Abstract
A subset of beryllium-exposed workers develop beryllium sensitisation (BeS) which precedes chronic beryllium disease (CBD). We conducted an in-depth analysis of differentially expressed candidate genes in CBD.
We performed Affymetrix GeneChip 1.0 ST array analysis on peripheral blood mononuclear cells (PBMCs) from 10 CBD, 10 BeS and 10 beryllium-exposed, nondiseased controls stimulated with BeSO4 or medium. The differentially expressed genes were validated by high-throughput real-time PCR in this group and in an additional group of cases and nonexposed controls. The functional roles of the top candidate genes in CBD were assessed using a pharmacological inhibitor. CBD gene expression data were compared with whole blood and lung tissue in sarcoidosis from the Gene Expression Omnibus.
We confirmed almost 450 genes that were significantly differentially expressed between CBD and controls. The top enrichment of genes was for JAK (Janus kinase)–STAT (signal transducer and activator of transcription) signalling. A JAK2 inhibitor significantly decreased tumour necrosis factor-α and interferon-γ production. Furthermore, we found 287 differentially expressed genes overlapped in CBD/sarcoidosis. The top shared pathways included cytokine–cytokine receptor interactions, and Toll-like receptor, chemokine and JAK–STAT signalling pathways.
We show that PBMCs demonstrate differentially expressed gene profiles relevant to the immunnopathogenesis of CBD. CBD and sarcoidosis share similar differential expression of pathogenic genes and pathways.
Introduction
Chronic beryllium disease (CBD), a granulomatous lung disease caused by beryllium exposure in the workplace, is characterised by the accumulation of beryllium-specific CD4+ T-cells in the lung. Depending on genetic susceptibility and exposure, CBD occurs in up to 20% of exposed workers [1]. Beryllium sensitisation (BeS) precedes CBD and is characterised by a positive beryllium lymphocyte proliferation test (BeLPT) [2]. BeS develops as a peripheral immune response to beryllium, but with no evidence of lung disease and/or granulomas on lung biopsy. The prevalence of BeS ranges from 2 to 20% of exposed workers [2–4]. BeS progresses to CBD at a rate of approximately 8% year−1 [5].
Once CBD CD4+ lung T-cells are activated, these cells clonally proliferate and secrete T-helper 1 (Th1)-type cytokines such as interleukin (IL)-2, interferon-γ and tumour necrosis factor (TNF)-α [6–8], which results in macrophage activation, accumulation, aggregation and the development of granulomatous inflammation [9]. Beryllium exposure stimulates a variety of cellular responses, including cell migration [10], cytokine regulation [11] and growth inhibition [12]. These immunomodulatory activities support the notion that beryllium triggers an innate immune response in non-CBD individuals as well as an acquired immune response.
The role of other genes and the regulation of their expression in BeS and CBD is unknown, and only limited beryllium-related studies have been conducted to date [13, 14]. Genome-wide expression patterns have been studied in sarcoidosis, a disease of unknown aetiology that manifests as noncaseating granulomas, predominantly in the lungs [15–18]. As CBD and sarcoidosis share clinical, radiological and histological similarities, we hypothesised that CBD would share common pathogenetic pathways and disease-specific patterns with sarcoidosis.
Our goal was to investigate the gene expression profile of CBD peripheral blood mononuclear cells (PBMCs) to define genes and patterns relevant to CBD and that overlap with sarcoidosis, which would provide insights into disease pathogenesis and assist in clinical phenotyping.
Materials and methods
Study population and design
All participants gave informed written consent in accordance with the Declaration of Helsinki and the study was approved by National Jewish Health Institutional Review Board for Human Subjects (HS#2435). 20 CBD patients, 20 BeS patients and 20 controls (Con), including 10 beryllium-exposed nondiseased controls (ND-Con, beryllium-exposed controls without CBD or BeS) and 10 nonexposed nondiseased controls (NE-Con, nonexposed controls without CBD or BeS), were enrolled in this study (figure 1). Attempts were made to frequency-match the controls and cases by age and gender.
FIGURE 1.
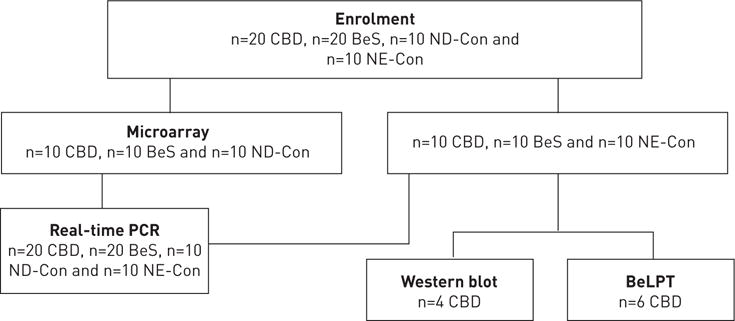
Flow diagram and study utilisation of the subjects enrolled in the study. CBD; chronic beryllium disease; BeS: beryllium sensitisation; ND-Con: beryllium-exposed nondiseased controls; NE-Con: nonexposed nondiseased controls; BeLPT: beryllium lymphocyte proliferation test.
A two-stage study design was undertaken with 10 subjects per group enrolled in the first phase. Those transcripts associated with CBD versus BeS and controls were validated in a second cohort of 20 subjects per group, including the initial 10 subjects per group and 10 new subjects per group, using high-throughput real-time PCR.
Methods
Disease diagnosis and exclusion
The diagnosis of CBD was established using previously defined criteria, including a history of beryllium exposure, the presence of granulomatous inflammation on lung biopsy, and a positive proliferative response of blood and/or bronchoalveolar lavage (BAL) cells to BeSO4 in vitro. The diagnosis of BeS was established based on a history of beryllium exposure, a positive proliferative response of PBMCs to BeSO4 in vitro on the BeLPT and the absence of granulomatous inflammation or other abnormalities on lung biopsy. We excluded subjects being treated for CBD with systemic immunosuppressants as this may impact gene expression. Some were treated with inhaled corticosteroids. We excluded subjects if they used tobacco (smoking or otherwise) in the past 6 months.
PBMC collection and beryllium treatment
Blood samples were drawn and PBMCs were isolated from heparinised blood by Ficoll-Hypaque separation. PBMCs (1×106 mL−1) were stimulated with 100 μM BeSO4 or medium alone and cultured for 24 h [19].
RNA preparation
Total RNA was extracted from PBMCs with an RNeasy Mini Kit according to the manufacturer’s protocol (Qiagen).
Probe preparation and hybridisation
Probe preparation and hybridisation were performed at the Genomics and Microarray Core at the University of Colorado Denver using a GeneChip 1.0 ST array (Affymetrix, High Wycombe, UK).
High-throughput real-time PCR
High-throughput real-time PCR was performed to verify the candidate genes using a TaqMan™ OpenArray© NT Cycler System (Applied Biosystems, Foster City, CA, USA) [20].
Western blotting
PBMCs were exposed to 100 μM BeSO4 for 0, 1, 2, 6 and 24 h, and then harvested for assay [21].
Beryllium lymphocyte proliferation test
Cells were incubated in the absence or presence of 1 μM TG101348 for 30 min and treated without or with 10 or 100 μM BeSO4 for 4 days. Proliferation was measured as described elsewhere [19].
Statistical analysis
Data (apart from microarray experiments) were analysed by one-way ANOVA, followed by Scheffe’s test for post hoc comparisons. The statistical analysis was performed using commercially available software (Statview version 5.0.1; SAS Institute, Cary, NC, USA). A p-value <0.05 was taken as statistically significant and all tests were two-sided.
Microarray analysis
The intensity probes of the microarray samples were log2-transformed, and expression normalisation and estimation were applied to the samples using the robust multiarray analysis method to adjust for experimental variability using R version 2.13.0 (https://cran.r-project.org). Microarray samples were grouped by CBD-stimulated/CBD-unstimulated, BeS-stimulated/BeS-unstimulated and Con-stimulated/Con-unstimulated. We adjusted for multiple comparisons using the Benjamini–Hochberg procedure. For each of the outcomes, we first rank-ordered the differentially expressed genes according to the p-value, then evaluated those genes that were upregulated or downregulated by >1.5-fold following the analysis of expression intensity [22–24]. The nominal significance level of each univariate test was p≤0.05 and the confidence level of the false discovery rate (FDR) was 90%.
Comparison of blood transcriptome with sarcoidosis
The genome expression levels for each comparison based on adjusted p-values were compared with the differentially expressed transcripts from patients with sarcoidosis as compared with control from KOTH et al. [17], which were downloaded from Gene Expression Omnibus (GEO). (http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS4587). Cases were confirmed with biopsy.
Comparison of lung tissue transcriptome with sarcoidosis
The expression levels for each comparison based on adjusted p-value were compared with the 386 differentially expressed transcripts in the analysis performed by CROUSER et al. [18], which were downloaded from GEO (http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS3580). Cases were confirmed with biopsy.
Gene set enrichment analysis was performed as described elsewhere [25, 26] using web-based software (http://www.broad.mit.edu/gsea/).
Functional classification and pathway analyses were evaluated with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis in the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatic Resources (http://david.abcc.ncifcrf.gov) [27].
Pathway analysis and network generation for overlapping genes in CBD and sarcoidosis were performed with Ingenuity Pathways Analysis software (http://www.ingenuity.com).
Results
Patient demographics
The demographics of the subjects are shown in table 1. No difference was seen in the age of the BeS and CBD patients enrolled in this study. The majority of both subject groups were male, typical of beryllium industries. No significant difference in the estimated duration from first beryllium exposure was observed between BeS and CBD subjects. CBD subjects had a statistically significant increase in the percentage of BAL lymphocytes compared with BeS patients (17.9±13.0 versus 7.6±5.9, p<0.01). The clinical characteristics of patients with sarcoidosis are noted in online supplementary table S1a. On average, lung function was similar between CBD and sarcoidosis cases. Sarcoidosis cases from KOTH et al. [17] had normal and low lung function, and were on systemic therapy and prednisone (online supplementary table S1a). Sarcoidosis cases in KOTH et al. [17] and our CBD cases showed similar usage of inhaled corticosteroids.
TABLE 1.
Clinical characteristics of control, beryllium sensitisation (BeS) and chronic beryllium disease (CBD) subjects
| Controls
|
BeS | CBD | ||
|---|---|---|---|---|
| ND-Con | NE-Con | |||
| Subjects | 10 | 10 | 20 | 20 |
| Age years | 55.5 (50–62) | 62 (55–73) | 56 (39–68) | 62.5 (50–81) |
| Male/female | 8/2 | 8/2 | 17/3 | 16/4 |
| Race White/African-American/other | 9/0/1 | 10/0/0 | 19/0/1 | 19/0/1 |
| Duration since first exposure years | 27.8±14.6 | 24.7±7.8 | ||
| Smoking status current/ex/never | 0/1/9 | 0/0/10 | 0/3/17 | 0/5/15 |
| FEV1 L | 2.9±0.74 | 2.71±0.57 | ||
| FEV1 % pred | 83.47±16.63 | 84.80±17.46 | ||
| FVC L | 3.78±0.89 | 3.51±0.74 | ||
| FVC % pred | 83.11±12.58 | 82.85±16.67 | ||
| TLC L | 6.69±1.46 | 6.47±1.15 | ||
| TLC % pred | 105.42±14.42 | 106.30±17.78 | ||
| DLCO | 26.41±7.03 | 24.41±9.23 | ||
| DLCO % pred | 79.84±18.53 | 76.63±23.04 | ||
| Inhaled corticosteroids | 0 (0) | 6 (30)# | ||
| BAL cells | ||||
| WBC count ×106 | 36.9±22.1 | 43.1±28.4 | ||
| Lymphocytes % | 7.6±5.9 | 17.9±13.0# | ||
Data are presented as n, median (range), mean±SE or n (%). ND-Con: beryllium-exposed nondiseased control group; NE-Con: nonexposed nondiseased control group; FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; TLC: total lung capacity; DLCO: diffusing capacity of the lung for carbon monoxide; BAL: bronchoalveolar lavage; WBC: white blood cell.
p<0.01.
Gene expression profiles demonstrate a prominent response from CBD compared with BeS, and similarities between BeS and control PBMCs
Our analyses among the three groups of PBMCs (CBD-stimulated, BeS-stimulated and Con-stimulated) revealed 472 significant genes (FDR <10% and p<0.05) (online supplementary table S1). Comparing CBD-stimulated versus BeS-stimulated PBMCs revealed 797 significant genes, including 661 upregulated and 136 downregulated genes. Only 47 genes were differentially expressed >1.5-fold, with 42 upregulated and five downregulated (online supplementary table S1b and c). We observed 735 significant differentially expressed genes between CBD-stimulated and Con-stimulated PBMCs. Interestingly, 543 genes were upregulated and 191 were downregulated, with 237 differentially expressed >1.5-fold (233 upregulated and four downregulated genes; online supplementary table S2a and b). The CBD-stimulated PBMCs demonstrated significantly higher expression levels of cytokine/chemokine genes than Con-stimulated PBMCs, including CXCL10, CXCL9, CCL8, CCL3, TNF, STAT2 and SOCS3. Only three cytokine/chemokine genes (CCL3, CCL3L1 and SOCS1) were significantly higher in CBD-stimulated versus BeS-stimulated PBMCs.
Comparing BeS-stimulated with Con-stimulated PBMCs, 37 genes were differentially expressed >1.5-fold, with 18 upregulated and 19 downregulated genes (data not shown). Both CBD-stimulated and BeS-stimulated PBMCs demonstrated a Th1 immune response with increased STAT1, TNF, IRF1, IRF4, IRF8 and ICAM1 expression. Figure 2 shows gene expression patterns based on KEGG pathways for CBD-stimulated versus Con-stimulated and BeS-stimulated PBMCs (FDR <5% and p<0.001). The CBD-stimulated gene expression was strikingly distinct from BeS-stimulated and Con-stimulated gene expression, with BeS-stimulated most similar to Con-stimulated. In our study, we did not find activation of the IL-17 pathways, which was confirmed by our T-cell immunophenotype study in CBD (data under review). We have characterised the expression of Th17 from the lungs and peripheral blood of patients with CBD in our study. There was no significant increase in IL-17+ CD4+ T-cells detected on the memory T-cells in CBD, which is consistent with our current gene expression data.
FIGURE 2.
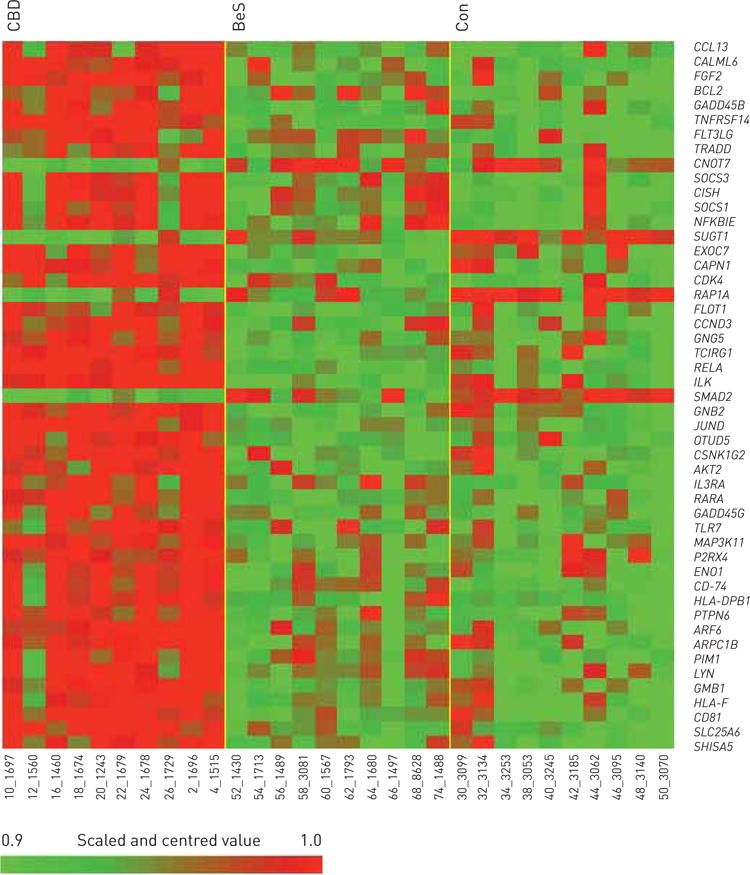
Heatmap representing expressing profiles of beryllium-stimulated beryllium sensitisation (BeS), chronic beryllium disease (CBD) and control (Con) peripheral blood mononuclear cells determined by microarray analysis. Red indicates increased expression; green indicates decreased expression relative to the nonfailing population mean. Gradients indicate the level of gene expression change over time. Data range before thresholding: −2.7 to 2.9.
Comparing the CBD-unstimulated with BeS-unstimulated PBMCs, only nine genes (three upregulated and six downregulated) were differentially expressed >1.5-fold (online supplementary table S3a and b). There were 55 genes (27 upregulated and 28 downregulated) differentially expressed >1.5-fold in the CBD-unstimulated versus Con-unstimulated PBMCs, while no genes were differentially expressed >1.5-fold in BeS-unstimulated versus Con-unstimulated PBMCs (online supplementary table S4a and b).
Analysis of gene sets and functional analyses revealed new and previously identified candidate genes in CBD-stimulated versus Con-stimulated PBMCs
Based on the findings above, 161 functional gene sets were identified, representing 117 molecular functions or 766 biological processes, comparing CBD-stimulated with Con-stimulated PBMCs. GO analysis revealed that these genes are mainly involved in functional or biological processes related to leukocyte-mediated cytotoxicity, T-cell-mediated cytotoxicity, cell killing, the cytokine-mediated signalling pathway and regulation and antigen/protein/translation factor activity nucleic acid/major histocompatibility complex (MHC) protein binding (figure 3). Using DAVID to compare CBD-stimulated with Con-stimulated expression levels, we found more immune system processes, immune responses, defence responses, responses to stress, inflammation responses, and responses to external stimulus, wounding, other organisms, viruses and bacteria (p<0.0001). The most significant of 18 enriched KEGG pathways was the JAK (Janus kinase)–STAT (signal transducer and activator of transcription) pathway, containing 15 genes (p=0.000003; table 2).
FIGURE 3.
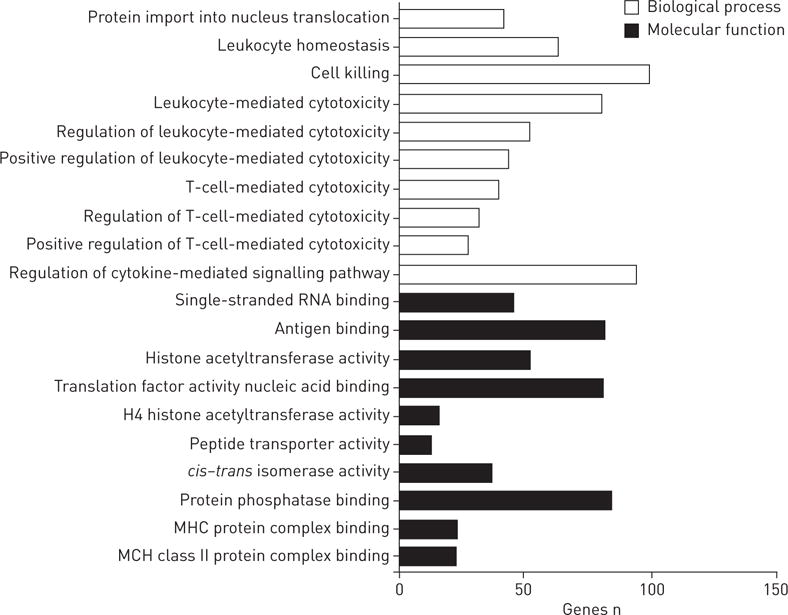
Gene Ontology category analysis related to the candidate genes found to differ between the beryllium-stimulated chronic beryllium disease and control groups. MHC: major histocompatibility complex. These 161 functional gene sets were identified to represent a total of 117 molecular functions or 766 biological processes. The top 10 biological processes and the top 10 molecular functions are shown.
TABLE 2.
Pathways identified to be altered in beryllium-stimulated chronic beryllium disease peripheral blood mononuclear cells (PBMCs) compared with beryllium-stimulated control PBMCs
| KEGG term | Genes | Pathway fold enrichment | False discovery rate | p-value |
|---|---|---|---|---|
| JAK–STAT signalling pathway | PTPN6, IL2RA, SOCS2, SOCS3, SOCS1, PIM1, BCL2L1, STAT1, CISH, STAT2, CCND2, CSF2RB, IL15RA, SPRED1, IL3RA | 4.599 | 0.003 | 0.000003 |
| Cytokine–cytokine receptor interaction | CCL3, TNF, CCL2, IL2RA, CCR1, CXCL9, CCL8, CD40, CXCL11, CXCL10, INHBA, TNFSF10, TNFSF13B, CCL3L1, CCL3L3, CSF2RB, IL15RA, IL3RA | 3.084 | 0.097 | 0.000087 |
| Asthma | TNF, FCER1G, HLA-DPB1, CD40, HLA-DOB, HLA-DRA | 9.832 | 0.318 | 0.000285 |
| Antigen processing and presentation | PSME1, PSME2, TAP2, TAP1, HLA-DPB1, HLA-DOB, CD74, TAPBP, HLA-DRA | 5.153 | 0.329 | 0.000295 |
| Intestinal immune network for IgA production | TNFSF13B, CD80, IL15RA, HLA-DPB1, CD40, HLA-DOB, HLA-DRA | 6.789 | 0.534 | 0.000481 |
| Allograft rejection | TNF, CD80, HLA-DPB1, CD40, HLA-DOB, HLA-DRA | 7.921 | 0.895 | 0.000807 |
| Toll-like receptor signalling pathway | CCL3, TNF, CD80, CXCL9, CD40, CXCL11, STAT1, TLR7, CXCL10 | 4.235 | 1.231 | 0.001111 |
| Chemokine signalling pathway | CCL3, CCL2, LYN, NCF1, CCR1, NCF1C, CXCL9, CCL8, STAT1, CXCL11, CXCL10, STAT2, CCL3L1, CCL3L3 | 3.050 | 1.801 | 0.001630 |
| Viral myocarditis | ICAM1, CD80, HLA-DPB1, CD40, ABL2, HLA-DOB, HLA-DRA | 4.685 | 3.725 | 0.003402 |
| Systemic lupus erythematosus | TNF, CD80, FCGR1C, FCGR1A, HLA-DPB1, CD40, HLA-DOB, TRIM21, HLA-DRA | 3.840 | 4.721 | 0.004331 |
| B-cell receptor signalling pathway | PTPN6, DAPP1, LYN, NFKBIE, LILRB3, CD81, PIK3AP1 | 4.436 | 4.872 | 0.004473 |
| Cell adhesion molecules | ICAM1, CD80, CD274, HLA-DPB1, CD40, SDC4, HLA-DOB, PDCD1LG2, HLA-DRA | 3.240 | 6.432 | 0.005950 |
| Graft-versus-host disease | TNF, CD80, HLA-DPB1, HLA-DOB, HLA-DRA | 6.093 | 8.946 | 0.008377 |
| Apoptosis | TNFSF10, TNF, BCL2, CSF2RB, BCL2L1, IL3RA, TRADD | 3.824 | 9.764 | 0.009179 |
| Type I diabetes mellitus | TNF, CD80, HLA-DPB1, HLA-DOB, HLA-DRA | 5.658 | 11.464 | 0.010870 |
| Proteasome | PSMB10, PSME1, PSME2, PSMB8, PSMB9 | 5.056 | 16.452 | 0.016005 |
| Autoimmune thyroid disease | CD80, HLA-DPB1, CD40, HLA-DOB, HLA-DRA | 4.659 | 21.105 | 0.021053 |
| NOD-like receptor signalling pathway | TNF, CCL2, MEFV, CCL8, RIPK2 | 3.833 | 36.208 | 0.039548 |
KEGG: Kyoto Encyclopedia of Genes and Genomes; JAK: Janus kinase; STAT: signal transducer and activator of transcription; IgA: immunoglobulin A; NOD: nucleotide-binding oligomerisation domain.
CBD and sarcoidosis share transcriptional profiles, pathways and networks
The top differentially expressed genes in CBD and sarcoidosis were compared by Entrez gene number, with 287 differentially expressed genes shared between CBD and sarcoidosis PBMCs, including 271 upregulated and 16 downregulated genes (online supplementary table S5a and b). The top regulators were NFKBIE, ICAM1 and LILRB4, while the top pathways were cytokine–cytokine receptor interactions, and Toll-like receptor, chemokine and JAK–STAT signalling pathways. There were 14 genes regulated in opposite directions (online supplementary table S5c and d). Evaluating these differentially expressed genes/pathways in CBD and sarcoidosis revealed overlapping networks, involved in the inflammatory response, immune response, lipid metabolism and molecular transport. The top overlapping networks (figure 4) included STAT, CISH, SOCS1, SOCS2, SOCS3, TNF and others. The STAT transcription factors serve as a hub for molecular connections.
FIGURE 4.
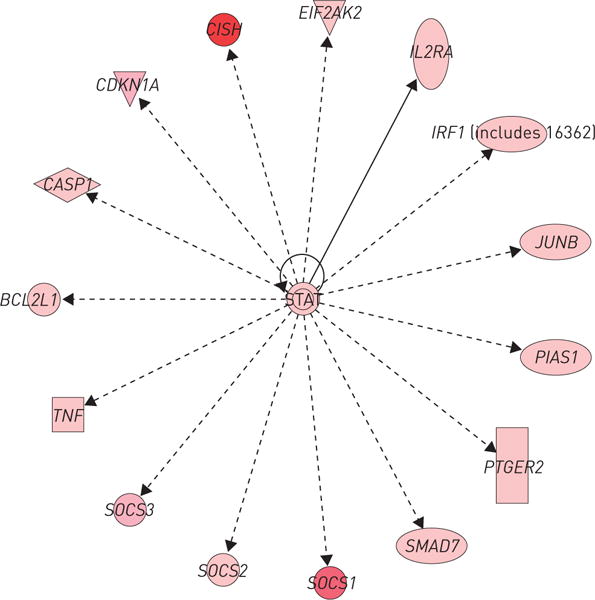
Ingenuity Pathway Analysis of the top overlapping networks in chronic beryllium disease and sarcoidosis. The network is displayed graphically as nodes (genes/gene products) and edges (biological relationship between nodes). The node colour intensity indicates the fold change expression of genes (red representing upregulation).
Furthermore, we examined differentially expressed genes from sarcoidosis lung tissue in our CBD profiles. We found that 33 differentially expressed genes overlapped in CBD PBMCs and sarcoidosis lung, which included 13 upregulated genes (including CXCL9, FAM26F, TAP1, CXCL11, GBP4, CCL8 and STAT1) and one downregulated gene. There were 19 genes regulated in opposite directions (online supplementary table S5e–h).
The top differentially expressed candidate genes were validated in a separate set of CBD, BeS and control groups
We confirmed almost 450 of the 526 genes that we found above using real-time PCR. The genes with the highest fold change included CXCL9, HCAR3, HCAR2, CD274, CLEC6A and EGR2, which were statistically significantly different between CBD-stimulated and Con-stimulated groups (figure 5). Some samples had similar values, resulting in the overlapping symbols in the “Con” groups (e.g. figure 5a). As above, BeS subjects were not statistically different from CBD or controls, but displayed a more variable response than controls.
FIGURE 5.
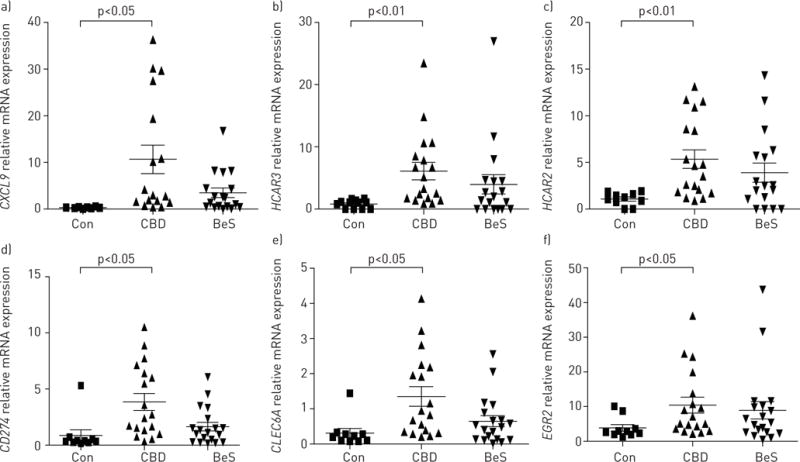
Validation of the top chronic beryllium disease (CBD) candidate genes using the OpenArray system: a) CXCL9, b) HCAR3, c) HCAR2, d) CD274, e) CLEC6A and f) EGR2. Con: control-stimulated group; CBD: CBD-stimulated group; BeS: beryllium sensitisation-stimulated group. These top six genes overexpressed in CBD are shown as representative of our findings for our validation. Each symbol represents an individual subject; bars show mean±SEM.
The JAK2–STAT1 pathway is functional in CBD
To assess the functional role of the top pathway and genes, we evaluated JAK2 protein phosphorylation in four CBD PBMCs by Western blotting. Expression of phospho (p)-JAK2/p-STAT1 was increased after 1 h of beryllium stimulation, while SOCS3 was increased after 6 h (figure 6a). The JAK2 inhibitor TG101348 significantly decreased the proliferation of PBMCs in the BeLPT after both 10 and 100 μM beryllium treatment (n=6), suggesting that the JAK pathway and JAK2 gene contribute to ongoing inflammation in the lung in response to beryllium (figure 6b).
FIGURE 6.
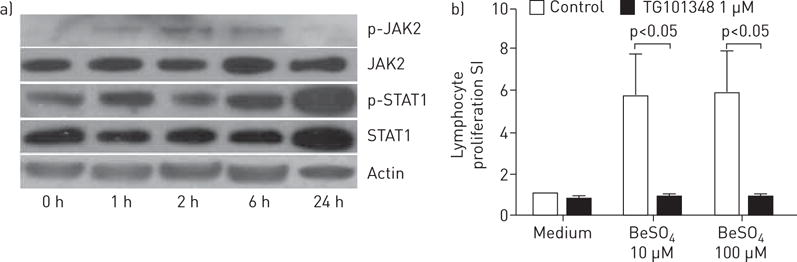
a) Janus kinase (JAK)2/signal transducer and activator of transcription (STAT)1 phosphorylation was stimulated by beryllium in peripheral blood mononuclear cells (PBMCs) from chronic beryllium disease (CBD) patients. PBMCs were treated with 100 μM BeSO4 for the indicated period of time and cell lysates containing equal amounts of total proteins were analysed by Western blot using antibodies against phospho (p)-JAK2, total JAK2, p-STAT1, STAT1 and actin. Total JAK2 and STAT1 served as a loading control. b) The JAK2 inhibitor TG101348 downregulated lymphocyte proliferation in CBD (n=6). PBMCs were incubated in the absence (“Medium”) or presence of 1 μM TG101348 for 30 min, and treated with without or with 10 and 100 μM BeSO4 for 4 days. Data are presented as mean±SEM stimulation index (SI).
Discussion
In this study, we showed that BeS and CBD demonstrate a Th1 genomic transcriptional profile, with cytokines and chemokines at higher levels in CBD than controls. Specifically, the genes differentially expressed in CBD were mainly involved in functional or biological processes related to leukocyte-mediated cytotoxicity, T-cell-mediated cytotoxicity, cell killing, the cytokine-mediated signalling pathway and their regulation. The most significant pathway was the JAK–STAT pathway, which is important in cytokine regulation. Not surprisingly, the top genes associated with CBD included cytokine/chemokine and signalling genes, such as CXCL10, CXCL9, TNF, STAT1, STAT2 and SOCS3. Furthermore, these genes/pathways were apparent in another granulomatous lung disease, i.e. sarcoidosis, with almost 300 genes concordantly regulated in both diseases. Finally, we confirmed the functional aspects of JAK2, demonstrating that altering these genes impacts beryllium-stimulated cytokine production and cell proliferation. The data provide important information regarding the pathogenesis of CBD and, with similar findings noted in sarcoidosis, have implications for other exposure-mediated immunological diseases.
We identified enrichment of genes involved in the JAK–STAT, cytokine–cytokine receptor interaction, antigen processing and presentation, Toll-like receptor, and NOD (nucleotide-binding oligomerisation domain)-like receptor signalling pathways as well as genes and pathways not previously known to be involved in CBD, such as SOCS3 and STAT1. Abnormal activation of the JAK–STAT pathway has been implicated in autoimmune and inflammatory diseases, such as rheumatoid arthritis [28, 29], systemic lupus erythematosus [30] and Crohn’s disease [31], and other granulomatous lung diseases, including tuberculosis [32] and sarcoidosis [32, 33]. Furthermore, the JAK2 inhibitor TG101348 decreased beryllium-induced proliferation in CBD, suggesting that the JAK pathway and the JAK2 gene contribute to the ongoing inflammation in the lung in response to beryllium (figure 6b). Inhibition of JAK genes has been proposed as treatment for autoimmune inflammation, with JAK inhibitors among the first successful agents reaching clinical application [34, 35]. Tofacitinib (tasocitinib), a JAK3 inhibitor, is used for psoriasis, while TG101348, a JAK2 inhibitor, is in phase 1 trial for myelofibrosis [36]. Our results suggest the potential of the JAK–STAT pathway as a target for immunomodulatory therapy in CBD and potentially other granulomatous lung diseases.
Moreover, our data identify a CBD blood transcriptional signature including TNF-α and cytokine/chemokine genes, which are concordantly regulated in blood and likely contribute to BeS and CBD pathogenesis. TNF-α is a potent pro-inflammatory cytokine that is essential for the formation and maintenance of granulomas in tuberculosis and sarcoidosis [37–40]. MAIER et al. [41] identified CBD patients with increased TNF-α levels associated with worse clinical markers of disease severity. The TNF-α*02 allele in the promoter region was a functional polymorphism that correlated with CBD severity [42]. Our study indicates that a CBD blood transcriptional signature is a powerful tool to: 1) gain insight into different pathways involved in the pathogenesis of CBD that can be studied further on a molecular level; 2) define potential therapeutic targets based on additional study; and 3) explore differentially expressed genes such as CCL3, CCL3L1 and SOCS1 that may be important in BeS progression.
The expression profiles of beryllium-stimulated BeS are more similar to controls than to CBD (figure 2), which is consistent with findings from our previous studies [43]. While BeS individuals demonstrate an immune response to beryllium based on the BeLPT, they do not show a significant Th1 immune response in the blood or lung. In addition, this is likely a spectrum with beryllium-exposed workers (normal healthy controls) developing sensitisation, some of whom are likely to progress to CBD, or may even be in the early stages of this transition.
Among the GO terms related to biological process, regulation of leukocyte- or T-cell-mediated cytotoxicity was present. Almost all of the genes included in the term encoded MHC class I molecules, such as HLA-A, -B, -C, -E, -F and -G, which are involved in the presentation of antigen to CD8+ cytotoxic T-cells in CBD. This evidence suggests that CD8+ T-cells may also play a role in the development of CBD, although this has not been evaluated to date. We did not find increases in Th17 genes, which is not surprising. We have conducted a T-cell immunophenotype study in CBD (data under review), and characterised the expression of Th17 from the lungs and peripheral blood of patients with CBD; we found no significant increase in IL-17+ CD4+ T-cells detected on the memory T-cells in CBD, which is consistent with our current gene expression data.
Evaluation of differentially expressed genes/pathways shared by CBD and sarcoidosis showed that the top overlapping networks were involved in inflammation, immune responses, lipid metabolism and molecular transport. Of the almost 300 differentially expressed genes shared between CBD and sarcoidosis, STAT transcription factors (STAT1 and STAT2) serve as a hub on which biologically important molecular connections concentrate [44]. Interestingly, in another study, sarcoidosis lymph node granulomas expressed abundant STAT1 and p-STAT1, suggesting its relevance in sarcoidosis [45]. While we found STAT1 gene overexpression in PBMCs, we also found it increased in alveolar macrophages (data not shown). In addition, STAT1-regulated chemokines, i.e. CXCL9, CXCL10, CXCL11 and CXCL16, were markedly upregulated in CBD and sarcoidosis patients (online supplementary table S5a), suggesting the importance of STAT1 in these diseases. STAT1 may also be a potential novel target for pharmacotherapy of chronic granulomatous disease. Furthermore, the only gene upregulated in sarcoidosis but downregulated in CBD was RNASE3, encoding for a ribonuclease which exhibits antimicrobial activity against pathogenic bacteria. Sarcoidosis may be triggered by the immune system responding to foreign substances, such as viruses, bacteria or chemicals, which maybe a reason for the upregulated RNASE3. It is possible that the differences found between CBD and sarcoidosis could be due to differences in the diseases themselves (as we postulate for the findings for RNASE3 above), differences in the compartment studied (PBMCs versus lung tissue), the use of beryllium stimulation, and/or differences in aetiology and/or study design.
In addition to comparing CBD PBMCs with those of sarcoidosis PBMCs, we compared the PBMC transcriptional profiling with lung gene expression and found a number of differentially expressed genes between the two. Most of the concomitantly regulated genes were related to the immune response (e.g. CCL8, CXCL9, CXCL11, TAP1, STAT1, CXCL9 and CXCL11). These genes have been implicated in granuloma formation in primates [46]. Interestingly, there were 19 genes which were divergent between these two diseases (online supplementary table S5g–f). Genes that were upregulated in sarcoidosis but downregulated in CBD were related to cellular macromolecule metabolic processes (e.g. FYB, PSIP1, SH2D1A, CCNG2, IFNGR1 and LPAR6). The only gene downregulated in sarcoidosis but upregulated in CBD was RHBDD2, which functions in signalling pathways related to cell proliferation/survival in breast cancer [47]. These differences could be related to the two different compartments tested, as some genes are known to be upregulated in the lung or blood and decreased in the other [17]. Some of these differences could be due to differences in disease severity, although on average the lung function was similar between groups, or differences in treatment (as KOTH et al. [17] included individuals on treatment). Alternatively, these findings might suggest that the two diseases demonstrate some difference in aspects of the immune response and/or proliferation, which should be evaluated further. Furthermore, our blood experiments with beryllium stimulation reveal higher gene expression levels than some of the sarcoidosis blood studies to date. In addition, we only obtained transbronchial tissue for CBD on diagnosis, which is not sufficient to evaluate gene expression. We have planned studies to obtain BAL cell gene expression beyond the scope of this study.
In summary, our study shows that BeS and CBD PBMCs demonstrate differentially expressed profiles relevant to immune function and pathogenesis. In addition, CBD shares pathogenic genes/pathways with sarcoidosis. This study fills a crucial gap in our current knowledge, allowing for the identification of new regulatory genes and networks that dictate CBD and/or perpetuation of BeS. This important translational information will likely have implications for other granulomatous and immune-mediated diseases. Most importantly, this study provides the groundwork for further studies to explore biomarkers and investigate the function of the genes and networks discovered herein, ultimately increasing our understanding of the pathogenesis of CBD and other similar diseases.
Supplementary Material
Acknowledgments
Support statement: Supported by NIH P01 ES11810-06A1, K12 HL090147-01, K01 ES020857-01 and 1 UL1 TR001082 from NCRR/NIH. Funding information for this article has been deposited with FundRef.
Footnotes
This article has supplementary material available from erj.ersjournals.com
Conflict of interest: Disclosures can be found alongside this article at erj.ersjournals.com
References
- 1.Henneberger PK, Cumro D, Deubner DD, et al. Beryllium sensitization and disease among long-term and short-term workers in a beryllium ceramics plant. Int Arch Occup Environ Health. 2001;74:167–176. doi: 10.1007/s004200100237. [DOI] [PubMed] [Google Scholar]
- 2.Newman L, Mroz MM, Balkissoon R, et al. Beryllium sensitization progresses to chronic beryllium disease: a longitudinal study of disease risk. Am J Respir Crit Care Med. 2005;171:54–60. doi: 10.1164/rccm.200402-190OC. [DOI] [PubMed] [Google Scholar]
- 3.Saltini C, Richeldi L, Losi M, et al. Major histocompatibility locus genetic markers of beryllium sensitization and disease. Eur Respir J. 2001;18:677–684. doi: 10.1183/09031936.01.00106201. [DOI] [PubMed] [Google Scholar]
- 4.Infante P, Newman L. Beryllium exposure and chronic beryllium disease. Lancet. 2004;363:415–416. doi: 10.1016/S0140-6736(04)15523-2. [DOI] [PubMed] [Google Scholar]
- 5.Henneberger P, Miller W, Doney B, et al. Industries in the United States with airborne beryllium exposure and estimates of the number of current workers potentially exposed. J Occup Environ Hyg. 2004;1:648–659. doi: 10.1080/15459620490502233. [DOI] [PubMed] [Google Scholar]
- 6.Maier L, Mroz M, Rossman M, et al. Infliximab therapy modulates an antigen-specific immune response in chronic beryllium disease. Respir Med. 2012;106:1810–1813. doi: 10.1016/j.rmed.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontenot A, Canavera S, Gharavi L, et al. Target organ localization of memory CD4+ T cells in patients with chronic beryllium disease. J Clin Invest. 2002;110:1473–1482. doi: 10.1172/JCI15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maier L, Sawyer R, Tinkle S, et al. IL-4 fails to regulate in vitro beryllium-induced cytokines in berylliosis. Eur Respir J. 2001;17:403–415. doi: 10.1183/09031936.01.17304030. [DOI] [PubMed] [Google Scholar]
- 9.Fontenot A, Palmer B, Sullivan A, et al. Frequency of beryllium-specific, central memory CD4+ T cells in blood determines proliferative response. J Clin Invest. 2005;115:2886–2893. doi: 10.1172/JCI24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer R, Doherty D, Schumacher B, et al. Beryllium-stimulated in vitro migration of peripheral blood lymphocytes. Toxicology. 1999;138:155–163. doi: 10.1016/s0300-483x(99)00101-8. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary A, Sauer N, Gupta G. Beryllium-specific immune response in primary cells from healthy individuals. Toxicology. 2004;201:9–19. doi: 10.1016/j.tox.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Lehnert N, Gary R, Marrone B, et al. Inhibition of normal human lung fibroblast growth by beryllium. Toxicology. 2001;160:119–127. doi: 10.1016/s0300-483x(00)00439-x. [DOI] [PubMed] [Google Scholar]
- 13.Gaede K, Mamat U, Schlaak M, et al. Analysis of differentially regulated mRNAs in peripheral blood monocytes of berylliosis patients after figure stimulation. J Mol Med. 2000;78:293–299. doi: 10.1007/s001090000105. [DOI] [PubMed] [Google Scholar]
- 14.Hong-Geller E, Pardington PE, Cary RB, et al. Chemokine regulation in response to beryllium exposure in human peripheral blood mononuclear and dendritic cells. Toxicology. 2006;218:216–228. doi: 10.1016/j.tox.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Zhou T, Zhang W, Sweiss N, et al. Peripheral blood gene expression as a novel genomic biomarker in complicated sarcoidosis. PLoS One. 2012;7:e44818. doi: 10.1371/journal.pone.0044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su R, Li M, Bhakta N, et al. Longitudinal analysis of sarcoidosis blood transcriptomic signatures and disease outcomes. Eur Respir J. 2014;44:985–993. doi: 10.1183/09031936.00039714. [DOI] [PubMed] [Google Scholar]
- 17.Koth L, Solberg O, Peng J, et al. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am J Respir Crit Care Med. 2011;184:1153–1163. doi: 10.1164/rccm.201106-1143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crouser E, Culver D, Knox K, et al. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am J Respir Crit Care Med. 2009;179:929–938. doi: 10.1164/rccm.200803-490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Hamzeh N, Gillespie M, et al. Beryllium increases the CD14dimCD16+ subset in the lung of chronic beryllium disease. PLoS One. 2015;10:e0117276. doi: 10.1371/journal.pone.0117276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forreryd A, Johansson H, Albrekt AS, et al. Evaluation of high throughput gene expression platforms using a genomic biomarker signature for prediction of skin sensitization. BMC Genomics. 2014;15:379. doi: 10.1186/1471-2164-15-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Zheng H, Gillespie M, et al. p38 mitogen-activated protein kinase in beryllium-induced dendritic cell activation. Human Immunol. 2014;75:1155–1162. doi: 10.1016/j.humimm.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry R, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 23.Li C, Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:0032.1–0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha V, Lindgren C, Eriksson K, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Malemud C, Miller A. Pro-inflammatory cytokine-induced SAPK/MAPK and JAK/STAT in rheumatoid arthritis and the new anti-depression drugs. Expert Opin Ther Targets. 2008;12:171–183. doi: 10.1517/14728222.12.2.171. [DOI] [PubMed] [Google Scholar]
- 29.Walker JG, Ahern MJ, Coleman M, et al. Expression of Jak3, STAT1, STAT4, and STAT6 in inflammatory arthritis: unique Jak3 and STAT4 expression in dendritic cells in seropositive rheumatoid arthritis. Ann Rheum Dis. 2006;65:149–156. doi: 10.1136/ard.2005.037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivashkiv L. Type I interferon modulation of cellular responses to cytokines and infectious pathogens: potential role in SLE pathogenesis. Autoimmunity. 2003;36:473–479. doi: 10.1080/08916930310001605882. [DOI] [PubMed] [Google Scholar]
- 31.Glas J. Evidence for STAT4 as a common autoimmune gene: rs7574865 is associated with colonic Crohn’s disease and early disease onset. PLoS One. 5:e10373. doi: 10.1371/journal.pone.0010373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maertzdorf J. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A. 109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou T, et al. Peripheral blood gene expression as a novel genomic biomarker in complicated sarcoidosis. PLoS One. 7:e44818. doi: 10.1371/journal.pone.0044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mesa R, Yasothan U, Kirkpatrick P. Ruxolitinib. Nat Rev Drug Discov. 2012;11:103–104. doi: 10.1038/nrd3652. [DOI] [PubMed] [Google Scholar]
- 35.Vaddi K, Sarlis N, Gupta V. Ruxolitinib, an oral JAK1 and JAK2 inhibitor, in myelofibrosis. Expert Opin Pharmacother. 2012;13:2397–2407. doi: 10.1517/14656566.2012.732998. [DOI] [PubMed] [Google Scholar]
- 36.Pardanani A, Gotlib JR, Jamieson C, et al. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. J Clin Oncol. 2011;29:789–796. doi: 10.1200/JCO.2010.32.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagata K, Maruyama K, Uno K, et al. Simultaneous analysis of multiple cytokines in the vitreous of patients with sarcoid uveitis. Invest Ophthalmol Vis Sci. 2012;53:3827–3833. doi: 10.1167/iovs.11-9244. [DOI] [PubMed] [Google Scholar]
- 38.Ziegenhagen M, Muller-Quernheim J. The cytokine network in sarcoidosis and its clinical relevance. J Intern Med. 2003;253:18–30. doi: 10.1046/j.1365-2796.2003.01074.x. [DOI] [PubMed] [Google Scholar]
- 39.Plessner H, Lin PL, Kohno T, et al. Neutralization of tumor necrosis factor (TNF) by antibody but not TNF receptor fusion molecule exacerbates chronic murine tuberculosis. J Infect Dis. 2007;195:1643–1650. doi: 10.1086/517519. [DOI] [PubMed] [Google Scholar]
- 40.Lin P, Myers A, Smith L, et al. Tumor necrosis factor neutralization results in disseminated disease in acute and latent Mycobacterium tuberculosis infection with normal granuloma structure in a cynomolgus macaque model. Arthritis Rheum. 2010;62:340–350. doi: 10.1002/art.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maier L, Sawyer R, Bauer R, et al. High beryllium-stimulated TNF-α is associated with the −308 TNF-α promoter polymorphism and with clinical severity in chronic beryllium disease. Am J Respir Crit Care Med. 2001;164:1192–1199. doi: 10.1164/ajrccm.164.7.2012123. [DOI] [PubMed] [Google Scholar]
- 42.Sato H, Silveira L, Fingerlin T, et al. TNF polymorphism and bronchoalveolar lavage cell TNF-alpha levels in chronic beryllium disease and beryllium sensitization. J Allergy Clin Immunol. 2007;119:687–696. doi: 10.1016/j.jaci.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Martin AK, Mack DG, Falta MT, et al. Beryllium-specific CD4+ T cells in blood as a biomarker of disease progression. J Allergy Clin Immunol. 2011;128:1100–1106. doi: 10.1016/j.jaci.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brierley M, Fish E. Stats: multifaceted regulators of transcription. J Interferon Cytokine Res. 2005;25:733–744. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- 45.Rosenbaum J, Pasadhika S, Crouser E, et al. Hypothesis: sarcoidosis is a STAT1-mediated disease. Clin Immunol. 2009;132:174–183. doi: 10.1016/j.clim.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuller C, Flynn J, Reinhart T. In situ study of abundant expression of proinflammatory chemokines and cytokines in pulmonary granulomas that develop in cynomolgus macaques experimentally infected with Mycobacterium tuberculosis. Infect Immun. 2003;71:7023–7034. doi: 10.1128/IAI.71.12.7023-7034.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abba M, Lacunza E, Nunez M, et al. Rhomboid domain containing 2 (RHBDD2): a novel cancer-related gene over-expressed in breast cancer. Biochim Biophys Acta. 2009;1792:988–997. doi: 10.1016/j.bbadis.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


