The 2-to-3-fold increased risk of multiple myeloma (MM) within families, and among African Americans vs. Whites support a role of genetic factors in disease development(1, 2). HLA proteins initiate immune surveillance through peptide presentation to T-cell receptors(3). Each HLA allele has the capability of presenting a differing limited repertoire of peptides derived from self and non-self proteins, therefore HLA polymorphim has been associated with numerous immune-mediated diseases(4).
A genome wide association study (GWAS) of MM identified a risk variant within the major histocompatibility complex (MHC) but the associated SNP (rs228580) was not directly characterized with classical HLA typing(5). GWAS has identified loci in the major histocompatibility region as the strong associations in other B cell malignancies, with p-values of up to 10−60(6). Previous MM association studies with HLA are limited by sample size(7). Building on work in chronic lymphocytic leukemia (CLL), where a similar analysis identified several HLA alleles associated with susceptibility(8), we comprehensively analyzed HLA associations with MM risk among White, African American, Hispanic, and Asian US populations.
Cases:US MM patients who searched National Marrow Donor Program (NMDP) for potential donors for hematopoietic cell transplantation (HSCT) or had HLA data collected by the Center for International Blood and Marrow Transplant Research (CIBMTR) after transplant (Table 1). HLA typing data were available on 2897 White, 569 African American, 184 Hispanic and 74 Asian / Pacific Islander patients. Independent association studies were conducted with 50,000 controls for each ethnic group. Controls were selected randomly from the NMDP volunteer adult donor registry recruited since 2005 and frequency-matched for patient age quartiles and gender proportions.. Cases and controls were HLA typed using DNA-based methods (Sanger sequence-based typing (SBT) or sequence-specific oligonucleotide (SSO))(9).
Table 1.
Haplotype Associations:
| Table 1A: Extended HLA Haplotype Associations from Factor Analysis for Whites | ||||||
|---|---|---|---|---|---|---|
| Variant | Control Frequency | Odds Ratio | Lower 95% CI | Upper 95% CI | FDR P | Factor Analysis |
| A*03~C*07~B*07~DRB5*01~DRB1*15~DQB1*06 | 0.030 | 1.27 | 1.01 | 1.61 | 3.00E-02 | Group 1 |
| A*02~C*05~B*44~DRB4*01~DRB1*04~DQB1*03 | 0.024 | 0.67 | 0.45 | 0.97 | 2.15E-02 | Group 2 |
| A*03:01g~C*07:02g~B*07:02g | 0.051 | 1.33 | 1.10 | 1.61 | 4.68E-05 | Group 3 |
| C*05~B*44~DRB4*01~DRB1*04~DQB1*03 | 0.031 | 0.71 | 0.52 | 0.96 | 9.82E-03 | Group 4 |
| C*05:01g~B*44:02g | 0.075 | 0.80 | 0.66 | 0.96 | 4.42E-03 | Group 5 |
| C*07:02g~B*07:02g~DRB4*01:01g~DRB1*04:01~DQB1*03:02g | 0.003 | 1.85 | 1.00 | 3.43 | 4.97E-02 | Group 6 |
| C*06:02g~B*57:01g~DRB4*01:01g~DRB1*07:01g~DQB1*02:01g | 0.005 | 0.41 | 0.17 | 1.03 | 4.97E-02 | Group 7 |
| C*05:01g~B*44:02g~DRB4*01:01g~DRB1*04:01 | 0.027 | 0.71 | 0.52 | 0.99 | 3.00E-02 | Group 8 |
| DRB1*15:01~DQB1*06:03g | 0.001 | 5.31 | 1.75 | 16.15 | 5.00E-05 | Group 9 |
| C*02:02g | 0.043 | 1.20 | 1.01 | 1.43 | 3.91E-02 | Group 10 |
| DQB1*02:01g | 0.220 | 0.87 | 0.77 | 0.98 | 6.66E-03 | Group 11 |
| C*02:02g~DRBX*NNNN | 0.007 | 1.50 | 1.01 | 2.24 | 4.20E-02 | Group 12 |
| Table 1B: Extended HLA Haplotype Associations from Factor Analysis for African Americans | ||||||
|---|---|---|---|---|---|---|
| Variant | Control Frequency | Odds Ratio | Lower 95% CI | Upper 95% CI | FDR P | Factor Analysis |
| C*04~B*15 | 0.003 | 2.87 | 1.01 | 8.19 | 4.57E-02 | Group 1 |
| DRB3*02~DRB1*11~DQB1*02 | 0.001 | 4.61 | 1.15 | 18.46 | 1.59E-02 | Group 2 |
| Table 1C: Extended HLA Haplotype Associations from Factor Analysis for Hispanics | ||||||
|---|---|---|---|---|---|---|
| Variant | Control Frequency | Odds Ratio | Lower 95% CI | Upper 95% CI | FDR P | Factor Analysis |
| A*02~C*04~B*35~DRB3*01~DRB1*14~DQB1*03 | 0.0035 | 5.43 | 1.88 | 15.69 | 2.74E-04 | Group 1 |
| A*24:02g~C*07:02g~B*39:06~DRB3*01:01~DRB1*14:06~DQB1*03:01g | 0.0053 | 3.93 | 1.57 | 9.83 | 1.63E-03 | Group 2 |
| DRBX*NN NN~DRB1*08:04~DQB1*03:01g | 0.0052 | 3.37 | 1.09 | 10.40 | 2.45E-02 | Group 3 |
| A*68:02~C*07:01g~B*57:03 | 0.0040 | 2.94 | 1.04 | 8.26 | 3.92E-02 | Group 4 |
| Table 1D: Extended HLA Haplotype Associations from Factor Analysis for Asians | ||||||
|---|---|---|---|---|---|---|
| Variant | Control Frequency | Odds Ratio | Lower 95% CI | Upper 95% CI | FDR P | Factor Analysis |
| C*03:02g~B*58:01g~DRB3*02:02g~DRB1*03:01~DQB1*02:01g | 0.0269 | 2.40 | 1.04 | 5.58 | 3.91E-02 | Group 1 |
| DRB5*01~DRB1*15~DQB1*05 | 0.0497 | 2.98 | 1.26 | 7.08 | 3.85E-03 | Group 2 |
| B*27:05g | 0.0074 | 4.14 | 1.21 | 14.17 | 2.29E-03 | Group 3 |
| A*34:01~DRB1*15:02 | 0.0074 | 7.68 | 2.66 | 22.12 | 3.17E-05 | Group 4 |
| C*07~B*38~DRB5*01~DRB1*15~DQB1*05 | 0.0135 | 3.91 | 1.30 | 11.82 | 9.38E-03 | Group 5 |
| DRB5*01~DRB1*15~DQB1*06 | 0.1251 | 0.39 | 0.16 | 0.97 | 3.90E-02 | Group 6 |
| C*07:02g~B*15:35 | 0.0060 | 8.99 | 2.53 | 31.97 | 2.23E-05 | Group 7 |
| DRBX*NN~DRB1*01~DQB1*05 | 0.0268 | 2.51 | 1.04 | 6.06 | 3.46E-02 | Group 8 |
Predisposing or protective associations of HLA polymorphisms with MM were identified at: (1) the individual allele level (HLA-A,-C,-B,-DRB3/4/5,-DRB1,-DQB1 loci); (2) haplotype combinations of these loci; and (3) genotypes of the loci. Associations were tested at two levels of HLA typing resolution – allele family and high resolution. DRBX*NNNN designates absence of any DRB3/4/5 gene on the chromosome. Groups of alleles with identical amino acid sequence in the antigen recognition site, designated with a ‘g’ notation, were not distinguished(10).
We developed a novel statistical methodology for fine-mapping of HLA associations, previously applied in CLL(8). We incorporated HLA typing ambiguity into our statistical model, allowing reporting of high-resolution haplotype-level associations. We also employed factor analysis to group associated HLA variants that are highly correlated by linkage disequilibrium(11). Multivariate logistic regression on independent sets of cases and controls for each ethnic subgroup identifies HLA that are associated with MM. P-values were adjusted for multiple testing using False Discovery Rate (FDR) with a 5% threshold; associations for age and gender interactions were unadjusted(12).
Cases and Controls Were Younger than Typical MM Patients: Baseline demographics are summarized in Table 1. Cases and controls are younger (49 years) than the typical median age for MM incidence (69 years). Generalizability of our findings may be limited to younger MM patients and possibly higher risk disease where allogeneic HSCT was considered.
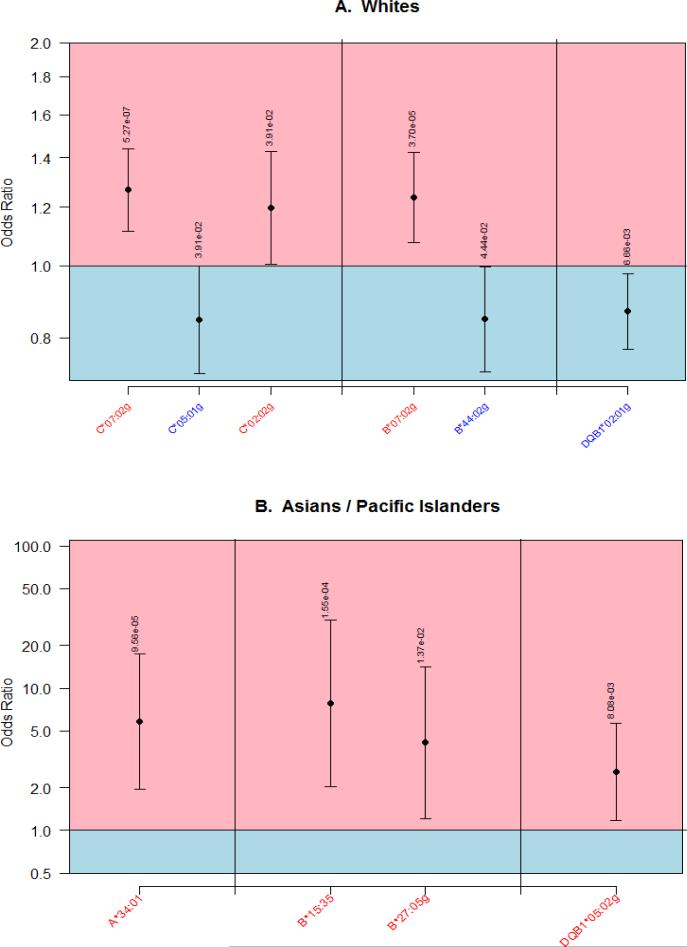
Allele-Level HLA Associations Identified in Whites and Asians: Figure 1 depicts the high resolution allele-level associations for Whites (3 predisposing or positively associated, 3 protective or negatively associated with MM) and Asians (4 predisposing). While no significant high resolution allelic associations were identified among Africans Americans and Hispanics, several allele-family-level and haplotypic associations were detected.
Figure 1.
High resolution HLA alleles associated with Multiple Myeloma among Whites (A) and Asian / Pacific Islanders (B)
We list all associated haplotypes and alleles at both high-resolution and allele family level for each population, with correlated associations grouped by factor analysis in Supplementary Table 1. The longest extended haplotypic association within each factor analysis group is provided in Table 1 for each population. Suggestive associations that were no longer significant after correction for multiple testing are listed in Supplementary Table 2.
C*07 Positively Associated with MM Risk: An increased MM risk was observed for both C*07:02g (OR=1.27,P=5.27×10−7) and B*07:02g (OR=1.24,P=3.70×10−5) in Whites. Because these two alleles are in extremely high linkage disequilibrium (D'>0.98)(10), it was not possible to disentangle effects of these alleles in Whites. We found three haplotypes containing these alleles independently associated with MM: A*03~C*07~B*07~DRB5*01~DRB1*15~DQB1*06 (OR=1.27,P=0.03), A*03:01g~C*07:02g~B*07:02g (OR=1.33,P=4.68×10−5), and C*07:02g~B*07:02g~DRB4*01:01g~DRB1*04:01~DQB1*03:02g (OR=1.85,P=0.0497) (Table 1A).
While neither single allele C*07:02g nor B*07:02g was associated with MM in other populations, several extended haplotype associations containing C*07 were positively associated with MM in Hispanics and Asians also. Haplotypes A*24:02g~C*07:02g~B*39:06~DRB3*01:01~DRB1*14:06~DQB1*03:01g (OR=3.93,P=0.0016) and A*68:02~C*07:01g~B*57:03 (OR=2.94,P=0.039) in Hispanics (Table 1C) and haplotypes C*07~B*38~DRB5*01~DRB1*15~DQB1*05 (OR=3.91,P=0.0094) and C*07:02g~B*15:35 were associated in Asians (OR=8.99,P=2.23×10−5) (Table 1D). Because C*07, but not B*07, was associated across populations, we infer that C*07 most likely drives the association with increased risk.
C*05:01g Negatively Associated with MM Risk: C*05:01g was negatively associated with MM in Whites (OR=0.85,P=0.039) along with B*44:02g (OR=0.85,P=0.044) on the same haplotype (Table 1A). The negative association of MM risk with C*05:01g, but not B*44:02g, was statistically significant in Hispanics and African Americans (Supplementary Table 2).
Many Other HLA Associations Were Unique to Single Populations: Differences in the frequency of HLA alleles, their underlying LD with other genes, and interactions with other genes that differ in LD and frequency across populations may account for heterogeneous HLA associations across populations14. For example, positively associated alleles A*34:01, B*15:35, B*27:05g, and DQB1*05:02g were not replicated across populations. C*02:02g was predisposing in Whites (OR=1.20,P=0.039) (Figure 1), with higher risk observed when C*02:02g occurred on a haplotype lacking a DRB3/4/5 gene, C*02:02g~DRBX*NNNN (OR=1.50,P=0.042) (Table 1A). Among African Americans, two independent haplotype associations (C*04~B*15 and DRB3*02~DRB1*11~DQB1*02) were associated with increased risk of MM (Table 1B).
We tested for interactions of HLA associations with age (Supplementary Table 3) and gender (Supplementary Table 4). None of the associations had a gender effect. Genotypic associations, grouped by factor analysis, are listed in Supplementary Table 5.
With the large numbers of cases/controls across four population groups, this study represents the most extensive examination of associations of HLA variants with MM risk. . Our results using classical HLA typing and haplotype inference clarify findings from a recent GWAS in Northern European Whites (4,692 MM cases and 10,990 controls) where HLA alleles were imputed from MHC region SNPs(5). The most significant GWAS association was DRB5*01 (OR 1.22,P=1.42×10−5), while C*07:02g and B*07:02g were also listed as apparently independent MM risk factors in the GWAS. We confirmed these three risk alleles, and illustrate their high correlation by linkage disequilibrium in Whites. Cross-population analysis suggests C*07 as the independent risk allele, while B*07:02 and DRB5*01 are likely surfacing on the same haplotype rather than contributing to MM risk. Similarly confirmed were, the protective association with MM of C*05:01g and B*44:02g alleles occurring on the same haplotype in Whites and the predisposing associations of C*12:03g~B*38:01 haplotype and B*58:01g in Whites and Asians respectively (Supplementary Table 2). While the cause of the positive risk association for C*07:02 is unclear from our data, it is known that C*07 group alleles have the lowest levels of expression among HLA-C alleles, which reduces immune control of HIV and may likewise reduce control of MM(13). C*07:02 is also a C1 group ligand for inhibitory KIR2DL2/2DL3 receptors on natural killer cells, and this KIR/HLA genotype combination has been associated with increased risk of melanoma(14).
The HLA associations observed here and in GWAS demonstrate that the HLA region is a key risk locus for mature B-cell malignancies(15, 16). MM etiology often involves clonal immunoglobulin production directed against post-translationally modified human proteins, such as hyperphosphorylated paratarg-7 (P-7) or sumoylated Hsp90(17, 18). HLA presentation of altered self-antigens derived from these modified proteins within B cells to T cell receptors could stimulate production of autoantibodies. Additional research is needed to characterize differences among HLA alleles in presentation of these altered self-antigens and to identify additional antigenic targets of MM clones(19). While inherited variation in immunologic response to protein-derived peptides may explain some of the increased risk of MM, because odds ratios for HLA were modest, genetic differences within oncogenes probably account for most of this differing risk. Interactions between HLA variants and myeloma subtypes / somatic mutations may also impact efficacy of emerging immunotherapy in MM(20). Identification of MM associated peptides bound by HLA alleles would be important next steps in developing immune oncologic therapies in MM.
Supplementary Material
Key Points.
Inherited HLA polymorphisms influence risk for developing multiple myeloma
HLA variation impacts immune surveillance of cancer
Acknowledgements
Dr. Beksac's work was supported by the Turkish Academy of Sciences. The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research; and grants from Allos, Inc.; Amgen, Inc.; Angioblast; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Buchanan Family Foundation; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Children's Leukemia Research Association; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Genzyme Corporation; GlaxoSmithKline; HistoGenetics, Inc.; Kiadis Pharma; The Leukemia & Lymphoma Society; The Medical College of Wisconsin; Merck & Co, Inc.; Millennium: The Takeda Oncology Co.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Optum Healthcare Solutions, Inc.; Osiris Therapeutics, Inc.; Otsuka America Pharmaceutical, Inc.; RemedyMD; Sanofi; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; StemCyte, A Global Cord Blood Therapeutics Co.; Stemsoft Software, Inc.; Swedish Orphan Biovitrum; Tarix Pharmaceuticals; Teva Neuroscience, Inc.; THERAKOS, Inc.; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the U.S. Government. Dr. Cozen's work was supported by the following grants: 2P50CA100707-06, 1R01CA134786-02, N01CP-67010, HHSN261201000035C.
Footnotes
Authorship Contributions:
All authors participated and co-wrote the manuscript. In addition MB and PH designed the study, analyzed the data and interpreted the results. LG, SF, MM, MA, XZ, MZ designed the methods for analysis and analyzed the data. WC, AD and SL interpreted the results and reviewed and edited the paper. MB and LG's contributions to the manuscript were equal.
Disclosure of Conflicts of Interest:
There are no conflicts of interest relevant to the content of this manuscript.
References
- 1.Vachon CM, Kyle RA, Therneau TM, Foreman BJ, Larson DR, Colby CL, et al. Increased risk of monoclonal gammopathy in first-degree relatives of patients with multiple myeloma or monoclonal gammopathy of undetermined significance. Blood. 2009 Jul 23;114(4):785–790. doi: 10.1182/blood-2008-12-192575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landgren O, Gridley G, Turesson I, Caporaso NE, Goldin LR, Baris D, et al. Risk of monoclonal gammopathy of undetermined significance (MGUS) and subsequent multiple myeloma among African American and white veterans in the United States. Blood. 2006 Feb 1;107(3):904–906. doi: 10.1182/blood-2005-08-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JH, Reinherz EL. Structural basis of T cell recognition of peptides bound to MHC molecules. Mol Immunol. 2002 May;38(14):1039–1049. doi: 10.1016/s0161-5890(02)00033-0. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Forsti A, et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nature genetics. 2013 Oct;45(10):1221–1225. doi: 10.1038/ng.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smedby KE, Foo JN, Skibola CF, Darabi H, Conde L, Hjalgrim H, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 2011 Apr;7(4):e1001378. doi: 10.1371/journal.pgen.1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludwig H, Mayr W. Genetic aspects of susceptibility to multiple myeloma. Blood. 1982 Jun;59(6):1286–1291. [PubMed] [Google Scholar]
- 8.Gragert L, Fingerson S, Albrecht M, Maiers M, Kalaycio M, Hill BT. Fine-mapping of HLA associations with chronic lymphocytic leukemia in US populations. Blood. 2014 Oct 23;124(17):2657–2665. doi: 10.1182/blood-2014-02-558767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012 Jul;80(1):1–11. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- 10.Gragert L, Madbouly A, Freeman J, Maiers M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Human immunology. 2013 Oct;74(10):1313–1320. doi: 10.1016/j.humimm.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Fabrigar LR, Wegener DT. Exploratory Factor Analysis (Understanding Statistics) Oxford University Press; 2011. [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 13.Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011 Apr 28;472(7344):495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naumova E, Mihaylova A, Stoitchkov K, Ivanova M, Quin L, Toneva M. Genetic polymorphism of NK receptors and their ligands in melanoma patients: prevalence of inhibitory over activating signals. Cancer Immunol Immunother. 2005;54(2):172–178. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcoceba M, Sebastian E, Marin L, Balanzategui A, Sarasquete ME, Chillon MC, et al. HLA specificities are related to development and prognosis of diffuse large B-cell lymphoma. Blood. 2013 Aug 22;122(8):1448–1454. doi: 10.1182/blood-2013-02-483420. [DOI] [PubMed] [Google Scholar]
- 16.Wang SS, Abdou AM, Morton LM, Thomas R, Cerhan JR, Gao X, et al. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood. 2010 Jun 10;115(23):4820–4823. doi: 10.1182/blood-2010-01-266775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preuss KD, Pfreundschuh M, Fadle N, Regitz E, Raudies S, Murwaski N, et al. Hyperphosphorylation of autoantigenic targets of paraproteins is due to inactivation of PP2A. Blood. 2011 Sep 22;118(12):3340–3346. doi: 10.1182/blood-2011-04-351668. [DOI] [PubMed] [Google Scholar]
- 18.Preuss KD, Pfreundschuh M, Weigert M, Fadle N, Regitz E, Kubuschok B. Sumoylated HSP90 is a dominantly inherited plasma cell dyscrasias risk factor. J Clin Invest. 2015 May;125(5):2179. doi: 10.1172/JCI82091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar MV. Clonal Immunoglobulin against Lysolipids in the Origin of Myeloma. N Engl J Med. 2016 Feb 11;374(6):555–561. doi: 10.1056/NEJMoa1508808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar S, van Gelder M, Noort W, Xu Y, Rouschop KM, Groen R, et al. Optimal selection of natural killer cells to kill myeloma: the role of HLA-E and NKG2A. Cancer Immunol Immunother. 2015 Aug;64(8):951–963. doi: 10.1007/s00262-015-1694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.