Abstract
Mechanical forces play key roles in regulating cellular pathways but are challenging to study using standard biological approaches. In a recent issue of Cell, Seo et al. (2016) present a platform for in vivo single-molecule manipulation, using magnetoplasmonic nanoparticles capable of imaging, localizing, and force-loading receptor proteins at high spatiotemporal resolution.
Cells use a diverse set of membrane receptors to sense mechanical, chemical, spatial, and temporal cues from their environment to take appropriate action. Pioneering work over the past couple of decades has highlighted how mechanosensitive receptors respond to mechanical stimuli at the molecular level to activate downstream signaling pathways (Iskratsch et al., 2014; Vogel and Sheetz, 2006). These studies have benefitted from emerging technologies for measuring and applying mechanical forces, including traction force microscopy, optical traps, magnetic tweezers, atomic force spectroscopy, and pillar arrays that were developed and implemented through intense interdisciplinary collaboration (Iskratsch et al., 2014; Vogel and Sheetz, 2006). One approach involves the use of nanotechnology to manipulate individual receptors in vivo. Using this method, Mannix et al. (2008) demonstrated that magnetic nanoparticles can control the spatial organization of cell membrane receptors to activate cellular signal transduction. Seo et al. (2016), in a recent issue of Cell, now extend this work, describing a versatile magnetoplasmonic nanoparticle-based platform that allows imaging, localizing, and force-loading receptor proteins in vivo at high spatiotemporal resolution. Using magnetoplasmonic nanoparticles (MPNs) in combination with optical microscopy and force-generating magnetic tweezers, the authors demonstrate that mechanical force loading of single Notch receptors in vivo is sufficient for inducing proteolytic cleavage and investigate how distinct E-cadherin responses to spatial and mechanical cues impact cytoskeleton complex formation.
The foundation of this platform is the use of uniformly sized MPNs. These nanoparticles have a magnetic core, which enables them to deliver piconewton levels of force in the presence of an applied magnetic field, as well as a plasmonic gold shell, which allows their imaging using dark-field microscopy (Seo et al., 2016). Furthermore, MPNs can be readily functionalized with 5′-thiolated oligonucleotides to enable the specific targeting of cell surface receptors labeled with the complementary sequence. Critically, the authors demonstrate that MPNs can achieve monovalent targeting of receptors (i.e., one MPN per receptor)—a challenge for micron-size force-probe techniques that often exhibit multivalent binding, which can disrupt the spatial organization of receptors and perturb cellular structures (Figure 1). Seo et al. (2016) demonstrate both dynamic control of receptor organization and single-receptor force loading with a well-defined calibrated force (from 1 to 50 pN) on MPN targeted receptors and use this system to study mechanical signaling pathways for Notch and E-cadherin.
Figure 1. Mechanically Probing Cell-Signaling Pathways with Magnetoplasmonic Nanoparticles.
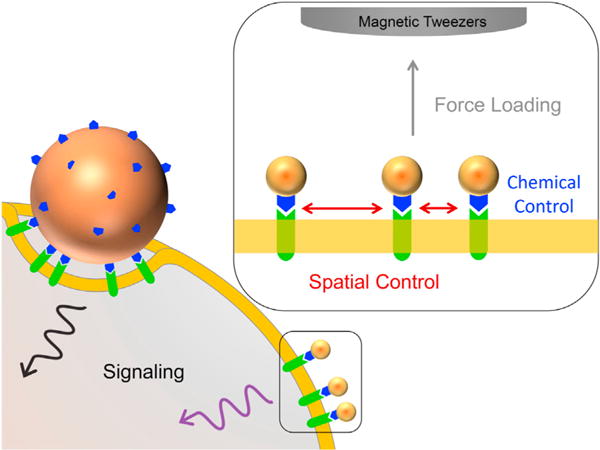
Conventional micron-size probes can form multivalent interactions with cell surface receptors, perturbing cellular structure and immobilizing receptors, whereas magnetoplasmonic nanoparticles (MPNs) have the ability to bind monovalently (one-to-one), providing precise chemical and physical control of ligand-receptor binding. In the presence of a magnetic field, the MPNs can spatially localize and apply controlled mechanical forces to the targeted receptors.
The Notch signaling pathway is a highly conserved mechanism for intercellular communication that regulates gene expression in a wide variety of settings, from neurogenesis during embryonic development to disease onset in adults (Kopan and Ilagan, 2009). Previously, force loading was shown to be required for ligand-induced activation of the Notch signaling pathway, with force inducing the unfolding of the negative regulatory region (NRR) in the extracellular domain (Gordon et al., 2015). The unfolding exposes the buried proteolytic site S2 to ADAM metalloproteases, which leads to dissociation of the extracellular domain, a necessary step in Notch activation (Gordon et al., 2007; Stephenson and Avis, 2012; Gordon et al., 2015). Seo et al. (2016) utilized the MPN platform to further address whether receptor clustering and spatial immobilization can contribute to Notch activation. Using monovalent single-molecule binding of MPNs to Notch in an in vivo pulling assay, Seo et al. (2016) show that 9 pN of force is sufficient to promote Notch extracellular domain (NECD) dissociation in the absence of receptor clustering. This result is consistent with previous single-molecule in vitro experiments performed by Gordon et al. (2015), which found that the force sensitivity of NRR cleavage is around 3.5–5.4 pN, force levels comparable to the force generated by single cytoskeletal motors such as myosin. Systematic examination of Notch receptor clustering and Delta-like 1 ligand binding with the MPN platform further demonstrate that force loading is required for the proteolytic activation of Notch.
Seo et al. (2016) also investigated E-cadherin-mediated mechanical signaling, a system that allowed the authors to study the cooperativity between spatial receptor organization and force loading (Figure 1). With functionalized MPNs that target E-cadherin on the surface of the cell, actin within the cell formed circular assemblies upon clustering of E-cadherin. When the loading force on the cluster was increased to 9 pN, recruitment of vinculin and other molecular changes occurred, which further stabilized the E-cadherin-cytoskeletal complex (Lecuit and Yap, 2015).
These examples illustrate the potential of magnetic nanoparticle-based force spectroscopy to extend single-molecule manipulation from the domain of in vitro assays performed with purified components to in vivo studies that illuminate how living systems respond to force at the nanoscale. By combining mechanical perturbation with engineered cells designed to report the activation of force-dependent pathways, insights into cell signaling and mechanotransduction have already been gained. The incorporation of additional reporter signals, such as patch clamps, or fluorescent reporters of voltage, calcium, and so on would extend this “mechanogenetic toolbox” to a wider variety of cellular and biological processes. In addition to uncovering natural force-regulated processes, one could also envision genetically engineering force regulation into cellular processes that are not natively force dependent, creating biological systems that can be regulated with magnetic fields. When combined with the “action-at-a-distance” enabled by magnetic manipulation, this approach opens up a wide range of possibilities, both for basic research and for clinical applications. In a basic research context, single-molecule force studies could be performed below the surface of cells or even tissues, enabling, for example, detailed studies of the mechanics of phagocytosis or the immune response. At the level of tissues and organisms, activation of mechanotransduction pathways with magnetic fields has already been used to influence the differentiation of stem cells in developing organisms (Pankhurst et al., 2009). These types of studies point to the wide variety of applications for magnetic nanoparticles, from magnetically regulated transfection and tissue engineering to hyperthermia therapies and selective ion channel activation (Pankhurst et al., 2009). Further development of these techniques, including the work of Seo et al. (2016), will continue to open research and clinical possibilities.
References
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Zimmerman B, He L, Miles LJ, Huang J, Tiyanont K, McArthur DG, Aster JC, Perrimon N, Loparo JJ, Blacklow SC. Dev Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T, Wolfenson H, Sheetz MP. Nat Rev Mol Cell Biol. 2014;15:825–833. doi: 10.1038/nrm3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Yap AS. Nat Cell Biol. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- Mannix RJ, Kumar S, Cassiola F, Montoya-Zavala M, Feinstein E, Prentiss M, Ingber DE. Nat Nanotechnol. 2008;3:36–40. doi: 10.1038/nnano.2007.418. [DOI] [PubMed] [Google Scholar]
- Pankhurst QA, Thanh NTK, Jones SK, Dobson J. J Phys D Appl Phys. 2009;42:224001. [Google Scholar]
- Seo D, Southard KM, Kim JW, Lee HJ, Farlow J, Lee JU, Litt DB, Haas T, Alivisatos AP, Cheon J, et al. Cell. 2016;165 doi: 10.1016/j.cell.2016.04.045. S0092-8674(16)30490-1, in press. Published online May 12, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson NL, Avis JM. Proc Natl Acad Sci USA. 2012;109:E2757–E2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]


