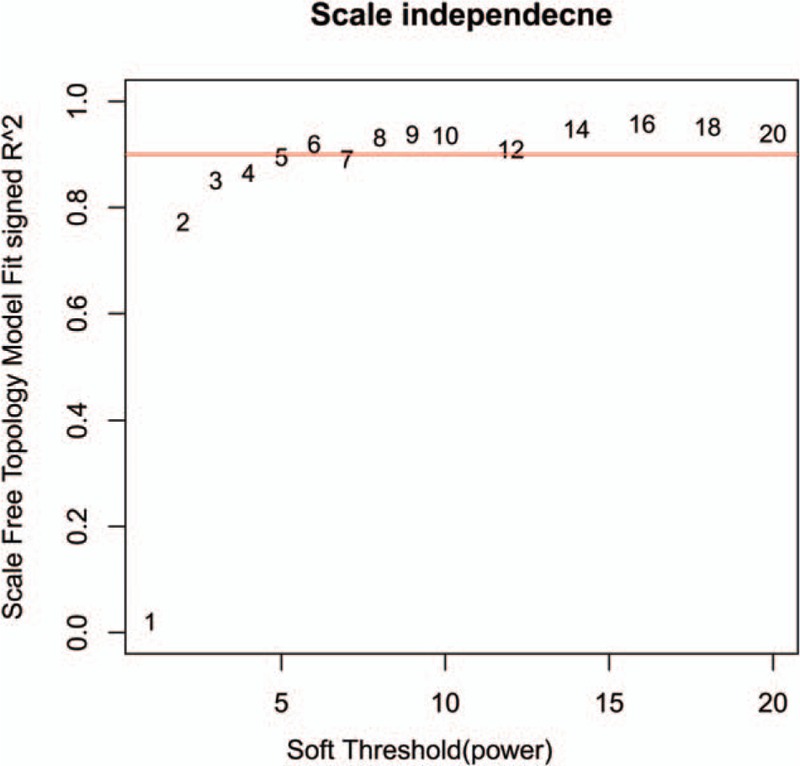
Power value for the adjacency matrix in weighted gene coexpression network analysis. The red line signals 0.9 on the vertical axis.
Figure 2.
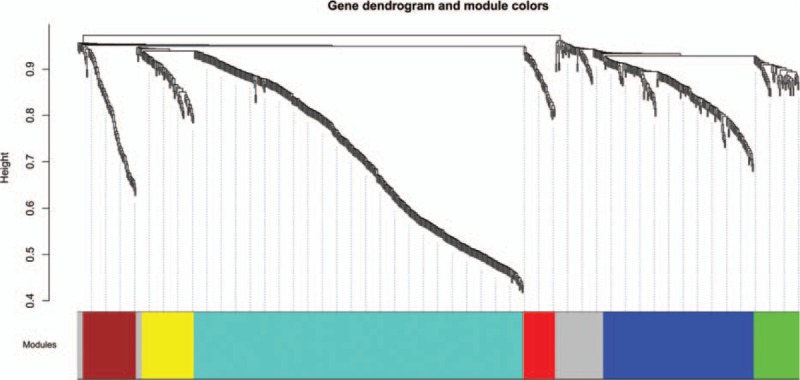
Gene dendrogram and modules from weighted gene coexpression network analysis. Horizontal axis represents modules in different colors. Vertical axis represents height of the gene clustering tree.
Figure 3.
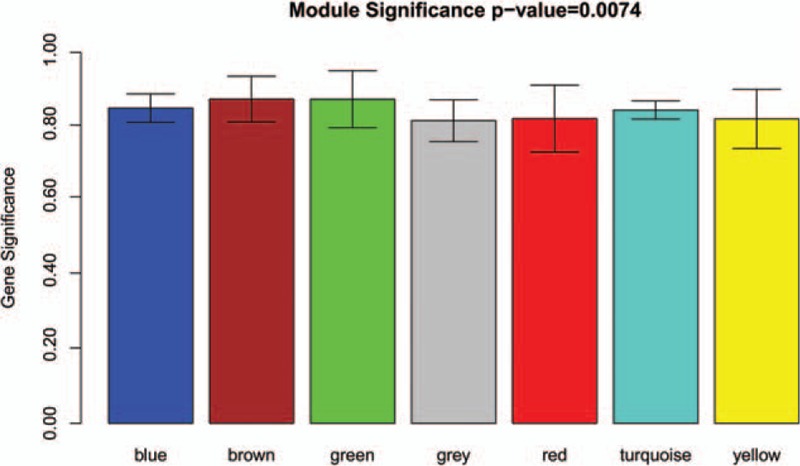
Module significance of different modules. Different modules are marked in different colors.
3.3. Prediction of target genes of diabetes-related miRNAs
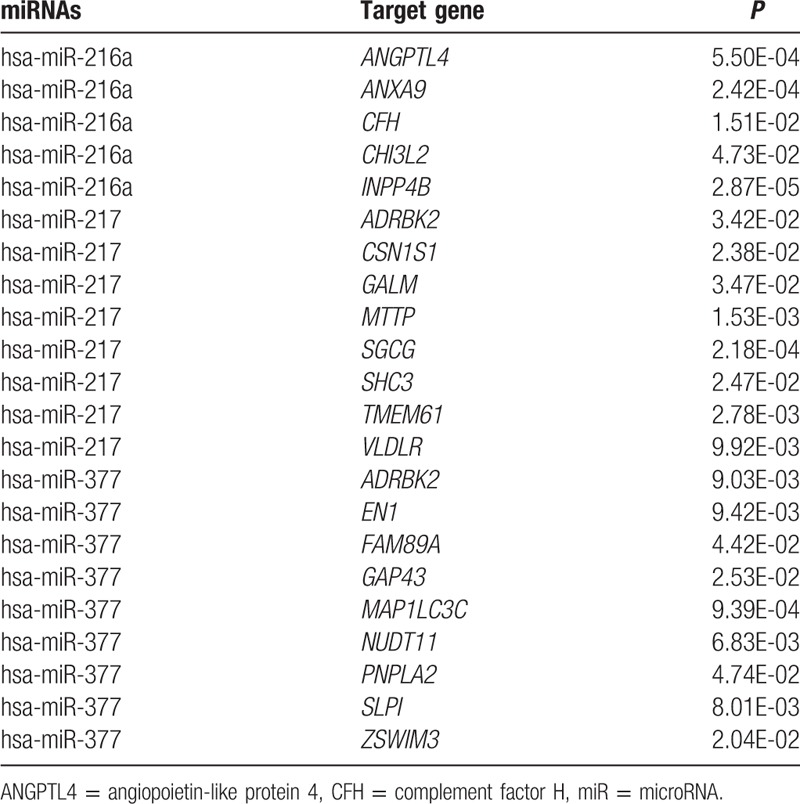
Using miR2Disease database, we obtained 3 miRNAs associated with diabetes: has-miR-377,[23] has-miR-216a,[24] and has-miR-217.[24] With the help of miRanda software and TargetScan software, the study predicted 1052 target genes of has-miR-377, 962 target genes of has-miR-261a, and 1025 target genes of has-miR-217 (Table 1).
Table 1.
Diabetes-related microRNAs and their target genes.
3.4. Construction of the miRNA–target gene network
Has-miR-377, has-miR-216a, has-miR-217 and their target genes were mapped to the DEGs in the blue module, brown module, and green module. Consequently, a miRNA–target gene network was built, and shown in Fig. 4.
Figure 4.
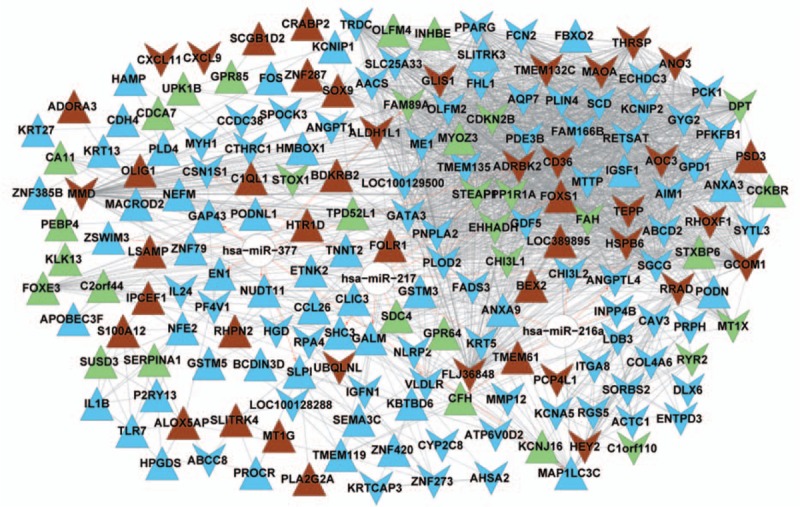
A miRNA–target gene network. In the network, upper triangles represent upregulated genes and downward triangles represent downregulated genes. White nodes stand for miRNAs. Blue, brown, and green nodes are genes extracted from the blue module, the brown module, and the green module, irrespectively. A dotted line suggests that the two genes are negatively correlated. A solid line suggests the positive correlation between two genes. A red arrow-headed line linking a gene and a miRNA suggests that the gene is regulated by the miRNA. miRNA = microRNA.
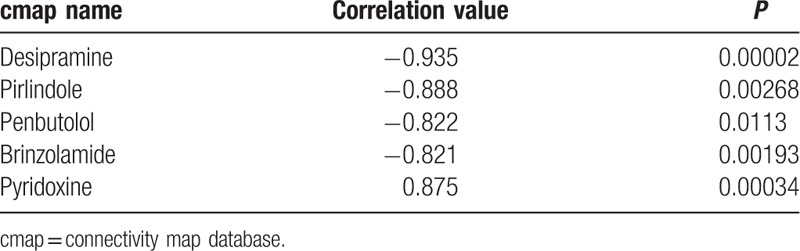
3.5. Selection of potential drugs against diabetic neuropathy
Based on the Connectivity map database, 5 small molecules with P value <0.05 and |connectivity score| >0.8 were selected to be potential drugs for diabetic neuropathy: desipramine, pirlindole, penbutolol, brinzolamide and pyridoxine (Table 2). The connectivity score of desipramine was lowest (−0.935).
Table 2.
Promising drugs predicted for diabetic neuropathy.
3.6. GO and KEGG pathway enrichment analysis
GO enrichment analysis revealed that the DEGs in the miRNA–target gene network were significantly related to 14 GO terms (Fig. 5A). The most significant GO term was significantly enriched with 13 genes, such as adenosine A3 receptor (ADORA3), chemokine (C-X-C motif) ligand (CXCL)9, bradykinin receptor B2 (BDKRB2), and complement factor H (CFH). CFH was a target gene of has-miR-216a. As for KEGG pathway enrichment analysis (Fig. 5B) there were 8 significant KEGG pathways associated with DEGs. The most significant KEGG pathway was peroxisome proliferator-activated receptor (PPAR) pathway significantly enriched with 8 genes: cluster of differentiation (CD)36, malic enzyme (ME)1, enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase (EHHADH), angiopoietin-like protein 4 (ANGPTL4), PPAR gamma (PPARG), aquaporin (AQP)7, stearoyl-CoA desaturase (SCD), and phosphoenolpyruvate carboxykinase 1 (PCK1). ANGPTL4 was a target gene of has-miR-216a. Adenosine monophosphate (AMP)-activated protein kinase (AMPK) signaling pathway was significantly enriched with PPARG, SCD, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 (PFKFB1), CD36, and PCK1.
Figure 5.
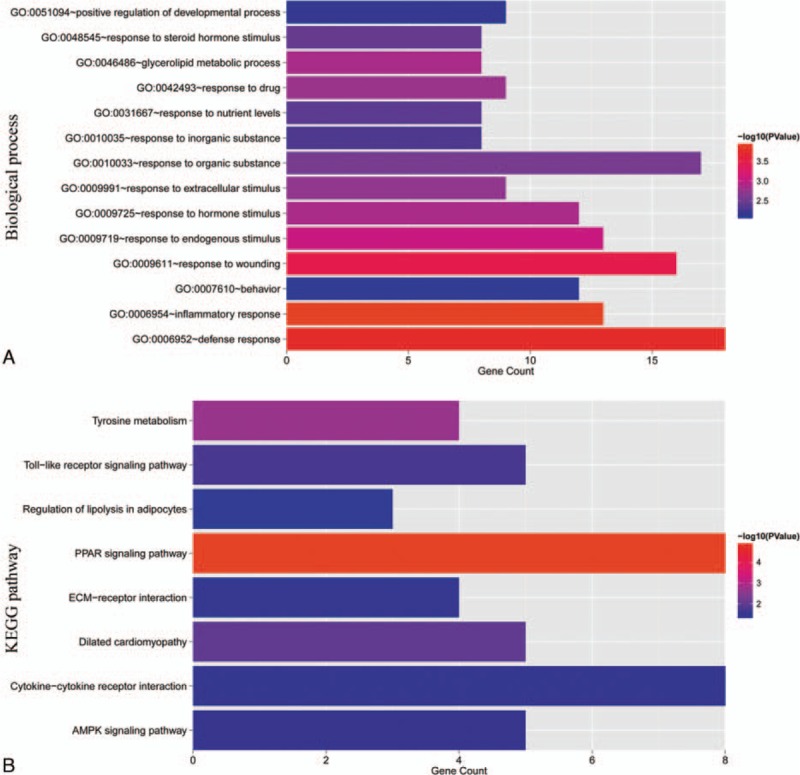
Significant GO terms and KEGG pathways. A, significant GO terms. Horizontal axis represents the number of genes significantly enriched in each GO term. Vertical axis represents GO terms. P values are marked in different colors. B, significant KEGG pathways. Horizontal axis represents the number of genes significantly enriched in each KEGG pathway. Vertical axis represents GO terms. P values are marked in different colors. GO = gene ontology, KEGG = Kyoto Encyclopedia of Genes and Genomes.
4. Discussion
Diabetic neuropathy is the most common complication occurred in diabetic patients. In order to unravel the molecular mechanisms underlying the progression of diabetic neuropathy, the study carried out WGCNA analysis to identify gene clusters associated with progression of diabetic nephropathy. The study further found that the DEGs in nonprogressing and progressing diabetic neuropathy were significantly enriched with several GO terms and KEGG pathways, such as inflammation, PPAR, and AMPK signaling pathways.
Increasing studies have demonstrated that inflammation-related pathways play critical roles in the progression of diabetic nephropathy.[25,26] The study confirmed the key role of inflammation, as evidenced by the finding that inflammation was the most significant GO term associated with DEGs in the miRNA–gene network. Hu et al[27] have found that PPARα expression is decreased in diabetic neuropathy rat model, and that PPARα overexpression inhibits diabetic neuropathy-related inflammation. Moreover, increasing evidence has suggested that PPARα and PPARγ agonists may be promising agents for management of diabetic nephropathy.[28,29] Similarly, the study also unveiled that PPAR pathway was the most significant signaling pathway enriched with several DEGs, including ANGPTL4, a target gene of has-miR-216a. ANGPTL4 is a member of the angiopoietin-like gene family produced from fat tissue and liver, involving in energy metabolism and insulin sensitivity. It is also a target of PPARs. It has been reported that ANGPTL4 overexpression modulates mitochondria function and methionine metabolic cycles in the liver of diabetic mice.[30] The study indicates that ANGPTL4 regulated by has-miR-216a and PPAR signaling pathway might participate in progression of diabetic neuropathy.
AMPK is an important enzyme in regulating cellular and whole-body energy homeostasis.[31] AMPK which plays critical roles in key metabolically relevant tissues is considered as an attractive drug target for diabetes.[32] The present study suggests that AMPK signaling pathway might be implicated in progression of diabetic neuropathy. Moreover, both AMPK and PPAR signaling pathways were significantly enriched with PPARG, SCD, CD36, and PCK1 in the present study. PPARG encodes PPARγ protein. SCD gene encodes SCD protein, which is a critical enzyme involved in fatty acid metabolism. There is evidence that loss of SCD1 promotes glucose utilization in the heart of mice model.[33] CD36 is an integral membrane protein located on the surface of many cell types, participating in several process, such as inflammation and oxidative stress.[34] It has been reported that CD36 is involved in proximal tubular epithelial cell apoptosis of diabetic nephropathy.[35] Moreover, Shiju et al[36] have implied that soluble CD36 in plasma and urine might be a prognostic biomarker for diabetic neuropathy. PCK1 encodes phosphoenolpyruvate carboxykinase 1 protein, which is a key enzyme in gluconeogenesis.[37] Findings of this study indicate that PPARG, SCD, CD36, and PCK1 might participate in progression of diabetic neuropathy, partly by regulating AMPK and PPAR signaling pathways.
The study predicted that desipramine, pirlindole, penbutolol, brinzolamide, and pyridoxine might be promising agents for treatment of diabetic neuropathy. In consistence with this finding, it has been established that desipramine is effective in relieving pain in painful diabetic neuropathy.[38] However, the other 4 drugs have not been reported in management of diabetic neuropathy. In addition, there are some limitations in the study, including the single platform of dataset, the dataset is not sufficiently robust and the reproducibility has not been determined, and the sample size is small. In addition, the study only included microarray data analysis. Therefore, further experimental studies are needed to verify the findings of this study.
5. Conclusion
The study suggests that PPARG, SCD, CD36, PCK1, AMPK pathway, and PPAR pathway may be implicated in progression of diabetic neuropathy. The genes might be recommended as possible biomarkers for progression of diabetic neuropathy. Further experimental studies are necessary to verify the findings of this study.
Acknowledgments
We would like to thank the patients and investigators of the study group for participating in this trial. We want to express our immeasurable thanks to Prof Taixiang Wu (Chinese Cochrane Center, Chinese Evidence-Based Medicine Center and Regional Clinical Epidemiology Re, West China Hospital, Sichuan University, Chengdu, China) for his assistance with statistical data analysis.
Footnotes
Abbreviations: ADORA3 = adenosine A3 receptor, AMP = adenosine monophosphate, AMPK = AMP-activated protein kinase, ANGPTL4 = angiopoietin-like protein 4, AQP7 = aquaporin 7, BDKRB2 = bradykinin receptor B2, CD = cluster of differentiation, CEL = cell Intensity, CFH = complement factor H, cmap = Connectivity Map, CXCL9 = C-X-C motif chemokine ligand 9, DEGs = differentially expressed genes, EHHADH = enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase, FC = fold change, GEO = Gene Expression Omnibus, GO = gene ontology, GS = gene significance, KEGG = Kyoto Encyclopedia of Genes and Genomes, ME = malic enzyme, miR = microRNA, MS = module significance, NCBI = National Center for Biotechnology Information, NLRP3 = NOD-like receptor 3, NOD = nucleotide-binding oligomerization domain, PCK1 = phosphoenolpyruvate carboxykinase 1, PFKFB1 = 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1, PPAR = peroxisome proliferator-activated receptor, PPARG = peroxisome proliferator-activated receptor gamma, SCD = stearoyl-CoA desaturase, TNF= tumour necrosis factor, WGCNA = weighted gene coexpression network analysis.
Funding/support: This work was supported by the Natural Science Foundation of Shaanxi Province, China (Grant No. 2012sp2-08). This work was supported by the Shaanxi Province Department of Education Fund, China (Grant No. 2013JK0781).
All authors declare that they have no conflicts of interest to state.
References
- [1].Callaghan BC, Cheng HT, Stables CL. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol 2012;11:521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Edwards JL, Vincent AM, Cheng HT, et al. Diabetic neuropathy: mechanisms to management. Pharmacol Ther 2008;120:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maser RE, Steenkiste AR, Dorman JS. Epidemiological correlates of diabetic neuropathy. Report from Pittsburgh Epidemiology of Diabetes Complications Study. Diabetes 1989;38:1456–61. [DOI] [PubMed] [Google Scholar]
- [4].Tesfaye S, Boulton AJ, Dyck PJ, et al. Diabetic neuropathies: update on definitions, diagnostic criteria, estimation of severity, and treatments. Diabetes Care 2010;33:2285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Omote K, Gohda T, Murakoshi M, et al. Role of the TNF pathway in the progression of diabetic nephropathy in KK-A(y) mice. Am J Physiol Renal Physiol 2014;306:F1335–47. [DOI] [PubMed] [Google Scholar]
- [6].Zenker J, Dan Z, Chrast R. Novel pathogenic pathways in diabetic neuropathy. Trends Neurosci 2013;36:439–49. [DOI] [PubMed] [Google Scholar]
- [7].Chen K, Zhang J, Zhang W, et al. ATP-P2X4 signaling mediates NLRP3 inflammasome activation: a novel pathway of diabetic nephropathy. Int J Biochem Cell Biol 2013;45:932–43. [DOI] [PubMed] [Google Scholar]
- [8].Long J, Wang Y, Wang W, et al. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem 2011;286:11837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lina W, Qingzhu W, Feng G, et al. MicroRNA-27a induces mesangial cell injury by targeting of PPARγ, and its in vivo knockdown prevents progression of diabetic nephropathy. Sci Rep 2016;6:26072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chien HY, Chen CY, Chiu YH, et al. Differential microRNA profiles predict diabetic nephropathy progression in Taiwan. Int J Med Sci 2016;13:457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hur J, Sullivan KA, Pande M, et al. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain 2011;134:3222–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17:520–5. [DOI] [PubMed] [Google Scholar]
- [15].Liu J, Yang XY, Shi WJ. Identifying differentially expressed genes and pathways in two types of non-small cell lung cancer: adenocarcinoma and squamous cell carcinoma. Genet Mol Res 2014;13:95–102. [DOI] [PubMed] [Google Scholar]
- [16].Warnes GR, Bolker B, Bonebakker L, et al. gplots: Various R programming tools for plotting data. R Package Version 2009;2: [Google Scholar]
- [17].Jiang Q, Wang Y, Hao Y, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res 2009;37:D98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 2010;11:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Garcia DM, Baek D, Shin C, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol 2011;18:1139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929–35. [DOI] [PubMed] [Google Scholar]
- [21].Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 2004;20:1464–5. [DOI] [PubMed] [Google Scholar]
- [22].Wu J, Mao X, Cai T, et al. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res 2006;34:W720–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Q, Wang Y, Minto AW, et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 2008;22:4126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kato M, Putta S, Wang M. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nature Cell Biology 2009;11:881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wada J, Makino H. Inflammation and the pathogenesis of diabetic nephropathy. Clin Sci (Lond) 2013;124:139–52. [DOI] [PubMed] [Google Scholar]
- [26].Kanasaki K, Taduri G, Koya D. Diabetic nephropathy: the role of inflammation in fibroblast activation and kidney fibrosis. Front Endocrinol (Lausanne) 2013;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hu Y, Chen Y, Ding L, et al. Pathogenic role of diabetes-induced PPAR-α down-regulation in microvascular dysfunction. Proc Natl Acad Sci U S A 2013;110:15401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Balakumar P, Arora MK, Singh M. Emerging role of PPAR ligands in the management of diabetic nephropathy. Pharmacol Res 2009;60:170–3. [DOI] [PubMed] [Google Scholar]
- [29].Park C, Zhang Y, Zhang X, et al. PPARα agonist fenofibrate improves diabetic nephropathy in db/db mice. Kidney Int 2006;69:1511–7. [DOI] [PubMed] [Google Scholar]
- [30].Wang Y, Lam KS, Lam JB, et al. Overexpression of angiopoietin-like protein 4 alters mitochondria activities and modulates methionine metabolic cycle in the liver tissues of db/db diabetic mice. Mol Endocrinol 2007;21:972–86. [DOI] [PubMed] [Google Scholar]
- [31].Viollet B, Lantier L, Devin-Leclerc J, et al. Targeting the AMPK pathway for the treatment of type 2 diabetes. Front Biosci (Landmark Ed) 2009;14:3380–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab 2009;9:407–16. [DOI] [PubMed] [Google Scholar]
- [33].Dobrzyn P, Sampath H, Dobrzyn A, et al. Loss of stearoyl-CoA desaturase 1 inhibits fatty acid oxidation and increases glucose utilization in the heart. Am J Physiol Endocrinol Metab 2008;294:E357–64. [DOI] [PubMed] [Google Scholar]
- [34].Okamura DM, Pennathur S, Pasichnyk K, et al. CD36 regulates oxidative stress and inflammation in hypercholesterolemic CKD. J Am Soc Nephrol 2009;20:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Susztak K, Ciccone E, Mccue P, et al. Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med 2007;2:152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shiju TM, Mohan V, Balasubramanyam M, et al. Soluble CD36 in plasma and urine: A plausible prognostic marker for diabetic nephropathy. J Diabetes Complications 2014;29:400–6. [DOI] [PubMed] [Google Scholar]
- [37].Pilz A, Willer E, Povey S, et al. The genes coding for phosphoenolpyruvate carboxykinase-1 (PCK1) and neuronal nicotinic acetylcholine receptor α4 subunit (CHRNA4) map to human chromosome 20, extending the known region of homology with mouse chromosome 2. Ann Hum Genet 1992;56:289–93. [DOI] [PubMed] [Google Scholar]
- [38].Max MB, Kishore-Kumar R, Schafer SC. Efficacy of desipramine in painful diabetic neuropathy: a placebo-controlled trial. Pain 1991;45:3–9. [DOI] [PubMed] [Google Scholar]


