Abstract
Previously, we determined that enhanced disease activity in patients with systemic lupus erythematosus (SLE) was associated with dramatic increases in numbers of B lymphocytes expressing the transcription factor ARID3a. Our data now indicate ARID3a is important for interferon alpha (IFNa) expression and show a strong association between ARID3a expression and transcription of genes associated with lupus IFN signatures. Furthermore, both ARID3a and IFNa production were elicited in healthy control B cells upon stimulation with the TLR 9 agonist, CpG. Importantly, secretion of IFNa from ARID3a+ healthy B lymphocytes stimulated increased IFNa production in plasmacytoid dendritic cells. These data identify ARID3a+ B cells as a novel type of effector B cell, and link ARID3a expression in B lymphocytes to IFN-associated inflammatory responses in SLE.
Keywords: Lupus, ARID3a, effector B lymphocyte, interferon alpha, inflammation
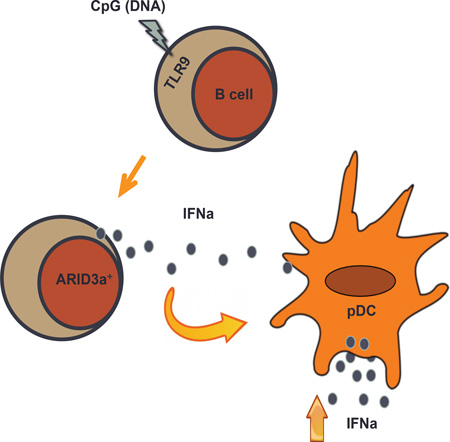
Graphical Abstract
1. Introduction
Previously, we found that constitutive expression of the transcription factor A-T rich interacting domain 3a (ARID3a) in murine B cells led to the development of anti-nuclear antibodies [1, 2], a common phenotype of the autoimmune disease systemic lupus erythematosus (SLE). While most circulating peripheral blood B lymphocytes fail to express ARID3a in healthy persons, we found that numbers of ARID3a+ B lymphocytes were dramatically increased (up to >40-fold) in SLE patients [3]. Importantly, elevated numbers of ARID3a-expressing B cells were associated with increased disease activity and varied over time in association with SLE disease activity index scores [3]. These data suggested a potential role for ARID3a in SLE disease pathogenesis. To date, there is not a unifying biomarker predictive of susceptibility or disease activity among SLE patients [4].
Approximately half of SLE patients exhibit increased levels of the cytokine interferon alpha (IFNa), and these levels have been associated with inflammation and disease activity in SLE [5–7]. Elevated IFNa levels result in increased expression of multiple IFN-responsive genes in SLE peripheral blood cells, collectively referred to as an “interferon signature” [8, 9]. Although the mechanisms which initiate IFN signatures in SLE remain elusive, plasmacytoid dendritic cells (pDCs) are thought to be the primary cells responsible for secreting IFNa [reviewed in [10]].
In the last decade, several investigators reported that B lymphocytes can act as cytokine producing effector B cells that modulate activities of other cell types during an immune response [reviewed in [11, 12]]. B cell effector functions were highlighted when SLE patients receiving B cell depleting therapy lessened disease activity without changes in autoantibody levels [13]. B cells responding to toll-like receptors (TLR) ligands produce an array of cytokines as B effector cells, including IFNγ, IL-12, IL-2, TNF-α, and IL-6 [14]. However, IFNa production by B cells has been understudied. Although B cells infected with the Epstein-Barr virus (EBV) express IFNa [15], little is known regarding IFNa production by human primary B cells.
Because our data indicated a strong association between numbers of ARID3a+ B lymphocytes and increased disease activity, we asked how ARID3a expression contributes to autoimmunity in SLE. Intriguingly, we observed no direct correlation between ARID3a expression in human SLE B cells and autoantibody production [16]. Therefore, we hypothesized that the relationship between ARID3a+ B cells and disease activity was antibody-independent, and might be associated with inflammatory responses that lead to “interferon signatures” observed in SLE [5].
2. Materials and methods
2.1. Participants
Peripheral blood mononuclear cells from a total of 22 SLE patients and 11 healthy controls were analyzed for ARID3a expression. SLE samples were defined as ARID3aH if numbers of ARID3A+ B cells > 2 standard deviations above the average numbers of ARID3a+ B cells in healthy controls (9,830 ARID3a+ B cells/ml), as previously described [3].
2.2. Plasma assessment
Due to low IFNa levels in peripheral blood, ELISA-based methods for quantitation can be unreliable [5, 17]. WISH endothelial cells express the IFNa receptor and have low/− endosomal Toll-like receptor (TLR) expression, and do not trigger endogenous IFN pathways [17]. Patient plasma was measured for the ability to elevate expression of IFNa-inducible genes using the human WISH reporter cell line (ATCC, CCL-25; gift from S. Kovats) by qRT-PCR, as previously described [17]. Briefly, WISH epithelial cells (50,000 cells/well) were cultured 1:2 with SLE patient or control plasma and RPMI supplemented with 10% FBS, for 6 hrs at 37°C prior to lysis for RNA isolation. Data were normalized to HPRT1 or GAPDH. Each value was expressed relative to the average normalized IFIT1 expression induced by 6 healthy control plasma samples.
2.3. RT-PCR, quantitative RT-PCR, and BioMark HD
Total RNA was isolated using Tri-Reagent (MRC, Inc). For RT-PCR, cDNA synthesis was performed at 37°C for 1 hour with M-MLV reverse transcriptase (Promega) and random primers (Promega), and amplification was for 40 cycles: 57–60°C for 30 s, 72°C for 1 min, and 95°C for 30 sec, according to the gene of interest. Real-Time qPCR was preformed according to the manufacturer’s protocol, using the 7500 Real-Time PCR System (Applied Biosystems) with SYBR Green PCR Master Mix (Qiagen) and gene-specific primers. Technical replicates were performed in duplicate or triplicate. Target gene expression was normalized to GAPDH or HPRT1, and expressed relative to gene expression in positive (EBV-transformed B cells or 293T) or B cells from healthy individuals for the Biomark HD data. The IFNa primers used for qRT-PCR of SLE B cells amplified ~70% of the IFNa subtype genes and quantified the collective expression of those genes. For BioMark HD assays, fluorescence activated cell sorted (FACS) CD20+ B lymphocytes were isolated using a FACSAria II (BD Biosciences). Isolated RNA was quantified, assessed for integrity using Agilent Total RNA Pico chips on a 2100 Bioanalyzer (Agilent Technologies, Boblingen, Germany). Analyses and qRT-PCR were all performed as previously described [18]. Data sets showing fold-increases and statistical relevance are available in Tables 1 and 2 in [19]. A list of primers for the genes assessed is available elsewhere (see Table 3 in [19]).
Table 1.
Summary of B cell array data
| Upregulated* genes in ARID3aH vs. ARID3aN SLE B cells |
|---|
|
ARID3a, OAS1, OAS3, HERC5, ISGI5, Ly6E, MX1, USP18, IFI44L, IFI6, IFI27, EPSTI1, IRF3, IRF5, IRF7, TLR7, BCL2L1, and BCL2 |
| Upregulated* genes in SLE vs. Control B cells |
|
IFNA2, IFNB1, IFI44, IFIT3, IFI44L, IFI6, IFI27, EPSTI1, SIGLEC1, and TLR7 |
| Upregulated* genes in Stim. vs. Unstim. Control B cells |
|
IFNA2, IFNRA1, IFNB1, EPSTI1, HERC5, IFI44L, IFIT3, MX1, Ly6E, OAS1, PLSCR1, OAS3, IRF3, TLR7, and TLR9 |
| Downregulated genes in Stim. vs. Unstim. Control B cells |
| BCL2L1 |
(> 2 fold); IFN signature genes are in bold
2.4. Methyl-seq
Purified genomic DNA was isolated from frozen aliquots containing PBMCs obtained from two ARID3a high and two ARID3a low expressing SLE patients using standard phenol/chloroform extraction protocols. Sample gDNA was fragmented on a Covaris S2 sonicator (Covaris, Woburn, MA) to an average size of ~350bp in length. Fragmented DNA was subjected to MethylMiner Methylated DNA Enrichment Kit (Life Technologies, Carlsbad, CA), according to the manufacturers protocol, and were then sequenced and analyzed by the Genomic Sequencing Core at Oklahoma Medical Research Foundation. The data base and additional details regarding its preparation are available elsewhere [19].
2.5. ARID3a knockdown
EBV-transformed lymphoblastoid (LCLs) B cell lines were plated into triplicate wells (40,000 cells/well) and transfected with 3 lentiviral constructs (MOI 3) containing ARID3a shRNA and co-expressing GFP (1–3), or a control vector expressing a scrambled sequence and GFP, as previously described [20]. Briefly, cells were cultured in RPMI 1640 supplemented with 4% FBS for 36 hrs at 37° C, and lysed for RNA isolation, cDNA synthesis, and qPCR after 36 hours (n=3).
2.6. Flow cytometry
PBMCs were isolated from heparinized peripheral blood (~15 ml) with Ficoll-Paque Plus (GE Healthcare), and stained with the following fluorochrome-labeled antibodies: CD19 PE-Cy5, CD10 Pacific Blue (BioLegend), IgD PerCP-Cy5.5, CD27 PE-Cy7, CD38 Alexa Fluor 700 (BD Pharmingen), and IgM APC (Southern Biotech). PBMCs were fixed (3% paraformaldehyde) and permeabilized (0.1% Tween-20) prior to staining with goat anti-human ARID3a antibody [21] and a rabbit anti-goat IgG FITC secondary (Invitrogen). Gating for individual B cell subsets was described previously [3] and used with the following B (CD20+) cell subset markers: transitional (IgD+CD27−CD10+), naïve (IgD+CD27−CD10+), MZ-like Memory (IgD+CD27+), Memory (IgD+CD27+), and double-negative (DN) (IgD−CD27−) B cells. Non-B cells were excluded using the following markers on the fluorochrome, APC: T cells (CD3), Monocytes, macrophages, and granulocytes (CD14), NK cells, neutrophils, macrophage and dendritic cells (CD16), and NK and NKT cells (CD56). Isotype controls (Caltag, BD Pharmingen, and eBioscience) were used for gating. Data (500,000 events per sample) were collected using an LSRII (BD Biogenics) and FACSDiva (BD Biosciences) software version 4.1 and were analyzed using FlowJo (Tree Star) software version 9.5.2.
2.7. Cell culture, B cell isolation and transfections
Healthy control and SLE B cells from PBMCs were enriched for B lymphocytes via negative selection using magnetic beads (Stem Cell Technologies), and were then fluorescence activated cell sort (FACS) purified using doublet exclusion to isolate single cells of > 99% purity. Post-sort analyses indicated a range of 97–99.8 percent CD20+ cells. For stimulation experiments, control B cells were cultured (40,000 cells/well) for 18–24 hrs in complete RPMI media (RPMI 1640, 5 x 10−5 M β-mercaptoethanol, 100 U/ ml penicillin, 100 ug/ ml streptomycin, 2 mM glutamine and 1 mM sodium pyruvate) supplemented with 4% heat inactivated fetal bovine serum (FBS) +/− the following agonists: lipoteichoic acid (50 ng/ml) (Sigma), PAM3CSK4 (50 ng/ml), Staphylococcus aureus peptidoglycan (10 µg/ml) and Bacillus anthracis peptidoglycan (10 µg/ml) [gifts from M. Coggeshall [22]], Poly I:C (0.1–0.3 µg/ml) and Imiquimod (10 µg/ml) [gifts from D. Farris], or Class C CpG oligonucleotide (1, 3, 5, or 10 µg/ml) and 5 µg/ml control ODN oligonucleotide (HPLC purified to >85% purity, and without detectable lipoproteins or endotoxins, as determined by the manufacturer, InvivoGen, San Diego, CA). Brefeldin A (BFA, eBiosciences) was added the final 4 hrs of culture to prevent protein secretion. Human EBV-transformed B cell lines were cultured in complete RPMI media supplemented with 8% or 4% FBS, at 37° C in a CO2 incubator. To assess secretion of IFNa, CpG-stimulated healthy control or SLE (40,000–100,000 cells/well), FACS-purified B cells (99% purity) were coated with an interferon capture antibody using the IFN-a Secretion Assay Kit (Miltenyi Biotec). Cells were cultured for 20 min., and then stained for flow cytometry. For B cell and plasmacytoid dendritic cell (pDC) coculture, B cells were enriched by negative selection, FASC purified, and stimulated with CpG (3–5 µg/ml) for 24 hrs. Autologous pDCs were positively selected with a CD304 MicroBead Kit (Miltenyi Biotec) using MACS 25 LS columns, FACS-purified, and cultured for 24 hrs in RPMI 1640 supplemented with 5% FBS. Following stimulation, B cells were cocultured with pDCs (3:1) pre-treated with an FcR block for 20 hrs, and stained for flow cytometry. BFA was added the final 5 hrs of culture to prevent secretion.
2.8. Statistics
GraphPad Prism 6 was used for all statistical analyses. A two-tailed Student’s T test or the nonparametric Mann-Whitney test was used for data comparing 2 groups. A one-way ANOVA was used for comparisons between 3 groups, followed by Turkey or Dunn’s posttest to correct for multiple comparisons. All statistical tests, and corresponding P values, are indicated in the figure legend. P values < 0.05 were considered significant and are indicated with the following symbols in the figures: *P < 0.05, **P < 0.01, ***P < 0.001.
2.9. Study Approval
Healthy controls (n=7) and patients (n=22) who met a minimum of four American College of Rheumatology Classification Criteria for SLE [23] were recruited after informed consent from the Oklahoma Medical Research Foundation Clinical Pharmacology clinic at as part of the Oklahoma Lupus Cohort (IRB compliance #09-07 and #06–19), in accordance with the Declaration of Helsinki.
3. Results
3.1. ARID3a is associated with IFNa expression
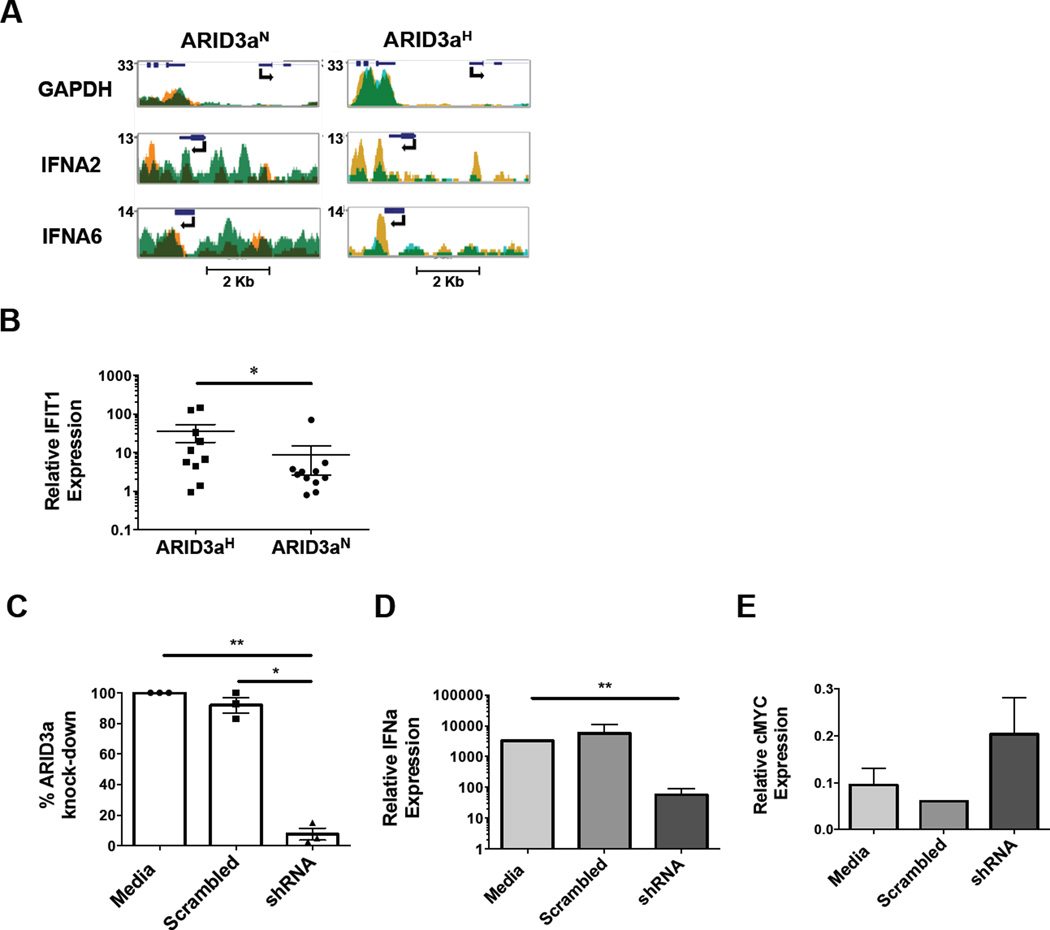
We postulated that ARID3a over-expression in SLE might be associated with differential gene regulation in total PBMCs. Because we found that numbers of cells expressing ARID3a in individuals vary over time, division of SLE samples based on total numbers of ARID3a+ B cells allowed us to better evaluate phenotypes directly associated with ARID3a expression [3]. Others have shown differential methylation patterns in SLE PBMCs compared to PBMCs from healthy controls [24, 25]. We hypothesized that ARID3a expression might ultimately affect the methylation status of multiple promoters, providing clues regarding which genes might be dysregulated in patient samples with increased numbers of ARID3a+ B cells (ARID3aH) versus samples with normal numbers of ARID3a-expressing B cells (ARID3aN). Genome-wide methyl-seq analyses of total PBMC samples from 2 ARID3aH and 2 ARID3aN designated SLE patient samples indicated methylation was globally higher across all chromosomes in the ARID3aH samples compared to ARID3aN samples (total data set available in ref. [19]). Promoter hypermethylation is typically correlated with gene repression [26,27]. However, PBMCs from ARID3aH SLE patients showed hypomethylation of several IFNa promoters, including IFNA 2, 5, 6, 8, 10, 14, 16, and 21, compared to ARID3aN SLE PBMCs (Figure 1A), implying that PBMCs from samples with increased numbers of ARID3a+ B cells express IFNa. Additionally, a review of data from the ENCODE group indicated potential ARID3a binding sites in promoters of IFNa subtype genes in some human cell lines [28], suggesting ARID3a could participate in regulation of those genes.
Figure 1. ARID3a is associated with IFNa expression.
A) Profiles show methylation patterns of two IFNa genes from PBMCs of 2 ARID3aN (orange and light green) and 2 ARID3aH (gold and blue) SLE patients. Dark green regions are positions methylated in both samples. Gene positions and transcription direction are indicated with arrows. The housekeeping gene, GAPDH, promoter served as a hypomethylated control. B) Plasma from ARID3aH (n= 10) and ARID3aN (n= 11) SLE patients (symbols) was tested for the ability to elicit expression of the interferon-signature gene, IFIT1, by qRT-PCR using the WISH reporter cell line. Means, standard errors, and significance (Mann-Whitney, *p< 0.05) are shown. C) Percentages of ARID3a expression with scrambled shRNA or ARID3a shRNA-treatment relative to media control, and corresponding D) IFNa and E) cMyc expression were assessed by qRT-PCR of EBV-transformed B cell lines. Means, standard errors, and significance (one-way ANOVA, with Turkey’s multiple comparison test, *p< 0.05, **p< 0.01) are shown.
To determine if increased numbers of ARID3a+ SLE B lymphocytes were associated with elevated circulating IFNa, ARID3aH and ARID3aN plasma samples were assessed for IFNa levels. Using a standard reporter assay allowing measurement of interferon-responsive genes by qRT-PCR, we found that ARID3aH SLE plasma samples showed significantly higher expression (p< 0.05) of the IFNa response gene, IFIT1, compared to ARID3aN SLE plasma samples (Figure 1B). These data indicate that plasma from patients with increased numbers of ARID3a+ B cells was more effective at inducing IFNa-stimulated gene expression than plasma from ARID3aN SLE samples, suggesting that IFN levels were higher in these samples.
Human lymphoblastoid B cell lines generated by infection with the Epstein Barr virus (EBV), express IFNa [15]. Moreover, we observed that all EBV-transformed cell lines express ARID3a [21], and others showed that ARID3a is necessary for expression of EBV latency proteins [29]. To test the requirement of ARID3a for IFNa expression, EBV-transformed B cells were infected with lentivirus expressing ARID3a shRNA, or scrambled control shRNA. Primers designed to amplify 6 of 12 highly homologous IFNa subtype gene products were demonstrated to yield appropriate sized products by RT-PCR using several EBV lines (see Figure 1 [19]). Thirty-six hours post-infection, expression of ARID3a and IFNa was determined via qRT-PCR (Figure 1C–D). While scrambled control shRNA did not significantly alter expression of ARID3a or IFNa, ARID3a knockdown decreased IFNa levels by more than 95%. However, expression of cMyc (Figure 1E), a gene previously determined to increase after ARID3a knockdown in 293T cells [20], was increased, suggesting that RNA expression was not globally affected by ARID3a inhibition. These data indicate expression of IFNa in EBV-transformed B cells requires ARID3a.
3.2. SLE B lymphocytes produce IFNa
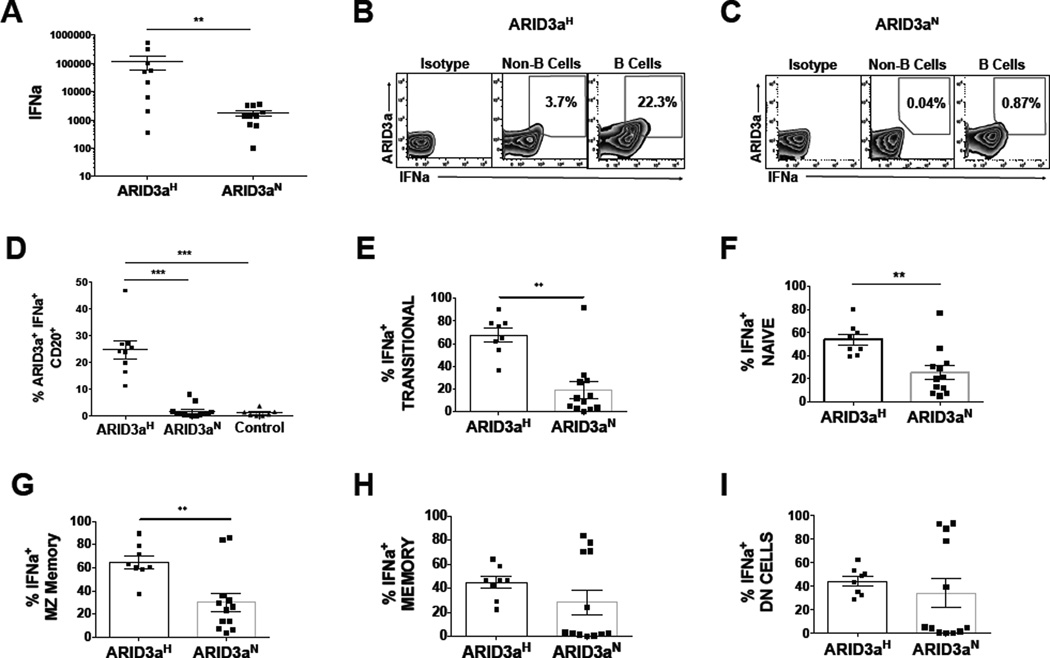
Typically, IFNa production is not attributed to B lymphocytes. To directly assess if primary B lymphocytes from SLE patients express IFNa, IFNa transcripts were quantified by qRT-PCR from peripheral blood B cells of 19 SLE patients segregated by total numbers of total ARID3a+ B cells. ARID3aH SLE B cells expressed an average of 100-fold higher levels of IFNa mRNA, compared to ARID3aN B cells (p< 0.01) (Figure 2A). To determine if IFNa mRNA levels reflected IFNa protein synthesis in SLE B cells, CD20+ B lymphocytes were evaluated for intracellular IFNa and ARID3a by flow cytometry. Representative flow cytometry plots for ARID3aH (Figure 2B) and ARID3aN (Figure 2C) SLE patient samples are shown. Numbers of ARID3a+ and IFNa-expressing B cells in ARID3aH samples averaged 25% of total CD20+ B cells. These cells were sorted to >99% purity for CD20, making it unlikely that the IFN-producing cells were the result of contaminating plasmacytoid dendritic cells. Co-expression of ARID3a and IFNa was significantly higher (p< 0.001) in ARID3aH versus ARID3aN and healthy control B cells (Figure 2D), but approximately 30% of ARID3aN B cells also showed IFNa staining, some without ARID3a. These data indicate ARID3a+ primary B cells from SLE patients produce IFNa.
Figure 2. IFNa is expressed in SLE B cells in association with ARID3a.
A) IFNa was measured by qRT-PCR in CD19+ B cells from 9 ARID3aH and 10 ARID3aN SLE patient samples. Data were normalized to GAPDH. Symbols represent averages of two replicates from individual samples (Mann-Whitney, **p<0.01). Representative flow cytometry plots of an ARID3aHB) and an ARID3aNC) SLE sample, showing percentages of IFNa and ARID3a within CD20+ B cells. D) Data show percentages of B cells expressing ARID3a+ and IFNA+ in PBMCs from ARID3aH (n=9) and ARID3aN (n=13) SLE patients, and healthy controls (n=6) analyzed by flow cytometry. Means, standard errors, and significance [ANOVA Kruskal Wallis (p < 0.0001) with Dunn’s multiple comparison correction (***p< 0.001)] are shown. E–I) Flow cytometry shows percentages of IFNa+ total B cells within transitional (IgD+CD27−CD10+), naïve (IgD+CD27−CD10−), MZ memory (IgD+CD27+), memory (IgD−CD27+), and DN (IgD−CD27−) B cells from ARID3aH (n=8–9) and ARID3aN (n=12–13) defined SLE samples. Means, standard errors, and significance (Mann-Whitney, **p < 0.01) are shown.
Our previous data showed that ARID3a expression in SLE B cells within individual peripheral blood B cell subsets [3]. Therefore, we hypothesized that IFNa and ARID3a expression might correlate more strongly in specific B cell subpopulations. In healthy controls, ARID3a is not expressed in early naïve B cells, yet in SLE naïve B cells, the ARID3a protein is abundantly expressed [3]. Intriguingly, examination of early transitional and naïve B cells, as well as marginal zone-like B cells, showed strong correlations between ARID3a and IFNa expression (Figure 2 E–G), while more mature memory and double-negative (DN) B cell subsets from some ARID3aN patient samples showed IFNa expression without ARID3a (Figure 2 H–I). One explanation for these data is that mature B cell subsets may express IFNa subtypes that do not require ARID3a. Alternatively, continued expression of ARID3a may not be required for maintenance of IFNa expression in mature B cell subsets. Nonetheless, these data indicate ARID3a and IFNa expression are strongly associated in early SLE B lineage cells.
3.3. Expression of IFNa signature genes is elevated in B cells with increased ARID3a
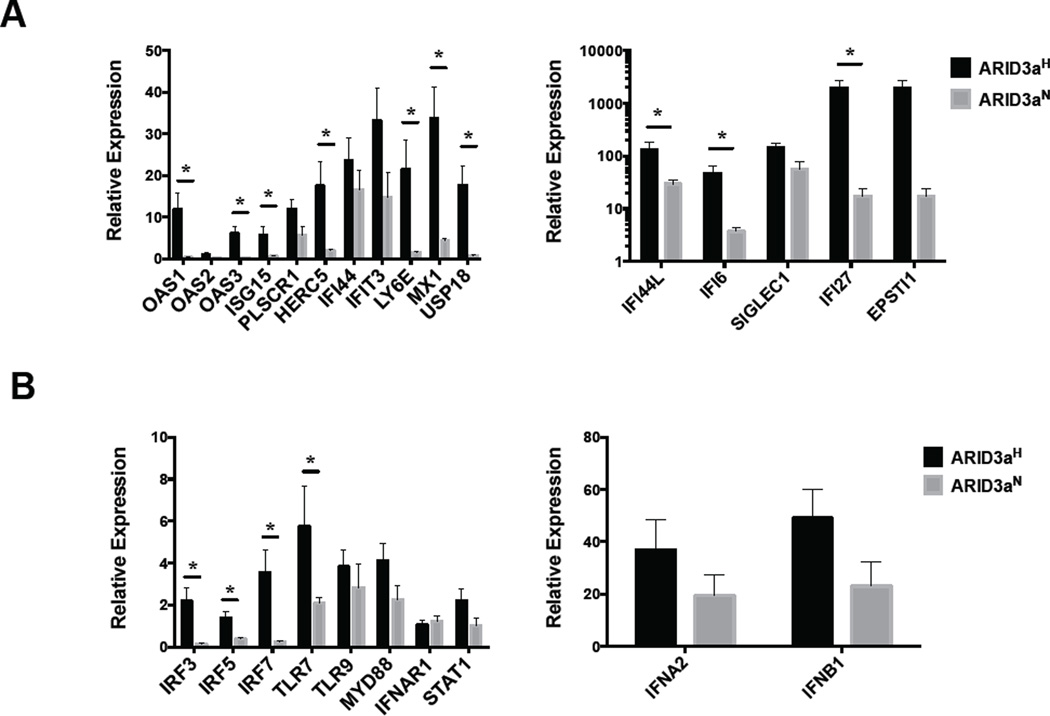
IFN can exert autocrine effects on cells that produce it [30]. To determine if IFNa-responsive genes typically assessed in SLE were upregulated in B lymphocytes from ARID3aH versus ARID3aN B cells, FACS-purified ARID3aH (n=3) and ARID3aN (n=3) CD20+ cells were evaluated for expression of IFNa pathway genes via BioMark HD qRT-PCR. As expected, ARID3aH B cells showed high expression (> 10-fold) of ARID3a mRNA, versus ARID3aN samples, and had significant expression (>2 fold) of 11 IFN signature genes versus ARID3aN B cells (Figure 3A and Table 1 in [19]). One IFNa subtype gene, IFNA2a, was included on the array; however, there were no significant differences in expression between ARID3a-based patient groups (Figure 3B). Additional genes involved in the IFNa pathway, including IRF3, IRF5, and IRF7, showed increased expression in ARID3aH versus ARID3aN B cells (Figure 3B). These data indicate genes involved in the IFNa pathway are upregulated in ARID3a+ SLE B cells, associating these cells with responses to IFNa, suggesting those cells may have been previously exposed to IFN.
Figure 3. Innate and IFNa signaling pathway genes are upregulated in association with ARID3a in SLE patient B cells.
RNA from isolated CD20+ SLE patient B cells (3 each ARID3aH and ARID3aN) was evaluated by Biomark HD qRT-PCR for interferon signature genes A) and other innate signaling pathway genes B). Gene expression was normalized to HPRT1. Means, standard errors and significance of genes upregulated > 2-fold by Student’s T test (*p) are shown.
3.4. TLR 9 signals induce ARID3a and IFNa expression in healthy control B cells
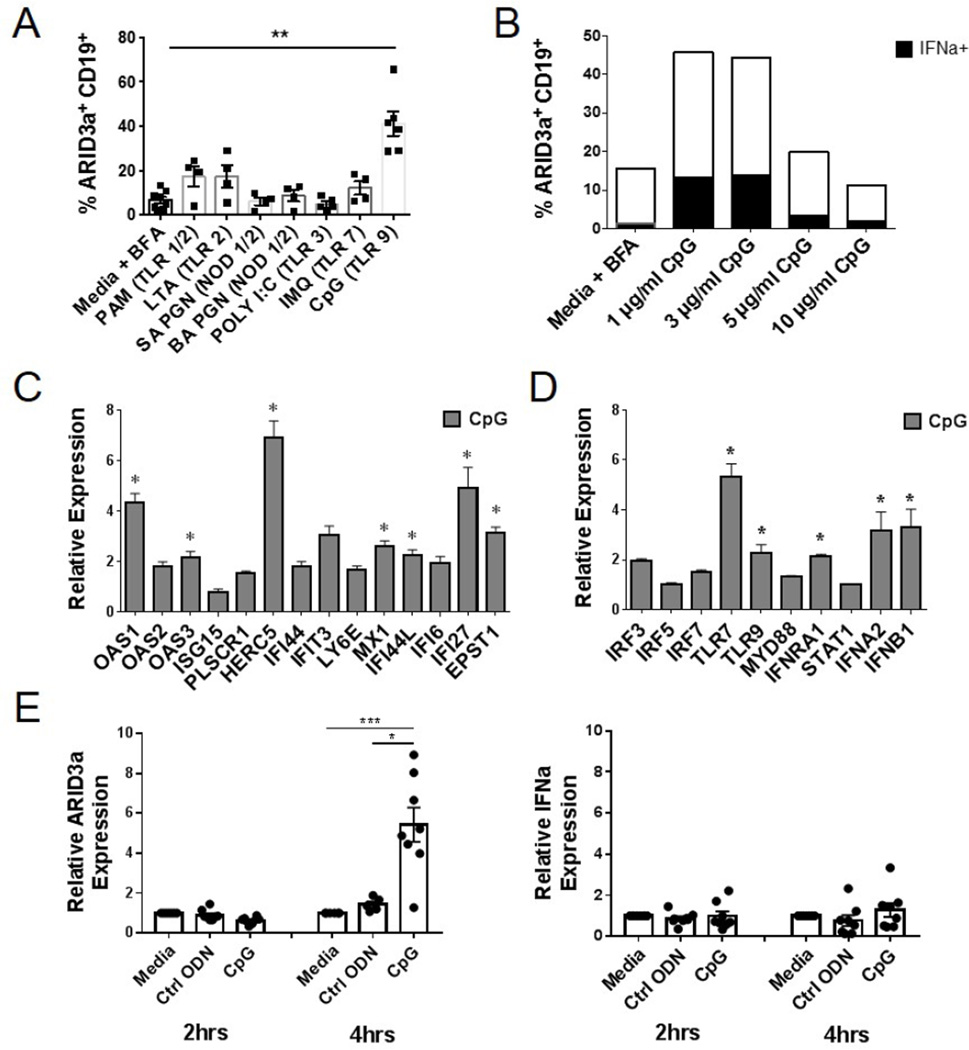
The initiating signals that induce ARID3a expression in SLE B cells are unknown. Although multiple stimuli induce ARID3a expression in mouse B cells [31–33], induction of ARID3a in healthy human B lymphocytes has been difficult [21]. Viral pathogens, microbial products, and self-antigens trigger innate immune responses in cell types expressing endosomal TLRs, including B cells, leading to the production of IFNa [34–37]. We hypothesized that ARID3a expression would be induced by stimuli that lead to IFNa expression (i.e., microbial products or self-antigens). Therefore, we assessed a variety of TLR and NOD-like receptor agonists for their ability to induce ARID3a expression in B lymphocytes from healthy donors. Although, expression of ARID3a was modestly stimulated by several of these agonists, stimulation of TLR 9 led to robust increases (p=0.0015) in numbers of ARID3a+ B cells (Figure 4A). Therefore, we further evaluated effects of increasing concentrations of CpG on ARID3a and IFNa expression in healthy B cells from 3 donors after 24 hours. Representative percentages of IFNa+ARID3a+ B cells induced by CpG stimulation, versus media control are shown in Figure 4B. These data indicate that ARID3a expression is robustly stimulated through activation of TLR 9 in healthy control B cells, and titrates with expression of IFNa.
Figure 4. Healthy control B cells express ARID3a upon stimulation via TLR 9.
A) Flow cytometry was used to assess percentages of ARID3a+ CD19+ B cells from 4–6 healthy controls after stimulation (18–24 hrs) with PAM, LTA, SA PGN, BA PGN, Poly I:C, IMQ, and CpG. Means, standard errors and significance (Paired T test, **p< 0.01) are shown. B) Representative percentages of ARID3a+ and IFNa-expressing B cells in response to increasing concentrations of CpG are indicated. C) Biomark HD qRT-PCR analyses of CD20+ B cells from CpG-treated (n=3) and unstimulated (n=3) control samples were assessed for IFN signature and D) TLR pathway-associated genes. All genes were normalized to HPRT1. Means, standard errors and significance of genes upregulated > 2-fold by Student’s T test (*p< 0.05) are shown. E) Healthy control B cells stimulated with ODN control or CpG were evaluated for ARID3a and IFNa transcription via qRT-PCR at 2 and 4 hours compared to media controls. Symbols are independent values determined from two individuals. Student’s T test (*p< 0.05, ***p=0.0007)
We further evaluated ARID3a+ healthy B cells for expression of IFNa signature genes, as we did for SLE ARID3aH and ARID3aN B cells, by BioMark HD qRT-PCR. CpG-stimulated healthy B cells showed increased expression of many of the same IFNa signature genes associated with ARID3a expression in SLE B cells (Figure 4C). CpG-stimulated B cells showed increased expression of several IFN signature genes in comparison to control B cells, including IFNA2 and IFNB1 (Figures 4 C–D, and Table 2 in [19]). In line with previous studies, several IFN signature genes were highly expressed in SLE versus healthy control B cells (Table 1). Furthermore, comparison of upregulated genes from CpG-stimulated B cells and ARID3aH SLE B cells showed up regulation of TLR7, as well as 7 IFN signature genes, in both array sets (Table 1). Interestingly, expression of BCL2L1 was increased in ARID3aH SLE B cells, but downregulated in healthy control B cells upon stimulation with CpG (Table 1, and Table 2 [19]). While most genes we analyzed were similarly regulated in SLE ARID3a+ versus healthy CpG-stimulated B cells, these data suggest that ARID3a+ SLE B cells may differ from healthy ARID3a+ B cells and may become dysregulated in healthy versus SLE inflammatory responses.
To further investigate the effects of CpG on healthy control B cells, we stimulated FACS-purified B cells with 5 µg/ml CpG, or the ODN control, and evaluated cells for ARID3a and IFNa transcription via qRT-PCR after 2 and 4 hours of stimulation. Control ODN showed little effect on ARID3a or IFNa expression at any time point evaluated, including 20 hours (Figure 4E and data not shown). However, CpG-stimulated B cells exhibited 5-fold increases in ARID3a expression without detectable increases in IFNa expression (Figure 4E). These data, along with supporting data in Figure 1C–E, indicate ARID3a expression occurs prior to expression of IFNa, and are consistent with the idea that ARID3a is required for IFNa expression.
3.5. IFNa is secreted from SLE B cells
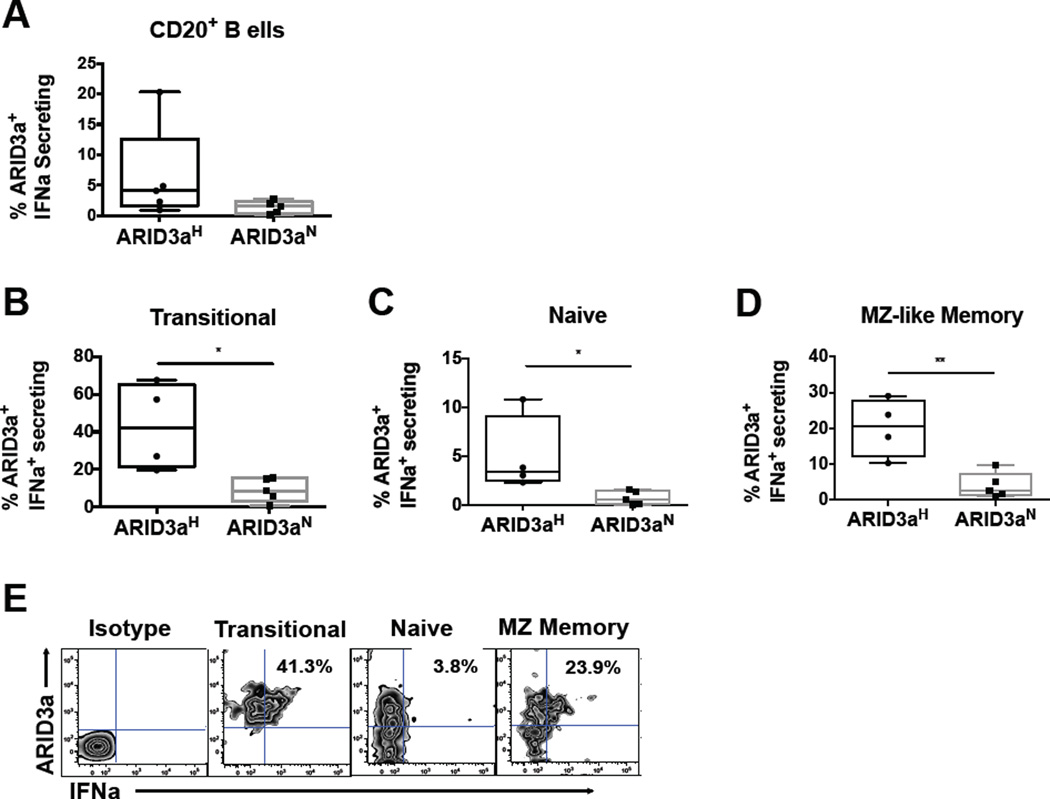
Although our data demonstrated IFN protein expression in ARID3a+ B cells, we determined if IFNa was secreted from SLE B cells. Using an IFNa capture antibody, we found that both ARID3aH and ARID3aN B cell samples showed evidence of IFNa secretion (Figure 5A). Examination of individual B cell subsets indicated that ARID3aH B cell samples had significantly higher percentages of ARID3a+ IFNa-secreting in transitional (p= 0.0159), naïve (p= 0.0159) and MZ memory (p= 0.0047) B cells, versus ARID3aN samples (Figures 5B–D). A representative flow plot for an ARID3aH B cell sample, showing percentages of ARID3a+ IFNa-secreting naïve, MZ-like memory, and transitional B cells is presented in Figure 5E. These data indicate that SLE ARID3a+ B cells can secrete IFNa at multiple stages of differentiation, including the early transitional B cell stage.
Figure 5. ARID3a+ SLE B cells secrete IFNa.
A) Results of flow cytometry show percentages of ARID3a+ IFNa-secreting B cells in ARID3aH (n=4) and ARID3aN (n=5) SLE patient samples. Secretion of IFNa in individual B cell subsets was evaluated using an extracellular capture antibody for B) transitional (IgD+CD27−CD10+), C) naïve (IgD+CD27−CD10−), and D) MZ memory (IgD+CD27+) B cell subsets as shown. Means, standard errors, and significance (Mann-Whitney or Student’s T test, *p< 0.05, **p< 0.01) are shown. E) Representative flow plots from one ARID3aH individual showing percentages of ARID3a+ IFNa-secreting cells in multiple subsets.
3.6. ARID3a+ healthy B lymphocytes act as effector B cells
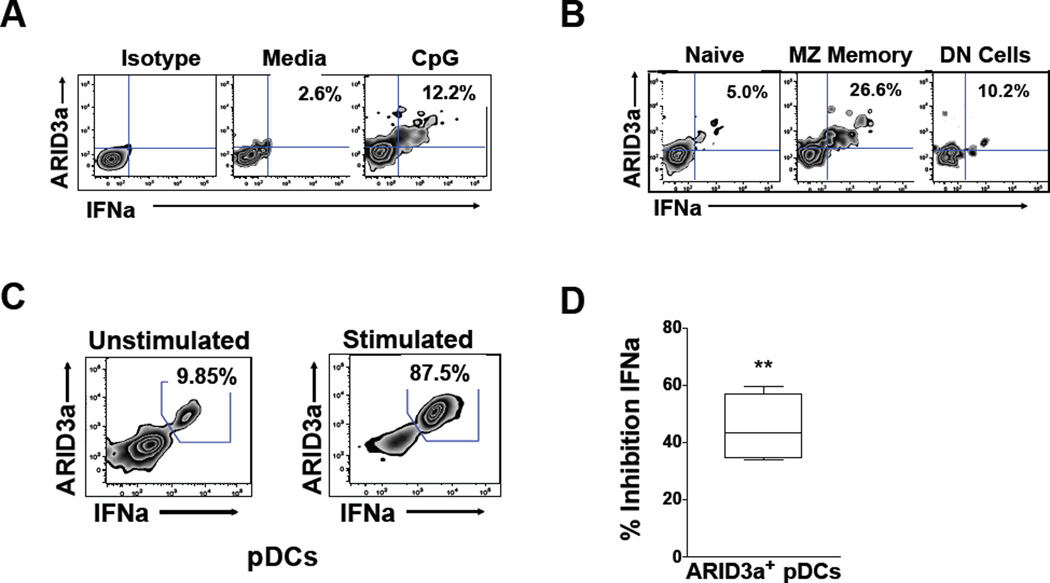
To determine if IFNa secretion is a property of healthy ARID3a+ B cells, healthy donor cells were stimulated with CpG (3 µg/ml) and assessed for secreted IFNa as described above. Healthy ARID3a+ B cells also secreted IFNa (Figure 6A). However, consistent with our previous findings that healthy naïve B cells do not typically express ARID3a, even after CpG stimulation, there was little IFNa secretion from those cells, or from the more mature DN subset. Interestingly, healthy MZ-like ARID3a+ B cells showed robust IFNa secretion (Figure 6B). These data suggest that healthy MZ-like ARID3a+ B cells secrete IFNa.
Figure 6. Healthy donor ARID3a+ B cells secrete IFNa and act as effector cells.
A) IFNa-secretion was analyzed by flow cytometry for percentages of ARID3a+ IFNa-secreting total B cells with and without CpG stimulation (n=6). B) Representative secretion of IFNa in individual naïve (IgD+CD27−CD10−), MZ memory (IgD+CD27+) and DN (IgD−CD27−) B cell subsets are shown. C) Representative autologous pDCs cocultured with CpG-stimulated or unstimulated B cells were evaluated by flow cytometry for ARID3a and IFNa expression (n=6). D) Autologous pDCs from 4 healthy controls were cocultured with CpG-stimulated B cells, with or without an IFNa blocking antibody, and the percent inhibition of IFNa expression by the blocking antibody was determined in ARID3a+ pDCs by flow cytometry. Means, standard errors and significance (paired Student’s T test, **p< .01) are shown.
We found it unlikely that secretion of IFNa from B lymphocytes could account for the association between ARID3a expression and IFNa plasma levels (Figure 1A). Plasmacytoid dendritic cells (pDCs) are the most notable human IFNa producers, secreting 10–100X more IFNa than other cell type upon activation [10]. Because IFNa can act as an autocrine factor to induce additional IFNa secretion [30], we hypothesized that ARID3a+ IFNa-secreting B cells were effector cells that would stimulate increased IFNa production in pDCs. Therefore, healthy control (n=6) CpG-stimulated or unstimulated B cells were cultured with autologous pDCs (3:1) for 20 hours prior to assessment of intracellular ARID3a and IFNa expression in gated pDCs. Interestingly, IFNa+ pDCs also expressed ARID3a when cocultured with CpG-treated control B cells, which stimulated (p=0.0409) a 3.49 fold increase in IFNa+ARID3a+ pDCs relative to unstimulated B cells on average (range 1.34–8.84). The most dramatic increase in IFNa+ARID3a+ pDCs was approximately 9-fold (87.5%), versus coculture with unstimulated B cells (9.85%) (Figure 6C). Stimulation of pDCs with CpG alone did not induce ARID3a or IFNa expression under these conditions (not shown), suggesting that results were not due to small numbers of contaminating pDCs present in the initial B cell cultures. Further, addition of polyclonal IFNa blocking antibodies to pDC cocultures (n=4) with autologous CpG-stimulated B cells inhibited production of IFNa+ARID3a+ pDCs (p= 0.0026) (Figure 6D). Together these data indicate that ARID3a+ IFNa-secreting B cells are innate effector B cells capable of secreting IFNa and influencing IFNa production in other cells.
4. Discussion
We now show that expanded numbers of ARID3a+ B cells in SLE are associated with increased plasma levels of IFNa. SLE B cells that express ARID3a synthesize IFNa, and express increased levels of IFNa signature genes previously associated with lupus disease activity [44]. In addition, our data indicate that the TLR 9 agonist CpG induces ARID3a expression and IFNa production in healthy donor B lymphocytes. Furthermore, both SLE ARID3a+ B cells and healthy donor ARID3a+ MZ-like B cells secrete IFNa. Moreover, healthy ARID3a+ B cells can act as effector cells to enhance IFNa secretion in pDCs. Together, these data suggest that ARID3a expression in B lymphocytes, and perhaps other cell types, including pDCs, is associated with IFNa production. These data identify ARID3a as a marker for a new type of IFNa-secreting effector B lymphocyte that is enriched in SLE patient samples.
While lipopolysaccharide (LPS), CD40 ligand, interleukin-5 (IL-5) plus antigen, and agonistic monoclonal antibodies against CD38 or RP105 (CD180) induced ARID3a expression in mouse B cells [31, 33, 39], the stimuli that induce ARID3a expression in human B cells have not been described. Interestingly, EBV infection resulted in upregulation of ARID3a expression [40] where it is required for expression of EBNA latency genes [29]. Our data now show that the TLR 9 agonist CpG is particularly effective inducing ARID3a expression in healthy B lymphocytes. We speculate ARID3a expression in SLE B cells may result from exposure to DNA and/or stimulation by TLR agonists. Further, others have shown associations between SLE pathogenesis and demethylation of gene promoter regions [24]. In this study, SLE PBMCs with increased ARID3a expression revealed hypomethylation of several IFNa subtype genes. Therefore, it will be important to determine if ARID3a regulates expression of IFNa genes directly, or via epigenetic changes, in response to DNA agonists in B cells.
ARID3a expression in EBV-transformed B cell lines, CpG-stimulated healthy B lymphocytes, and SLE B cells was associated with IFNa production, and our data indicate that ARID3a+ IFNa-secreting B cells can stimulate elevated IFNa production in pDCs. Others showed that supernatants from Class A CpG-stimulated B cells resulted in increased IFNa production in pDCs, but did not observe detectable IFNa in the immunoassays they used [41]. In addition, no differences were observed in the ability of CpG-stimulated memory or naïve B cells to induce IFNa production in pDCs in those studies [41]. Class A-CpG (ODN 2216/type “D”) was previously shown to be a poor B cell stimulator [42, 43], yet effective in stimulating pDC IFNa secretion by 48 hours [44, 45]. Our studies used class C-CpG, which is an effective B cell stimulator [44], and may account for the IFNa production we observed in B cells, and the ability to enhance IFN expression in pDCs.
Although our data show a strong correlation between IFNa expression and ARID3a expression, it should be noted that the association was not 100% in all SLE patient-derived B cells (Figure 2). IFNa belongs to the type I interferon subclass, which includes 13 subtypes that are 75% homologous encoded by individual genes on chromosome 9, as well as 4 additional IFN genes [10]. Our methyl- and ChIP-seq data suggested the possibility of ARID3a-mediated regulation of nine IFNa subtype genes. Therefore, one explanation for the expression of IFNa without ARID3a is that some subtypes of IFNa may not require ARID3a for their production. Usage of different IFNa subtypes in B lymphocytes, and individual B lymphocyte subsets, remains unexplored, and is complicated by the high homology among the subtypes, each of which utilize the same IFNa receptor. It is unclear how these subtypes generate differential IFN responses; however, this may be partially explained by cell-type specific expression of some IFNa subtypes [10]. Further studies to elucidate IFNa subtype expression in B lymphocytes may be informative and could account for some of the differences in IFNa expression we observed. In addition, our data also showed expression of ARID3a without IFNa in some B cells. Because our data was a snapshot of ARID3a and IFNa expression at a single time point, one explanation is that ARID3a expression occurs prior to IFNa production. Indeed, time course expression studies from healthy CpG stimulated B lymphocytes indicated that ARID3a transcription was apparent earlier than expression of IFNa (Fig. 4E). These data are supported by studies in mouse hematopoietic stem cells which also found a correlation between ARID3a and IFNa expression [46]. In those studies, ARID3a knockouts were also deficient in IFNa signaling, suggesting that ARID3a functions upstream of IFNa in that system. Irrespective of the possibility that some IFNa subtypes may not require ARID3a, knockdown of ARID3a in EBV-transformed cells also greatly decreased IFNa transcript levels, indicating it is required for expression of at least a subset of IFNa subtypes.
ARID3a expression was not ubiquitously expressed in all B cell subsets [3]. Interestingly, ARID3a+ MZ-like memory B cell subpopulations were most highly associated with increased disease activity in our previous studies (p= 0.0225) [3], and with IFNa production in these experiments. ARID3aH SLE samples had nearly 2.5-fold more IFNa+ MZ-like B cells than ARID3aN samples (Figure 2). Intriguingly, the most robust IFNa secretion in CpG-stimulated healthy donors was also the MZ-like subset. ARID3a is abnormally expressed in naïve B cells in SLE patients [3], and somewhat surprisingly, we also observed IFNa secretion in early naïve and transitional ARID3a+ SLE B cells in this study. This is contrary to what we observed in CpG stimulated healthy donor B cells, where ARID3a and IFNa were not induced in naïve and transitional B cells. The reasons for this are unclear, but could indicate that other stimuli are necessary to induce ARID3a expression in early B lineage cells. Alternatively, SLE transitional and naïve B cells may differ intrinsically from healthy donor B cells. Our limited BioMark HD analyses indicated some genes are differentially regulated in healthy stimulated versus SLE B cells (Table 1).
Multiple IFN-stimulated genes were induced in both healthy control B cells using the TLR 9 agonist, CpG, and in SLE ARID3a+ B cells, suggesting autocrine stimulation of the B cells by IFNa [30]. Importantly, two anti-apoptotic molecules, Bcl-2 (>3.5 fold) and Bcl-2-like protein 1 (Bcl2I1) [47] were increased (>3 and >19 fold, respectively) in ARID3aH versus ARID3aN SLE B cells (Table 1 in [19]), suggesting the interesting possibility of increased survival of ARID3aH SLE B cells. Intriguingly, Bcl2I1 was downregulated in CpG-stimulated healthy control B cells (Table 1), suggesting expression may be dysregulated in lupus B cells. Further identification of genes that are differentially expressed in healthy ARID3a+ versus SLE ARID3a+ B cells may be informative regarding mechanisms that contribute to inflammation and enhanced IFNa levels in lupus.
In this study, we have identified a previously undefined ARID3a+ innate effector B cell that secretes IFNa in SLE patient samples previously associated with increased disease activity. Human innate B cell functions have been understudied; therefore, interrogation of how ARID3a affects coregulatory processes associated with TLR 9 and other DNA sensors [48] will be important for future studies. Little is known regarding expression of IFNa expression in human B cells. These studies are the first to show the B cell subset distribution of IFNa secreting B cells, and highlight new mechanisms for innate B cells. It will be important to determine if ARID3a+ B cells have additional effector functions, and if they influence cell types other than pDCs. A recent phase II study indicated that targeting IFNa in SLE patients with low IFN signatures lessened disease activity [49], suggesting that the ability to define patients with early symptoms of inflammation might be therapeutically useful. The possibility that ARID3a can be used as a marker for IFNa-producing effector B cells may open new avenues for therapeutic approaches in autoimmunity.
Highlights.
ARID3a inhibition reduces IFNa expression
ARID3a+ human B lymphocytes secrete IFNa
ARID3a is induced by TLR agonists in healthy B cells
ARID3a expressing B lymphocytes enhance IFNa production in pDCs
Ward et al. identified a new human effector B lymphocyte characterized by expression of the transcription factor ARID3a and type I interferon. Increased numbers of ARID3a+ B lymphocytes in lupus patients were associated with inflammatory responses and interferon production. These data implicate ARID3a+ B cells as therapeutic targets in SLE.
Acknowledgments
The authors wish to thank T. D. Templeton and V. Roberts for technical assistance, Drs. S. Kovats, D. Farris, and M. Coggeshall for reagents, and Drs. TF Tedder and MB Humphrey for helpful discussions. Funding was provided by the Lupus Foundation of America (CFW), and grants from the National Institutes of Health: AI090343 and AI044215 (CFW), and AR053483, GM103510, AI101934, AI082714, GM104938 (JMG, JAJ), and AR056360, AR063124, and GM110766 (PMG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors declare that there are no competing financial interests.
References
- 1.Oldham AL, Miner CA, Wang HC, Webb CF. The transcription factor Bright plays a role in marginal zone B lymphocyte development and autoantibody production. Mol Immunol. 2011;49:367–379. doi: 10.1016/j.molimm.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankar M, Nixon JC, Maier S, Workman J, Farris AD, Webb CF. Anti-nuclear antibody production and autoimmunity in transgenic mice that overexpress the transcription factor Bright. J Immunol. 2007;178:2996–3006. doi: 10.4049/jimmunol.178.5.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward JM, Rose K, Montgomery C, Adrianto I, James JA, Merrill JT, et al. Disease activity in systemic lupus erythematosus correlates with expression of the transcription factor AT-rich-interactive domain 3A. Arthritis Rheumatol. 2014;66:3404–3412. doi: 10.1002/art.38857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lateef A, Petri M. Unmet medical needs in systemic lupus erythematosus. Arthritis Res Ther. 2012;14(Suppl 4):S4. doi: 10.1186/ar3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 7.Bengtsson AA, Sturfelt G, Truedsson L, Blomberg J, Alm G, Vallin H, et al. Activation of type I interferon system in systemic lupus erythematosus correlates with disease activity but not with antiretroviral antibodies. Lupus. 2000;9:664–671. doi: 10.1191/096120300674499064. [DOI] [PubMed] [Google Scholar]
- 8.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155:109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niewold TB, Clark DN, Salloum R, Poole BD. Interferon alpha in systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:948364. doi: 10.1155/2010/948364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eloranta ML, Alm GV, Ronnblom L. Disease mechanisms in rheumatology--tools and pathways: plasmacytoid dendritic cells and their role in autoimmune rheumatic diseases. Arthritis Rheum. 2013;65:853–863. doi: 10.1002/art.37821. [DOI] [PubMed] [Google Scholar]
- 11.Lund FE, Randall TD. Effector and regulatory B cells: modulators of CD4+ T cell immunity. Nat Rev Immunol. 2010;10:236–247. doi: 10.1038/nri2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao Y, Cao X. The immune potential and immunopathology of cytokine-producing B cell subsets: a comprehensive review. J Autoimmun. 2014;55:10–23. doi: 10.1016/j.jaut.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Ramos-Casals M, Sanz I, Bosch X, Stone JH, Khamashta MA. B-cell-depleting therapy in systemic lupus erythematosus. Am J Med. 2012;125:327–336. doi: 10.1016/j.amjmed.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund FE. Cytokine-producing B lymphocytes-key regulators of immunity. Curr Opin Immunol. 2008;20:332–338. doi: 10.1016/j.coi.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickramasinghe SN, Hasan R, Smythe J. Reduced interferon-alpha production by Epstein-Barr virus transformed B-lymphoblastoid cell lines and lectin-stimulated lymphocytes in congenital dyserythropoietic anaemia type I. Br J Haematol. 1997;98:295–298. doi: 10.1046/j.1365-2141.1997.1993016.x. [DOI] [PubMed] [Google Scholar]
- 16.Ward JM, James JA, Zhao YD, Webb CF. Antibody reactivity of B cells in lupus patients with increased disease activity and ARID3a expression. Antibodies. 2015;4:354–368. doi: 10.3390/antib4040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54:1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 18.Ratliff ML, Mishra M, Frank M, Guthridge JM, Webb CF. The transcription factor ARID3a is important for in vitro differentiaion of human hematopoietic progenitors. J Immunol. 2015;192:614–623. doi: 10.4049/jimmunol.1500355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JM, Ratliff ML, Dozmorov MG, Wiley G, Guthridge JM, Gaffney PM, James JA, Webb CF. Data showing the correlation between ARID3a, interferon alpha, and IFN-stimulated gene expression in SLE and CpG-stimulated B lymphocytes. Data in Brief. 2016 submitted. [Google Scholar]
- 20.An G, Miner CA, Nixon JC, Kincade PW, Bryant J, Tucker PW, et al. Loss of Bright/ARID3a function promotes developmental plasticity. Stem Cells. 2010;28:1560–1567. doi: 10.1002/stem.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nixon JC, Rajaiya JB, Ayers N, Evetts S, Webb CF. The transcription factor, Bright, is not expressed in all human B lymphocyte subpopulations. Cell Immunol. 2004;228:42–53. doi: 10.1016/j.cellimm.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Iyer JK, Khurana T, Langer M, West CM, Ballard JD, Metcalf JP, et al. Inflammatory cytokine response to Bacillus anthracis peptidoglycan requires phagocytosis and lysosomal trafficking. Infect Immun. 2010;78:2418–2428. doi: 10.1128/IAI.00170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 24.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffries MA, Sawalha AH. Epigenetics in systemic lupus erythematosus: leading the way for specific therapeutic agents. Int J Clin Rheumtol. 2011;6:423–439. doi: 10.2217/ijr.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 27.Tribioli C, Tamanini F, Patrosso C, Milanesi L, Villa A, Pergolizzi R, et al. Methylation and sequence analysis around EagI sites: identification of 28 new CpG islands in XQ24-XQ28. Nucleic Acids Res. 1992;20:727–733. doi: 10.1093/nar/20.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borestrom C, Forsman A, Ruetschi U, Rymo L. E2F1, ARID3A/Bright and Oct-2 factors bind to the Epstein-Barr virus C promoter, EBNA1 and oriP, participating in long-distance promoter-enhancer interactions. J Gen Virol. 2012;93:1065–1075. doi: 10.1099/vir.0.038752-0. [DOI] [PubMed] [Google Scholar]
- 30.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb CF, Das C, Coffman RL, Tucker PW. Induction of immunoglobulin mu mRNA in a B cell transfectant stimulated with interleukin-5 and a T-dependent antigen. J Immunol. 1989;143:3934–3939. [PubMed] [Google Scholar]
- 32.Webb CF, Das C, Eaton S, Calame K, Tucker PW. Novel protein-DNA interactions associated with increased immunoglobulin transcription in response to antigen plus interleukin-5. Mol Cell Biol. 1991;11:5197–5205. doi: 10.1128/mcb.11.10.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb CF, Smith EA, Medina KL, Buchanan KL, Smithson G, Dou S. Expression of bright at two distinct stages of B lymphocyte development. J Immunol. 1998;160:4747–4754. [PubMed] [Google Scholar]
- 34.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 35.Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T, et al. Quantitative expression of toll-like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 36.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 37.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker AM, Dao KH, Han BK, Kornu R, Lakhanpal S, Mobley AB, et al. SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature. PLoS One. 2013;8:e67003. doi: 10.1371/journal.pone.0067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb CF, Das C, Eneff KL, Tucker PW. Identification of a matrix-associated region 5’ of an immunoglobulin heavy chain variable region gene. Mol Cell Biol. 1991;11:5206–5211. doi: 10.1128/mcb.11.10.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nixon JC, Rajaiya J, Webb CF. Mutations in the DNA-binding domain of the transcription factor Bright act as dominant negative proteins and interfere with immunoglobulin transactivation. J Biol Chem. 2004;279:52465–52472. doi: 10.1074/jbc.M403028200. [DOI] [PubMed] [Google Scholar]
- 41.Berggren O, Hagberg N, Weber G, Alm GV, Ronnblom L, Eloranta ML. B lymphocytes enhance interferon-alpha production by plasmacytoid dendritic cells. Arthritis Rheum. 2012;64:3409–3419. doi: 10.1002/art.34599. [DOI] [PubMed] [Google Scholar]
- 42.Verthelyi D, Ishii KJ, Gursel M, Takeshita F, Klinman DM. Human peripheral blood cells differentially recognize and respond to two distinct CPG motifs. J Immunol. 2001;166:2372–2377. doi: 10.4049/jimmunol.166.4.2372. [DOI] [PubMed] [Google Scholar]
- 43.Gursel M, Verthelyi D, Gursel I, Ishii KJ, Klinman DM. Differential and competitive activation of human immune cells by distinct classes of CpG oligodeoxynucleotide. J Leukoc Biol. 2002;71:813–820. [PubMed] [Google Scholar]
- 44.Vollmer J, Weeratna R, Payette P, Jurk M, Schetter C, Laucht M, et al. Characterization of three CpG oligodeoxynucleotide classes with distinct immunostimulatory activities. Eur J Immunol. 2004;34:251–262. doi: 10.1002/eji.200324032. [DOI] [PubMed] [Google Scholar]
- 45.Krug A, Rothenfusser S, Hornung V, Jahrsdorfer B, Blackwell S, Ballas ZK, et al. Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur J Immunol. 2001;31:2154–2163. doi: 10.1002/1521-4141(200107)31:7<2154::aid-immu2154>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 46.Kim PG, Canver MC, Rhee C, Ross SJ, Harriss JV, Tu HC, et al. Interferon-alpha signaling promotes embryonic HSC maturation. Blood. 2016;128:204–2016. doi: 10.1182/blood-2016-01-689281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moser K, Wong DM, Manz RA. Antibody memory: a question of life or death beyond the germinal center. Immunol Cell Biol. 2011;89:164–166. doi: 10.1038/icb.2010.159. [DOI] [PubMed] [Google Scholar]
- 48.Sharma S, Fitzgerald KA, Cancro MP, Marshak-Rothstein A. Nucleic Acid-Sensing Receptors: Rheostats of Autoimmunity and Autoinflammation. J Immunol. 2015;195:3507–3512. doi: 10.4049/jimmunol.1500964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalunian KC, Merrill JT, Maciuca R, McBride JM, Townsend MJ, Wei X, et al. A Phase II study of the efficacy and safety of rontalizumab (rhuMAb interferon-alpha) in patients with systemic lupus erythematosus (ROSE) Ann Rheum Dis. 2016;75:196–202. doi: 10.1136/annrheumdis-2014-206090. [DOI] [PubMed] [Google Scholar]