Marchantia polymorpha, like all liverworts, accumulates a large array of terpenes, and this process depends on a unique family of terpene synthases.
Abstract
Marchantia polymorpha is a basal terrestrial land plant, which like most liverworts accumulates structurally diverse terpenes believed to serve in deterring disease and herbivory. Previous studies have suggested that the mevalonate and methylerythritol phosphate pathways, present in evolutionarily diverged plants, are also operative in liverworts. However, the genes and enzymes responsible for the chemical diversity of terpenes have yet to be described. In this study, we resorted to a HMMER search tool to identify 17 putative terpene synthase genes from M. polymorpha transcriptomes. Functional characterization identified four diterpene synthase genes phylogenetically related to those found in diverged plants and nine rather unusual monoterpene and sesquiterpene synthase-like genes. The presence of separate monofunctional diterpene synthases for ent-copalyl diphosphate and ent-kaurene biosynthesis is similar to orthologs found in vascular plants, pushing the date of the underlying gene duplication and neofunctionalization of the ancestral diterpene synthase gene family to >400 million years ago. By contrast, the mono- and sesquiterpene synthases represent a distinct class of enzymes, not related to previously described plant terpene synthases and only distantly so to microbial-type terpene synthases. The absence of a Mg2+ binding, aspartate-rich, DDXXD motif places these enzymes in a noncanonical family of terpene synthases.
INTRODUCTION
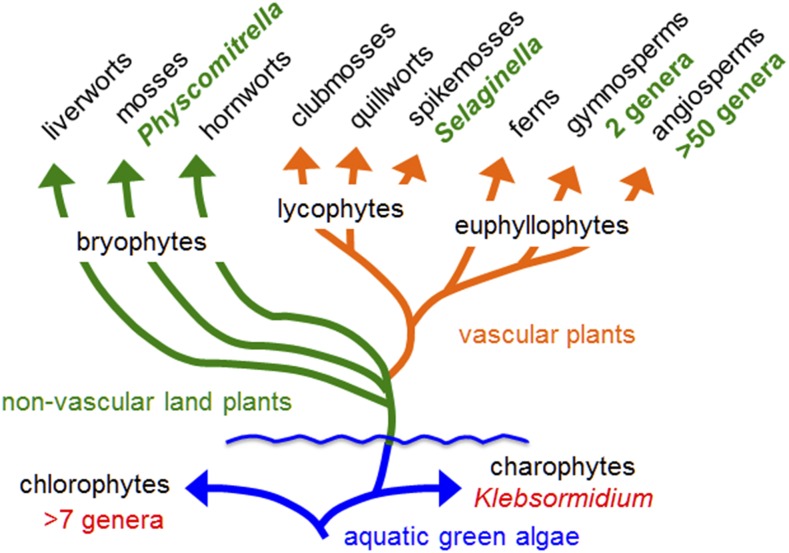
Although there is no universal agreement on the phylogenetic relationships of terrestrial plant clades, it is generally accepted that liverworts with 6000 to 8000 extant species were among the first and diverged earliest from other land plant lineages based on morphology, fossil evidence, and some molecular analyses (Figure 1) (Mishler et al., 1994; Kenrick and Crane, 1997; Qiu et al., 2006). Similar to other bryophytes, liverworts have a haploid gametophyte-dominant life cycle, with the diploid sporophyte generation nutritionally dependent upon the gametophyte. Lacking sophisticated vascular systems like xylem and phloem typical of true vascular plants for the long distance transport of water, minerals, and the distribution of photosynthate restricts the growth habits and the ecological niches liverworts occupy. The liverwort vegetative gametophyte body plan is either a prostrate thallus, whose complexity varies between taxa, or a leafy prostrate or erect frond-like structure. A defining feature of liverworts is the presence of oil bodies, which are present in every major lineage of liverworts, implying the last common ancestor of extant liverworts possessed oil body cells (Schuster, 1966; He et al., 2013). In some species, several oil bodies are found in nearly all differentiated cells of both gametophyte and sporophyte. By contrast, in the Treubiales and Marchantiopsida, oil bodies are large and usually found in only specialized cells referred to as idioblasts. In early studies, it was noted that the fragrance of crushed liverworts was associated with prevalence of oil bodies and it was proposed that they contain ethereal oils (Gottsche, 1843; von Holle, 1857; Lindberg, 1888). The contents of oil bodies were later shown to be of a terpenoid nature (Lohmann, 1903), with modern analyses demonstrating that oil bodies contain a mixture of mono-, sesqui-, di-, and triterpenoids, as well as constituents such as bibenzyl phenylpropanoid derivatives (Asakawa, 1982; Huneck, 1983). Interestingly, the chemical diversity of terpenes found in liverworts appears to rival that found altogether in bacteria, fungi, and vascular plants (Paul et al., 2001). For instance, Marchantia polymorpha has been documented to constitutively accumulate thujopsene, a sesquiterpene common to the seed plant genus Thujopsis (Oh et al., 2011), and cuparene, a sesquiterpene common to the fungus Fusarium verticillioides (Dickschat et al., 2011). Many of the terpene compounds associated with liverworts have been reported to have various biological activities, including antimicrobial (Gahtori and Chaturvedi, 2011), and serve as a deterrence to insect predation (Asakawa, 2011).
Figure 1.
A Phylogenetic Tree to Illustrate the Evolutionary Relationship between the First Land Plants (Bryophytes) and the Most Evolutionarily Diverged Forms of Plants (Angiosperms) (Adapted from Banks et al., 2011).
The annotations in red and green refer to genera whose genomes have been sequenced and published. All the terrestrial plant genera genomes examined to date appear to harbor diterpene synthase genes for kaurene biosynthesis, a precursor to gibberellins and gibberellin-like growth regulators. For mono- and sesquiterpene synthase genes, the angiosperms Arabidopsis and Vitis harbor 27 and 66 annotated genes, respectively (Chen et al., 2011; Vaughan et al., 2013; Schwab and Wüst, 2015). In the case of non-seed plants like Selaginella, 48 mono- and sesquiterpene synthase genes were predicted (Li et al., 2012), while Physcomitrella was found to contain only one functional diterpene synthase gene (Hayashi et al., 2006).
A common tenet about the biological fitness of plants is that they have evolved a variety of mechanisms to suit their sessile life style to cope with ever-changing environmental conditions and their abilities to occupy specific environmental niches. In fact, the dizzying array of specialized metabolites found in plants has been suggested to serve an equally diverse range of functions. For example, many of the over 210,272 identified alkaloid, phenylpropanoid, and terpene natural products (Marienhagen and Bott, 2013; Buckingham, 2013) have been proposed to provide protection against biotic (i.e., microbial pathogens and herbivores; Davis and Croteau, 2000) and abiotic (i.e., UV irradiation; Gil et al., 2012) stresses, as well as to serve in attracting beneficial insect/microbe/animal associations (Gershenzon and Dudareva, 2007) and mediating plant-to-plant communication (Aharoni et al., 2003). Consistent with these notions is that the mechanisms responsible for these adaptations have been subject to evolutionary processes yielding, in the case of specialized metabolism, molecular changes in the genes encoding for the relevant biosynthetic enzymes and thus providing for changes in catalytic outcomes and chemical diversification. Hence, it has seemed reasonable to explore molecular comparisons of enzymes for specialized metabolism between plant species as a means for possibly revealing structural features underpinning the biochemical diversification of these enzymes. Alternatively, and possibly equally instructive, has been to broaden this comparison of key biosynthetic enzymes to those from some of the earliest land plants. In fact, Li et al. (2012) recently reported on the characterization of the mono-, sesqui-, and diterpene synthase gene families uncovered from the DNA sequence of the Selaginella moellendorffii genome. These investigators found evidence that while the diterpene synthase genes in S. moellendorffii appear consistent with a plant phylogeny, the mono- and sesquiterpene synthase genes appear to be evolutionarily related to microbial terpene synthases.
In earlier work aimed at the functional characterization of putative terpene synthase genes in Arabidopsis thaliana, Wu et al. (2005) and Tholl et al. (2005) described a sesquiterpene synthase catalyzing the biosynthesis of α-barbatene, thujopsene, and β-chamigrene. While the observation of terpene synthases capable of generating more than one reaction product was not unusual (Degenhardt et al., 2009), the particular mix of sesquiterpene reaction products was unexpected. These particular sesquiterpenes are commonly found in liverworts rather than in vascular plants (Wu et al., 2005), leaving the impression that at least some of the vascular plant terpene synthases could have arisen from an ancestral gene in common with liverworts. Hence, the aim of this work was to first identify the terpene synthase genes within the model liverwort species M. polymorpha and then to assess the phylogenetic relationships between the vascular plants and liverwort genes with hopes of uncovering peptide residues or domains that might mediate various facets of their catalytic specificity. Unexpectedly, our results suggest that the mono- and sesquiterpene synthase genes of M. polymorpha are only very distantly related to terpene synthase genes in common with fungi and bacteria and are not related to those previously described from land plants. Thus, despite the presence of functional diterpene synthases genes in M. polymorpha clearly related to other vascular plant terpene synthases and the suggestions that these genes could have functionally diversified to give rise to mono- and sesquiterpene biosynthesis (Trapp and Croteau, 2001), alternative gene families appear to have been recruited for the production of these important metabolites in liverworts.
RESULTS
Developmental Profile of Terpenes in M. polymorpha
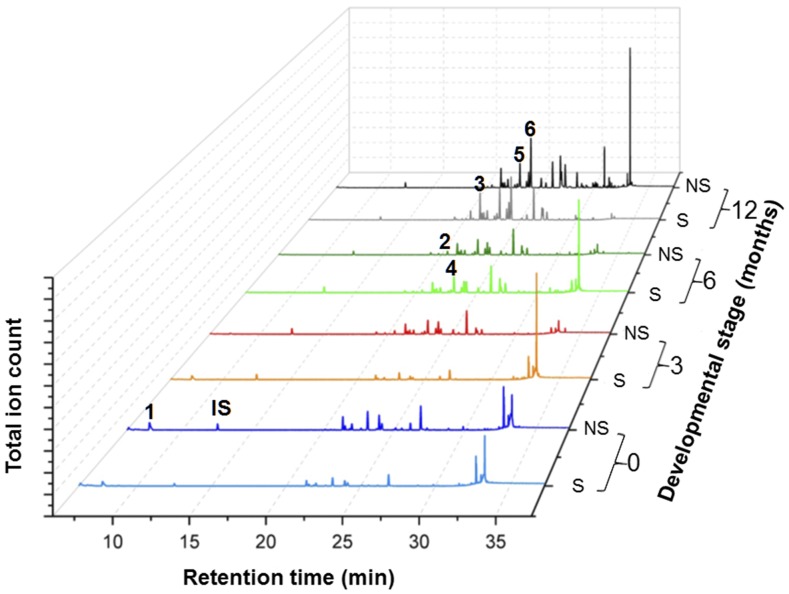
M. polymorpha can reproduce vegetatively via gemmae, multicellular propagules produced from single cells at the base of specialized receptacles referred to as gemmae cups, produced on the dorsal surface of the thallus. Once displaced from the confines of the cup, gemmae germinate to produce new thalli. Oil body cells differentiate both in developing gemmae and later produced thallus tissue. To determine how terpene accumulation might be associated with these development events, organic extracts of temporally staged thallus material were profiled by gas chromatography-mass spectroscopy (GC-MS) (Figure 2). Although many of the compounds present in these extracts have not been structurally identified, their MS patterns (parent ions and fragmentation patterns) are fully consistent with mono- and sesquiterpene classes of isoprenoids. Nonetheless, a few of the compounds have been characterized previously (Suire et al., 2000) or their retention time and MS patterns could be matched with available standards. For instance, limonene, a monoterpene, was evident during the early growth stage (i.e., gemma stage), yet decreased thereafter. By contrast, sesquiterpenes, such as (−)-α-gurjunene, (−)-thujopsene, β-chamigrene, β-himachalene, and (+)-cuparene, tended to accumulate over developmental time, increasing 2-fold (β-chamigrene) to >20-fold [(+)-cuparene] over a 12-month period (Supplemental Figure 1). While no significant differences in the chemical entities between extracts of axenic plants versus those propagated under greenhouse conditions (Figure 2) were noted, the absolute level of a constituent between replicates varied up to 50% or more. Moreover, close inspection of the GC-MS data indicated multiple, coeluting compounds, which might mask differences in the level of individual components contributing to a single peak. Our extraction and analytical procedure (GC-MS) was sufficient to detect diterpenes, but the only diterpene that we observed in the chemical profiles was phytol, a linear diterpene alcohol.
Figure 2.
Chemical Profiles of M. polymorpha Grown Axenically in Tissue Culture Conditions and in Soil under Greenhouse Conditions over a Developmental Time Course.
Organic extracts prepared from thallus material (or gemma for 0 months) collected at the indicated time points were profiled by GC-MS and the indicated peaks identified by reference to the NIST (2011) library and compound identifications according to Suire et al. (2000). Peaks 1 to6 correspond to d-limonene, (−)-α-gurjunene, (−)-thujopsene, β-chamigrene, β-himachalene, and (+)-cuparene, respectively. The dominant peak at ∼34 min was identified as phytol based on MS comparisons to NIST standards. IS, internal standard; S, sterile; NS, not sterile.
Identification of Putative M. polymorpha Terpene Synthases
To identify terpene synthase enzymes contributing to the terpene profiles in M. polymorpha, RNA was isolated from various staged thallus tissues, pooled for DNA sequencing, and the sequence information assembled into 42,617 contigs using the CLC Workbench software version 4.7. Validation of the assembled contigs was provided by confirming the presence of a previously characterized M. polymorpha gene (encoding a calcium-dependent protein kinase) (Nishiyama et al., 1999), plus two additional genes (actin MrActin and 18S rRNA). However, when the transcriptome was searched with a conventional BLAST search function using protein sequences for archetypical mono- and sesquiterpene synthases as the search queries, no contigs with significant TBLASTN sequence similarity scores (E-value < 10−3) were identified, except for four diterpene synthase-like genes (Supplemental Table 1). Similar assembly of contigs was performed with the M. polymorpha transcriptome sequence present in the NCBI SRA database (SRP029610).
To more thoroughly investigate the M. polymorpha transcriptome for terpene synthase-like contigs, the HMMER search software reliant on probabilistic rather than absolute sequence identity/similarity scoring was used in combination with conserved protein sequence domains (Pfams) associated with terpene synthases on the basis of sequence similarity and hidden Markov models. Pfams PF01397, PF03936, and PF06330 refer to a domain associated with N-terminal α-helices, a C-terminal domain associated with a metal cofactor binding function essential for initial ionization of the allylic diphosphate substrates by all terpene synthases, and a domain associated with the trichodiene synthase gene family responsible for initiating this biosynthetic pathway in Fusarium and Trichothecium species, respectively. The Pfam domain queries led to the identification of 17 putative terpene synthase transcripts, including four partial reading frames, which were not pursued further, and the four putative diterpene synthase genes identified in the conventional BLAST searches described above. When each of the contigs was then used as the query for a NCBI BLASTX search analysis, the transcripts clustered into two different groups (Supplemental Tables 2 to 5). M. polymorpha terpene synthases 1 to 9 exhibited low similarity to functionally characterized microbial-type terpene synthases and were hence designated as M. polymorpha microbial terpene synthase-like contigs MpMTPSL1 to MpMTPSL9 (Supplemental Table 6 and Supplemental Data Set 1). By contrast, contigs MpDTPS1 to MpDTPS4 demonstrated greater similarity to other plant diterpene synthases rather than to microbial forms and were thus designated as M. polymorpha diterpene synthases (MpDTPS). MpDTPS1 to 3 showed significant sequence similarity to an ent-kaurene synthase previously reported for another liverwort species Jungermannia subulata (Kawaide et al., 2011), while MpDTPS4 exhibit the greatest similarity to a gymnosperm ent-kaurene synthase (Supplemental Table 6 and Supplemental Data Set 1).
Among the Pfam search queries, the PF06330 search lead to identification of eight MpMTPSL contigs, with MpMTPSL6 and 7 exhibiting the highest similarity scores. However, only one of the putative diterpene synthase-like contigs (MpDTPS2) was recognized (Supplemental Table 4). Searches with PF01397 lead to only MpDTPS genes (Supplemental Table 2), while searches with PF03936 revealed all the MpMTPSLs, except 6 and 7 (Supplemental Table 3). To determine if these searches might be biased by the relative abundance of the MpMTPSL and MpDTPS contigs, the number of sequence reads observed for each contig (fragments per kilobase of exon per million fragments mapped [FPKM] analysis) was determined (Supplemental Figure 2). The relative abundance of all the terpene synthase contigs was directly comparable, except for MpMTPSL6, which was 5- to 10-fold more abundant than any of the other TPS-like contigs. Qualitative RT-PCR assays to measure the respective mRNA levels provided similar information (Supplemental Figure 3).
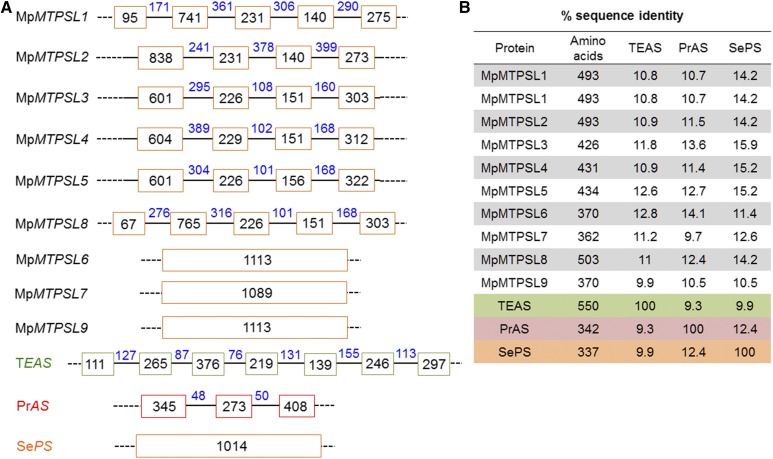
Given the low amino acid sequence similarity of MpMTPSL1-9 to microbial TPSs, additional evidence that these TPS genes are resident within the M. polymorpha genome and not derived from bacterial or fungal endophytes was sought. First, intron-exon mapping of the TPS genes was performed (Figure 3A). For comparison, vascular plant TPS genes contain six or more introns that are positionally conserved, as illustrated for the tobacco (Nicotiana tabacum) gene encoding for the TEAS enzyme (Trapp and Croteau, 2001). Fungal TPSs genes, which may contain a variable number of introns without any positional conservation, as exemplified by the two introns present in a Penicillium TPS gene, and bacterial TPS genes, of course, contain no introns, as depicted for the pentalene synthase from Streptomyces exfoliates (Figure 3A). No introns were evident within MpMTPSL6, MpMTPSL7, and MpMTPSL9, while MpMTPSL2-5 all appear to have a highly conserved intron-exon organization with three introns. MpMTPSL1 and 8 were found to have four introns with the position of the three downstream introns conserved with those of the other MpMTPSL genes. Interestingly, excluding MpMTPSL6, 7, and 9, the intron-exon organization of the MpMTPSL genes does not resemble that found in the plant or fungal TPS genes characterized to date.
Figure 3.
Structural and Organizational Comparison of M. polymorpha Terpene Synthase-Like Genes to Archetypical Plant and Microbial Terpene Synthases.
Comparison of intron-exon maps of M. polymorpha terpene synthase-like genes (MpMTPSL1-9) to the tobacco 5-epi-aristolochene synthase (TEAS), aristolochene synthase from Penicillium roqueforti (PrAS), and pentalene synthase from S. exfoliates (SePS) genes (A). Colored blocks represent exons that are connected by introns (black line). The numbers within the boxes and above the lines represent the respective nucleotide lengths. Total number of amino acids and percentage of sequence identity between the deduced amino acid sequences of the MpMTPSLs to the TEAS, PrAS, and SePS protein sequences are provided in the table in (B).
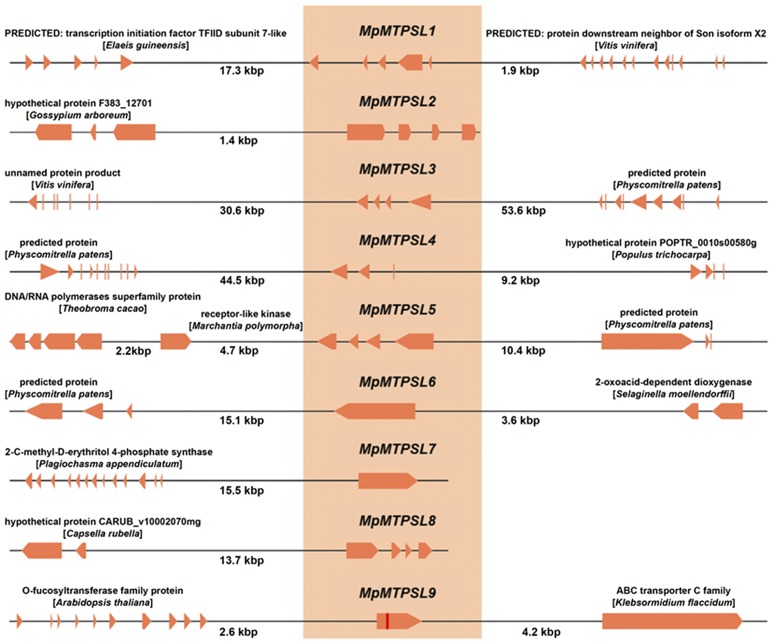
Additional evidence for the residence of the MpMTPSL genes in the M. polymorpha genome was obtained by searching the assembled reference genome of M. polymorpha (version 2.0; http://genome.jgi.doe.gov). SNAP trained for Arabidopsis and FGENESH trained for Physcomitrella patens were used to predict and annotate the genome. The resulting protein sequences of predicted genes were then searched individually using each of the nine MpMTPSLs as queries. As the protein encoding sequences of MpMTPSL2 and MpMTPSL6 are identical to genomic loci, and the sequence identities between MpMTPSL1, 5, 7, and 8 and their corresponding genomic loci are between 95 and 99% identical, we designated these as alleles of the respective genomic loci (Figure 4). By contrast, MpMTPSL3 and MpMTPSL4 are only 91 and 79% identical to genomic loci, and it remains to be determined whether these sequence differences represent allelic diversity or gene content diversity in the different M. polymorpha accessions. The genes neighboring each of the individual MpMTPSLs from the reference genome were also analyzed. Using the same gene annotation described above, the nearest genes flanking each of individual MpMTPSL genes were also identified and annotated (Figure 4). The deduced protein sequences for each of the neighboring genes were then searched against the nonredundant database of NCBI. The best hits for the neighboring genes are indicated in Figure 4, but it is worth noting that all of the neighboring genes appear to be orthologous to plant genes.
Figure 4.
Schematic Diagram of the Nearest Gene Neighbors to the Nine MpMTPSL Genes Based on the Available M. polymorpha Genomic DNA Information.
The assembled M. polymorpha genome was queried with each of the MpMTPSL gene sequences and the best matches are depicted relative to the nearest 5′ and 3′ annotated genes. Contiguous genomic sequence information was not always available on both sides of the MpMTPSL genes. Orange boxes with arrowheads represent exons, with direction of transcription indicated by the arrows, and the black lines between exons represent introns. The nearest neighbor functional annotations are based on the best BLASTp hits, with distances between genes shown below the lines. For visualization purposes, this figure is not drawn to scale.
Apart from intron-exon mapping and genome localization, the amino acid sequence comparisons between the M. polymorpha, vascular plant, fungal, and bacterial terpene synthases also suggest that the MpMTPSL genes are only distantly related to the microbial and other plant genes (Figure 3B). First, the amino acid comparisons show a low sequence identity between these proteins, where the maximum identity was ∼16% between MpMTPSL3 and the pentalene synthase from Streptomyces. Second, while there is variability in the number of amino acids associated with any family of terpene synthases, the range of amino acids associated with MpMTPSL1-5 and 8 (426 to 493) lies outside the range typical for vascular plant or bacterial TPS proteins, while the amino acid sequence range in MpMTPSL6, 7, and 9 (∼370 to 362) is quite similar to the lengths of fungal TPS. Comparisons were also performed against the recently identified microbial (SmMTPSL) and nonmicrobial (SmTPS) type terpene synthases of S. moellendorffii (Li et al., 2012), and this too showed low similarity and identity scores of <28 and <15%, respectively (Supplemental Table 6 and Supplemental Data Set 1).
Another difference between the predicted terpene synthase-like proteins of M. polymorpha and all other functionally characterized terpene synthases is the absence of the canonical DDXXD motif, with the exception of MpMTPSL2 and the plant-like diterpene synthases MpDTPS1-4 (Supplemental Figures 4 and 5). However, a second conserved metal binding motif, NDXXSXXXE, which is characteristic of nearly all class I terpene synthases (Aaron and Christianson, 2010), is completely conserved in five out of eight sequences, MpMTPSL1-4 and 6. The corresponding sequence is apparent in the other MpMTPSL proteins, but missing or having substitutions for highly conserved residues (MpMTPSL5 and 8, G for S substitution; MpMTPSL7, S for N substitution).
Functional Characterization of the MpMTPSL Genes
To initially characterize the synthases encoded by the MpMTPSL genes, the respective cDNAs were heterologously expressed in Escherichia coli and crude lysates screened for substrate preference (Table 1). Among the substrate preferences, only lysates from bacteria expressing MpMTPSL3 and MpMTPSL4 appeared capable of converting the unusual all-cis configuration of farnesyl diphosphate (FPP) (Z,Z-FPP) to hydrocarbon products detectable by GC-MS at activity levels barely above background levels of 1 ρmol h−1 µg−1. Lysates from bacteria overexpressing MpMTPSL1 and 8 exhibited only modest terpene synthase activity without any clear substrate preference. MpMTPSL2 demonstrated an unusual substrate preference for neryl diphosphate (NPP), the cis-form of geranyl diphosphate (GPP). While most monoterpene synthases catalyze the cyclization of GPP with good fidelity, NPP might be an intermediate and hence readily used. MpMTPSL2 showed a 25-fold preference for NPP over GPP, achieving a specific activity of 108.1 ± 12.7 ρmol h−1 µg−1. The highest specific activity for any of the synthases examined was 525.17 ± 14.05 ρmol h−1 µg−1 for MpMTPSL3 followed by MpMTPSL9 (250.5 ± 39.07 ρmol h−1 µg−1) using all-trans FPP as its preferred substrate, the most common physiological configuration of FPP. MpMTPSL4, 5, and 7 also exhibited a substrate preference for the all-trans FPP substrate but with much more modest activities of 41 to 56 ρmol h−1 µg−1. MpMTPSL6 was found to be the most versatile synthase because of its ability to use both mono- and sesquiterpene substrates. Interestingly, lysate from bacteria overexpressing MpMTPSL6 was found to be slightly more catalytically active with NPP as substrate than GPP (52.38 ± 1.98 versus 43.48 ± 3.64 ρmol h−1 µg−1) and possessed a comparable catalytic activity with all-trans FPP. The catalytic specificities of the MpMTPSL enzymes were confirmed with purified synthase proteins (Supplemental Figure 6), all exhibiting steady state kinetic constants (Km and kcat) (Supplemental Figure 7) comparable to those reported previously for plant and microbial terpene synthases (Supplemental Table 7) (Cane et al., 1997; Mathis et al., 1997).
Table 1. Substrate Preferences of the M. polymorpha Terpene Synthase-Like Enzymes.
| Enzyme | NPP | GPP | (2E,6E)-FPP | (2Z,6Z)-FPP | GGPP |
|---|---|---|---|---|---|
| MpMTPSL1 | 0.50 ± 0.01 | 1.60 ± 0.95 | 2.41 ± 0.63 | – | 0.18 ± 0.07 |
| MpMTPSL2 | 108.1 ± 12.7 | 3.82 ± 0.70 | 0.22 ± 0. 07 | – | 0.17 ± 0.04 |
| MpMTPSL3 | 43.9 ± 5.89 | 18.04 ± 3.54 | 525.17 ± 14.05 | + | 0.45 ± 0.10 |
| MpMTPSL4 | 2.10 ± 0.33 | 0.58 ± 0.11 | 46.59 ± 8.64 | + | 1.76 ± 0.04 |
| MpMTPSL5 | 2.18 ± 0.34 | 0.86 ± 0.19 | 56.23 ± 15.62 | – | 0.20 ± 0.05 |
| MpMTPSL6 | 52.38 ± 1.98 | 43.48 ± 3.64 | 41.48 ± 0.88 | – | 1.59 ± 0.61 |
| MpMTPSL7 | 16.94 ± 1.92 | 5.28 ± 0.13 | 49.54 ± 8.31 | – | 4.69 ± 0.23 |
| MpMTPSL8 | 0.01 ± 0.064 | 1.04 ± 0.12 | 0.30 ± 0.04 | – | 0.10 ± 0.03 |
| MpMTPSL9 | 1.8 ± 1.18 | 0.77 ± 0.27 | 250.5 ± 39.07 | + | 1.37 ± 0.79 |
Each of the respective genes was expressed in E. coli and bacterial lysates assayed for conversion of 30 µM NPP, GPP, all-trans FPP [(2E,6E)-FPP], all-cis FPP [(2Z,6Z)-FPP], or GGPP to hexane extractable (hydrocarbon) products. Assays were performed with radiolabeled substrates except for (2Z,6Z)-FPP, and activities were determined by scintillation counting or as peak areas determined by GC-MS [(2Z,6Z)-FPP] of the hexane extracts. Activities are expressed as pmol of product(s) formed/µg of lysate protein/h, with preferred substrate activities in bold, or “–” for no product detected and “+” for product detected by GC-MS.
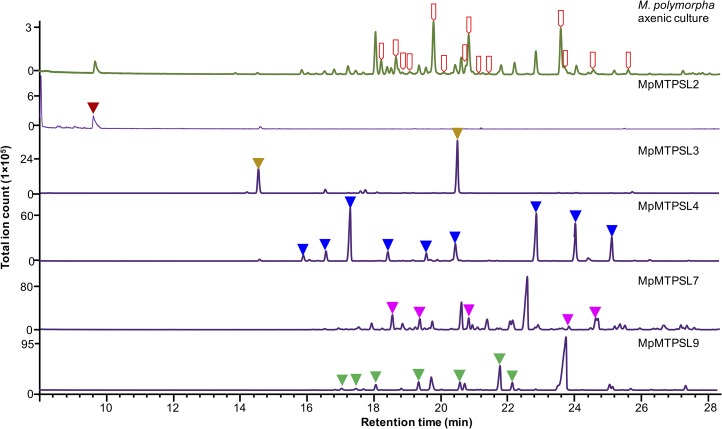
To qualify and identify the reaction products generated by the various MpMTPSL enzymes, in vitro and in vivo generated products were profiled by GC-MS relative to the terpene profile found in extracts prepared from M. polymorpha (Figures 5 and 6). The in vitro product profiles were generated by incubating the purified MpMTPSL enzymes with their preferred substrates (Table 1), while the in vivo products were produced by heterologous expression of the respective cDNAs in E. coli and Saccharomyces cerevisiae engineered for high-level accumulation of FPP (Supplemental Figures 8 to 11). Importantly, no qualitative or quantitative differences were noted in rigorous comparisons between the in vitro versus in vivo reaction profiles, as illustrated by that for MpMTPSL3 and 4 (Figure 5; Supplemental Figure 8). However, some of the MpMTPSL reaction products were not observed in the extracts prepared from M. polymorpha thallus material. For instance, of the 22 in vitro reaction products generated by MpMTPSL7, only five are evident in extracts prepared from axenic plant material. This may arise because specific metabolites are metabolized to other products or because the specific terpene is volatile and does not accumulate. Likewise, MpMTPSL6 catalyzed the conversion of GPP to cis-β-ocimene (Supplemental Figure 12), yet no ocimene was detected in extracts prepared from plant material (Figure 2). By contrast, MpMTPSL2 encodes a limonene synthase capable of using either NPP or GPP for limonene biosynthesis (Supplemental Figure 13), and limonene is a common component found during the early development stages of our propagation platform (Figure 2; Supplemental Figure 1).
Figure 5.
Comparison of the Terpene Profile of M. polymorpha to Those Generated in Vitro by the Indicated MpMTPSL Enzymes.
Equal amounts of M. polymorpha thallus tissue cultured axenically for 3, 6, and 12 months were pooled, extracted, and profiled by GC-MS, along with extracts prepared from in vitro assays of the indicated MpMTPSL enzymes purified from bacteria expressing the corresponding genes. In all cases, the preferred substrates noted in Table 1 were used. The solid arrowheads indicate those in vitro products found in the plant extract. The open red arrows indicate terpenes found in the M. polymorpha extract but not observed in any of the in vitro-synthesized reactions.
Figure 6.
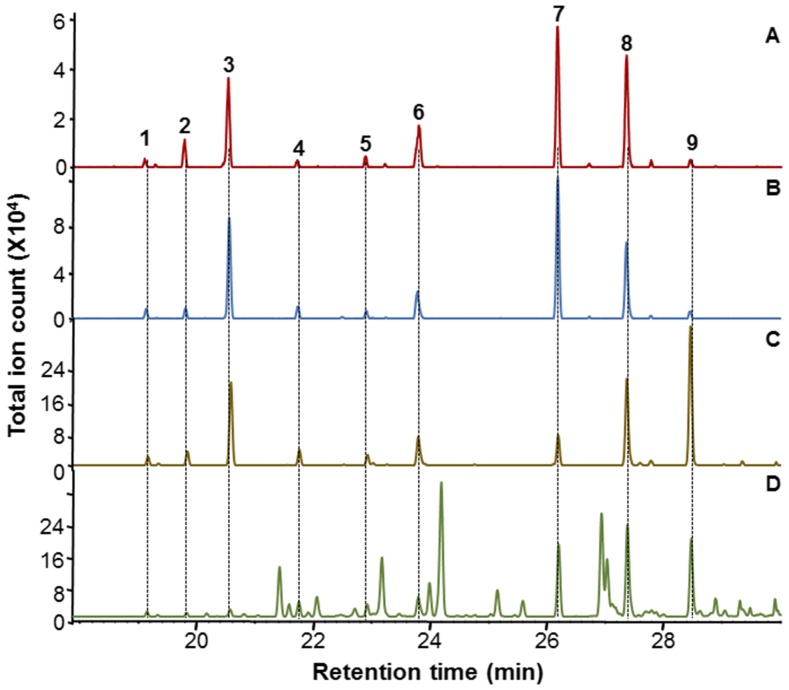
Functional Characterization of the MpMTPSL4 Sesquiterpene Synthase.
Chromatograms show the GC-MS analysis of sesquiterpenes produced by the recombinant MpMTPSL4 using all trans-farnesyl diphosphate [(2E,6E)-FPP] as substrate (A) and the metabolites produced by the heterologous expression of MpMTPSL4 in E. coli (B) and yeast (C) engineered for high-level availability of FPP in comparison to the profile of extractable terpenes from M. polymorpha (D). The in vitro products are labeled 1 to 9. MS provided in Supplemental Figure 14.
While the identified MpMTPSLs can account for approximately one-half of the terpene products observed in planta, we have not elucidated the chemical structure for the majority of these reaction products, nor have we identified all of the terpene synthases responsible for the biosynthesis of a number of the most abundant terpenes accumulating in planta. Nonetheless, among the multiple compounds generated by the MpMTPSL4 synthase were two sesquiterpene alcohols, which are dominant in the chemical profile of M. polymorpha tissue extracts and for which no reports of compounds with comparable MS patterns in the literature were found (Figures 5 and 6; Supplemental Figure 14). Because of the possible chemical novelty of these compounds, we sought their identification. One additional observation that became informative upon closer inspection of the reaction product profile of MpMTPSL4, products generated either in vitro or in vivo, was that the compound corresponding to peak 3 did not appear to accumulate in planta (Figure 6). The MS pattern for the compound corresponding to peak 3 was fully consistent with that for a sesquiterpene hydrocarbon with a parent ion of 204 [M]+ and a MS pattern consistent with that for a known sesquiterpene. Full confirmation of peak 3 corresponding to (−)-α-gurjunene was obtained by direct GC-MS and 1H-NMR comparisons with an authentic standard (Sigma-Aldrich).
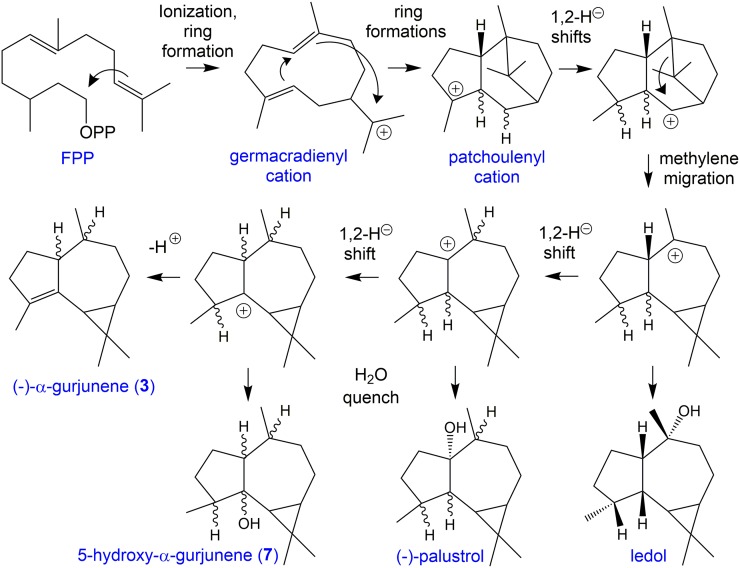
The identification of gurjunene as a biosynthetic product of MpMTPSL4 has important implications for the sesquiterpene alcohols corresponding to peaks 7 to 9 because they share MS fragments in common with gurjunene (Figure 7; Supplemental Figure 14). This leads to the possibility that the sesquiterpene alcohols might arise from carbocation reaction intermediates along the catalytic cascade to gurjunene being quenched by a water molecule, yielding formation of the alcohols. The compound corresponding to peak 7 was hence isolated and its structure determined to be 5-hydroxy-α-gurjunene according to NMR (Supplemental Tables 8 and 9). Based on this evidence and inferences from MS data (Supplemental Figure 14), we suggest that the structures of the other two alcohols corresponding to peaks 8 and 9 might be C10 hydroxylated gurjunene isomers [e.g., (+)-ledol, (+)-globulol, and (−)-viridiflorol] or C1 hydroxylated products like (+/−)-palustrol (Figure 7).
Figure 7.
Proposed Biosynthetic Mechanism for Formation of (−)-α-Gurjunene (3), 5-Hydroxy-α-Gurjunene (7), and Related Sesquiterpene Alcohols Generated by MpMTPSL4 from All-trans Farnesyl Diphosphate [(2E,6E)-FPP].
Blue lettering refers to chemical names and black lettering to biochemical processes. Adapted from Schmidt et al. (1999).
Functional Characterization of the MpDTPS Genes
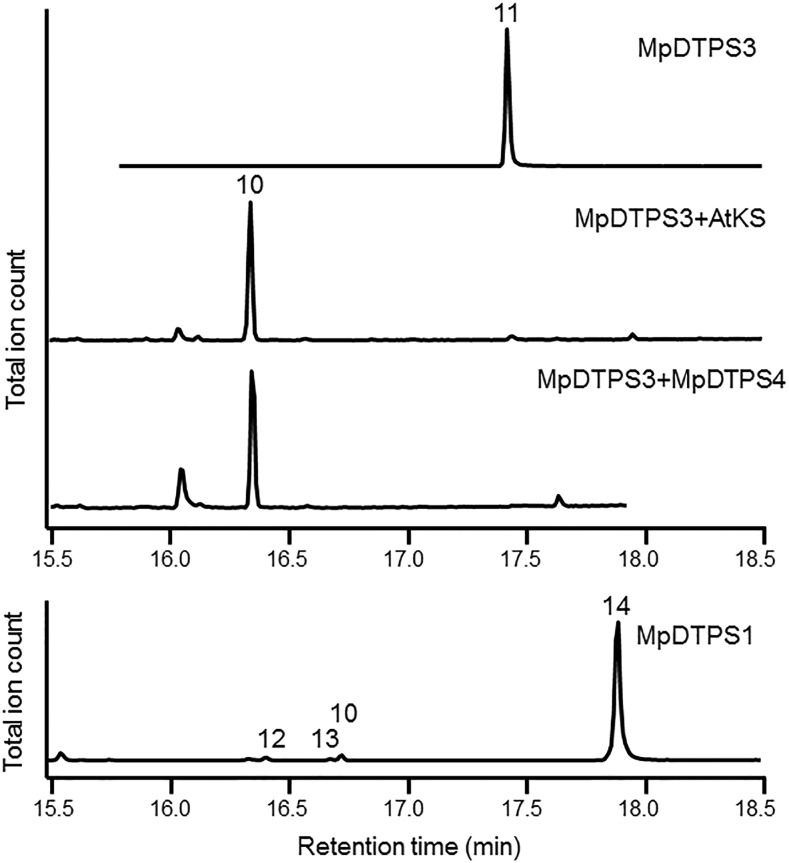
The putative M. polymorpha diterpene synthase genes, MpDTPS1, 3, and 4, were functionally characterized using an in vivo bacterial expression platform (Cyr et al., 2007). We were unable to functionally characterize MpDTPS2 because we were not able to recover a full-length cDNA for this gene and observed a frame-shift mutation within the coding sequence in any case. The system relies on the generation of the general diterpene precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) by coexpression of a GGPP synthase (GGPS) using a previously described metabolic engineering system (Cyr et al., 2007), plus one or more of the putative diterpene synthases. Bacterial cultures are then simply extracted and the organic extract containing the diterpene synthase product analyzed by GC-MS. When MpDTPS3 was evaluated in this manner, copalol was the only diterpene produced, indicating the production of copalyl diphosphate (CPP), with dephosphorylation to the primary alcohol by endogenous phosphatases (Figure 8). The stereochemical configuration of this CPP was determined by coexpression of a subsequently acting diterpene synthase possessing stereochemical selectivity for ent-, syn-, or normal CPP, much as previously described (Gao et al., 2009). In particular, MpDTPS3 was coexpressed with the GGPS plus either the ent-CPP specific kaurene synthase from Arabidopsis (AtKS), a rice (Oryza sativa) kaurene synthase-like gene (OsKSL4) specific for syn-CPP, or a variant of the Abies grandis abietadiene synthase (AgAS:D404A) that can only react with normal CPP (Supplemental Figure 15). Only when MpDTPS3 was coexpressed with AtKS was copalol no longer observed; instead, stoichiometric production of ent-kaurene was observed, thus establishing MpDTPS3 as an ent-CPP synthase (Figure 8).
Figure 8.
Functional Characterization of the M. polymorpha Diterpene Synthases.
Truncated, soluble forms of the indicated M. polymorpha and Arabidopsis enzymes were expressed in bacteria engineered for high-level production of GGPP (Cyr et al. 2007) and the extractable diterpenes profiled by GC-MS. Note that different chromatographic conditions were used for the upper and lower chromatograms. Compound identification was afforded by reference to authentic standards (10, kaurene; 11, copalol; 13, atiserene) and confirmation by NMR (14, atiseran-16-ol). Spectra are provided in Supplemental Figure 15 and Supplemental Table 10. A minor, uncharacterized diterpene product (12) was also reproducibly detected in extracts from cells expressing MpDTPS1.
Coexpression of MpDTPS4 with just the GGPS did not result in any observable diterpene product accumulation. Assuming that MpDTPS4 might act specially on CPP, the MpDTPS4 gene was coexpressed with the GGPS and with the gene encoding one of three CPP synthases (CPSs), either that from maize (Zea mays; An2/ZmCPS2) (Harris et al., 2005), rice (OsCPS4) (Xu et al., 2004), or a variant of AgAS (D621A) (Peters et al., 2001), which produces ent-, syn-, or normal CPP, respectively. Only upon coexpression with the ent-CPP producing An2/ZmCPS2 did a further elaborated diterpene appear, specifically ent-kaurene (Supplemental Figure 15). Not surprising then, when the MpDTPS3 gene was coexpressed with that for MpDTPS4, ent-kaurene was produced, supporting the identification of MpDTPS4 as a stereospecific ent-kaurene synthase (Figure 8).
Similar to MpDTPS4, coexpression of MpDTPS1 with just the GGPS also did not yield any diterpene synthase product. Again assuming that this specifically acts on CPP, MpDTPS1 was further coexpressed with each of the stereochemically differentiated CPSs. Similarly, only when coexpressed with the maize An2/ZmCPS2/ent-CPP synthase were further elaborated products observed (one dominant and three very minor; Figure 8). While two of the minor products could be identified as ent-kaurene and ent-atiserene by comparison to authentic standards by GC-MS, the major product did not match any of the available diterpene standards. To isolate a sufficient quantity for structural analysis, further metabolic engineering to increase flux to terpenoid metabolism, as previously described (Morrone et al., 2010), was employed, along with increased volumes of recombinant culture. Upon isolation and subsequent analysis by NMR, the dominant product was identified as atiseran-16-ol (Supplemental Table 10 and Supplemental Figure 16).
DISCUSSION
The impetus behind this effort was to uncover what we had hoped would be the vestiges of the first terpene synthase genes, via examination of an early diverging lineage of land plants, the liverworts. The rationale being that because liverworts are known to accumulate large amounts of structurally diverse terpenes, the forms of the genes associated with this biochemical capacity within an extant liverwort might provide insight into what structural features and what peptide domains might have helped to drive the molecular and biochemical evolution leading to the diversity of terpene synthases found in the evolutionarily derived gymnosperm and angiosperm species. Although we have functionally characterized a range of mono-, sesqui-, and diterpene synthase genes in M. polymorpha, the results suggest a much more complex picture for the evolution of terpene synthase genes within land plant species than expected.
Diterpene Synthases Provide an Anchor for Assessment
Although the diterpene synthases found in fungi and vascular plants can be functionally analogous, they are phylogenetically distinct from one another. Kaurene biosynthesis in fungi, for example, is mediated by a single bifunctional diterpene synthase enzyme converting GGPP to ent-kaurene via ent-CPP (Kawaide et al., 1997). While land plant diterpene synthases also appear to have arisen from an ancestral bifunctional enzyme, this appears to have a separate, potentially bacterial, origin (Morrone et al., 2009; Gao et al., 2012). In land plants, this bifunctional ancestral gene was presumably involved in the generation of ent-kaurene via ent-CPP for production of derived signaling molecules such as the gibberellin hormones in vascular plants. At some point, this gene underwent a duplication and subfunctionalization, with one copy retaining the class II diterpene cyclase activity for production of ent-CPP (i.e., become a CPS), while the other retained the class I diterpene synthase activity for production of ent-kaurene from ent-CPP (i.e., served as a kaurene synthase [KS]). Through additional gene duplication events, that CPS then gave rise to all plant class II diterpene cyclases, while the KS is hypothesized to have given rise not only to all plant class I diterpene synthases, but to have undergone another gene duplication and neofunctionalization event, with subsequent loss of the N-terminal domain in the non-KS paralog, which then gave rise to all the terpene synthases found in angiosperms (Gao et al., 2012). Bifunctional CPS-KSs have been found in nonvascular plants, although that from P. patens, a non-seed ancestral moss, produces largely 16α-hydroxy-ent-kaurane (Hayashi et al., 2006), while that from the liverwort J. subulata produces exclusively ent-kaurene (Kawaide et al., 2011). S. moellendorffii, a lycophyte occupying an intermediate position in the evolutionary landscape (Figure 1) between bryophytes and the euphyllophytes also has been reported to possess bifunctional diterpene synthases, although neither of those characterized to date produces ent-kaurene (Mafu et al., 2011; Sugai et al., 2011). S. moellendorffii also contains monofunctional CPSs (Li et al., 2012), one of which produces ent-CPP, as well as a monofunctional KS (Shimane et al., 2014). Given that it has been shown that gymnosperms (Keeling et al., 2010), as well as angiosperms (Hedden and Thomas, 2012), have separate CPS and KS for gibberellin biosynthesis, it seems most reasonable to assume that the underlying gene duplication and subfunctionalization of the ancestral CPS-KS to separate CPSs and KSs occurred prior to or soon after the split between the bryophyte and vascular plant lineages.
However, our results demonstrate that the diterpene synthase genes of M. polymorpha, MpDTPS1, 3, and 4, encode discrete monofunctional enzymes responsible for the production of ent-atiseranol, ent-CPP, and ent-kaurene, respectively (Figure 8; Supplemental Figure 15). Accordingly, this liverwort/bryophyte contains separate CPS (MpDTPS3) and KS (MpDTPS4) enzymes. Phylogenetic analyses suggest that these are related to other plant monofunctional CPSs and KSs (Figure 9; Supplemental Figure 17), which form the TPS-c and TPS-e/f subfamilies, respectively (Chen et al., 2011). Interestingly, in this analysis, MpCPS/DTS3 groups with previously reported bryophyte bifunctional CPS-KSs, as well as other class II CPSs, rather than the bifunctional diterpene synthases from gymnosperms or class I KSs. MpDTPS1 groups with MpKS/DTPS4, indicating homologous origins for these two class I diterpene synthases, which are most similar to the KS from S. moellendorffii. By contrast, the bifunctional diterpene synthases from S. moellendorffii are related to gymnosperm (di)terpene synthases, along with a previously reported monofunctional 16α-hydroxy-ent-kaurane synthase (Shimane et al., 2014), suggesting that this arose independently from a bifunctional ancestor, much as has been shown for monofunctional class I diterpene synthases involved in more specialized diterpene metabolism in gymnosperms (Hall et al., 2013). Equally intriguing to consider is the possibility that gene duplication and neofunctionalization of the ancestral CPS-KS to form separate monofunctional CPS and KS could have occurred multiple times, prior to and after the split between the bryophyte and vascular plant lineages. Unfortunately, the available sequence information and comparisons are not sufficient to resolve direct lineages for either of these events. Nonetheless, this split of the catalytic functions of diterpene synthase has not been uniformly retained. For example, the moss P. patens and the liverwort J. subulata appear to rely on a single bifunctional CPS-KS (Rensing et al., 2008). This may reflect the less essential role of ent-kaurene production in these nonvascular plants, which do not produce gibberellins (Hirano et al., 2007). Although there does appear to be a physiological role for ent-kaurene-derived metabolites in P. patens (Anterola et al., 2009; Hayashi et al., 2010; Miyazaki et al., 2014, 2015), its CPS-KS catalyzes the biosynthesis of mostly 16α-hydroxy-ent-kaurane (Hayashi et al., 2006), which is extruded (Von Schwartzenberg et al., 2004). In any case, given the ability of M. polymorpha to give rise to functionally distinct diterpene synthases (i.e., MpDTPS1, 3, and 4), it is unclear why this liverwort recruited separate gene families to catalyze mono- and sesquiterpene cyclization rather than to rely on evolution of these genes from diterpene synthase genes as suggested by Trapp and Croteau (2001), but this could represent a fascinating case of adaptive gene evolution by horizontal transfer.
Figure 9.
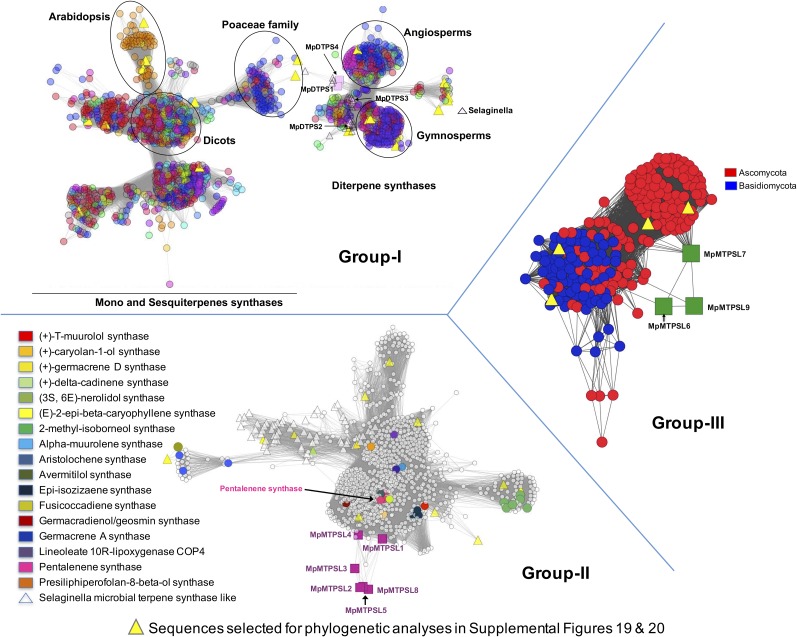
Classification of Terpene Synthase-Like Proteins Using Sequence Similarity Networks of Related Proteins.
Group I: Diterpene synthase-like proteins from M. polymorpha (MpDTPS; pink squares) in the context of their closest relatives from the SFLD, the terpene synthase-like 1 C-terminal domain subgroup, at a –log (E-value) cutoff of 110, colored by genus. Selaginella sequences are represented as magenta triangles. Group II: MpMTPSL1-5 and -8 from M. polymorpha (magenta squares) in the context of their closest relatives from the SFLD, the terpene cyclase-like 2 subgroup, at a –log (E-value) cutoff of 19, colored by Swiss-Prot annotation. S. moellendorffii sequences are represented as triangles. Note that the M. polymorpha sequences connect to other fungal and bacterial genes via pentalenene synthase. Group III: MpMTPSL6-7 and -9 from M. polymorpha (green squares) in the context of their closest relatives from the SFLD trichodiene synthase-like subgroup at a –log (E-value) cutoff of 17, colored by phylum. The sequences chosen for construction of phylogenetic trees are identified by yellow triangle nodes and represent synthases that have been functionally characterized and are evenly dispersed across the groups making up the networks.
Recruitment of Novel Gene Families Encoding Terpene Synthases in M. polymorpha
While the MpDTPS genes resemble diterpene synthases found in gymnosperms and a liverwort, the mono- and sesquiterpene synthases characterized here appear to be rather unique. This became apparent when conventional BLAST search functions of the M. polymorpha transcriptome were unsuccessful when a wide range of plant and microbial terpene synthases were used as the search queries. It was not until we employed the HMMER search algorithm with PFAM domains conserved across all microbial and plant terpene synthases that nine putative target contigs were uncovered. The complete functional characterization of these genes included heterologous expression in bacterial and yeast hosts, detailed in vitro and in vivo characterization of the encoded enzyme activities, and careful molecular documentation that these genes actually reside within the M. polymorpha genome and are expressed within the thallus tissue. It is within this suite of information that several distinguishing features became apparent for the MpMTPSLs.
First, none of the MpMTPSLs show more than 20% sequence identity to any of the archetypical plant, fungal, or bacterial terpene synthases. Second, the total number of amino acids for any of the MpMTPSLs is distinctly different from any plant or microbial TPSs. Third, exon size and intron number associated with the MpMTPSLs differ from any plant or microbial TPS genes characterized to date, features previously used to infer the molecular events underpinning the evolution of gymnosperm and angiosperm TPSs (Trapp and Croteau, 2001).
Given such low sequence similarities between the MpMTPSLs and all other TPS proteins, building statistically significant (bootstrap values greater that 50%) phylogenetic trees was not possible and could be misleading. Instead, we used sequence similarity networks (Atkinson et al., 2009; Barber and Babbitt, 2012) to place the MpMTPSLs into association with 2700 other TPS sequences housed within the Structure-Function Linkage Database (SFLD) database (Akiva et al., 2014) (Figure 9). These sequence similarity networks differ from conventional phylogenetic tree building programs in that all-by-all BLAST pairwise sequence alignments are performed rather than relying on multiple sequence alignments, and the significance of clustering can be controlled by selecting the BLAST E-value cutoff required for inclusion of an edge (line) between two sequences (nodes). In Figure 9, −log E-values for the three groups were 110, 19, and 17, respectively, with group I representing mono-, sesqui-, and diterpene synthases found in the most evolutionarily diverged plants; group II representing bacterial TPSs; and group III depicting fungal TPSs.
The edges between particular MpMTPSL proteins and others within a group indicate pairwise sequence similarities better than the specified e-value cutoff, but edge length is not directly weighted by e-value. Single edges suggest limited sequence similarities to other members within a cluster. For instance, MpMTPSL6 exhibits significant sequence similarity to a single Basidiomycota fungal TPS (POSPLDRAFT_106438) and to two other Marchantia TPSs, MpMTPSL7 and 9. By contrast, MpMTPSL7 shares sequence similarity with more than a dozen Ascomycota fungal TPSs, as well as with MpMTPSL6 and 9. For MpMTPSL6 and 9, one of their three connections to the Ascomycota TPS cluster, is mediated via their sequence similarity to MpMTPSL7.
While the connection of MpMTPSL6, 7, and 9 to the Ascomycota and Basidiomycota fungal TPS cluster might appear to be somewhat tenuous, these connections are found at a statistically significant −log E-value of 17 and thus support our hypotheses about possible origins of the Marchantia genes encoding for these proteins. The MpMTPSL6, 7, and 9 gene family could have arisen from an ancestral gene in common with fungi that duplicated and radiated within liverwort species. Or, this particular Marchantia gene family could have arisen by some convergent mechanism wherein particular domains essential for catalysis evolved in common with those found in the Ascomycota fungal TPS genes. Differentiating between these possibilities will require more information, such as 3D resolution of the TPS proteins and detailed examination of the catalytic role of the amino acid residues driving these particular sequence similarity relationships.
MpMTPSLs 1 to 5 and 8 cluster with the bacterial TPS group II that also includes many of the previously identified Selaginella enzymes (Li et al., 2012). However, it is clear that the Selaginella mono- and sesquiterpene synthases are quite distantly related to those from M. polymorpha. Moreover, the M. polymorpha bacterial-like enzymes appear to exhibit more sequence similarity among themselves than do the Selaginella enzymes, which has important implications about the evolutionary origin of the MpMTPSL1-5 and 8 genes. Recalling that these Marchantia genes possess a conserved intron-exon organization, it is relatively easy to envision that a bacterial-like TPS gene acquired by horizontal gene transfer or convergent evolution acquired introns prior to amplification and dispersal within this liverwort lineage. Precedence for such a mechanism was predicted previously when catalytic functions were associated with exon domains of sesqui- and diterpene synthases from Solonaceae and Euphorbiaceae plants (Mau and West, 1994; Back and Chappell, 1996).
Given the intron-exon organization of the M. polymorpha genes encoding MpDTPS1, 3, and 4, it is not surprising that these M. polymorpha diterpene synthases are related to similarly functioning diterpene synthases found in Angiosperms and Gymnosperms, the group I TPSs (Supplemental Figure 18). However, the importance of finding these genes in Marchantia suggests that this class of genes encoding diterpene synthases evolved prior to or just after the split of the earliest bryophytes and euphyllophytes, and based on fossil records for liverworts (Kenrick and Crane, 1997), pushes that date to ∼430 million years ago. Equally intriguing is that the genes found in angiosperms and gymnosperms encoding mono- and sesquiterpene synthases are suspected of arising from progenitor, multi-intronic diterpene synthase genes (Trapp and Croteau, 2001). Because M. polymorpha does not appear to harbor any such mono- or sesquiterpene biosynthetic enzymes clustering with similar functioning enzymes in the group I TPSs, the mechanism(s) propelling the evolutionary events for mono- and sesquiterpene synthase development must have occurred after the divergence of liverworts from the lineage leading to derived land plants and serves to further differentiate these two lineages.
Figure 9 is also highlighted with yellow triangles to identify sequences that were used to construct neighbor joining and maximum likelihood phylogenetic trees (Supplemental Figures 19 and 20 and Supplemental Data Sets 2 and 3). The overall topologies of these trees are similar to one another. That is, the MpDTPSs associate with the angiosperm and gymnosperm clade for mono-, sesqui-, and diterpene synthases, while the MpMTPSLs 1-9 associate with fungal and bacterial TPS clades. Although these trees are not statistically robust based on their bootstrap values, they too are consistent with the inferences derived from the sequence similarity networks described above.
Much More Remains Underappreciated
The GC-MS traces of Figures 2 and 5 illustrate how the profile and abundance of mono- and sequiterpene hydrocarbons and mono-oxygenate forms changes over the course of normal growth and development of M. polymorpha. Some terpenes are more abundant during the early phase of growth, while others tend to accumulate at later developmental stages. These accumulation profiles might also be correlated with the expression profile of the terpene synthase genes associated with their biosynthesis (Supplemental Figure 3). For instance, the level of MpMTPSL2 mRNA, which encodes an enzyme with limonene synthase activity, is more abundant during the early stages of development when limonene levels are high, while MpMTPSL4, which encodes an enzyme catalyzing the biosynthesis of multiple sesquiterpene products, including the dominant product palustrol, exhibits essentially the opposite trend (Figure 5; Supplemental Figures 3 and 21). MpMTPSL4 mRNA levels increase over the developmental time course, as does the accumulation of palustrol. Such associations are indicative of a possible role of a gene in the accumulation pattern of a specific terpene, but certainly far from a rigorous proof of such. More evidence, such as loss-of-function and gain-of-function alleles of specific genes, is required.
Equally important to recognize is that the profiles presented are biased by the type of analyses performed. Our analysis is specific for terpene hydrocarbons and their mono-oxygenated forms. This means that further metabolism to more oxygenated forms or conjugates with carbohydrates and other substituent groups will be missing in this analysis. This also means that we are unable to account for the possible metabolism of terpene products identified by the in vitro biochemistry. Hence, while we have identified mono- and sesquiterpene products generated by MpMTPSL enzymes fed substrates in vitro, we have only been able to document the presence of about one-half of these products in vivo. This does not mean these products are not generated in vivo because we have complemented the in vitro product profiles with in vivo profiles generated by the heterologous expression of the MpMTPSL genes in bacteria and yeast. While we cannot exclude the possibility that in vivo conditions in M. polymorpha might be different, we would suggest that at least some of the products are subject to further metabolic transformations that are not visible with the current analytic assessments, and this might include those terpene hydrocarbons that would be lost because of their volatility.
METHODS
Plant Materials and Chemical Profiling
Cultures of Marchantia polymorpha were obtained from D.N. McLetchie (Department of Biological Sciences, University of Kentucky) and B.J. Candall-Stotler (Department of Plant Biology, Southern Illinois University-Carbondale) and propagated via vegetative cuttings in standard greenhouse and growth room conditions. Axenic cultures of M. polymorpha were obtained from greenhouse-grown materials by surface sterilization in 10% bleach for 2 to 4 min, washing extensively with sterile water, and plating onto T-tissue culture media (4.2 g MS salts [Phytotechnology Laboratories], 0.112 g B5 vitamins [Phytotechnology Laboratories], 30 g sucrose, and 8 g agar without any growth regulators added) at 25°C with a 16-h-light/8-h-dark cycle provided by standard fluorescent lamps. Cultures were periodically assessed for bacterial and fungal contaminants by plating macerated thallus material onto nutrient rich microbiological mediums and microscopy examinations using vital stains. The axenic cultures have been maintained for more than 5 years. The same culture lines were also maintained under nonaxenic conditions grown in greenhouse and growth room facilities.
Chemical profiling of plant material (axenic versus nonaxenic) was performed at four developmental stages. Stages 1 (gemma still in gemma cups), 2, 3, and 4 correspond to 0, 3, 6, and 12 months of gemma development, respectively. The plant material was harvested in three replicates, stored at −80°C, and processed for chemical profiling. Frozen plant material was ground into fine powder and mixed with an equal amount (w/v) of 5 mM NaCl followed by the addition of (w/v) 100% acetone. An internal standard of 1 µg of dodecane per gram of plant material was also added, and this mixture was incubated at room temperature on an incubator shaker at 120 rpm for 3 h. Then, an equal amount of hexane:ethyl acetate (7:3 v/v) mixture was added and the samples were centrifuged at 100g for 5 min at room temperature. The extracted organic layer was passed through a hexane saturated silica gel column. One microliter of elute was analyzed by GC-MS. This method was validated for detection of mono-, sesqui-, and diterpene hydrocarbons and mono-oxidized products of each.
RNA-Seq Analysis
For RNA-seq analysis, samples of axenic M. polymorpha cultures were harvested from three biological replicates representing three different developmental stages (3, 6, and 12 months old). RNA was extracted separately from the tissue samples and equal aliquots of RNA from each stage pooled and used for paired-end sequencing on an Illumina GAIIx, followed by sequence filtering, trimming, and RNA-seq analyses according to Góngora-Castillo et al. (2012). All the sequence read data are available at the NCBI website under SRA accession number SRP074621.
Annotation of M. polymorpha MpMTPSL and MpDTPS Genes
The HMMER3.0 (http://www.hmmer.org/) algorithm was used to search the M. polymorpha assembled contigs (42,617) obtained from RNA-seq analysis and retrieve gene models encoding proteins containing the conserved terpene synthase PFAM motif sequences, N-terminal domain (PF01397), metal binding domain (PF03936), and Trichodiene synthase (TRI5) (PF06330). A relatively low stringent HMM cut off E-value of 10.0 was used for the initial searches to assure capture of all possible terpene synthase-like genes. The resulting gene models were then manually curated to consider alternative splice variants and translation start/stop sites. Differential expression among MpMTPSL and MpDTPS genes was calculated based on the FPKM method of Trapnell et al. (2012).
Isolation of Full-Length MpMTPSL and MpDTPS cDNAs
The computationally predicted M. polymorpha terpene and diterpene synthase genes (MpMTPSL and MpDTPS, respectively) were confirmed by DNA sequencing of RT-PCR amplification products from RNA isolated from axenic and nonaxenic M. polymorpha plant material. RNA was isolated and converted to single-stranded cDNA according to Yeo et al. (2013). The PCR amplification conditions were 30 cycles of 10 s at 98°C, followed by 30 s at 60°C, and 30 s at 72°C using gene-specific primers (Supplemental Tables 11 and 12). Amplification products were cloned into the pGEM-T Easy vector (Promega) and subjected to standard DNA sequencing protocols using sequencing primers (Supplemental Table 13).
Heterologous MpMTPSL Expression in Bacteria and Enzyme Characterization
The putative MpMTPSL genes were reamplified from their pGEM-T Easy vectors and ligated into the pET28a vector based on restriction sites introduced via their PCR amplification primers (Supplemental Table 14) in order to append a hexa-histidine purification tag at the N or C terminus of the encoded protein. DNA sequence confirmed clones were transformed into Escherichia coli BL21 (DE3). Bacterial expression and enzyme assays were performed as described previously by Yeo et al. (2013). Cultures initiated from single colonies were cultured at 37°C until an optical density (600 nm) of 0.8 and then protein induction of MpMTPSL genes was initiated by the addition of 1.0 mM IPTG. The cultures were incubated for an additional 7 h at room temperature with shaking. Cells were then collected by centrifugation at 4000g for 10 min, resuspended in lysis buffer of 50 mM NaH2PO4, pH 7.8, 300 mM NaCl, 5 mM imidazole, 1 mM MgCl2, 1mM PMSF, and 1% glycerol (v/v), and sonicated six times for 20 s. The cleared supernatants (16,000g for 10 min at 4°C) were used directly for enzyme assays (screening for substrate preference) as well as for affinity purification (to be used for kinetic activity measurements) according to Niehaus et al. (2011). Typical enzyme assays were initiated by mixing aliquots of the cleared supernatants with 250 mM Tris-HCl, pH 7.0, 5 mM MgCl2, and 30 µM allylic diphosphate substrates (NPP, GPP, FPP, and GGPP), and each 30 µM radiolabeled (i.e., [1-3H]NPP, [1-3H]GPP, [1-3H]FPP, and [1-3H]GGPP) allylic diphosphate substrate was prepared in such a manner that the final specific activity for each reaction was 250 dpm per ρmole. All nonradioactive allylic diphosphates substrates were purchased from Echelon Bioscience, while [1-3H] radiolabeled substrates were from American Radiolabeled Chemicals. Reactions were performed in a total reaction volume of 50 µL, incubated at 37°C for 5 min, and stopped upon addition of 50 μL of 0.2 M EDTA, pH 8.0, and 0.4 M NaOH. Reaction products were extracted with 200 μL of hexanes. Unreacted prenyl diphosphates and prenyl alcohols were removed with silica gel, and incorporation of radioactivity was measured by scintillation counter. For kinetic analyses, purified enzyme was incubated with preferred allylic diphosphate substrates (NPP, GPP, FPP, and GGPP) ranging from 0.1 to 100 μM in 50-μL reaction volumes. Each kinetic analysis was performed in three biological replicates and these three biological replicates include three technical replicates. In order to confirm reaction products, nonradioactive assays were performed using purified enzyme with substrates (NPP, GPP, FPP, and GGPP), and reaction products were profiled by GC-MS. These assays were performed in 500 μL of reaction mixture (250 mM Tris-HCl, pH 7.0, and 5 mM MgCl2) containing 10 µg of purified protein and initiated with 100 µM of the indicated allylic diphosphate substrate. After incubating for 0.5 h at 37°C, the reactions were stopped with 500 μL of stop solution (0.5 M EDTA, pH 8.0, and 0.4 M NaOH), extracted twice with an equal volume of hexanes, concentrated 2-fold under nitrogen gas, and analyzed by GC-MS. The substrates included the all-trans configurations of GPP, FPP (E,E-FPP), and GGPP for conventional mono-, sesqui-, and diterpene synthase activity measurements, as well as the cis-isomer NPP for monoterpene in radiolabeled and nonradiolabeled forms, while all cis-FPPs (Z,Z-FPP) for sesquiterpene biosynthesis only in nonradiolabeled forms were provided. For the radiolabeled assays, activity was measured as hexane extractable products quantified by scintillation counting. For the nonradiolabeled substrate Z,Z-FPP, aliquots of the hexane extracts were profiled by GC-MS.
Heterologous MpMTPSL Expression in Yeast
A modified yeast expression system was used for in vivo characterization of the MpMTPSL genes and generation of sufficient terpenes for chemical characterizations. The yeast line used for this work was ZX 178-08, derived from Saccharomyces cerevisiae 4741 by a series of genetic and molecular genetic selections for ergosterol auxotrophy, yet high FPP production (Zhuang and Chappell, 2015). The M. polymorpha terpene synthase-like genes (MpMTPSL) were cloned into modified yeast expression vectors using primers and restriction sites as noted in Supplemental Table 15. The modified yeast vectors contained the GPD promoter (Pgpd) amplified from the PYM-N14 plasmid described by Janke et al. (2004). These pESC-vectors with MpMTPSL genes were expressed in the ZX178-08 host for the production of sesquiterpenes.
For NMR studies of the compound produced by MpMTPSL4 (sesquiterpenes and sesquiterpene alcohol), a 5.0-liter fermentation of S. cerevisiae ZX17808/(pES-XHIS-TPS4) was grown for 10 d in SCE media lacking histidine before harvesting, as previously described (Yeo et al., 2013). The culture was covered in 5.0 liters of acetone, and the cells were extracted by shaking at 200 rpm for 8 h. The sesquiterpene components were extracted with 5.0 liters of hexane and dried to a volume of 10 mL under nitrogen. The 10-mL extract was spotted onto preparative TLC plates (silica gel G60; Sigma-Aldrich) and developed with cyclohexane:acetone (7:3). A portion of the plate was developed with vanillin stain, giving a characteristic blue/green color for terpene components. Zones corresponding to well-isolated sesquiterpenes were scraped and eluted in n-hexane before evaluating purity via GC-MS. (1) (−)-α-Gurjunene was isolated as a band corresponding to an Rf of 0.9 in this separation system and (2) was isolated as a band corresponding to Rf of 0.73 in the same separation system. Approximately 5 mg of α-gurjunene and 1 mg of sesquiterpene alcohol were recovered for NMR studies. α-Gurjunene was authenticated by comparison to genuine standard (Sigma-Aldrich) as determined by identical retention time and mass spectral properties, as well as identical 1H-NMR spectra. The new sesquiterpene alcohol was identified by 1H-NMR, 13C-NMR, 1H,13C-gHSQC, and 1H,1H-gCOSY spectroscopy methods.
(2S,4R,7R,8R)-3,3,7,11-Tetramethyltricyclo[6.3.0.02.4]undec-1(11)-ene, also known as (-)-α-gurjunene, was the isolated compound. The NMR spectral studies were performed using the following parameters: 1H-NMR (400.1 MHz, CDCl3); MS (EI, 70 eV), m/z (rel. int.): 204 [M]+ (100), 189 (77), 175 (10), 161 (90), 147 (33), 133 (48), 119 (74), 105 (94), 91 (71), 77 (33), 67 (18), 55 (26). The isolated compound is compared to standard compounds with the following physical and spectral properties: colorless oil; 1H-NMR (400.1 MHz, CDCl3); MS (EI, 70 eV), m/z (rel. int.): 204 [M]+ (100), 189 (76), 175 (10), 161 (88), 147 (37), 133 (51), 119 (69), 105 (97), 91 (72), 77 (35), 67 (17), 55 (25).
Terpene GC-MS Analysis
Metabolites were detected using GC-MS. One microliter of sample was injected using a splitless injection technique into a GC-MS instrument. This instrument consisted of a 7683 series autosampler, a 7890A GC system, and a 5975C inert XL mass selective detector with a Triple-Axis Detector (Agilent Technologies). The inlet temperature was set at 225°C. The compounds were separated using a HP-5MS (5%-phenyl)-methylpolysiloxane (30 m × 0.25 mm × 0.25 µm film thickness) column. Helium was used as the carrier gas at a flow rate of 0.9 mL/min. GC parameters were as follows: initial oven temperature was set at 70°C for 1 min followed by ramp 1 of 20°C/min to 90°C; ramp 2 of 3°C/min to 170°C; ramp 3 of 30°C/min to 280°C, hold at this temperature for 5 min; then final 20°C/min to 300°C. The mass selective detector transfer line and the MS quadrupole were maintained at 270°C and 150°C, respectively, whereas the MS source temperature was 230°C. Compounds were detected using the scan mode with a mass detection range of 45 to 400 atomic mass units. Retention peaks for the various terpenes were recorded using Agilent ChemStation software, and quantifications were performed relative to a dodecane internal standard.
Experimental Procedures for MpDTPS Characterization
Unless otherwise noted, chemicals were purchased from Fisher Scientific and molecular biology reagents from Invitrogen. Recombinant expression was performed with the OverExpress C41 strain of E. coli. Gas chromatography was performed with a Varian 3900 GC with Saturn 2100 ion trap mass spectrometer in electron ionization (70 eV) mode. Samples (1 µL) were injected in splitless mode at 250°C and, after holding for 3 min at 250°C, the oven temperature was raised at a rate of 15°C/min to 300°C, where it was held for an additional 3 min. MS data from 90 to 600 mass-to-charge ratio (m/z) were collected starting 15 min after injection until the end of the run.
MpDTPS Expression Constructs
The diterpene synthases from M. polymorpha were transferred to the Gateway vector system via PCR amplification and directional topoisomerization insertion into pENTR/SD/D-TOPO. Simultaneously, the diterpene synthases were truncated to remove the plastid targeting sequence from its N terminus (MpDTPS1 after residue 40 and MpDTPS3 after residue 131). MpDTPS4 was reconstructed to residue 52 (using predicted sequence) as the cloned fragment was shorter than expected. The clones were subsequently transferred via directional recombination to destination vectors pGG-DEST, which is a pACYC-Duet (Novagen /EMD)-derived plasmid with an upstream GGPP synthase and an inserted DEST cassette as previously described and/or pDEST15 (Cyr et al., 2007; Morrone et al., 2009).
Functional Characterization of MpDTPS Genes
Functional characterization of MpDTPS genes was performed using of a previously described modular metabolic engineering system (Cyr et al., 2007). For analysis of MpDTPS3 (class II), this gene was coexpressed with an upstream GGPP synthase that was carried on the pACYC-Duet (Novagen/EMD) derived plasmid as previously described (Cyr et al., 2007; Morrone et al., 2009). Stereochemistry of the CPP product was determined via coexpression of GGPPs and CPS on pACYC-Duet with KS(L) genes (carried on the pDeST15 plasmid) that selectively react with ent-, syn-, and normal CPP (AtKS, OsKSL4, and AgAs, D404A, respectively). Likewise, to assess the function and stereochemistry of KS(L) with the characteristic class I motif, the GGPP synthase and CPS were coexpressed with the pACYC-Duet-derived plasmid, where pGGeC produces ent-CPP, pGGsC produces syn-CPP, and pGGnC produces normal CPP (Peters et al., 2000; Xu et al., 2004; Harris et al., 2005; Cyr et al., 2007).
Diterpene activity was assessed by cotransformation described previously into the E. coli strain C41 (Lucigen). The recombinant bacterium was grown in a culture of TB medium (50 mL) to the mid-log phase (OD600 ∼0.6) at 37°C, then at 16°C for 1 h prior to induction with IPTG (0.5 mM). Thereafter, it was grown for an additional 72 h, after which the culture was then extracted with an equal volume of hexanes. The extract was dried under a stream of N2 and then resuspended with hexane (500 µL) for analysis by GC-MS as previously described (Wu et al., 2012; Zhou et al., 2012). Diterpene products were identified by comparison of retention time and mass spectra to that of authentic samples.
Diterpene Production
For MpDTPS3, the novel enzymatic product was obtained in sufficient amounts for NMR analysis by both increasing flux into isoprenoid metabolism and scaling up the culture volumes. Flux toward isoprenoid biosynthesis was increased using the pIRS plasmid, which encodes the methylerythritol pathway (Morrone et al., 2010). This enabled increased production of the isoprenoid precursors isopentenyl diphosphate and dimethylallyl diphosphate, while feeding 50 mM pyruvate significantly increased diterpenoid production as described previously. The resulting bacterial culture was grown in 2 × 1 liter cultures and extracted, as described above. The extract was dried by rotary evaporation, resuspended in 10 mL hexanes, and passed over a column of silica gel (10 mL), which was then eluted with ethyl acetate in hexanes (10 mL, 20% v/v) The resulting residue was dissolved in acetonitrile and the hydroxylated diterpenoid purified by HPLC. This was performed using an Agilent 1200 series instrument equipped with autosampler, fraction collector, and diode array UV detection, over a Zorbax Eclipse XDB-C8 column (4.6 × 150 mm, 5 μm) at a 0.5 mL/min flow rate. The column was preequilibrated with 50% acetonitrile/distilled water and the sample loaded. The column then washed with 50% acetonitrile/distilled water (0 to 2 min) and eluted with 50 to 100% acetonitrile (2 to 7 min), followed by a 100% acetonitrile wash (7 to 30 min). Following purification, the compound was dried under a gentle stream of N2 and then dissolved in 0.5 mL deuterated chloroform (Sigma-Aldrich), with this evaporation-resuspension process repeated two more times to completely remove the protonated acetonitrile solvent.
Diterpene Structure Identification
NMR spectra for the diterpenoids were recorded at 25°C on a Bruker Avance 700 spectrometer equipped with a cryogenic probe for 1H and 13C. Structural analysis was performed using 1D 1H, 1D DQF-COSY, HSQC, HMQC, HMBC, and NOESY experiment spectra acquired at 700 MHz and 13C (175 MHz) spectra using standard experiments from the Bruker TopSpin v1.3 software. All samples were placed in NMR tubes purged with nitrogen gas for analyses, and chemical shifts were referenced using known chloroform (13C 77.23 1H 7.24 ppm) signals offset from tetramethylsilane and compared with those previously reported (Moraes and Roque, 1988).
qRT-PCR Analysis of Terpene Synthase Gene Expression
Total RNA (2.5 µg) was extracted from M. polymorpha axenic culture (fresh weight of 200 mg), which was harvested at different developmental stages viz. 0, 3, 6, and 12 months using the RNeasy plant mini kit (Qiagen). The RNA was reverse-transcribed with SuperScript II reverse transcriptase (Invitrogen), and the PCR reaction was performed using Takara ExTaq DNA polymerase (Takara Bio) with RT product as a template. The PCR reaction was performed for 25 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s using gene-specific primer for terpene synthase as well as housekeeping genes listed in Supplemental Table 16.
Genomic DNA Extraction and Terpene Synthase Intron-Exon Mapping
Template genomic DNA was extracted using a DNeasy plant mini kit (Qiagen) and 50 ng of each sample was used per assay. Sexual determination of thalli was determined by PCR using gene-specific primers as described previously (Okada et al., 2001). To determine intron and exon boundaries, PCR amplification was performed using genomic DNA as template with gene-specific primers for MpMTPSLs (Supplemental Table 6). Amplified products were cloned in pGEMT-Easy vector and sequenced.
Phylogenetic Analysis
The phylogenetic tree for the M. polymorpha diterpene synthase genes (MpDTPS) was constructed according to the description associated with Supplemental Figure 17. Maximum likelihood and neighbor-joining phylograms of the M. polymorpha mono- and sesquiterpene synthase genes were constructed using the amino acid alignment presented in Supplemental Data Sets 2 and 3. These terpene synthase sequences were downloaded from the SFLD (Pegg et al., 2006) through the links http://sfld.rbvi.ucsf.edu/django/subgroup/1019/, http://sfld.rbvi.ucsf.edu/django/subgroup/1020/, and http://sfld.rbvi.ucsf.edu/django/subgroup/1021/. The sequences were selected based on sequence similarity network groupings, as shown by yellow triangles in Figure 9. Sequences were chosen such that each major cluster in the networks was represented, with functionally characterized proteins and proteins closely related to M. polymorpha preferentially selected. Additionally, we selected our in house-predicted terpene synthase-like genes from another liverwort, Pellia endiviifolia, based on its reported transcriptomes (Alaba et al., 2015) and recently reported hypothetical M. polymorpha genes having similarity with terpene synthases genes from our study (Proust et al., 2016). Multiple alignment was performed on the amino acid sequences of the 59 selected genes (including M. polymorpha) using PROMALS3D with default parameters (Pei et al. 2008). The aligned sequences were trimmed manually to remove the gaps using Jalview as alignment editor (Clamp et al., 2004). After removing gaps, the sequences were aligned again using PROMALS3D. The aligned amino acid sequences (Supplemental Data Set 2) were used to build maximum likelihood and neighbor-joining phylogenetic trees using MEGA 7.0 (Kumar et al., 2016) using the LG matrix-based substitution model only for maximum likelihood method (Le and Gascuel, 2008). The selection of the substitution model was based on the ProtTest 2.4 model search program at http://darwin.uvigo.es/software/prottest2_server.html with default criteria (Abascal et al., 2005). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a LG model. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 10.1109)). The rate variation model allowed for some sites to be evolutionarily invariable [+I] (JTT matrix-based model; (ones et al., 1992). The tree was further annotated manually based on a description of terpene synthase genes reported in the Uniprot database (Supplemental Data Set 4).
Sequence Similarity Networks
Full sequence similarity networks for the Terpene Cyclase Like 2 and Trichodiene Synthase Like subgroups, a representative set of sequences for the Terpene Cyclase Like 1 C-terminal domain subgroup, along with their closest relatives from M. polymorpha, were obtained from the SFLD (Akiva et al., 2014). SFLD networks were created using Pythoscape (Barber and Babbitt, 2012). Each node (circle) represents a unique sequence from the appropriate subgroup, and each edge (line) between two nodes indicates that the sequences represented by the connected nodes had a BLAST similarity score with an E-value at least as significant as the specified cutoff. The nodes were arranged using the yFiles organic layout provided with Cytoscape version 2.8 (Smoot et al., 2011). Lengths of edges are not meaningful except that sequences in tightly clustered groups are relatively more similar to each other than sequences with few connections.
Accession Numbers
Sequence data from this article can be found in the GenBank/NCBI databases under the following accession numbers: KU664188, MpMTPSL1; KU664189, MpMTPSL2; KU664190, MpMTPSL3; KU664191, MpMTPSL4; KU664192, MpMTPSL5; KU664193, MpMTPSL6; KU664194, MpMTPSL7; KU664195, MpMTPSL8; KU886240, MpMTPSL9; KU664196, MpDTPS1; KU664197, MpDTPS3; and KU664198, MpDTPS4.
Supplemental Data
Supplemental Figure 1. Major terpenes present in axenic (sterile) cultures of Marchantia polymorpha harvested after 0, 3, 6, and 12 months of growth.
Supplemental Figure 2. Measurement of terpene synthase-like gene expression based on the fragments per kilobase of exon per million fragments mapped.
Supplemental Figure 3. Relative expression of the terpene synthase-like genes (MpMTPSL) from axenic as well as nonaxenic M. polymorpha tissue grown for 0, 3, 6, and 12 months.
Supplemental Figure 4. Web logo-based consensus sequence pattern for Marchantia terpene synthase-like genes and aspartate-rich substrate binding motif description using ClustalW alignment.
Supplemental Figure 5. ClustalW alignment of the diterpene synthase-like genes present in M. polymorpha (MpDTPS1-4) centered on class I (DXDD) and class II (DDXXD) divalent metal binding motifs.
Supplemental Figure 6. Purification of recombinant MpMTPSL terpene synthases.
Supplemental Figure 7A. Enzyme kinetic determinations for MpMTPSL2, 3, 4, 5, 6, and 7.
Supplemental Figure 7B. Enzyme kinetic determinations for MpMTPSL4 and MpMTPSL9 for total (nonscrubbed) and all hydrocarbon (scrubbed) reaction products.
Supplemental Figure 8. GC chromatogram of terpene reaction product(s) generated by MpMTPSL3 in vitro and in vivo.
Supplemental Figure 9. GC chromatogram of terpene reaction product(s) generated by MpMTPSL5 in vitro and in vivo.
Supplemental Figure 10. GC chromatogram of terpenes generated by MpMTPSL7 in vitro and in vivo.
Supplemental Figure 11. GC chromatogram of terpenes generated by MpMTPSL9 in vitro and in vivo.
Supplemental Figure 12. GC chromatogram of terpene reaction product(s) generated in vitro by MpMTPSL6.
Supplemental Figure 13. GC chromatograms of terpene reaction product(s) generated by MpMTPSL2 in vitro using NPP as substrate.
Supplemental Figure 14. Mass spectra of selected compounds produced by MpMTPSL4 as shown in Figure 6 and for (−)-α-gurjunene standard.
Supplemental Figure 15. GC-MS analysis of diterpenes products generated by coexpression of MpDTPS4 with GGPP synthase plus An2, or OsCPS2 or AgAS:D621A, which are copalyl diphosphate synthases that produce ent-, syn-, and normal CPP from GGPP, respectively.
Supplemental Figure 16. Numbering and selected 1H-1H COSY, HMBC, and NOESY correlations for atiseranol.
Supplemental Figure 17. Phylogenetic relationships of MpDTPS1, 3, and 4 to other plant monofunctional CPSs and KSs and a bifunctional KS from Physcomitrella as inferred using the neighbor-joining method (Saitou and Nei, 1987).
Supplemental Figure 18. Intron-exon organization of MpDTPS1 to 4 in comparison to a typical monofunctional diterpene synthase (CPS) found in Arabidopsis (AT4g02780) (Sun and Kamiya, 1994).
Supplemental Figure 19. Phylogenetic analysis of the terpene synthase-like and diterpene synthase-like proteins from M. polymorpha in relationship to bacterial, fungal, and plant terpene synthase proteins (Supplemental Data Sets 2 and 4).
Supplemental Figure 20. Evolutionary relationships of Marchantia terpenes synthase proteins to other terpene synthase proteins presented in Supplemental Data Sets 2 and 4.
Supplemental Figure 21. Developmental time course for the amounts of the major MpMTPSL4 products found in M. polymorpha.
Supplemental Table 1. TBLASTN search of Marchantia transcriptome database (42,617 contigs) using archetypical mono- and sesquiterpene synthases.
Supplemental Table 2. Pfam domain search of the M. polymorpha assembled contigs with PF01397.
Supplemental Table 3. Pfam domain search of the M. polymorpha assembled contigs with PF03936.
Supplemental Table 4. Pfam domain search of the M. polymorpha assembled contigs with PF06330.
Supplemental Table 5. Summary of Pfam domain searches of the M. polymorpha transcriptome (45,309 contigs, assembled from the NCBI SRA database SRP029610 according to Sharma et al., 2013) for PF01397, PF03936, and PF06330 motifs.
Supplemental Table 6. References sequences used for similarity and identity comparisons.
Supplemental Table 7. Kinetic constants for the terpene synthase-like enzymes from M. polymorpha.
Supplemental Table 8. 1H-NMR for compound 7 (from Figure 6) in CDCl3.
Supplemental Table 9. 13C-NMR data for compound 7 (from Figure 6), (+)-globulol, and (+)-ledol in CDCl3.
Supplemental Table 10. 1H- and 13C-NMR data for atiseranol.
Supplemental Table 11. Primers used for cloning full-length MpMTPSL genes.
Supplemental Table 12. Primers used for cloning full length MpDTPS genes.
Supplemental Table 13. Primers used for DNA sequencing.
Supplemental Table 14. Primers used for cloning of MpMTPSL genes into bacterial (E. coli) protein expression vectors and their restriction site.
Supplemental Table 15. Primers used for cloning MpMTPSL genes into specific restriction sites within yeast expression vectors.
Supplemental Table 16. Primers used qualitative RT-PCR.
Supplemental Data Set 1. Sequence comparison (similarity and identity) of Marchantia terpene synthase-like proteins to selected terpene synthase proteins from primitive plants like Selaginella (SmMTPSL and SmTPS) and Physcomitrella (PhyTPS), diverged plants such as gymnosperm (TASY_TAXBR), and angiosperm (TEAS from tobacco and 4LS from peppermint), fungal (TRI5_FUSSP), and bacterial (ScGS) terpene synthases.
Supplemental Data Set 2. Multiple sequence alignment of Marchantia terpene synthase genes (MpMTPSL and MpDTPS) with unique terpene synthase sequences from the Structure Function Linkage Database and chosen on the basis of their relationship to one another as illustrated in Figure 9.
Supplemental Data Set 3. Multiple sequence alignment of Marchantia diterpene synthase proteins (MpCPS1, MpKS, and MpDTS; MpDTPS3, 4, and 1, respectively) with other characterized monofunctional diterpene synthase sequences (CPSs and KSs) and the bifunctional KSs from Physcomitrella, Jungermannia, and Selaginella.
Supplemental Data Set 4. Correspondence between symbols used in protein sequence alignment (Supplemental Data Set 2) and phylogenetic trees (Supplemental Figures 19 and 20) to their identifiers in UniProt and NCBI databases.
Supplementary Material
Acknowledgments
We thank Tobias G. Köllner and Jonathan Gershenzon for their contributions in the early stage of this study. This work was supported, in part, by Grants 1RC2GM092521 from the National Institutes of Health to JC, GM076324 from the National Institutes of Health to RJP, an Innovation Grant from the University of Tennessee Institute of Agriculture to F.C., GM60595 from the National Institutes of Health to P.C.B., and DP160100892 from the Australian Research Council to J.L.B. We also thank our colleagues at Joint Genome Institute for access to the M. polymorpha genome sequence information, work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02-05CH11231.
AUTHOR CONTRIBUTIONS
S.K., C.K., S.A.B., S.E.N., X.Z., S.E.K., Z.J., S.G., K.B.L., A.N., X.C., Q.J., S.M., J.Z., and S.D.B. designed and conducted the experiments. S.K., F.C., R.J.P., P.C.B., J.L.B., and J.C. designed the project, directed the experimental work, guided the discussions, and wrote the article.
Glossary
- GC-MS
gas chromatography-mass spectroscopy
- FPP
farnesyl diphosphate
- GPP
geranyl diphosphate
- NPP
neryl diphosphate
- GGPP
geranylgeranyl diphosphate
- GGPS
GGPP synthase
- CPP
copalyl diphosphate
- KS
kaurene synthase
- FPKM
fragments per kilobase of exon per million fragments mapped
- SFLD
Structure-Function Linkage Database
Footnotes
Articles can be viewed without a subscription.
References
- Aaron J.A., Christianson D.W. (2010). Trinuclear metal clusters in catalysis by terpenoid synthases. Pure Appl. Chem. 82: 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Aharoni A., Giri A.P., Deuerlein S., Griepink F., de Kogel W.J., Verstappen F.W., Verhoeven H.A., Jongsma M.A., Schwab W., Bouwmeester H.J. (2003). Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiva E., et al. (2014). The structure-function linkage database. Nucleic Acids Res. 42: D521–D530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaba S., Piszczalka P., Pietrykowska H., Pacak A.M., Sierocka I., Nuc P.W., Singh K., Plewka P., Sulkowska A., Jarmolowski A., Karlowski W.M., Szweykowska-Kulinska Z. (2015). The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol. 206: 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anterola A., Shanle E., Mansouri K., Schuette S., Renzaglia K. (2009). Gibberellin precursor is involved in spore germination in the moss Physcomitrella patens. Planta 229: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Asakawa Y. (1982). Chemical constituents of the hepaticae. In Progress in the Chemistry of Organic Natural Products, W.Herz, H. Grisebach, and G.W. Kirby, eds (Vienna, Austria: Springer Verlag), pp. 269–285. [Google Scholar]
- Asakawa Y. (2011). Bryophytes: Chemical diversity, synthesis and biotechnology. A review. Flavour Fragr. J. 26: 318–320. [Google Scholar]
- Atkinson H.J., Morris J.H., Ferrin T.E., Babbitt P.C. (2009). Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4: e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back K., Chappell J. (1996). Identifying functional domains within terpene cyclases using a domain-swapping strategy. Proc. Natl. Acad. Sci. USA 93: 6841–6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J.A., et al. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber A.E. II, Babbitt P.C. (2012). Pythoscape: a framework for generation of large protein similarity networks. Bioinformatics 28: 2845–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham J. (2013). Dictionary of Natural Products. (London: Taylor and Francis Group). [Google Scholar]
- Cane D.E., Chiu H.T., Liang P.H., Anderson K.S. (1997). Pre-steady-state kinetic analysis of the trichodiene synthase reaction pathway. Biochemistry 36: 8332–8339. [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., Bohlmann J., Pichersky E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66: 212–229. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J. (2004). The Jalview Java alignment editor. Bioinformatics 20: 426–427. [DOI] [PubMed] [Google Scholar]
- Cyr A., Wilderman P.R., Determan M., Peters R.J. (2007). A modular approach for facile biosynthesis of labdane-related diterpenes. J. Am. Chem. Soc. 129: 6684–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E.M., Croteau R. (2000). Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. In Topics in Current Chemistry: Biosynthesis—Aromatic Polyketides, Isoprenoids, Alkaloids, F.J. Leeper and J.C. Vederas, eds (Heidelberg, Germany: Springer-Verlag; ), pp. 53–95. [Google Scholar]
- Degenhardt J., Köllner T.G., Gershenzon J. (2009). Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637. [DOI] [PubMed] [Google Scholar]
- Dickschat J.S., Brock N.L., Citron C.A., Tudzynski B. (2011). Biosynthesis of sesquiterpenes by the fungus Fusarium verticillioides. ChemBioChem 12: 2088–2095. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gahtori D., Chaturvedi P. (2011). Antifungal and antibacterial potential of methanol and chloroform extracts of Marchantia polymorpha L. Arch. Phytopathol. Pflanzenschutz 44: 726–731. [Google Scholar]
- Gao W., Hillwig M.L., Huang L., Cui G., Wang X., Kong J., Yang B., Peters R.J. (2009). A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 11: 5170–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Honzatko R.B., Peters R.J. (2012). Terpenoid synthase structures: a so far incomplete view of complex catalysis. Nat. Prod. Rep. 29: 1153–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J., Dudareva N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3: 408–414. [DOI] [PubMed] [Google Scholar]
- Gil M., Pontin M., Berli F., Bottini R., Piccoli P. (2012). Metabolism of terpenes in the response of grape (Vitis vinifera L.) leaf tissues to UV-B radiation. Phytochemistry 77: 89–98. [DOI] [PubMed] [Google Scholar]
- Góngora-Castillo E., Fedewa G., Yeo Y., Chappell J., DellaPenna D., Buell C.R. (2012). Genomic approaches for interrogating the biochemistry of medicinal plant species. Methods Enzymol. 517: 139–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottsche C.M. (1843). Anatomische-physiologische Untersuchungen uber Haplomitrium Hookeri N. v. E. mit Vergleichung anderer Lebermoose. Nov. Actorum Acad. Caes. Leop. Carol. Nac. Cur. 20: 267–289. [Google Scholar]
- Hall D.E., Zerbe P., Jancsik S., Quesada A.L., Dullat H., Madilao L.L., Yuen M., Bohlmann J. (2013). Evolution of conifer diterpene synthases: diterpene resin acid biosynthesis in lodgepole pine and jack pine involves monofunctional and bifunctional diterpene synthases. Plant Physiol. 161: 600–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L.J., Saparno A., Johnston A., Prisic S., Xu M., Allard S., Kathiresan A., Ouellet T., Peters R.J. (2005). The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol. Biol. 59: 881–894. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Horie K., Hiwatashi Y., Kawaide H., Yamaguchi S., Hanada A., Nakashima T., Nakajima M., Mander L.N., Yamane H., Hasebe M., Nozaki H. (2010). Endogenous diterpenes derived from ent-kaurene, a common gibberellin precursor, regulate protonema differentiation of the moss Physcomitrella patens. Plant Physiol. 153: 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Kawaide H., Notomi M., Sakigi Y., Matsuo A., Nozaki H. (2006). Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 580: 6175–6181. [DOI] [PubMed] [Google Scholar]
- He X., Sun Y., Zhu R.-L. (2013). The oil bodies of liverworts: Unique and important organelles in land plants. CRC Crit. Rev. Plant Sci. 32: 293–302. [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Hirano K., et al. (2007). The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorffii but not in the bryophyte Physcomitrella patens. Plant Cell 19: 3058–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huneck S. (1983). Chemistry and biochemistry of bryophytes. In New Manual of Bryology, Vol. 1, Schuster R.M., ed (Nichinan, Japan: Hattori Botanical Laboratory; ), pp. 1–116. [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kawaide H., Hayashi K., Kawanabe R., Sakigi Y., Matsuo A., Natsume M., Nozaki H. (2011). Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens. FEBS J. 278: 123–133. [DOI] [PubMed] [Google Scholar]
- Kawaide H., Imai R., Sassa T., Kamiya Y. (1997). Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J. Biol. Chem. 272: 21706–21712. [DOI] [PubMed] [Google Scholar]
- Keeling C.I., Dullat H.K., Yuen M., Ralph S.G., Jancsik S., Bohlmann J. (2010). Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiol. 152: 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P., Crane P.R. (1997). The origin and early evolution of plants on land. Nature 389: 33–39. [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Li G., Köllner T.G., Yin Y., Jiang Y., Chen H., Xu Y., Gershenzon J., Pichersky E., Chen F. (2012). Nonseed plant Selaginella moellendorffi [corrected] has both seed plant and microbial types of terpene synthases. Proc. Natl. Acad. Sci. USA 109: 14711–14715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg S.O. (1888). Bidrag till nordens mossflora. Meddelanden af Societas pro Fauna et Flora Fennica 14: 63–77. [Google Scholar]
- Lohmann C.E.J. (1903). Beitrag zur chemie und biologie der leber moose. Botan. Centrallbl. Beih. 15: 215–256. [Google Scholar]
- Mafu S., Hillwig M.L., Peters R.J. (2011). A novel labda-7,13e-dien-15-ol-producing bifunctional diterpene synthase from Selaginella moellendorffii. ChemBioChem 12: 1984–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienhagen J., Bott M. (2013). Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 163: 166–178. [DOI] [PubMed] [Google Scholar]
- Mathis J.R., Back K., Starks C., Noel J., Poulter C.D., Chappell J. (1997). Pre-steady-state study of recombinant sesquiterpene cyclases. Biochemistry 36: 8340–8348. [DOI] [PubMed] [Google Scholar]
- Mau C.J., West C.A. (1994). Cloning of casbene synthase cDNA: evidence for conserved structural features among terpenoid cyclases in plants. Proc. Natl. Acad. Sci. USA 91: 8497–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishler B.D., Lewis L.A., Buchheim M.A., Renzaglia K.S., Garbary D.J., Delwiche C.F., Zechman F.W., Kantz T.S., Chapman R.L. (1994). Phylogenetic relationships of the “Green Algae” and Bryophytes. Ann. Mo. Bot. Gard. 81: 451–483. [Google Scholar]
- Miyazaki S., Nakajima M., Kawaide H. (2015). Hormonal diterpenoids derived from ent-kaurenoic acid are involved in the blue-light avoidance response of Physcomitrella patens. Plant Signal. Behav. 10: e989046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S., Toyoshima H., Natsume M., Nakajima M., Kawaide H. (2014). Blue-light irradiation up-regulates the ent-kaurene synthase gene and affects the avoidance response of protonemal growth in Physcomitrella patens. Planta 240: 117–124. [DOI] [PubMed] [Google Scholar]
- Moraes M.P.L., Roque N.F. (1988). Diterpenes from the fruits of Xylopia aromatica. Phytochemistry 27: 3205–3208. [Google Scholar]
- Morrone D., Chambers J., Lowry L., Kim G., Anterola A., Bender K., Peters R.J. (2009). Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 583: 475–480. [DOI] [PubMed] [Google Scholar]
- Morrone D., Lowry L., Determan M.K., Hershey D.M., Xu M., Peters R.J. (2010). Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 85: 1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus T.D., Okada S., Devarenne T.P., Watt D.S., Sviripa V., Chappell J. (2011). Identification of unique mechanisms for triterpene biosynthesis in Botryococcus braunii. Proc. Natl. Acad. Sci. USA 108: 12260–12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R., Mizuno H., Okada S., Yamaguchi T., Takenaka M., Fukuzawa H., Ohyama K. (1999). Two mRNA species encoding calcium-dependent protein kinases are differentially expressed in sexual organs of Marchantia polymorpha through alternative splicing. Plant Cell Physiol. 40: 205–212. [DOI] [PubMed] [Google Scholar]
- Oh I., Yang W.Y., Park J., Lee S., Mar W., Oh K.B., Shin J. (2011). In vitro Na+/K+-ATPase inhibitory activity and antimicrobial activity of sesquiterpenes isolated from Thujopsis dolabrata. Arch. Pharm. Res. 34: 2141–2147. [DOI] [PubMed] [Google Scholar]
- Okada S., Sone T., et al. (2001). The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl. Acad. Sci. USA 98: 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C., König W.A., Muhle H. (2001). Pacifigorgianes and tamariscene as constituents of Frullania tamarisci and Valeriana officinalis. Phytochemistry 57: 307–313. [DOI] [PubMed] [Google Scholar]
- Pegg S.C., Brown S.D., Ojha S., Seffernick J., Meng E.C., Morris J.H., Chang P.J., Huang C.C., Ferrin T.E., Babbitt P.C. (2006). Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry 45: 2545–2555. [DOI] [PubMed] [Google Scholar]
- Pei J., Kim B.H., Grishin N.V. (2008). PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36: 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R.J., Flory J.E., Jetter R., Ravn M.M., Lee H.J., Coates R.M., Croteau R.B. (2000). Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry 39: 15592–15602. [DOI] [PubMed] [Google Scholar]
- Peters R.J., Ravn M.M., Coates R.M., Croteau R.B. (2001). Bifunctional abietadiene synthase: free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J. Am. Chem. Soc. 123: 8974–8978. [DOI] [PubMed] [Google Scholar]
- Proust H., Honkanen S., Jones V.A.S., Morieri G., Prescott H., Kelly S., Ishizaki K., Kohchi T., Dolan L. (2016). RSL Class I genes controlled the development of epidermal structures in the common ancestor of land plants. Curr. Biol. 26: 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y.L., et al. (2006). The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl. Acad. Sci. USA 103: 15511–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Schmidt C.O., Bouwmeester H.J., Bülow N., König W.A. (1999). Isolation, characterization, and mechanistic studies of (-)-alpha-gurjunene synthase from Solidago canadensis. Arch. Biochem. Biophys. 364: 167–177. [DOI] [PubMed] [Google Scholar]
- Schuster R.M. (1966). The Hepaticae and Anthocerotae of North America East of the Hundredth Meridian, Vol. 1 (New York: Columbia University Press; ). [Google Scholar]
- Schwab W., Wüst M. (2015). Understanding the constitutive and induced biosynthesis of mono- and sesquiterpenes in grapes (Vitis vinifera): A key to unlocking the biochemical secrets of unique grape aroma profiles. J. Agric. Food Chem. 63: 10591–10603. [DOI] [PubMed] [Google Scholar]
- Shimane M., Ueno Y., Morisaki K., Oogami S., Natsume M., Hayashi K., Nozaki H., Kawaide H. (2014). Molecular evolution of the substrate specificity of ent-kaurene synthases to adapt to gibberellin biosynthesis in land plants. Biochem. J. 462: 539–546. [DOI] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai Y., Ueno Y., Hayashi K., Oogami S., Toyomasu T., Matsumoto S., Natsume M., Nozaki H., Kawaide H. (2011). Enzymatic 13C labeling and multidimensional NMR analysis of miltiradiene synthesized by bifunctional diterpene cyclase in Selaginella moellendorffii. J. Biol. Chem. 286: 42840–42847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suire C., Bouvier F., Backhaus R.A., Bégu D., Bonneu M., Camara B. (2000). Cellular localization of isoprenoid biosynthetic enzymes in Marchantia polymorpha. Uncovering a new role of oil bodies. Plant Physiol. 124: 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., Chen F., Petri J., Gershenzon J., Pichersky E. (2005). Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 42: 757–771. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp S.C., Croteau R.B. (2001). Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158: 811–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan M.M., Wang Q., Webster F.X., Kiemle D., Hong Y.J., Tantillo D.J., Coates R.M., Wray A.T., Askew W., O’Donnell C., Tokuhisa J.G., Tholl D. (2013). Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 25: 1108–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holle G. (1857). Eine Pflanzen-Physiolog. (Heidelberg, Germany: Ueber die Zellenblaschen der Lebermoose). [Google Scholar]
- Von Schwartzenberg K., Schultze W., Kassner H. (2004). The moss Physcomitrella patens releases a tetracyclic diterpene. Plant Cell Rep. 22: 780–786. [DOI] [PubMed] [Google Scholar]
- Wu S., Schoenbeck M.A., Greenhagen B.T., Takahashi S., Lee S., Coates R.M., Chappell J. (2005). Surrogate splicing for functional analysis of sesquiterpene synthase genes. Plant Physiol. 138: 1322–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhou K., Toyomasu T., Sugawara C., Oku M., Abe S., Usui M., Mitsuhashi W., Chono M., Chandler P.M., Peters R.J. (2012). Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hillwig M.L., Prisic S., Coates R.M., Peters R.J. (2004). Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39: 309–318. [DOI] [PubMed] [Google Scholar]
- Yeo Y.S., et al. (2013). Functional identification of valerena-1,10-diene synthase, a terpene synthase catalyzing a unique chemical cascade in the biosynthesis of biologically active sesquiterpenes in Valeriana officinalis. J. Biol. Chem. 288: 3163–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Xu M., Tiernan M., Xie Q., Toyomasu T., Sugawara C., Oku M., Usui M., Mitsuhashi W., Chono M., Chandler P.M., Peters R.J. (2012). Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Chappell J. (2015). Building terpene production platforms in yeast. Biotechnol. Bioeng. 112: 1854–1864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.