Biosynthetic pathways for the terpenes DMNT and TMTT in maize (Zea mays) and their corresponding enzymes were identified by nested association mapping of herbivore-induced volatile terpenes.
Abstract
Plant volatiles not only have multiple defense functions against herbivores, fungi, and bacteria, but also have been implicated in signaling within the plant and toward other organisms. Elucidating the function of individual plant volatiles will require more knowledge of their biosynthesis and regulation in response to external stimuli. By exploiting the variation of herbivore-induced volatiles among 26 maize (Zea mays) inbred lines, we conducted a nested association mapping and genome-wide association study (GWAS) to identify a set of quantitative trait loci (QTLs) for investigating the pathways of volatile terpene production. The most significant identified QTL affects the emission of (E)-nerolidol, linalool, and the two homoterpenes (E)-3,8-dimethyl-1,4,7-nonatriene (DMNT) and (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT). GWAS associated a single nucleotide polymorphism in the promoter of the gene encoding the terpene synthase TPS2 with this QTL. Biochemical characterization of TPS2 verified that this plastid-localized enzyme forms linalool, (E)-nerolidol, and (E,E)-geranyllinalool. The subsequent conversion of (E)-nerolidol into DMNT maps to a P450 monooxygenase, CYP92C5, which is capable of converting nerolidol into DMNT by oxidative degradation. A QTL influencing TMTT accumulation corresponds to a similar monooxygenase, CYP92C6, which is specific for the conversion of (E,E)-geranyllinalool to TMTT. The DMNT biosynthetic pathway and both monooxygenases are distinct from those previously characterized for DMNT and TMTT synthesis in Arabidopsis thaliana, suggesting independent evolution of these enzymatic activities.
INTRODUCTION
Plant resistance against bacteria, fungi, nematodes, insects, and other herbivores depends to a large extent on defense-related metabolites that are produced in response to an attack (Harborne, 1991). With advances in analytical methods over the last three decades, an increasing number of volatile metabolites with important roles in plant defense have been identified. Many such volatiles were assigned functions in the defense against pathogens like lepidopteran larvae, aphids, fungi, and bacteria (Pichersky et al., 2006; Unsicker et al., 2009; McCormick et al., 2012). In addition, plant volatiles have been implicated in signaling within the plant, the coordination of defense responses, and interplant signaling (Yoneya and Takabayashi, 2013; Heil, 2014).
A better understanding of the major roles that volatiles have in plant defense should provide valuable strategies for controlling pests in cultivated crops (Degenhardt et al., 2003; Pickett et al., 2014). In maize (Zea mays), damage by lepidopteran larvae such as Spodoptera littoralis induces a complex volatile blend that attracts parasitic wasps (Turlings et al., 1990, 1991; Turlings and Tumlinson, 1991). These wasps are natural enemies of the lepidopteran larvae and can ultimately reduce damage to the plant (Hoballah et al., 2004). In field experiments, maize engineered to emit the volatile sesquiterpene (E)-β-caryophyllene displayed a higher resistance to western maize rootworm (Diabrotica virgifera virgifera), an important maize pest. Cocultivation of maize with an African grass (Melinis minutiflora) that releases abundant volatile compounds led to a significant reduction in damage by lepidopteran larvae as a result of increased parasitism by braconid wasps (Khan et al., 1997, 2000). Important volatiles in this cocultivation farming system are the C11 homoterpene (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT) and the corresponding C16 homolog (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT) (Tamiru et al., 2011). These homoterpenes are emitted by many angiosperms and gymnosperms in response to attack by both herbivores and pathogens (Su et al., 2009; Mumm and Dicke, 2010; Tholl and Lee, 2011). The functions of DMNT and TMTT in plant defense also have been characterized in lima bean (Phaseolus lunatus). A volatile blend with DMNT and TMTT is emitted after spider mite (Tetranychus urticae) attack and promotes the attraction of predatory mites (Dicke et al., 1990; De Boer et al., 2004; Kappers et al., 2005). In this tritrophic interaction, the presence of TMTT in the volatile blend can enable the predatory mite to select plants that are infested with suitable prey (De Boer et al., 2004). In Arabidopsis thaliana, homoterpenes are involved in the direct repulsion of aphids (Bruce et al., 2008).
Plant terpenes are formed from 5-carbon units that are fused to provide the 10-carbon (C10) geranyl diphosphate (GDP), the C15 farnesyl diphosphate (FDP), or the C20 geranylgeranyl diphosphate (GGDP), which are the precursors of mono-, sesqui-, and diterpenes, respectively. The biosynthesis of homoterpenes must proceed through the addition or elimination of carbons to form the C11 and C16 skeletons of DMNT and TMTT. Experiments with stable isotopes suggested that the formation of DMNT starts with C15 FDP and proceeds via the tertiary sesquiterpene alcohol (E)-nerolidol to form the C11 homoterpene DMNT after an oxidative degradation. Similarly, the C20 GGDP is converted to the tertiary diterpene alcohol (E,E)-geranyllinalool and subsequently oxidized to the C16 homoterpene TMTT (Donath and Boland, 1995; Boland et al., 1998). The latter pathway was identified in Arabidopsis, where a geranyllinalool synthase and a P450 enzyme (CYP82G1) catalyze the two respective steps in the biosynthesis of TMTT (Herde et al., 2008; Lee et al., 2010).
The corresponding formation of DMNT via (E)-nerolidol was not observed in Arabidopsis (Lee et al., 2010). Instead, DMNT is produced in the Arabidopsis roots via degradation of the triterpene arabidiol (Sohrabi et al., 2015). Although (E)-nerolidol synthases have been identified in many other plant species (Degenhardt et al., 2009), no enzymes converting this intermediate into DMNT have been found in planta. Geranyllinalool synthases with similarity to Arabidopsis geranyllinalool synthase were described in Nicotiana attenuata (NaGLS) and tomato (Solanum lycopersicum; SlGLS) (Falara et al., 2014). These enzymes convert GGDP more efficiently than FDP in vitro and their transcript levels correlate with the formation of TMTT (SlGLS) and hydroxylinalool glycosides (NaGLS). Two geranyllinallool synthases from Fabaceae, PlTPS2 from P. lunatus (Brillada et al., 2013) and MtTPS3 from Medicago truncatula (Gomez et al., 2005; Arimura et al., 2008), belong to a separate clade of terpene synthases possibly involved in volatile terpene production. These enzymes are expressed in the plastid and convert GDP and FDP with higher in vitro efficiencies than GGDP.
To elucidate the pathways of homoterpene biosynthesis in maize and identify the enzymes involved in the formation of these volatiles, we exploited the high variation in volatiles produced that occurs among maize inbred lines (Degen et al., 2004). The nested association mapping (NAM) design utilizes both ancient and recent recombination events to compensate for population structure but retains high mapping resolution (Yu et al. 2008). The United States NAM (US-NAM) maize population was constructed by crossing one common parental line (B73) with 25 diverse maize inbred lines to produce almost 6000 recombinant inbred line families (Yu et al., 2008; McMullen et al., 2009). This population has been successfully employed to map complex traits like quantitative resistance to insects and pathogens (Buckler et al., 2009; Poland et al., 2011; Tian et al., 2011; Cook et al., 2012; Meihls et al., 2013) and regulation of carotenoid biosynthesis (Owens et al., 2014).
In this study, we used the US-NAM population to map pathways and enzymes contributing to volatile terpene biosynthesis in maize leaves. Our results demonstrate that the C11 homoterpene DMNT is formed by the oxidative degradation of (E)-nerolidol. Similarly, TMTT is produced via oxidative degradation of (E,E)-geranyllinalool. The first step of both pathways, the formation of the tertiary terpene alcohols (E,E)-geranyllinalool and (E)-nerolidol, is catalyzed by the terpene synthase, TPS2, whereas the subsequent oxidative degradation to DMNT and TMTT is catalyzed by two specific P450 monooxygenases, CYP92C5 and CYP92C6.
RESULTS
Identification of QTLs for Production of Maize Volatiles
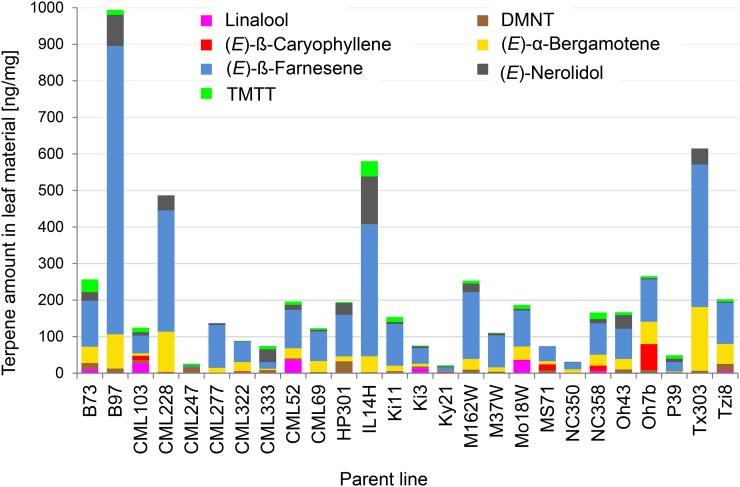
Maize volatiles released after herbivore damage display high levels of quantitative and qualitative variation (Degen et al., 2004). To utilize this diversity of terpene volatiles for quantitative trait locus (QTL) mapping, we characterized the volatile profiles of the 26 parental lines of the US-NAM population (Yu et al., 2008). As expected, the volatile profiles vary in both the quantity and composition of their terpene components (Figure 1). We continued to screen all 5982 RILs that comprise the US-NAM population. The hexane-soluble fraction of leaves was analyzed after treatment with an indanoyl isoleucine conjugate, which is a structural analog of the jasmonate isoleucine conjugate that mimics elicitation by herbivory (Schüler et al., 2001). The quantities of the terpene volatiles linalool, nerolidol, DMNT, and TMTT were mapped through joint-linkage analysis (Yu et al., 2008) using 1106 common parent-specific markers that were genotyped on all of the US-NAM RILs (Figure 2). The traits linalool, (E)-nerolidol, and DMNT were associated most significantly with the QTL606 (marker PZA02525.1, P values are 1.15E−04, 2.00E−09, and 1.91E−10, respectively), while TMTT correlated strongly with the QTL607 (marker PZA03677.1, P-value 1.09E−13). Combination of the four volatile traits resulted in a highly significant QTL at position 607 (P value 6.03−18), suggesting that a common locus is responsible for the formation of these volatiles. Candidate genes located in this region of chromosome 5 encode two putative terpene synthase genes, which were designated tps2 (at position 71.1 Mb, GRMZM2G046615 in AGPv3) and tps3 (at position 71.4 Mb, GRMZM2G064406 in AGPv3). Terpene synthases convert prenyl diphosphates into the basic carbon skeletons of terpenes and are therefore likely to be involved in the biosynthesis of the identified volatiles. The two terpene synthase genes share 92% amino acid sequence similarity and contain characteristic sequence motifs, including a DDxxD-motif (Starks et al., 1997; Degenhardt et al., 2009).
Figure 1.
Diversity of Volatile Terpene Production among the 26 Parent Lines of the NAM Population after Elicitor Induction.
Mono-, sesqui-, and diterpenes were extracted with hexane from each of 8 to 10 individual plants and were then analyzed by GC-MS as described in Methods. The diversity of the most common volatile terpenes is shown.
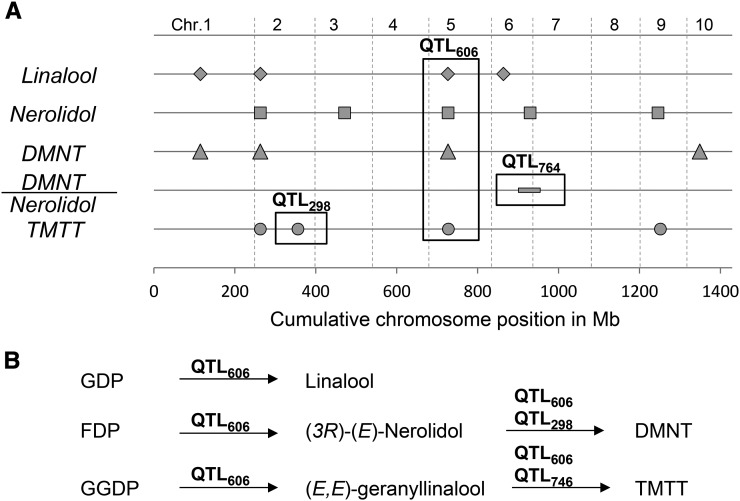
Figure 2.
Mapping of QTLs Involved in Plant Volatile Production.
(A) The production of linalool (diamonds), (E)-nerolidol (squares), DMNT (triangles), TMTT (circles), and the ratio of DMNT to (E)-nerolidol (rectangle) were determined across a set of 5982 RILs of the NAM population and mapped against 1106 common parent-specific markers. The cumulative chromosome positions of the major QTLs are indicated on the x axis. Prominent QTLs are highlighted with boxes and are described in Results.
(B) Suggested functions of the QTLs in the proposed biosynthetic pathways of these compounds. QTL606 is associated with a terpene synthase activity able to convert GDP, FDP, and GGDP to the respective tertiary alcohols. QTL298 and QTL764 are associated with an oxidase activity that forms DMNT and TMTT, respectively, depending on the substrate.
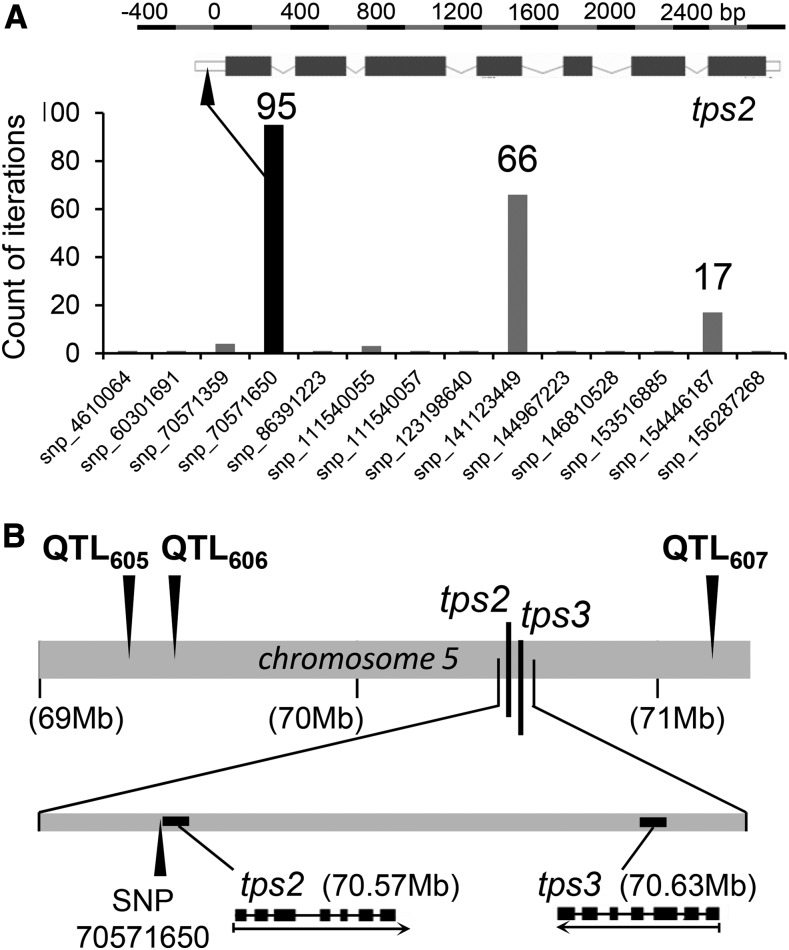
To provide further information about the loci corresponding to these two QTLs, we conducted a genome-wide association study (GWAS) using 1.6 million single nucleotide polymorphisms (SNPs) genotyped for the 26 founder lines that were projected on the US-NAM RILs (HapMap.v1 markers) (Gore et al., 2009). This procedure identified SNP70571650 as the most significant marker (resampling model inclusion probability [RMIP] = 0.95) located between QTL606 and QTL607 (Figure 3A). This SNP is located directly 5′ of tps2 and is only 160 bp upstream of the start codon (Figures 3A and 3B). The location of tps3 is ∼64.3kb downstream in a region covered by 15 SNPs that showed no significant association with the QTL. These analyses suggest that TPS2 might be responsible for the volatile products linalool, nerolidol, DMNT, and TMTT. Thus, this putative terpene synthase was predicted to convert GDP and FDP into their respective tertiary terpene alcohols: linalool and nerolidol. Because QTL606/607 also affects the trait DMNT, nerolidol is a likely intermediate in the formation of DMNT. In addition, QTL606/607 is strongly associated with TMTT production. This suggests that TPS2 also converts GGDP into a geranyllinalool intermediate, which is a substrate for TMTT formation. Unfortunately, the diterpene alcohol geranyllinalool only has low hexane solubility and could not be measured as a volatile trait in samples from the mapping population.
Figure 3.
Mapping of QTL606 for Nerolidol Production in a GWAS Using 1.6 Million SNP Markers.
(A) The bar graph presents the counts of iterations by GWAS for each of the SNPs located near QTL606. The counts of iterations are shown above the most prominent bars. The SNP with the most significant correlation to QTL606 (indicated by a black bar) is positioned in the promoter region of tps2 (arrow). Shown above is the pattern of exons (boxes) and introns (lines) of tps2 with the number of nucleotides from the translation start.
(B) Location of tps2, tps3, and the most relevant QTLs in a section of chromosome 5 (wide gray line). A close-up of this region (thin gray line) shows the position of SNP70571650 in relation to tps2 and tps3. For both tps2 and tps3, the respective pattern of exons (black boxes) and introns (black lines) is pictured below.
TPS2 Is an Active Mono-, Sesqui-, and Diterpene Synthase
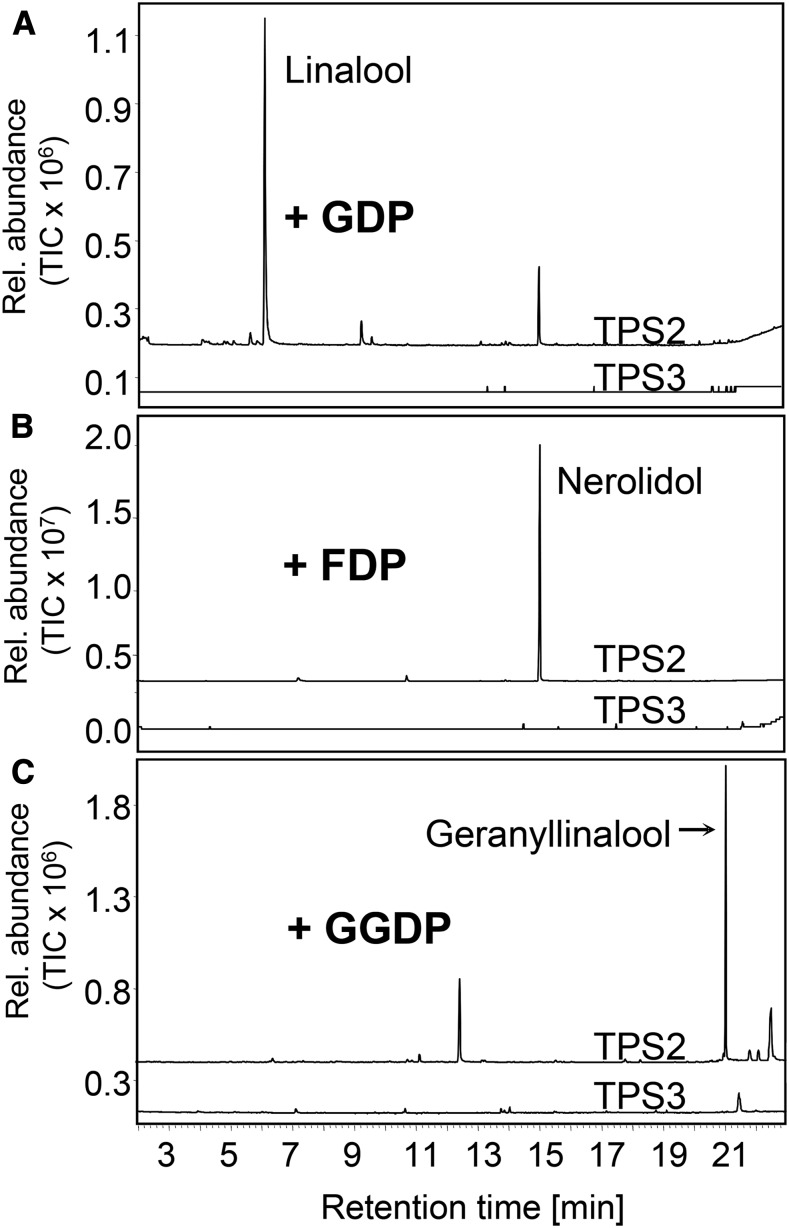
Characterization of QTL606 led us to hypothesize that terpene synthase TPS2, or less likely TPS3, is responsible for the production of volatile mono-, sesqui-, and diterpene alcohols. The open reading frames of both genes were cloned, and the enzymes were expressed in an Escherichia coli system to allow biochemical characterization in vitro. Both proteins were expressed at similar levels without the putative 21-amino acid signal peptide and were soluble (Supplemental Figures 1C and 1D). Whereas TPS2 converted GDP, the substrate for monoterpenes, into the terpene alcohol linalool, no activity was detected for TPS3 (Figure 4A). Also, TPS2 converted the sesquiterpene substrate FDP to nerolidol and the diterpene substrate GGDP to geranyllinalool (Figures 4B and 4C). Km values for the reactions with GDP, FDP, and GGDP were 16.9 μM (±6.1 μM), 18 μM (±7.8 μM), and 25 μM (±12.6 μM), respectively. These values are within the range expected for terpene synthases (Degenhardt and Gershenzon, 2000; Köllner et al., 2008) and indicate that the enzyme is functional in planta, as indicated by the QTL analysis. Again, no activity was detectable for TPS3. Stereospecific analysis of the TPS2 products on a chiral gas chromatography-mass spectrometry (GC-MS) column revealed that the enzyme only forms the S-isomers of the three tertiary terpene alcohols (Supplemental Figure 2).
Figure 4.
The Terpene Synthase TPS2 Encodes an Active Enzyme in the Inbred Line B73.
TPS2 and TPS3 were produced in E. coli, extracted, partially purified, and incubated with the substrates. The resulting terpene products were collected with a polydimethylsiloxane-coated SPME fiber and analyzed by GC-MS. The terpene synthase TPS2 produces the tertiary alcohols linalool from GDP (A), nerolidol from FDP (B), and geranyllinalool from GGDP (C). TPS3 formed no detectable products with any tested substrate. Products were identified by GC-MS. The products were identified by comparison of their retention times and mass spectra with those of authentic standards.
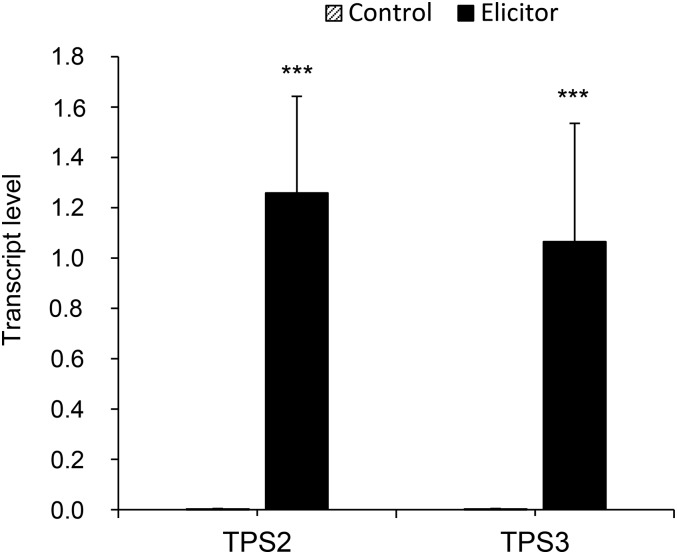
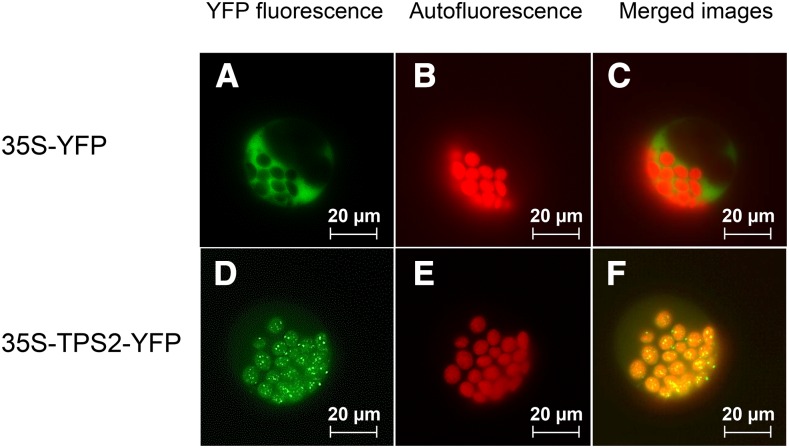
If TPS2 is involved in the formation of herbivore-induced compounds, its expression is likely to be induced by herbivory. To test this, we analyzed tps2 and tps3 transcript levels in leaves of the inbred line B73. Undamaged control plants showed very low expression of tps2 and tps3 (Figure 5). To induce volatile production, the leaves were wounded and treated with an indanone elicitor that mimics herbivory (Schüler et al., 2001). This elicitor treatment was used for the NAM population induction experiment because it is more reproducible than actual insect herbivory. After 24 h, a time of high volatile release, tps2 and tps3 both displayed elevated transcript levels (Figure 5). Treatment of maize seedlings with caterpillar oral secretions, in combination with wounding, resulted in a very similar induction of tps2 transcripts (Supplemental Figure 3). Transient expression of a TPS2-YFP construct in maize protoplasts localized the protein in globular structures within the plastids (Figure 6).
Figure 5.
Expression of the Terpene Synthases TPS2 and TPS3 Is Induced by Simulated Herbivory.
Leaves of 14-d-old plants of the inbred line B73 were wounded and treated with indanone derivative (elicitor) for 24 h or left undamaged (control). Levels of transcripts encoding TPS2 and TPS3 were determined relative to the APT1 reference gene. One-way ANOVA was used to calculate the significance based on three plants per treatment. Error bars represent se. Triple asterisks represent significant difference from the control at P < 0.01.
Figure 6.
Terpene Synthase TPS2 Is Localized in the Chloroplasts of Maize Mesophyll Protoplasts.
Images of maize mesophyll protoplasts transiently expressing 35S-YFP ([A] to [C]) or 35S-TPS2-YFP ([D] to [F]) showing YFP fluorescence ([A] and [D]), chlorophyll autofluorescence ([B] and [E]), and merged images ([C] and [F]). Bars = 20 µm.
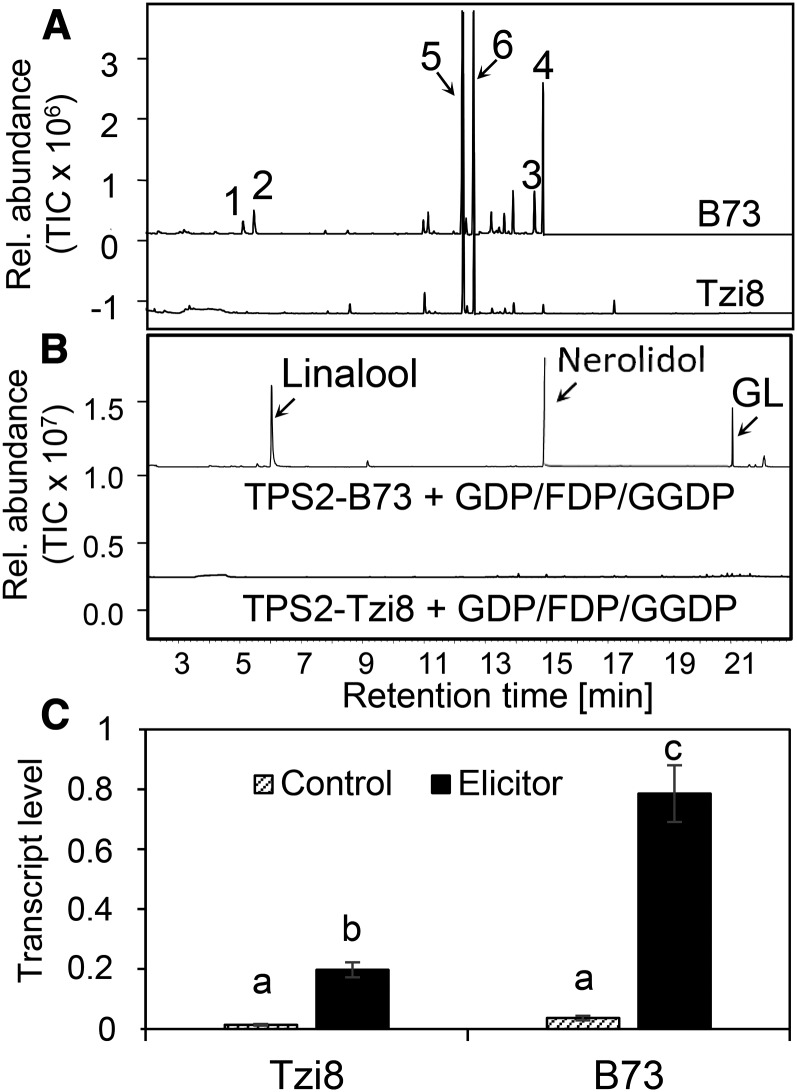
The Lack of Volatile Terpene Alcohol Production in the Parental Line Tzi8 Correlates with an Inactive tps2 Allele
To verify the association between QTL606 and tps2, we characterized a NAM parental line that contributes to the significance of the QTL. For the identification of such parental lines, we conducted comparative interval mapping with individual sets of recombinant inbred lines. The parental line Tzi8, which produces very low or undetectable amounts of linalool, nerolidol, DMNT, and TMTT (Figure 7A; Supplemental Figure 4), contributes to the significance of QTL606. In addition, the most significant SNP located in the promoter region of tps2, SNP70571650, is present in Tzi8. To determine whether the tps2-Tzi8 allele is inactive and thereby responsible for the Tzi8 phenotype, the gene was cloned and expressed in a bacterial system for biochemical characterization in vitro. Although the protein was soluble and expressed at levels similar to that of tps2-B73 (Supplemental Figure 1B), no activity could be observed with the FDP, GDP, or GGDP substrates (Figure 7B). Compared with the tps2-B73 allele, tps2-Tzi8 is altered at six amino acid positions in functionally important domains of the enzyme.
Figure 7.
TPS2 Is Inactive in the Inbred Line Tzi8.
(A) Volatiles found in leaves of the inbred lines B73 and Tzi8 after induction with indanone elicitor. The volatiles were analyzed by GC-MS. The major terpene compounds were identified as 1-linalool, 2-DMNT, 3-(E)-nerolidol, 4-TMTT, 5-(E)-α-bergamotene, and 6-(E)-β-farnesene.
(B) TPS2 alleles from B73 and Tzi8 were expressed in E. coli, and protein was extracted, partially purified, and incubated with GDP/FDP/GGDP substrates. The resulting terpene products were collected with a polydimethylsiloxane-coated SPME fiber and analyzed by GC-MS.
(C) Levels of transcripts encoding TPS2 in leaves of the lines Tzi8 and B73. Leaves of 14-d-old plants were wounded and treated with indanone derivative (Elicitor) for 24 h or left undamaged (Control). Transcript abundance was determined relative to the reference gene encoding APT1. One-way ANOVA was used to calculate the significance of three plants per treatment. Error bars represent se. Different letters denote significant differences at P < 0.01.
In addition to enzyme activity, tps2 expression was compared between Tzi8 and B73. In undamaged plants, very low transcript levels were detected in both lines (Figure 7C). After wounding and indanone treatment, tps2 transcripts were significantly lower in Tzi8, ∼25% of the B73 levels. This may be caused by a 690-bp insertion in the Tzi8 promoter that is found 566 bp upstream of the start codon. This insertion changes the promoter architecture. For example, the element SEBFCONSSTPR10A is found at −583 bp in B73 and at −1261 bp in Tzi8. This element is a binding site of the silencing element binding factor (Boyle and Brisson, 2001), which has been described in the promoter of several pathogenesis-related genes. Due to the inactive TPS2-Tzi8 allele, we would not expect any DMNT formation in this inbred line. The remaining, low levels of DMNT in Tzi8 (Supplemental Figure 4) may be caused by other terpene synthases that often produce minor amounts of (E)-nerolidol or the degradation of triterpenes (Sohrabi et al., 2015). Further evidence for the crucial role of TPS2 in the formation of linalool, nerolidol, DMNT, and TMTT was provided by a tps2/3::DS transposon insertion line in the W22 background (B.S08.0585; http://acdstagging.org/). After wounding and treatment with indanone-derivative, TPS2 products were not detected at all in this transposon insertion line (Figure 8).
Figure 8.
TPS2 Products Were Not Detected in a Maize tps2/3::DS Transposon Insertion Line.
Volatiles found in of leaves after wounding and treatment with indanone derivative (elicitor) of the inbred line W22 and a line with a tps2/3::DS transposon insertion in the W22 background. The volatiles were collected with a polydimethylsiloxane-coated SPME fiber and analyzed by GC-MS. Several terpene compounds were identified as 1-linalool, 2-DMNT, 3-(E)-nerolidol, and 4-TMTT.
Joint-Linkage Analysis of the Homoterpenes DMNT and TMTT Identified Two Cytochrome P450 Genes
Mapping of QTL606, which corresponds to tps2, showed significant effects not only on the formation of TPS2 products but also on the accumulation of the homoterpenes DMNT and TMTT (Figure 2). This suggests that two of the TPS2 products, (E)-nerolidol and (E,E)-geranyllinalool, are intermediates for the production of DMNT and TMTT, respectively. To identify the enzymes catalyzing the final biosynthetic steps, we searched the NAM population for QTLs that are significant for DMNT but do not affect nerolidol production. To facilitate detecting such QTLs, we determined the ratio of DMNT to (E)-nerolidol in each of the RILs of the NAM population. This ratio was treated as an independent trait and provided the single, highly significant QTL764 (NAM marker 764; PHM3078.12; P value 1.72E−06), at 6 Mb on chromosome 7 (AGPv3). This QTL is located close to a cytochrome P450 monooxygenase gene (GRMZM2G102079) that belongs to the CYP92 family. Such monooxygenases are likely to catalyze oxidative eliminations such as the conversion of (E)-nerolidol to DMNT. A previous study in Arabidopsis also found a P450 monooxygenase catalyzing this biosynthetic step, but the enzyme, CYP82G1, has been recruited from a different P450 family (Herde et al., 2008; Lee et al., 2010). Moreover, the amino acid identity between the maize and Arabidopsis enzymes is <30%. Because we were not able to measure levels of geranyllinalool, the precursor of TMTT, in the NAM population, we could not identify a QTL for the final biosynthetic step. However, one of the significant QTLs for the production of TMTT was QTL298 (NAM marker 298; PZA02170.1). This locus at 234.8 Mb (AGPv3) on chromosome 2 showed the second highest significance ranking (P value 9.99E−12) and appeared independent of other QTLs for volatile production (Figure 2). QTL298 is also located in the vicinity of a putative monooxygenase of the CYP92 group (GRMZM2G139467). The two monooxygenases, which were linked to DMNT and TMTT synthesis, respectively, share 72% amino acid identity.
The P450 Monooxygenases CYP92C5 and CYP92C6 Convert Tertiary Monoterpene Alcohols into Homoterpenes
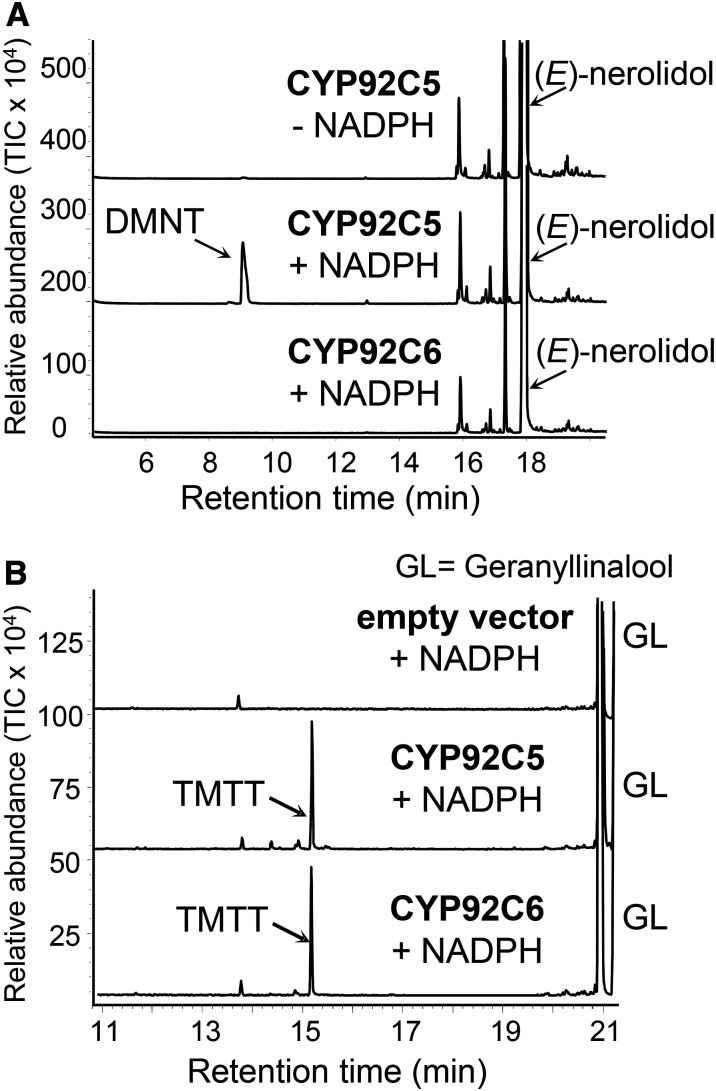
The putative P450 monooxygenase located at QTL764 for DMNT formation was named CYP92C5 and the closely related enzyme at QTL298 for TMTT was designated CYP92C6. To test whether these enzymes catalyze the reactions suggested by the US-NAM genetic mapping, they were expressed in a yeast (Saccharomyces cerevisiae) system to allow biochemical analysis in vitro. The activity assays were conducted with isolated yeast microsomes and included the substrates linalool, (E)-nerolidol, and (E,E)-geranyllinalool, as well as the cofactor NADPH, which is a common requirement for the function of P450 monooxygenases (Figure 9). Without the expressed protein and NADPH, no enzyme activity was detected. CYP92C5 converted (E)-nerolidol and (E,E)-geranyllinalool to DMNT and TMTT, respectively (Figure 9). The reaction has a Km value of 14.2 μM (±4.45 μM) for (E)-nerolidol and 5.6 μM (±1.26 μM) for (E,E)-geranyllinalool. CYP92C6 only converted (E,E)-geranyllinalool to TMTT, with a Km value of 10.4 μM (±2.6 μM). Neither of the enzymes accepted linalool as a substrate. The observed in vitro activities coincide exactly with those predicted by the QTLs. To confirm the role of both P450 monooxygenases in herbivore-induced volatile biosynthesis, we measured transcript levels in leaves that were undamaged or wounded and treated with the indanone elicitor (Figure 10). Both genes exhibited low transcript levels in undamaged leaves but were strongly induced in response to simulated herbivory.
Figure 9.
The P450 Monooxygenases CYP92C5 and CYP92C6 Convert Sesquiterpene Alcohols into Their Respective Homoterpenes.
CYP92C5 and CYP92C6 of B73 were expressed in yeast, extracted, partially purified, and incubated in the presence and absence of the substrates nerolidol (A), geranyllinalool (B), and the cosubstrate NADPH. The resulting terpene products were collected with a polydimethylsiloxane-coated SPME fiber and analyzed by GC-MS. The DMNT and TMTT products were identified by comparison of retention time and mass spectrum with those of authentic standards.
Figure 10.
Levels of Transcripts Encoding the P450 Monooxygenases CYP92C5 and CYP92C6 Are Induced by Simulated Herbivory.
Leaves of the inbred line B73 were wounded and treated with an indanone derivative elicitor (Elicitor) for 24 h or left undamaged (Control). Levels of transcripts encoding CYP92C5 (A) and CYP92C6 (B) were determined relative to the APT1 reference gene. One-way ANOVA was used to calculate the significance of three plants per treatment. Error bars represent se. Asterisks represent significant difference from the control at P < 0.01.
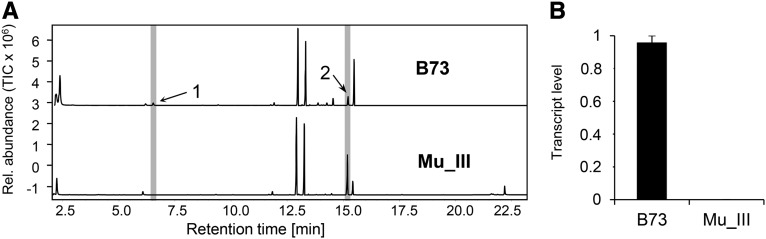
A Maize Mu Transposon Insertion into CYP92C5 Results in No DMNT Production and Reduced Levels of TMTT after Simulated Herbivory
We identified a cyp92C5 Mu transposon insertion (MuIII) in the B73 background, which results in undetectable DMNT levels not only in undamaged plants, but also in plants following induction by wounding and treatment with indanone (Figure 11A; Supplemental Figure 5). The precursor of DMNT, (E)-nerolidol, was present in higher concentrations in the volatile spectrum than in B73. This suggests that MuIII contains an active TPS2 enzyme but that the function of the P450 monooxygenase CYP92C5 responsible for the production of DMNT is compromised. TMTT abundance also was significantly reduced in MuIII, indicating that CYP92C5 also is involved in TMTT formation. To test whether cyp92C5 expression is affected by MuIII, we measured the transcript levels of this gene in induced plants. In comparison to B73, cyp92C5 transcript levels are strongly reduced (Figure 11B). Close examination of the cyp92C5 transcript in MuIII revealed that an mRNA with a length of 296 instead of 532 amino acids was detectable by RACE-PCR. This indicates that cyp92C5 inactivation affects the production of DMNT and to a lesser degree TMTT.
Figure 11.
CYP92C5 Is Inactive in the Transposon Insertion Mutant MuIII.
(A) Volatiles found in of leaves of the lines B73 and transposon insertion mutant MuIII after induction with the indanone elicitor. The volatiles were collected with a polydimethylsiloxane-coated SPME fiber and analyzed by GC-MS. The major terpene compounds that were identified are 1-DMNT, 2-(E)-nerolidol, and 3-TMTT.
(B) Levels of transcripts encoding CYP92C5 in leaves of the lines B73 and MuIII. Leaves of 14-d-old plants were wounded and treated with indanone derivate for 24 h. Transcript abundance was determined relative to the APT1 reference gene. One-way ANOVA was used to calculate the significance of three plants per treatment. Error bars represent se.
Association mapping of metabolic pathways not only characterizes the enzymes involved in a particular pathway but also provides the opportunity to determine which particular enzyme of a large gene family is responsible for catalysis in vivo. Only one terpene synthase, TPS2, produces the majority of tertiary mono-, sesqui-, and diterpene alcohols of the biosynthetic pathways studied here. The conversion of both (E)-nerolidol and (E,E)-geranyllinalool to their respective homoterpenes DMNT and TMTT is catalyzed by a cytochrome P450 monooxygenase in vitro. However, in planta DMNT was primarily produced by CYP92C5, whereas the majority of TMTT was produced by a related cytochrome P450 monooxygenase, CYP92C6, which is specific for the conversion of (E,E)-geranyllinalool to TMTT.
DISCUSSION
Association Mapping Identified the Pathways of Homoterpene Formation in Maize
We used QTL analysis of induced volatile production to characterize the biosynthetic pathways of the maize homoterpenes DMNT and TMTT. The C11 homoterpene DMNT is produced from C15 FPP via an (E)-nerolidol intermediate. Because transposon insertions in both TPS2 and CYP92C5 reduce DMNT production below detectable levels, it appears that this pathway is responsible for the bulk of DMNT formation in maize leaves. The biosynthetic pathway of DMNT presented here is consistent with previous feeding experiments utilizing deuterated nerolidol, which was converted to DMNT in nine dicotyledonous plant species (Donath and Boland, 1995). A very different pathway of DMNT biosynthesis was described for Arabidopsis roots, in which this compound is formed by degradation of the triterpene arabidiol (Sohrabi et al., 2015). In maize roots, the formation of DMNT by this or any other pathway was not observed (Köllner et al., 2008).
The analysis of QTLs for production of the C16 homoterpene TMTT identified a biosynthetic pathway similar to that of DMNT. In maize, TMTT is derived from C20 GGDP via the diterpene alcohol geranyllinalool. The same intermediate was identified in the biosynthesis of TMTT in Arabidopsis (Lee et al., 2010), indicating that this pathway is found in both mono- and dicotyledonous plants.
The QTLs most significant for DMNT, TMTT, and (E)-nerolidol biosynthesis were associated with the genes encoding the enzymes of the respective biosynthetic pathways. This indicates that the alteration of promoters or enzyme activities, rather than the modification of regulatory trans-factors, is responsible for the much of the variation of terpene metabolites within the US-NAM mapping population. Similarly, a study of eQTLs in Arabidopsis showed that most of the variation in gene expression maps to the affected genes, indicating cis-regulation rather than the effects of transcription factors or other trans-regulatory elements located elsewhere in the genome (Kliebenstein et al., 2006).
The Terpene Synthase TPS2 Is Involved in Biosynthesis of Both DMNT and TMTT
The QTL corresponding to TPS2 affected the production of several volatiles, including linalool, (E)-nerolidol, and (E,E)-geranyllinalool. This demonstrated that TPS2 not only is able to produce these compounds in vitro but also is involved in their formation in planta. Furthermore, a transposon insertion in TPS2 (Figure 8) and an inactive allele of TPS2 in line Tzi8 (Figure 7; Supplemental Figure 4) caused greatly reduced linalool, (E)-nerolidol, and (E,E)-geranyllinalool abundance. This indicates that TPS2 is involved in the production of mono-, sesqui-, and diterpenes in planta. The closely related enzymes MtTPS3 from M. truncatula (Gomez et al., 2005; Arimura et al., 2008) and PlTPS2 from P. lunatus (Brillada et al., 2013) accept the same range of substrates in vitro, but their activities in the respective plant species have not been determined. The products of TPS2 have previously been identified as some of the major herbivore-induced volatiles of maize. They are an important part of the indirect defense of maize against lepidopteran herbivores and direct defense against aphids (Degenhardt and Gershenzon, 2000; Pickett et al., 2014). The conversion of the three respective substrates, GDP, FDP, and GGDP, is thought to proceed by a classical carbocationic mechanism (Degenhardt and Gershenzon, 2000; Schnee et al., 2002).
The products formed by the plastidic TPS2 in planta are determined in part by the availability of its substrates. The methylerythritol phosphate pathway in plastids provides the enzyme with the precursors GDP for the formation of linalool and GGDP for the formation of geranyllinalool. The substrate FDP for (E)-nerolidol biosynthesis is commonly provided by the mevalonate pathway in the cytoplasm. Although some studies have indicated an interchange of precursors of terpene biosynthesis between the two compartments (Adam et al., 1999; Lichtenthaler, 1999; Hemmerlin et al., 2003), the import of FDP into the plastid was described only in a study of spinach chloroplasts (Bick and Lange, 2003). However, the analyses of QTLs and transposon insertion mutants in maize provide unambiguous evidence that TPS2 has access to all three substrates, GDP, FDP, and GGDP. Another indication for plastidic FDP production is found in the closely related monocot, rice (Oryza sativa), where the farnesyl diphosphate synthase FPPS1 is localized in the plastid (Sanmiya et al., 1999). In maize, a family of three farnesyl diphosphate synthases was characterized. One member of the maize farnesyl diphosphate synthases, FPPS3, is responsible for herbivore-induced production of FDP but has not been localized within the cell (Richter et al., 2015). Further studies will be required to determine whether FDP is available in plastids or whether TPS2 is localized also in the cytoplasm of some of the cells within the leaf tissue. Since genetic analyses of volatile terpene biosynthesis are not yet available in plants other than maize, we do not know whether production of sesquiterpene alcohols in plastids is a common phenomenon.
Maize Homoterpenes Are Formed by a Terpene Synthase of the TPS-g Subfamily
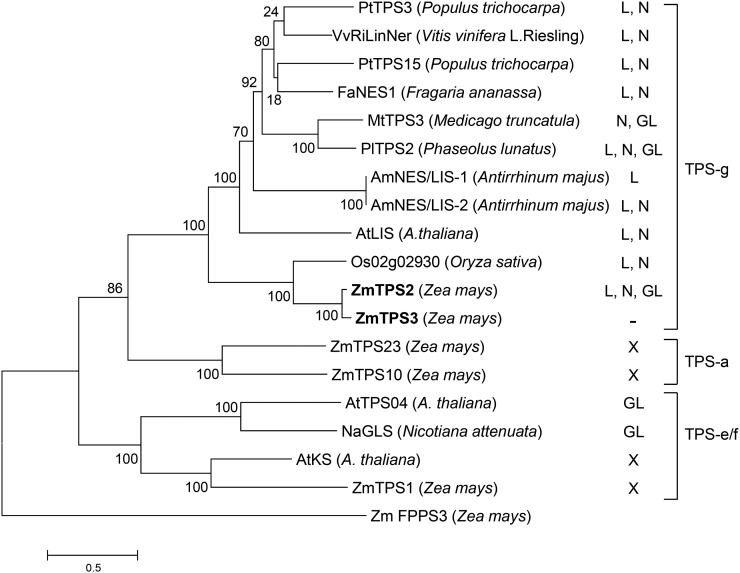
The amino acid sequence of TPS2 displays a high similarity to the terpene synthases of the TPS-g subfamily (Figure 12). Enzymes in this group were characterized as class I terpene synthases (Tholl and Lee, 2011) and include MtTPS3 from M. truncatula (Gomez et al., 2005; Arimura et al., 2008), PlTPS2 from P. lunatus (Brillada et al., 2013), FaNES1 from Fragaria ananassa (Aharoni et al., 2004), and Os02g022930 from rice (Yuan et al., 2008). Each of these enzymes produces one or more of the tertiary terpene alcohols linalool, (E)-nerolidol, and geranyllinalool. The legume enzymes MtTPS3 and PlTPS2 were less efficient in producing geranyllinalool and better at making linalool (PlTPS2) or nerolidol (MtTPS3) (Arimura et al., 2008; Brillada et al., 2013). However, maize TPS2 displayed similar Km values for the reactions with GDP, FDP, and GGDP. As geranyllinalool synthases were previously described only in dicots, it needs to be determined whether the similar acceptance of the three substrates is a feature common to enzymes of the TPS-g subgroup in monocots.
Figure 12.
Dendrogram Analysis of Maize TPS2 and TPS3 with Closely Related Linalool/Nerolidol/Geranyllinalool Synthases from Other Plant Species.
The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis includes TPS2 (NP_001105854.1) and TPS3 (NP_001105855.1), shown in bold; PtTPS3 (XM_006377207.1); MtTPS3 (AY766249); VvRiLinNer (JQ062931); FaNES1 (CAD57083); PtTPS15 (KF776502); PlTPS2 (KC012520); AtLIS (AAO85533); AmNES/LIS-1 (EF433761); AmNES/LIS-2 (ABR24418); Os02g02930 (Q6ZH94); ZmTPS23 (EU259634.1); ZmTPS10 (NM_001112380.2); AtTPS04 (AA085540); NaGLS (KJ755868); AtKS (NP_178064); and ZmTPS1 (NM_001111627.1). The FPP synthase ZmFPPS3 (BT085025) was used as an outgroup. Products of each terpene synthase are indicated by letters for linalool (L), (E)-nerolidol (N), (E,E)-geranyllinalool (GL), others (X), or inactivity (–).
Terpene alcohols also are formed by members of the TPS-e/f subgroup of terpene synthases, which differ from TPS2 in their domain structure and intracellular location (Figure 12). The geranyllinalool synthases of Arabidopsis (Herde et al., 2008) and two members of the Solanaceae, N. attenuata and tomato (Falara et al., 2014), do not contain a predicted signaling peptide and, in the case of Arabidopsis TPS04, were localized in both the cytoplasm and endoplasmic reticulum (Herde et al., 2008). Inactivation of TPS04 in Arabidopsis demonstrated that this gene is responsible for most of the TMTT formation in planta (Herde et al., 2008). A terpene synthase of the TPS-e/f subgroup was also identified in maize. Maize TPS1 produces linalool and (E)-nerolidol, but no geranyllinalool after heterologous expression in vitro (Degenhardt and Gershenzon, 2000). However, TPS1 is unlikely to have a function in volatile production because no linalool and (E)-nerolidol were detected in plants with inactive tps2 alleles. In addition, no QTL for the corresponding volatiles was observed at the location of TPS1 on chromosome 1. Third, the tps1 transcript levels did not correlate with the formation of linalool, (E)-nerolidol, or DMNT in some maize lines. Instead, TPS1 was shown to have an additional diterpene synthase activity producing ent-kaurene, a precursor for the nonvolatile gibberellins and other diterpenes (Fu et al., 2015). Taken together, the findings in maize indicate that only TPS2, an enzyme of the TPS-g subtype, is responsible for the formation of volatile homoterpenes in maize.
While geranyllinalool-producing enzymes of the TPS-e/f type have been identified in Arabidopsis, Solanum lycopersicum, and N. attenuata, no such enzymes of the TPS-g subtype have been identified in these plants (Falara et al., 2014). This suggests that Tps-e/f subtype enzymes are responsible for homoterpene production in dicots, while monocots may have recruited enzymes of the TPS-g subtype for this task. As a consequence, TMTT may originate most prominently from the mevalonate pathway in dicots, while monocots such as maize may generate both homoterpenes from the methylerythritol phosphate pathway of the plastid. The terpene synthases of the TPS-g subtype from maize and the TPS-e/f subtype from Arabidopsis belong to rather distant clades that may have separated during the early development of angiosperms or are the result of convergent evolution (Tholl and Lee, 2011; Falara et al., 2014). The characterization of more terpene synthases with a role in homoterpene formation will provide a better insight in the compartmentation of these biosynthetic pathways in monocots and dicots.
Two P450 Enzymes Are Responsible for the Emission of DMNT and TMTT in Maize
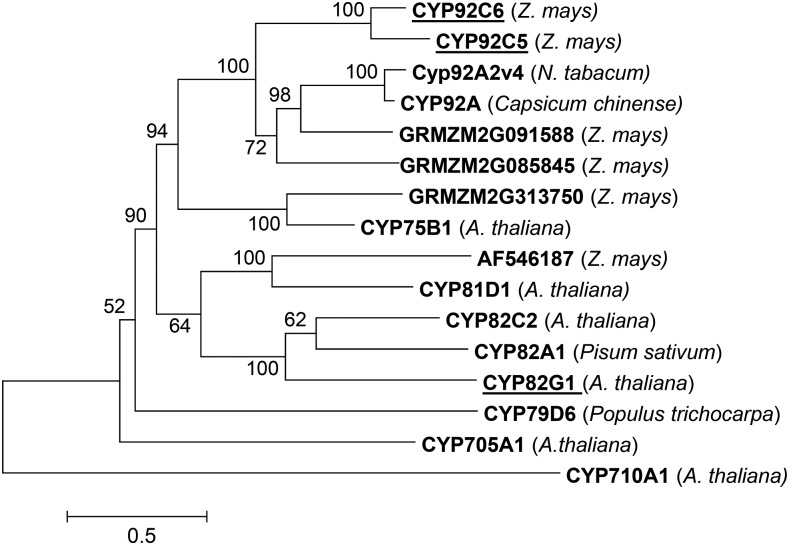
We were able to identify significant QTLs that were associated with cytochrome P450 monooxygenases involved in the formation of the homoterpenes DMNT and TMTT from their respective terpene alcohols (Figure 13). One QTL was associated with DMNT biosynthesis and corresponded to CYP92C5, an enzyme that produced both DMNT from (E)-nerolidol and TMTT from geranyllinalool after heterologous expression. The volatile profile of a plant with a transposon insertion in cyp92C5 demonstrated that CYP92C5 is not only responsible for DMNT biosynthesis but also participates in TMTT formation in planta. Despite this participation in TMTT formation, we did not observe a significant QTL for TMTT at the location of CYP92C5. This was probably due to the functional redundancy with the other identified cytochrome P450 monooxygenase, CYP92C6, which exclusively produces TMTT. The DMNT biosynthesis pathway is fundamentally different from that in Arabidopsis, where DMNT is formed via triterpene degradation (Sohrabi et al., 2015). The P450 enzyme involved in this alternate pathway, CYP705A1, has no apparent sequence similarity to maize CYP92C5 and CYP92C6 (Figure 14).
Figure 13.
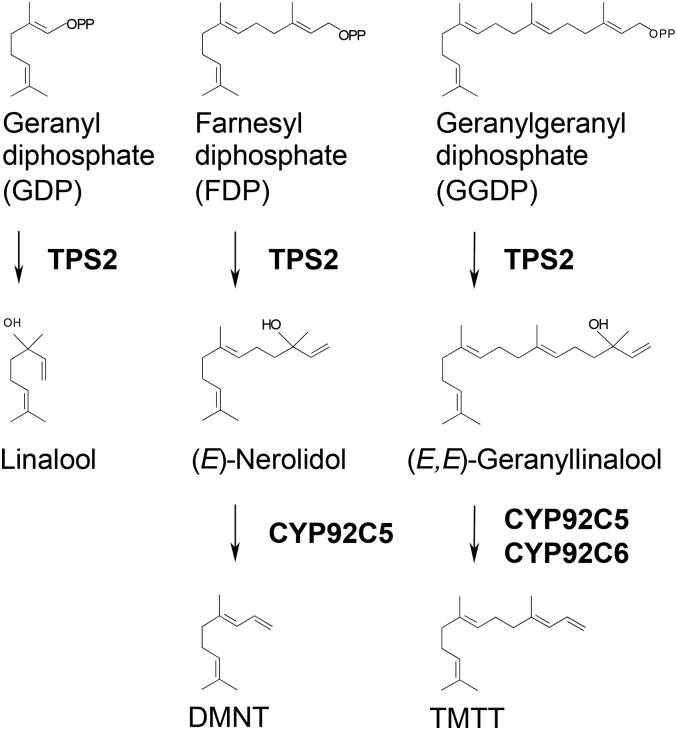
Overview of DMNT and TMTT Synthesis in Maize.
The first steps of the pathways for the synthesis of the maize homoterpenes DMNT and TMTT are catalyzed by the terpene synthase TPS2, which converts GDP, FDP, and GGDP to the corresponding tertiary alcohols. The second steps, which are the oxidative degradations, are catalyzed by two P450 monooxygenases, CYP92C5 and CYP92C6. OPP, diphosphate.
Figure 14.
Dendrogram Analysis of Maize CYP92C5 and CYP92C6 with Closely Related P450 Monooxygenases from Other Plant Species.
The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The P450 monooxygenase CYP710A1 was used as an outgroup. Underlined genes were shown to encode proteins producing homoterpenes upon in vitro expression. The analysis includes CYP92C5 (ACG28049), CYP92C6 (GRMZM2G139467), and CYP82G1 (AT3G25180.2), which are underlined. It also includes CYP92A2v4 (LOC107803700), CYP92A (ABG74350.1), CYP75B1 (AT5G07990), CYP81D1 (AT3G28740.1), CYP82C2 (AT4G31970.1), CYP82A1 (AAG09208.1), CYP705A1 (AT4G15330), CYP79D6 (AHF20913.1), and CYP710A1 (AT2G34500.1).
cyp92C5 and cyp92C6 are 78% identical in their DNA sequences. The proteins they encode both contain highly conserved domains typical of P450 enzymes, including the heme binding loop PFxxGxRxCxG/A motif, the WxxR motif, the PERF motif, and the proline-rich domain for proper folding (Kusano et al., 2001a, 2001b). In a dendrogram analysis, the enzymes fall into a clade with two additional, uncharacterized maize open reading frames (Figure 14). This clade is closely related to P450 enzymes that encode a putative flavonoid 3′ monooxygenase from tobacco (Nicotiana tabacum) and a β-butyric acid-induced gene from yellow lantern chili (Capsicum chinense). Surprisingly, maize CYP92C5 and CYP92C6 exhibit only 37% identity to the amino acid sequence of the Arabidopsis homoterpene synthase CYP82G1, which forms both DMNT and TMTT in vitro, but only TMTT in planta, likely due to the absence of the (E)-nerolidol substrate (Lee et al., 2010). CYP82G1 falls into a distant clade with maize AF546187, an uncharacterized gene that is not associated with a QTL of homoterpene biosynthesis. The separation of the maize and Arabidopsis homoterpene biosynthesis genes in distant clades suggests that the oxidative elimination reaction has evolved independently in both species and may be derived from hydroxylating enzymes of flavonoid biosynthesis. Since many plant volatile mixtures contain homoterpenes, one might have expected a highly conserved group of P450 monooxygenases catalyzing these activities. Also, the in vitro enzyme properties of these enzymes do not necessarily coincide with their in vivo roles. This creates many questions about the mechanisms of herbivore-induced gene regulation and the compartmentation and interplay of terpene biosynthetic pathways which have to be explored in the future. Further studies are necessary to elucidate the reaction mechanism and phylogeny of these apparently distantly related enzymes in monocots and dicots.
METHODS
Plant Material and Herbivore Induction
The 26 parent lines of the maize nested association mapping population, their 5982 RILS (Yu et al., 2008), and a dmnt-mutant line (MuIII) were used. For QTL analysis within the NAM population, plants grown in a field near Cornell University were chosen. All other plants were grown for 2 weeks in climate-controlled chambers under a 16/8-h and 22/18°C day/night cycle (1000 μmol m−2 s−1 PAR). The third leaf of 2-week-old seedlings was used to conduct the induction experiments. Plants were induced with a 6-substituted indanoyl isoleucine conjugate, which is an analog of jasmonic acid isoleucine that simulates the effect of caterpillar feeding (Schüler et al., 2001). For elicitation, leaves of 14-d-old plants were cut and placed in a vial of water (2 mL) containing the indanone derivative (2.3 µM) for 24 h.
Phenotypic Data Transformation
Five terpene compounds were measured in leaves from all 5982 RILs in the US-NAM population. For each trait, outliers were removed using Studentized deleted residuals obtained from mixed linear models with the plant line included as a random effect (Kutner et al., 2004). A Studentized residual is the quotient resulting from the division of a residual by an estimate of its sd. This determines whether or not an individual observation has outlying response variable values and/or outlying explanatory variable values (Kutner et al., 2004). For each of the tested traits, a best linear unbiased predictor of each line was obtained by fitting a mixed linear model across all replicates in ASREML version 3.0 (Gilmour et al., 2009). The model fitting procedure has been previously described (Chandler et al., 2012). The variance component estimates from these models were used to calculate heritabilities (Holland et al., 2003; Hung and Holland, 2012), and standard errors of these heritability estimates were approximated using the delta method (Holland et al., 2003). Pearson’s correlation coefficients were used to quantify the relationship between the untransformed trait BLUPs. Finally, the Box-Cox procedure (Box and Cox, 1964) was implemented to find the optimal transformation of each trait to correct for unequal error variances and non-normality of error terms. This procedure is critical for preventing violations in the statistical assumptions made in the joint-linkage and GWAS models. The transformation was implemented in SAS PROC TRANSREG (http://www.sas.com/).
QTL Mapping/Joint-Linkage Mapping
QTL mapping was conducted with joint-linkage analyses across 25 populations as described previously (Buckler et al., 2009). The analyses used a forward stepwise regression model to include genetic markers that have significant effects nested to populations. Permutation analysis was conducted to determine the threshold for markers entering the model. The analyses were conducted using SAS PROC GLMSelect (http://www.sas.com/).
NAM and GWAS
Following the previous procedure, the 1.6 million SNPs identified in the NAM founders (Gore et al., 2009) were imputed in the NAM RILs using pedigree and linkage marker information (Tian et al., 2011). GWAS was conducted on top of the joint linkage analysis. First, residuals for each chromosome were calculated from the full joint-linkage model with the removal of any QTL located on that chromosome. To obtain robust association results, 100 subsample data sets, each containing 80% of each family, were used for association mapping. SNPs were scored on the number of subsamples they were identified in the RMIP (Valdar et al., 2009). SNPs detected in at least five subsamples (RMIP ≥ 0.05) were considered as significant. The analyses were conducted by the TASSEL software package (Bradbury et al., 2007).
Sequence Isolation
To isolate the sequences of tps2 and tps3 from cDNA without getting chimeric versions of both genes, specific primers in the untranslated region were designed based on the B73 genomic sequence (www.gramene.org). The 5′ and 3′-RACE libraries from the inbred line Tzi8 were used to obtain the complete tps2 amplicon from this NAM parental line (Supplemental Table 1). The libraries were constructed with the SMARTer RACE cDNA amplification kit (Clontech). For the adjacent PCR reaction, the Advantage cDNA PCR kit (Clontech) was used as described in the manual. The open reading frames of the P450s could be amplified only with the Advantage GC Genomic LA PCR Polymerase (Takara Bio Europe/Clontech). All of the PCR fragments were cloned into pCR4-TOPO vector (TOPO TA cloning kit for sequencing; Invitrogen) and subsequently sequenced.
Transcript Quantification by qRT-PCR
For quantitative real-time PCR, total RNA was isolated from B73, the dmnt mutant line (MuIII), and Tzi8 using either the RNeasy Plant Mini Kit (Qiagen) or the GeneJET RNA purification kit (Thermo Scientific). Isolated RNA was treated with RQ1 RNase-free DNase (Promega) to remove genomic DNA. The Fermentas first-strand cDNA kit was used to synthesize cDNA from 1 µg purified RNA. For qRT-PCR, 10 μL Maxima SYBR Green/ROX qPCR Master Mix (Fermentas), 0.5 μL gene-specific forward primer, 0.5 μL reverse primer (10 pmol mL−1 each; Supplemental Table 1), 1 µg template, and 4 μL PCR-grade water were mixed. PCR was performed using a CFX96 real-time system (Bio-Rad) under the following conditions: 10 min at 95°C, 40 cycles of 30 s at 95°C, 30 s at 62°C, and 40 s at 72°C. APT1 (ADENINE PHOSPHORIBOSYLTRANSFERASE1) was used as a control for quantifying gene expression. For each gene analyzed, a cDNA pool from all plants was diluted from 3× to 27× to generate a standard curve. Using the program Bio-Rad CFX Manager, the ΔCT (Δ-threshold cycle) for each gene was calculated relative to APT1 expression, and CT averaging was used for three biological replicates, which each had three technical replicates. All experiments were conducted in independent triplicates and averages as well as the se of the mean are shown.
Protein Overexpression and Enzyme Assay of Terpene Synthases
The open reading frames of the putative terpene synthases tps2 and tps3 from B73 and tps2 from Tzi8 were cloned into the Escherichia coli expression vector pASK-IBA37plus (IBA). The cloning of the terpene synthase fragments into the vector yielded a fusion protein with a His-tag (6xHis) at the N terminus of each protein. For the determination of Km values, the putative 26-amino acid signaling sequence predicted by Signal P was removed from the N terminus. A starter culture of E. coli TOP10 cells in 3 mL Luria-Bertani medium (Sambrook et al., 1989) with 100 µg mL−1 ampicillin was grown overnight at 37°C and used to inoculate 100 mL of Luria-Bertani medium supplemented with 100 µg mL−1 ampicillin. Terpene synthase expression was induced by addition of anhydrotetracycline at 200 µg/L after the bacteria were grown to an optical density of 0.6 at 600 nm, and then the culture was shaken for 20 h at 18°C. Next, 100 mL of bacterial culture was incubated at 10°C at 70 rpm (benchtop incubation shaker; Certomat IS; Sartorius) with 1 mL Ni-NTA agarose (Qiagen) in a beaker to let the fusion protein bind to the nickel beads. After a 1-h incubation, the preparation was gently transferred into a 0.8 × 4-cm Poly-prep chromatography column (Bio-Rad) for the purification steps, which were done under native conditions as described in the Bio-Rad manual. Enzyme activity assays were performed with 38 μL extracted enzyme and 62 μL reaction mix (10 mM Tris-HCl, pH 7.5, 10% glycerol, 1 mM DTT, 60 μM GDP/FDP [Echelon Research Laboratories], and 10 mM MgCl2). Purified protein was quantified with Bradford reagent (Bradford, 1976) and separated under denaturing conditions using polyacrylamide electrophoresis with 12% sodium dodecyl sulfate-polyacrylamide gels. Protein probes were mixed with 5× buffer and denaturated for 10 min at 95°C and run at a constant 25 mA per gel. Following electrophoresis, proteins were stained with Coomassie Brilliant Blue solution. For the estimation of the protein sizes, a prestained protein marker (Invitrogen) was used. The assay for geranyl linalool was performed using 100 μL purified enzyme in a 400 μL reaction mix (10 mM Tris-HCl, pH 7.5, 10% glycerol, 1 mM DTT, 60 μM GGDP [Echelon Research Laboratories], and 10 mM MgCl2). Enzyme products were collected using a polydimethylsiloxane-coated SPME fiber (SUPELCO) that was exposed to the headspace above the assay mixture for 45 min at 35°C in a water bath to bind the volatile terpenes. Enzyme kinetics of TPS2 were determined with [1-3H]-labeled GDP, FDP, and GGDP substrates having an activity of 3000 Bq mmol−1. The enzyme products were extracted with 500 μL n-pentan and counted with 2 mL Lipoluma cocktail (Carl Roth) in a Tricarb TR2100 scintillation counter (Perkin-Elmer). The assays were repeated three times with five samples per substrate concentration. Km values were determined using a Lineweaver-Burk plot.
Isolation and Transformation of Maize Protoplasts
For chloroplast isolation, two leaves of 14-d-old plants (B73) grown under low light conditions were removed and cut into <0.5-mm strips with a razor blade. These leaf strips were transferred into 30 mL enzyme solution (0.6 M mannitol, 20 mM MES, pH 5.7, 20 mM KCl, 0.4% macerozyme R10 [Yakult Pharmaceuticals], 1.5% cellulase [Yakult Pharmaceuticals], 10 mM CaCl2, and 0.1% BSA) for a 30- to 60-min vacuum infiltration. Digestion was continued for 2 to 3 h with shaking at 30°C in the dark. The protoplasts were filtered through a nylon mesh (100 µM; Merck Millipore) and centrifuged for 1.5 min at 150g at 4°C. The pellet was washed twice with 5 mL wash solution (0.6 M mannitol, 4 mM MES, pH 5.7, and 20 mM KCl) and transferred into MMG buffer (0.6 M mannitol, 4 mM MES, pH 5.7, and 15 mM MgCl2) with a final concentration of one to two protoplasts per 10−6/mL. For transformation, 1 mL protoplast solution was incubated with 10 to 20 µg plasmid DNA for 20 min. Then, 1.1 mL PEG solution (40% [w/v] PEG4000, 0.6 M mannitol, and 0.1 M CaCl2) was added to the protoplasts and gently converted to mix the suspension. The transformation was stopped after 10 min by addition of 4.4 mL of wash solution. The protoplasts were washed and suspended in 1 mL incubation buffer (0.6 M mannitol, 4 mM MES, pH 5.7, and 4 mM KCl) overnight at 18°C in the dark.
Yeast Expression and Cytochrome P450 Activity Assays
For cloning the open reading frames of the two cytochrome P450 sequences, the pESC-Leu2d vector (Ro et al., 2008) was used and transformed into the Saccharomyces cerevisiae strains WAT11 and W(R) overexpressing an Arabidopsis thaliana P450 reductase and a yeast P450 reductase (Pompon et al., 1996) by chemical transformation as described (Gietz and Woods, 2002). Primers for cloning into pESC-Leu2d are shown in Supplemental Table 1. Microsomes were prepared according to Irmisch et al. (2013). Enzyme activity assays were performed in a glass vial with 70 μL extracted microsomes (0.3 to 3 µg protein) in a 230-μL reaction mix containing 75 mM sodium phosphate buffer, pH 6.8, 100 μM substrate (supplied in a maximum of 6 μL DMSO), and 1 mM NADPH. Using a polydimethylsiloxane-coated SPME fiber, the products were collected from the head space of the assay mixture for 1 h at 30°C and analyzed by GC-MS.
GC-MS Analysis of Leaf Volatiles and Products of Terpene Synthase and P450 Assays
For volatile terpene extraction, the induced leaf material was overlaid with 600 μL hexane and incubated for at least 7 d at 23°C. For solid-phase microextraction (SPME), a 100-μm polydimethylsiloxane fiber (Supelco) adsorbed the volatiles from the headspace of the plant material or enzyme assays within 1 h at 42°C. After injection of 1 μL hexane extract or the SPME into the GC (GC-2010; Shimadzu), terpenes were separated from the EC5-MS column (30-m length, 0.25-mm inner diameter, and 0.25-µm film) (Grace) based on the oven temperature and detected in the mass spectrometer (GCMS-QP 2010 Plus; Shimadzu). The injection was performed with a temperature of 220°C. Hydrogen was used for the carrier gas at a flow rate of 1 mL/min. To separate the volatiles, an EC5-MS column (30-m length, 0.25-mm inner diameter, and 0.25-µm film) (Grace Alltech) was used under following conditions: 80°C for 2 min, first ramp 7°C/min to 150°C, second ramp 100°C/min to 300°C, final 2 min hold. Total ion chromatograms with sums of intensities of all mass spectral peaks belonging to the same scan are shown for the volatile spectra. To identify the separated volatiles, the Shimadzu software GCMS Postrun Analysis was used with the mass spectra libraries Wiley8 (Hewlett Packard) and Adams (Adams, 2007). To determine the kinetics of the cytochrome P450 monooxygenases, assays were performed in triplicate and quantified by GC coupled to a FID detector. The assays were repeated twice.
Phylogenetic Analysis
The dendrogram analysis was conducted with the software Mega6 (Tamura et al., 2013). The loaded protein sequences were aligned by MUSCLE (Edgar, 2004) (gap open −2.9; gap extended 0; hydrophobicity multiplier 1.2; clustering method: UPGMB) (Supplemental Data Sets 1 and 2). Based on this alignment, the maximum likelihood method with the Le_Gascuel_2008 model (Le and Gascuel, 2008) was identified as the best protein model and used to reconstruct the tree. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. A discrete gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 1.7268)]. All positions with <80% site coverage were eliminated. That is, fewer than 20% alignment gaps, missing data, and ambiguous bases were allowed at any position. The percentages of trees were calculated by 1000 bootstrap trials.
Accession Numbers
The cDNA sequences obtained from B73 were deposited in GenBank/EMBL data libraries (http://www.ncbi.nlm.nih.gov) under accession numbers AY928081.1 (tps2), AY928082 (tps3), ACG28049 (cyp92C5), and GRMZM2G139467 (cyp92C6). See the legends to Figures 12 and 14 for accession numbers of sequences used in phylogenetic analyses.
Supplemental Data
Supplemental Figure 1. Analysis of purified maize TPS2 and TPS3 by SDS-PAGE.
Supplemental Figure 2. Chiral GC-MS analysis of the sesquiterpene and diterpene products from recombinant expressed TPS2.
Supplemental Figure 3. Expression of tps2 is induced by caterpillar oral secretions.
Supplemental Figure 4. Quantification of the major terpene compounds in the maize inbred lines B73 and Tzi8 after wounding and treatment with indanone derivative (elicitor).
Supplemental Figure 5. Quantification of the major terpene compounds in the maize inbred lines B73 and the transposon insertion mutant MuIII after wounding and treatment with indanone derivative (elicitor).
Supplemental Table 1. Sequences of nucleotide primers.
Supplemental Data Set 1. Text file of the amino acid sequences of TPS2 and TPS3 and linalool/nerolidol/geranyllinalool synthases from other plant species and the alignment used for the phylogenetic analysis shown in Figure 12.
Supplemental Data Set 2. Text file of the amino acid sequences of several plant P450 monooxygenases related to maize CYP92C5 and CYP92C6 and the alignment used for the phylogenetic analysis shown in Figure 14.
Supplementary Material
Acknowledgments
We thank Jens Wurlitzer for plant collection, Michael Gore for processing of samples from the field, and Peter Lindemann for help with enzyme characterization. We also thank KWS Seeds for maize B73 seeds. A.R. and J.D. were supported by project B7 of the Collaborative Research Center 648 of the German Research Foundation (DFG). Germplasm, field, and mapping analysis were supported by grants from the U.S. National Science Foundation (IOS-0820619 and IOS-1238014) and the USDA-Agricultural Research Service. G.J. was supported by U.S. National Science Foundation Awards IOS-1139329 and IOS-1339237.
AUTHOR CONTRIBUTIONS
A.R. and J.D. designed research. A.R., Z.Z., A.E.L., and E.S.B. designed bioinformatic analyses. A.R., C.S., T.G.K., S.I., and S.P. performed research. E.S.B. provided germplasm and performed the field trial. W.B. provided the elicitor. A.R., Z.Z., A.E.L., G.J., J.G., and J.D. wrote the article.
Glossary
- DMNT
(E)-4,8-dimethyl-1,3,7-nonatriene
- TMTT
(E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene
- GDP
geranyl diphosphate
- FDP
farnesyl diphosphate
- GGDP
geranylgeranyl diphosphate
- NAM
nested association mapping
- QTL
quantitative trait locus
- GWAS
genome-wide association study
- SNP
single nucleotide polymorphism
- RMIP
resampling model inclusion probability
- GC-MS
gas chromatography-mass spectrometry
- SPME
solid-phase microextraction
Footnotes
Articles can be viewed without a subscription.
References
- Adam K.-P., Thiel R., Zapp J. (1999). Incorporation of 1-[1–13 C] deoxy-d-xylulose in chamomile sesquiterpenes. Arch. Biochem. Biophys. 369: 127–132. [DOI] [PubMed] [Google Scholar]
- Adams R.P. (2007). Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry. (Carol Stream, IL: Allured Publishing).
- Aharoni A., Giri A.P., Verstappen F.W., Bertea C.M., Sevenier R., Sun Z., Jongsma M.A., Schwab W., Bouwmeester H.J. (2004). Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16: 3110–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura G., Garms S., Maffei M., Bossi S., Schulze B., Leitner M., Mithöfer A., Boland W. (2008). Herbivore-induced terpenoid emission in Medicago truncatula: concerted action of jasmonate, ethylene and calcium signaling. Planta 227: 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J.A., Lange B.M. (2003). Metabolic cross talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Arch. Biochem. Biophys. 415: 146–154. [DOI] [PubMed] [Google Scholar]
- Boland W.G.A., Gilbert M., Feng Z. (1998). Biosynthesis of C11 and C16 homoterpenes in higher plants: stereocheistry of C–C bond cleavage reaction. Tetrahedron 54: 725–736. [Google Scholar]
- Box G.E.P., Cox D.R. (1964). An analysis of transformations. J. R. Stat. Soc. B 26: 211–252. [Google Scholar]
- Boyle B., Brisson N. (2001). Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF. Plant Cell 13: 2525–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brillada C., Nishihara M., Shimoda T., Garms S., Boland W., Maffei M.E., Arimura G.i. (2013). Metabolic engineering of the C16 homoterpene TMTT in Lotus japonicus through overexpression of (E, E)-geranyllinalool synthase attracts generalist and specialist predators in different manners. New Phytol. 200: 1200–1211. [DOI] [PubMed] [Google Scholar]
- Bruce T.J., Matthes M.C., Chamberlain K., Woodcock C.M., Mohib A., Webster B., Smart L.E., Birkett M.A., Pickett J.A., Napier J.A. (2008). cis-Jasmone induces Arabidopsis genes that affect the chemical ecology of multitrophic interactions with aphids and their parasitoids. Proc. Natl. Acad. Sci. USA 105: 4553–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E.S., et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Chandler K., Lipka A.E., Owens B.F., Li H., Buckler E.S., Rocheford T., Gore M.A. (2012). Genetic analysis of visually scored orange kernel color in maize. Crop Science 53: 189–200. [Google Scholar]
- Cook J.P., McMullen M.D., Holland J.B., Tian F., Bradbury P., Ross-Ibarra J., Buckler E.S., Flint-Garcia S.A. (2012). Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 158: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer J.G., Posthumus M.A., Dicke M. (2004). Identification of volatiles that are used in discrimination between plants infested with prey or nonprey herbivores by a predatory mite. J. Chem. Ecol. 30: 2215–2230. [DOI] [PubMed] [Google Scholar]
- Degen T., Dillmann C., Marion-Poll F., Turlings T.C.J. (2004). High genetic variability of herbivore-induced volatile emission within a broad range of maize inbred lines. Plant Physiol. 135: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J., Gershenzon J. (2000). Demonstration and characterization of (E)-nerolidol synthase from maize: a herbivore-inducible terpene synthase participating in (3E)-4,8-dimethyl-1,3,7-nonatriene biosynthesis. Planta 210: 815–822. [DOI] [PubMed] [Google Scholar]
- Degenhardt J., Gershenzon J., Baldwin I.T., Kessler A. (2003). Attracting friends to feast on foes: engineering terpene emission to make crop plants more attractive to herbivore enemies. Curr. Opin. Biotechnol. 14: 169–176. [DOI] [PubMed] [Google Scholar]
- Degenhardt J., Köllner T.G., Gershenzon J. (2009). Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637. [DOI] [PubMed] [Google Scholar]
- Dicke M., Van Beek T., Posthumus M., Dom N.B., Van Bokhoven H., De Groot A. (1990). Isolation and identification of volatile kairomone that affects acarine predatorprey interactions Involvement of host plant in its production. J. Chem. Ecol. 16: 381–396. [DOI] [PubMed] [Google Scholar]
- Donath J., Boland W. (1995). Biosynthesis of acyclic homoterpenes: enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39: 785–790. [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V., Alba J.M., Kant M.R., Schuurink R.C., Pichersky E. (2014). Geranyllinalool synthases in Solanaceae and other angiosperms constitute an ancient branch of diterpene synthases involved in the synthesis of defensive compounds. Plant Physiol. 166: 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Ren F., Lu X., Mao H., Xu M., Degenhardt J., Peters R., Wang Q. (2015). A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metaolism. Plant Physiol. 170: 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A.R., Gogel B., Cullis B., Thompson R., Butler D. (2009). ASReml User Guide Release 3.0. (Hemel Hempstead, UK: VSN International; ). [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Gomez S.K., Cox M.M., Bede J.C., Inoue K., Alborn H.T., Tumlinson J.H., Korth K.L. (2005). Lepidopteran herbivory and oral factors induce transcripts encoding novel terpene synthases in Medicago truncatula. Arch. Insect Biochem. Physiol. 58: 114–127. [DOI] [PubMed] [Google Scholar]
- Gore M.A., Chia J.-M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., Peiffer A., McMullen M.D., Grills G.S., Ross-Ibarra J., Ware D.H., Buckler E.S. (2009). A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Harborne J.B. (1991). The chemical basis of plant defense. In Plant Defenses against Mammalian Herbivory, Palo R.T., Robbins C.T., eds (Boca Raton, FL: CRC Press; ), pp. 45–59. [Google Scholar]
- Heil M. (2014). Herbivore‐induced plant volatiles: targets, perception and unanswered questions. New Phytol. 204: 297–306. [Google Scholar]
- Hemmerlin A., Hoeffler J.-F., Meyer O., Tritsch D., Kagan I.A., Grosdemange-Billiard C., Rohmer M., Bach T.J. (2003). Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278: 26666–26676. [DOI] [PubMed] [Google Scholar]
- Herde M., Gartner K., Kollner T.G., Fode B., Boland W., Gershenzon J., Gatz C., Tholl D. (2008). Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C-16-homoterpene TMTT. Plant Cell 20: 1152–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoballah M.E., Degen T., Bergvinson D., Savidan A., Tamo C., Turlings T.C.J. (2004). Occurrence and direct control potential of parasitoids and predators of the fall armyworm (Lepidoptera: Noctuidae) on maize in the subtropical lowlands of Mexico. Agric. For. Entomol. 6: 83–88. [Google Scholar]
- Holland J.B., Nyquist W.E., Cervantes-Martínez C.T. (2003). Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 22: 9–112. [Google Scholar]
- Hung H.-Y., Holland J.B. (2012). Diallel analysis of resistance to Fusarium ear rot and fumonisin contamination in maize. Crop Sci. 52: 2173–2181. [Google Scholar]
- Irmisch S., McCormick A.C., Boeckler G.A., Schmidt A., Reichelt M., Schneider B., Block K., Schnitzler J.-P., Gershenzon J., Unsicker S.B., Köllner T.G. (2013). Two herbivore-induced cytochrome P450 enzymes CYP79D6 and CYP79D7 catalyze the formation of volatile aldoximes involved in poplar defense. Plant Cell 25: 4737–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappers I.F., Aharoni A., van Herpen T., Luckerhoff L.L.P., Dicke M., Bouwmeester H.J. (2005). Genetic engineering of terpenoid metabolism attracts, bodyguards to Arabidopsis. Science 309: 2070–2072. [DOI] [PubMed] [Google Scholar]
- Khan Z., Ampong-Nyarko K., Chiliswa P., Hassanali A., Kimani S., Lwande W., Overholt W.A., Picketta J.A., Smart L.E., Woodcock C.M. (1997). Intercropping increases parasitism of pests. Nature 388: 631–632. [Google Scholar]
- Khan Z.R., Pickett J.A., Berg J.d., Wadhams L.J., Woodcock C.M. (2000). Exploiting chemical ecology and species diversity: stem borer and striga control for maize and sorghum in Africa. Pest Manag. Sci. 56: 957–962. [Google Scholar]
- Kliebenstein D.J., West M.A., van Leeuwen H., Loudet O., Doerge R.W., St Clair D.A. (2006). Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics 7: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner T.G., Held M., Lenk C., Hiltpold I., Turlings T.C.J., Gershenzon J., Degenhardt J. (2008). A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20: 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Kagawa N., Sakaguchi M., Omura T., Waterman M.R. (2001a). Importance of a proline-rich sequence in the amino-terminal region for correct folding of mitochondrial and soluble microbial P450s. J. Biochem. 129: 271–277. [DOI] [PubMed] [Google Scholar]
- Kusano K., Sakaguchi M., Kagawa N., Waterman M.R., Omura T. (2001b). Microsomal P450s use specific proline-rich sequences for efficient folding, but not for maintenance of the folded structure. J. Biochem. 129: 259–269. [DOI] [PubMed] [Google Scholar]
- Kutner M.H., Nachtsheim C.J., Neter J., Li W. (2004). Applied Linear Statistical Models. 4th ed. (Boston: McGraw-Hill; ). [Google Scholar]
- Le S.Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Lee S., Badieyan S., Bevan D.R., Herde M., Gatz C., Tholl D. (2010). Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 21205–21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Biol. 50: 47–65. [DOI] [PubMed] [Google Scholar]
- McCormick A.C., Unsicker S.B., Gershenzon J. (2012). The specificity of herbivore-induced plant volatiles in attracting herbivore enemies. Trends Plant Sci. 17: 303–310. [DOI] [PubMed] [Google Scholar]
- McMullen M.D., et al. (2009). Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Meihls L.N., Handrick V., Glauser G., Barbier H., Kaur H., Haribal M.M., Lipka A.E., Gershenzon J., Buckler E.S., Erb M., Köllner T.G., Jander G. (2013). Natural variation in maize aphid resistance is associated with 2, 4-dihydroxy-7-methoxy-1, 4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25: 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm R., Dicke M. (2010). Variation in natural plant products and the attraction of bodyguards involved in indirect plant defense The present review is one in the special series of reviews on animal-plant interactions. Can. J. Zool. 88: 628–667. [Google Scholar]
- Owens B.F., et al. (2014). A foundation for provitamin A biofortification of maize: Genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Noel J.P., Dudareva N. (2006). Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett J.A., Woodcock C.M., Midega C.A., Khan Z.R. (2014). Push–pull farming systems. Curr. Opin. Biotechnol. 26: 125–132. [DOI] [PubMed] [Google Scholar]
- Poland J.A., Bradbury P.J., Buckler E.S., Nelson R.J. (2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D., Louerat B., Bronine A., Urban P. (1996). Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272: 51–64. [DOI] [PubMed] [Google Scholar]
- Richter A., Seidl-Adams I., Köllner T., Degenhardt J. (2015). A maize (Zea mays) herbivore-induced farnesyl diphosphate synthase supports the sesquiterpene synthesis in leaves. Planta 241: 1351–1361. [DOI] [PubMed] [Google Scholar]
- Ro D.-K., Ouellet M., Paradise E.M., Burd H., Eng D., Paddon C.J., Newman J.D., Keasling J.D. (2008). Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Sanmiya K., Ueno O., Matsuoka M., Yamamoto N. (1999). Localization of farnesyl diphosphate synthase in chloroplasts. Plant Cell Physiol. 40: 348–354. [DOI] [PubMed] [Google Scholar]
- Schnee C., Köllner T.G., Gershenzon J., Degenhardt J. (2002). The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-Î2-farnesene,(E)-nerolidol, and (E,E)-farnesol after herbivore damage. Plant Physiol. 130: 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler G., Görls H., Boland W. (2001). 6-Substituted indanoyl isoleucine conjugates mimic the biological activity of coronatine. Eur. J. Org. Chem. 2001: 1663–1668. [Google Scholar]
- Sohrabi R., Huh J.-H., Badieyan S., Rakotondraibe L.H., Kliebenstein D.J., Sobrado P., Tholl D. (2015). In planta variation of volatile biosynthesis: an alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell 27: 874–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starks C.M., Back K., Chappell J., Noel J.P. (1997). Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277: 1815–1820. [DOI] [PubMed] [Google Scholar]
- Su Y., Wang C.Z., Guo Y.L. (2009). Analysis of volatile compounds from Mentha hapioealyx Briq by GC-MS based on accurate mass measurement and retention index. Acta Chimi. Sin. 67: 546–554. [Google Scholar]
- Tamiru A., Bruce T.J., Woodcock C.M., Caulfield J.C., Midega C.A., Ogol C.K., Mayon P., Birkett M.A., Pickett J.A., Khan Z.R. (2011). Maize landraces recruit egg and larval parasitoids in response to egg deposition by a herbivore. Ecol. Lett. 14: 1075–1083. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Bradbury P.J., Brown P.J., Hung H., Sun Q., Flint-Garcia S., Rocheford T.R., McMullen M.D., Holland J.B., Buckler E.S. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43: 159–162. [DOI] [PubMed] [Google Scholar]
- Turlings T.C., Tumlinson J.H. (1991). Do parasitoids use herbivore-induced plant chemical defenses to locate hosts? Fla. Entomol. 74: 42–50. [Google Scholar]
- Turlings T.C., Tumlinson J.H., Lewis W.J. (1990). Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 250: 1251–1253. [DOI] [PubMed] [Google Scholar]
- Turlings T.C., Tumlinson J.H., Heath R.R., Proveaux A.T., Doolittle R.E. (1991). Isolation and identification of allelochemicals that attract the larval parasitoid, Cotesia marginiventris (Cresson), to the microhabitat of one of its hosts. J. Chem. Ecol. 17: 2235–2251. [DOI] [PubMed] [Google Scholar]
- Unsicker S.B., Kunert G., Gershenzon J. (2009). Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 12: 479–485. [DOI] [PubMed] [Google Scholar]
- Valdar W., Holmes C.C, Mott R., Flint J. (2009). Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneya K., Takabayashi J. (2013). Interaction–information networks mediated by plant volatiles: a case study on willow trees. J. Plant Interact. 8: 197–202. [Google Scholar]
- Yu J., Holland J.B., McMullen M.D., Buckler E.S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J.S., Köllner T.G., Wiggins G., Grant J., Degenhardt J., Chen F. (2008). Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J. 55: 491–503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.