Bamboo mosaic virus satellite RNA can move autonomously in a fibrillarin-dependent manner in the absence of its helper virus in Nicotiana benthamiana.
Abstract
RNA trafficking plays pivotal roles in regulating plant development, gene silencing, and adaptation to environmental stress. Satellite RNAs (satRNAs), parasites of viruses, depend on their helper viruses (HVs) for replication, encapsidation, and efficient spread. However, it remains largely unknown how satRNAs interact with viruses and the cellular machinery to undergo trafficking. Here, we show that the P20 protein of Bamboo mosaic potexvirus satRNA (satBaMV) can functionally complement in trans the systemic trafficking of P20-defective satBaMV in infected Nicotiana benthamiana. The transgene-derived satBaMV, uncoupled from HV replication, was able to move autonomously across a graft union identified by RT-qPCR, RNA gel blot, and in situ RT-PCR analyses. Coimmunoprecipitation experiments revealed that the major nucleolar protein fibrillarin is coprecipitated in the P20 protein complex. Notably, silencing fibrillarin suppressed satBaMV-, but not HV-, phloem-based movement following grafting or coinoculation with HV. Confocal microscopy revealed that the P20 protein colocalized with fibrillarin in the nucleoli and formed punctate structures associated with plasmodesmata. The mobile satBaMV RNA appears to exist as ribonucleoprotein (RNP) complex composed of P20 and fibrillarin, whereas BaMV movement proteins, capsid protein, and BaMV RNA are recruited with HV coinfection. Taken together, our findings provide insight into movement of satBaMV via the fibrillarin-satBaMV-P20 RNP complex in phloem-mediated systemic trafficking.
INTRODUCTION
RNA trafficking is essential for plant development, nutrient allocation, gene silencing, and stress responses (Lucas et al., 2001; Kehr and Buhtz, 2008; Turgeon and Wolf, 2009; Ursache et al., 2014). For efficient trafficking, plants have evolved complex networks of regulatory components that enable local and long-distance communication (Middleton et al., 2012). While systemic trafficking is enabled by phloem transport, local cell-to-cell communication relies on microchannels that traverse plant cell walls, known as plasmodesmata (PD) (Lucas et al., 2009; Kragler, 2013). Companion cell-sieve element PDs mediate the selective trafficking of RNAs through the phloem translocation stream (Aoki et al., 2005; Kehr and Buhtz, 2008; Turgeon and Wolf, 2009; Ursache et al., 2014). Recent findings on the movement of small RNAs, including microRNAs and small interfering RNAs (Dunoyer et al., 2010; Molnar et al., 2010), have greatly advanced our understanding of the intercellular signaling that coordinates gene expression during development.
RNAs can assemble with various proteins into ribonucleoprotein (RNP) complexes during cell-to-cell or long-distance trafficking through the phloem (Haywood et al., 2005; Bailey-Serres et al., 2009; Ham et al., 2009; Pallas and Gómez, 2013). Long-distance trafficking has been shown to be mediated by the binding of RNA to the RNA-interacting domains of phloem-mobile RNA binding proteins (RBPs) (Ham et al., 2009; Pallas and Gómez, 2013). For example, phloem RBP50 binds to the polypyrimidine-tract binding motif of GA-INSENSITIVE PHLOEM RNA (Ham et al., 2009) for phloem-mediated trafficking. In addition, phloem RBPs can selectively bind to small RNAs, as well as mRNAs or viral RNAs, to mediate their trafficking (Aoki et al., 2005; Kehr and Buhtz, 2008; Ham et al., 2009; Hipper et al., 2013). Thus, phloem RBPs are translocated with plant RNAs and are likely to be important determinants of plant RNA vascular trafficking (Lucas et al., 2001; Aoki et al., 2005; Kehr and Buhtz, 2008; Ham et al., 2009; Turgeon and Wolf, 2009; Pallas and Gómez, 2013).
Viral spread from infected cells to neighboring cells requires that the PD size exclusion limits be increased through the action of viral movement protein (MP) (Gopinath and Kao, 2007; Canetta et al., 2008; Harries et al., 2009; Hipper et al., 2013). Changes in PD permeability are also thought to enable movement into the vascular system during systemic phloem-mediated trafficking. Thus, viruses must have evolved various strategies to interact with cellular factors to be loaded into and unloaded from the vascular system (Chen et al., 2000; Kim et al., 2007; Harries et al., 2009; Raffaele et al., 2009; Taliansky et al., 2010; Semashko et al., 2012; Hipper et al., 2013). Host factors such as myosin are required for MP targeting to and virus movement through the PD (Amari et al., 2014; Harries et al., 2009); intercellular and long-distance trafficking of Tobacco mosaic virus (TMV) (Chen et al., 2000) and Groundnut rosette virus (GRV) (Kim et al., 2007; Taliansky et al., 2010; Semashko et al., 2012) were substantially delayed in plants silenced for PECTIN METHYLESTERASE and fibrillarin. However, remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, can interact physically with the MP from Potato virus X and negatively regulates Potato virus X intercellular and phloem-mediated trafficking (Raffaele et al., 2009). Some viral RNAs and viroids also contain important 3D RNA motifs required for intercellular movement (Takeda et al., 2011).
Satellite RNAs (satRNAs) are parasites of RNA viruses that are almost exclusively associated with plant viruses. These entities lack appreciable sequence similarity to the genomes of their helper viruses (HVs), but depend on HV-encoded proteins for replication and encapsidation (Hu et al., 2009). The mechanisms by which satRNAs undergo intracellular or intercellular trafficking and whether satRNA transport depends on HV replication remain unknown.
Bamboo mosaic virus (BaMV)-associated satRNA (satBaMV) has a single-stranded positive-sense RNA genome of ∼835 nucleotides (Lin and Hsu, 1994). SatBaMV encodes a 20-kD nonstructural RNA binding protein (P20) that is dispensable for replication (Lin et al., 1996) but is required for long-distance satBaMV transport in HV coinfected Nicotiana benthamiana (Vijaya Palani et al., 2006; Vijayapalani et al., 2012). The P20 protein has several MP features, including RNA binding activity (Tsai et al., 1999), strong self-interactions, and efficient cell-to-cell movement (Vijaya Palani et al., 2006). P20 accumulates in the cytoplasm and nuclei of HV and satBaMV coinfected cells (Palani et al., 2009) and can be phosphorylated to negatively regulate the formation of satBaMV RNP complexes (Vijayapalani et al., 2012). BaMV HV is a member of the Potexvirus genus that contains a single-stranded, positive-sense RNA genome with five open reading frames. These open reading frames encode a replicase, three MPs (TGBp1, TGBp2, and TGBp3, encoded by a triple gene block), and a capsid protein (CP) (Lin et al., 1994), each required for cell-to-cell movement (Lin et al., 2004, 2006; Lan et al., 2010; Lee et al., 2011; Wu et al., 2011; Chou et al., 2013). However, the factors involved in long-distance trafficking of BaMV remain to be determined.
Here, we provide the evidence that trafficking of P20-defective satBaMV can be restored in transgenic plants expressing P20 protein. Grafting experiments performed to uncouple the replication and trafficking events of satBaMV showed that satBaMV RNA alone can move systemically across a graft union into non-satBaMV-expressing tissue. Coimmunoprecipitation (co-IP) analysis revealed that a nucleolar protein, fibrillarin, was associated with the P20 protein complex. Thus, it can be suggested that movement of satBaMV requires P20 interaction with fibrillarin to form mobile satBaMV RNP complexes. TGBp1, TGBp2, TGBp3, and CP were also present in the fibrillarin-satBaMV-P20 RNP complex after coinfection with satBaMV and HV, indicating that viral proteins also play important roles in satRNP complex trafficking.
RESULTS
The P20 Protein Plays a Crucial Role in SatBaMV Systemic Movement in N. benthamiana
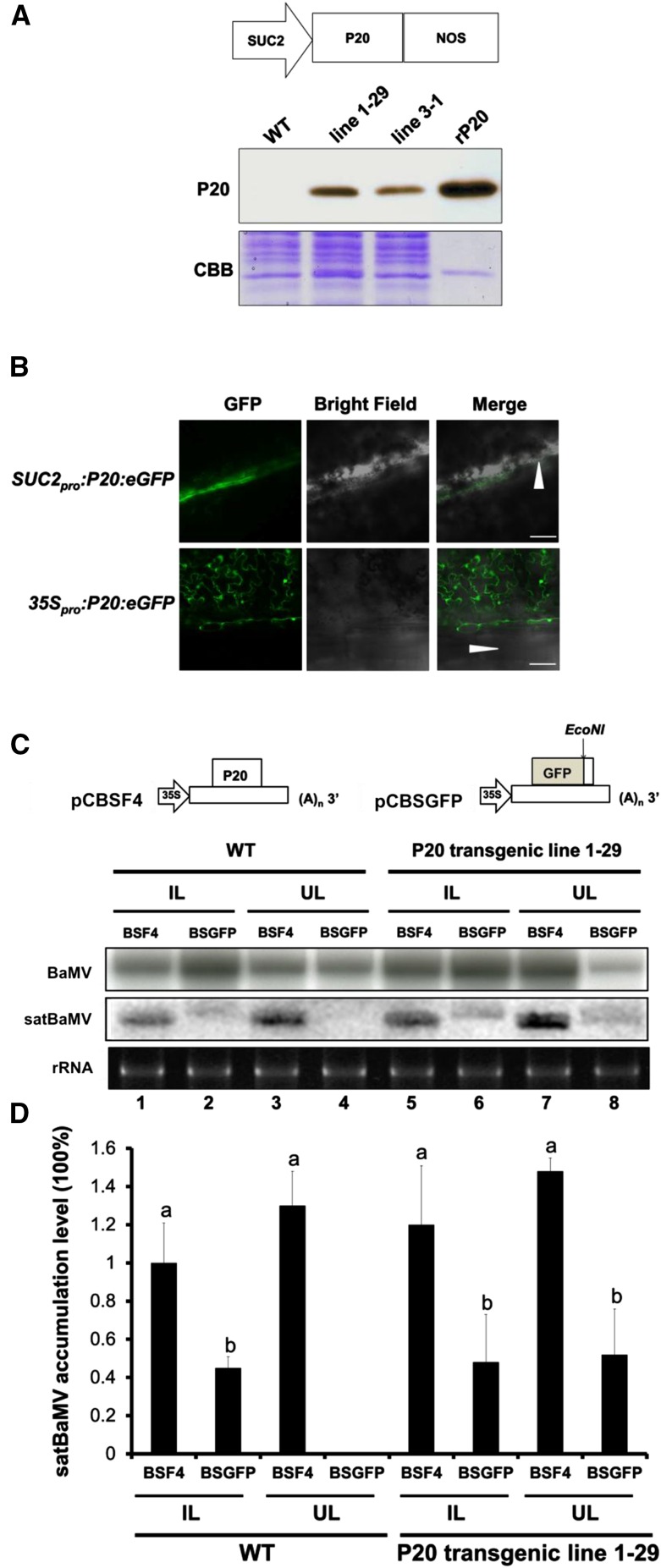
Previously, we showed that the trafficking of P20-defective satBaMV exhibits low efficiency in N. benthamiana coinfected with BaMV HV (Lin et al., 1996; Vijaya Palani et al., 2006; Vijayapalani et al., 2012). To provide further insights into the role of P20 in the systemic movement of satBaMV, we generated transgenic N. benthamiana expressing the P20 protein under the phloem companion cell-specific SUC2 promoter (Figure 1A). Immunoblot analysis confirmed that P20 was expressed in all 20 transgenic lines; we selected homozygous lines 1-29 and 3-1 for this study (Figure 1A). No abnormal phenotypes were observed for SUC2 promoter-driven P20 transgenic N. benthamiana plants. To verify the localization of SUC2 driven P20, we created the construct SUC2pro:P20:eGFP for transient assays. Confocal microscopy clearly showed that P20 expressed from the SUC2pro:P20:eGFP construct was present in the phloem along the leaf vein of agroinfiltrated leaves, whereas it localized to almost all cell types when it was expressed from 35Spro:P20:eGFP (Figure 1B).
Figure 1.
Trans-Complementation of the Systemic Movement of P20-Defective satBaMV BSGFP in P20 Transgenic N. benthamiana.
(A) Physical map of a SUC2 promoter-driven P20 expression plasmid and immunoblot analysis of P20 accumulation in leaves of SUC2pro:P20 transgenic N. benthamiana (lines 1-29 and 3-1). rP20, recombinant P20 protein purified from Escherichia coli. CBB, Coomassie Brilliant Blue staining.
(B) P20-eGFP localization under SUC2 promoter- and 35S promoter-driven expression in wild-type N. benthamiana leaves at 3 DPI. Arrowheads represent the leaf midrib.
(C) Schematic maps of satBaMV infectious clones and RNA accumulation of wild-type satBaMV (BSF4) and P20-defective satBaMV (BSGFP) in wild-type and P20 transgenic N. benthamiana (line 1-29). Leaves of wild-type and P20-transgenic N. benthamiana were coinoculated with pCB (Lin et al., 2004) and pCBSF4 (Lin et al., 2004) or pCBSGFP. Inoculated leaves (IL) were harvested at 10 DPI and uninoculated upper leaves (UL) at 20 DPI for RNA gel blot analysis of BaMV and satBaMV RNA accumulation. BaMV and satBaMV accumulation was detected using 32P-labeled RNA probes specific for the BaMV 3′ end and satBaMV 3′-untranslated region, respectively.
(D) Quantitative analysis of satBaMV accumulation in wild-type and P20-transgenic line 1-29 from four independent biological samples, each involving four plants. Values are normalized against BSF4 satBaMV in inoculated leaves of wild-type plants. Data are mean ± sd from four experiments and were analyzed by Student’s t test. Different letters indicate significant difference (P < 0.05).
For complementation assays, we first generated P20-defective satBaMV. The P20 gene of pCBSF4, the full-length cDNA clone of satBaMV (Lin et al., 2004), was replaced with GFP to generate a P20-defective satBaMV reporter plasmid (pCBSGFP); plants were mechanically inoculated with this plasmid and the BaMV infectious cDNA clone, pCB (Lin et al., 2004). BSGFP RNA accumulation was observed in the inoculated leaves of both wild-type and P20-transgenic lines, but BSGFP RNA was not detected in the upper uninoculated leaves of wild-type plants (Figure 1C, lane 4; Supplemental Figure 1A). Notably, a significant amount of BSGFP RNA (∼50% compared with wild-type BSF4 satBaMV in the wild-type inoculated leaves) was detected in the upper uninoculated leaves of two independent P20 transgenic lines at 20 d postinoculation (DPI) from four independent experiments, each involving four plants (Figures 1C, lane 8, and 1D; Supplemental Figure 1A). To further confirm the accumulation of BSGFP RNA in BaMV coinfected wild-type or P20 transgenic line 1-29, we performed confocal microscopy to visualize the accumulation of BSGFP protein after coinfection of N. benthamiana with pCB. Long-distance trafficking of BSGFP RNA from inoculated leaves to the systemic leaves was rescued in the P20 transgenic line 1-29 (Supplemental Figure 1Bd), which suggests that P20 in trans significantly contributes to long-distance trafficking of BSGFP RNA.
HV-Independent Long-Distance Trafficking of SatBaMV in N. benthamiana
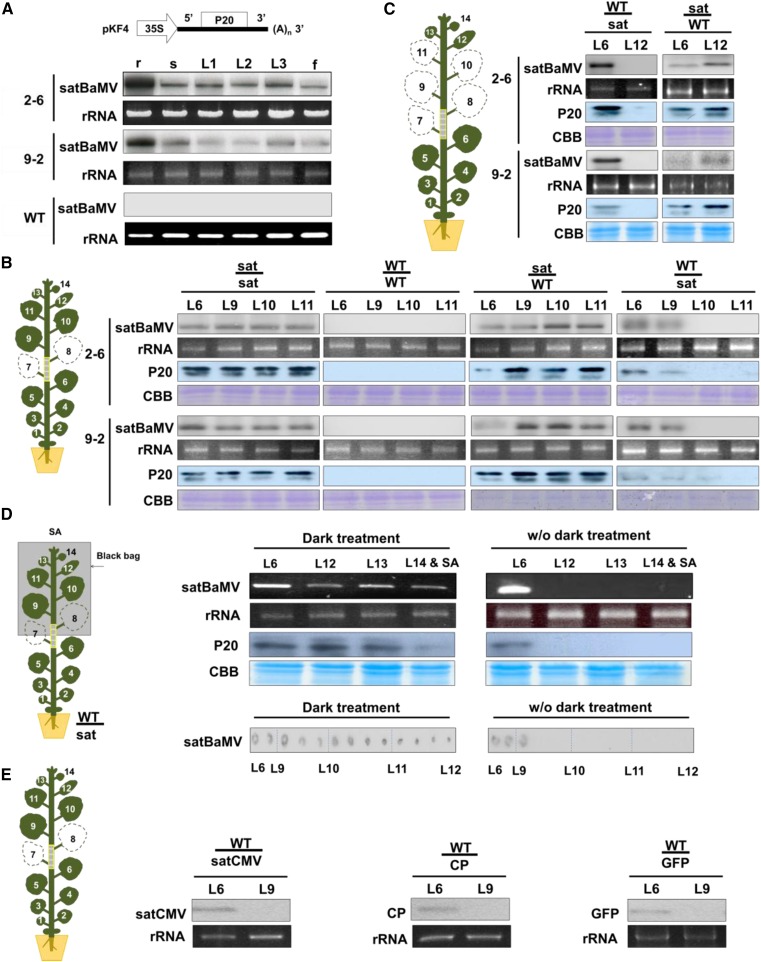
SatBaMV depends entirely on HV for replication, encapsidation, and efficient spread in plants (Lin and Hsu, 1994). To determine whether satBaMV can move systemically in the absence of HV, we generated five 35S promoter-driven satBaMV transgenic N. benthamiana lines. The satBaMV RNA of the transgene was expressed in all N. benthamiana tissues, including root, stem, leaves, and flowers, in lines 2-6 and 9-2 (Figure 2A). No abnormal phenotypes were observed in the 35S promoter-driven satBaMV transgenic N. benthamiana plants.
Figure 2.
HV-Independent Systemic Movement of SatBaMV.
(A) Physical map of pKF4 for generating satBaMV transgenic plants and satBaMV RNA accumulation in transgenic N. benthamiana lines 2-6 and 9-2 by RNA gel blot. r, root; s, stem; L1, 1st leaf; L2, 2nd leaf; L3, 3rd leaf; f, flower. rRNA of ethidium bromide served as loading control. Four independent biological samples, each involving four plants, generated similar results.
(B) and (C) Illustrations of grafting experiments with 40-d-old wild-type and satBaMV-transgenic N. benthamiana (sat). SatBaMV RNA and P20 protein accumulation were examined by RNA gel blot and immunoblot analysis, respectively. rRNA and Coomassie blue staining were used for loading controls. Four independent biological samples, each involving four plants, generated similar results.
(B) Leaf 7 (L7) and leaf 8 (L8) were detached before grafting. L6, L9, and L10 near the graft union were harvested at 12 DAG and L11 at 15 DAG.
(C) Leaves L7 to L11 were detached immediately after grafting; L6 and L12 were harvested at 15 DAG.
(D) RT-PCR and tissue blot detection of satBaMV RNA in grafting scions after dark treatment. The dark treatment of scions started at 12 DAG for 3 d. RNA and protein extracted from L6, L12-L14, and SA were sampled at 15 DAG and examined by RT-PCR and immunoblot analysis, respectively. Plants without dark treatment (w/o dark treatment) were used as controls. rRNA and Coomassie blue staining were used for loading controls. The tissue blots from left to right were prepared from grafting stem tissues between L6 and L12, followed by hybridization with satBaMV-specific probe. Four independent biological samples, each involving four plants, generated similar results.
(E) Detection of transgene RNA in transgenic stocks and wild-type scions after grafting. Wild-type plants were grafted onto transgenic N. benthamiana expressing GFP or BaMV CP or transgenic N. tabacum expressing satCMV. RT-PCR analysis of mRNA level at 15 DAG. Four independent biological samples, each involving four plants, generated similar results.
To determine whether satBaMV RNA alone can move systemically, we grafted satBaMV-transgenic N. benthamiana onto wild-type N. benthamiana and vice versa via cleft grafting (Figures 2B to 2D; Supplemental Figure 2). As shown in Figure 2B, the leaves L7 and L8 were detached before grafting, whereas leaves L6, L9, and L10 were harvested at 12 d after grafting (DAG) and L11 at 15 DAG. SatBaMV was detected in transgenic and wild-type leaves from chimeric grafts regardless of whether the wild-type plants served as scions or stocks (Figure 2B; Supplemental Figure 2A). When satBaMV transgenic lines served as scions and the wild type as stocks, satBaMV RNA and P20 protein were detected in all assayed leaves from L6 to L11; however, satBaMV RNA and P20 protein were detected only in L6 and L9 when satBaMV transgenic lines served as stocks and the wild type as scions (Figure 2B). Quantitative analyses of four independent experiments revealed that satBaMV RNA was undetectable in L10 and L11 by RNA gel blot (Supplemental Figure 2A). Moreover, RT-qPCR detected that accumulation of satBaMV progressively decreased in stem, petiole, and leaf L9, while satBaMV trafficked from satBaMV transgenic stocks to wild-type scion with levels ∼82, 41, and 14%, respectively, of those in L6 of the stocks (Supplemental Figure 3A). Our grafting experiments indeed showed that satBaMV RNA can move long distance alone across the graft union.
To investigate satBaMV long-distance trafficking in the presence of HV, L6 or L9 leaves near the graft union were coagroinfiltrated with the HV infectious cDNA clone pKB (Liou et al., 2014) at 9 DAG. As expected, satBaMV RNA accumulated in the leaves of both scions and stocks of satBaMV transgenic and wild-type plants after grafting with HV infection at 12 DAG (Supplemental Figure 2C, lanes 5 to 8).
To further determine whether satBaMV can undergo trafficking without HV in response to source-to-sink strength, we detached mature and young leaves (L7-L11) and left only the newly emerged youngest leaf L12 after grafting onto the scions (Figure 2C). RNA gel blot (Figure 2C), RT-PCR (Supplemental Figure 2B), and RT-qPCR (Supplemental Figure 3B) analyses revealed no satBaMV RNA accumulation in L12 of the wild type grafted onto satBaMV transgenic lines nor was there P20 protein accumulation based on immunoblot analysis (Figure 2C). However, satBaMV RNA and P20 protein were detected in L6 of wild-type stocks supported by satBaMV transgenic scions from four independent experiments (Figure 2C; Supplemental Figures 2B and 3B).
Since satBaMV could not move to the L12 (Figure 2C), we examined whether dark treatment could enhance satBaMV long-distance trafficking because previous studies have indicated that dark treatment may alter the source-sink relationship and increase virus susceptibility (Lemoin et al., 2013; Helms and McIntyre, 1967). Scions were subjected to dark treatment for 3 d before the leaves and stems were harvested for analysis (i.e., dark treatment started at 12 DAG), and RNA or protein was then extracted from L6 to L14 and shoot apex (SA) for analysis by RT-PCR or immunoblot, respectively; stem tissues were blotted onto nitrocellulose membrane by tissue blotting (Lin et al., 1990) at 15 DAG. RT-PCR or immunoblot analysis detected satBaMV or P20 protein accumulation in L12, L13, and SA after dark treatment but not in leaves without dark treatment (Figure 2D, upper panel). Stem tissues were harvested between L6 and L12 at 15 DAG (Figure 2D, lower panel). Tissue blotting further confirmed satBaMV RNA accumulation in wild-type scion stems under dark treatment, even in young stems close to L12. By contrast, only stem samples derived near L6 or L9 showed positive signals for satBaMV; satBaMV RNA failed to be detected in the upper stems near L10∼L12 without dark treatment (Figure 2D, lower panel; Supplemental Figure 3C). Therefore, dark treatment increased the long-distance trafficking of satBaMV to young leaves and stems, as well as to the SAs of wild-type plants (Figure 2D; Supplemental Figure 3C).
To determine whether other RNA is also mobile, we grafted wild-type plants onto transgenic Nicotiana tabacum plants expressing the satRNA of Cucumber mosaic virus (satCMV) or N. benthamiana expressing BaMV CP or GFP (16c) (Ruiz et al., 1998) as controls. However, none of these transgene RNAs was detectable in wild-type scions at 15 DAG (Figure 2E). These results indicate that satBaMV differs from satCMV in being dispensable for HV in systemic trafficking.
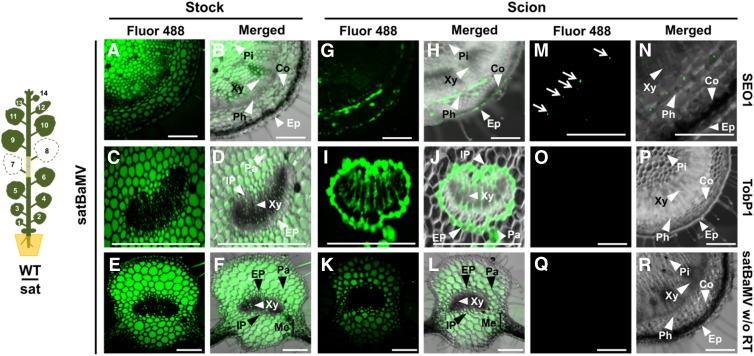
In Situ Localization of SatBaMV
To verify the phloem transportation and cell-to-cell movement of satBaMV, we used a modified method of in situ RT-PCR (Yoo et al., 2004). In situ RT-PCR allows the fluorescence associated with satBaMV to be visualized under confocal microscopy (Figures 3A to 3L). We performed the grafting experiment as for Figure 2B. Fresh sections were obtained from grafting stems, petioles, and leaves of wild-type scions at 12 DAG. In situ RT-PCR revealed strong fluorescence in the stem (Figures 3A and 3B), petiole (Figures 3C and 3D), and leaf (Figures 3E and 3F) of satBaMV transgenic stock, including pith, phloem, cortex, epidermis, parenchyma, and mesophyll cells, with relatively lower fluorescence in xylem. The fluorescence associated with satBaMV was mainly detected in phloem and some in the cortex of the wild-type scion stems (Figures 3G and 3H). In wild-type scion petioles, fluorescence was particularly located in internal phloem, external phloem, and parenchyma cells (Figures 3I and 3J). In addition, strong fluorescence was detected in phloem and parenchyma cells of the major veins of wild-type scion leaves, with little in the mesophyll and xylem (Figures 3K and 3L), indicating that satBaMV was able to move cell-to-cell from phloem and parenchyma to mesophyll cells in wild-type scion leaves. SIEVE ELEMENT OCCLUSION1 (SEO1) mRNA was detected as an internal control for in situ RT-PCR and was found only in phloem (Figures 3M and 3N), which is consistent with previous promoter assay findings (Ernst et al., 2012). Furthermore, we used another control, TOBACCO POLYPHENOL OXIDASE1 (TobP1), established as a flower-specific gene (Goldman et al., 1998); TobP1 mRNA was not detected in N. benthamiana stem tissues (Figures 3O and 3P). The absence of signal within stem and tissues in experiments with reverse transcriptase omitted from the reaction for satBaMV established the specificity of the protocol used in these studies (Figures 3Q and 3R). Taken together, these studies confirm that satBaMV can move from cell to cell and long distance via phloem.
Figure 3.
In Situ RT-PCR Detection of SatBaMV.
The grafting experiment was illustrated as in Figure 2B. Stems and petioles between L6 and L9 were harvested at 12 DAG for in situ RT-PCR detection.
(A) to (F) Presence of satBaMV RNA in the stem ([A] and [B]), petiole ([C] and [D]), and leaf midrib ([E] and [F]) of satBaMV transgenic N. benthamiana stock.
(G) to (L) Presence of satBaMV RNA in the stem ([G] and [H]), petiole ([I] and [J]), and leaf midrib ([K] and [L]) of wild-type scion.
(M) and (N) Detection of SEO1 mRNA in the wild-type scion stem. SEO1 mRNA was restricted to the sieve element (Ernst et al., 2012). Arrows in (M) indicate sieve element.
(O) and (P) Detection of TOBACCO POLYPHENOL OXIDASE1 mRNA in the wild-type scion stem, which was exclusively present in flower organs (Goldman et al., 1998), as a negative control.
(Q) and (R) Detection of satBaMV RNA without reverse transcriptase in the reaction in the wild-type scion stem as control for nonspecific amplification during RT-PCR.
The results of in situ RT-PCR were observed by confocal laser scanning microscopy. Green signal represents incorporation of Alexa Fluor 488-labeled nucleotides during specific amplification of target genes indicated beside images and the respective merged image, with bright-field image in the right panel. Four independent biological samples, each involving four plants, of in situ RT-PCR detection generated similar results. Bars = 100 μm. Pi, pith; Xy, xylem; Co, cortex; Ep, epidermis; Ph, phloem; IP, internal phloem; EP, external phloem; Pa, parenchyma; Me, mesophyll.
Host Factors Immunoprecipitated with P20 Protein
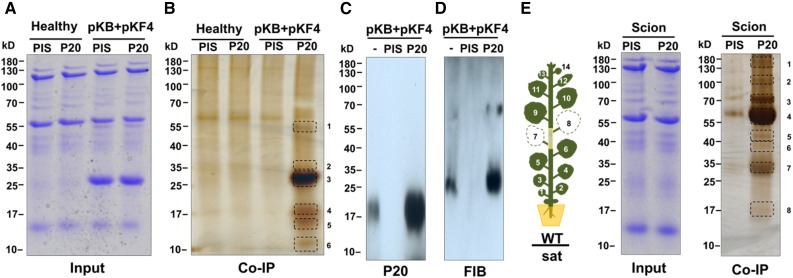
Since satBaMV is always associated with BaMV in the natural environment, we examined the host factors involved in movement of satBaMV in the presence of HV. Therefore, we used HV and satBaMV coinfected systemic leaves at 7 DPI to search for possible host factors involved in trafficking of satBaMV.
We performed co-IP experiments and analyzed the precipitated products by gel electrophoresis followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). To avoid interference from Rubisco, the most abundant protein in leaves, we prepared Rubisco-depleted fractions and then incubated those with anti-P20 or preimmune IgG, followed by Protein A Sepharose CL-4B. After washing, bound proteins were eluted and fractionated by gel electrophoresis. In mock-inoculated samples or in controls with preimmune IgG, only small numbers of nonspecific proteins were detected in silver-stained gels (Figure 4B). By contrast, numerous proteins were resolved after anti-P20 IgG precipitation (Figure 4B). LC-MS/MS analysis revealed several peptide sequences associated with anti-P20 coimmunoprecipitates after HV and satBaMV coinfection (Table 1). In three independent experiments, P20 was observed in band 4, which migrated slightly slower than the 17-kD standard; this band was absent in co-IP complexes from total proteins of coinfected leaves treated with preimmune IgG (Figure 4B), which indicates the specificity of the anti-P20 IgG preparation in the co-IP experiments. As shown in Table 1, the most abundant viral CP was detected in band 3, whereas TGBp1 was in band 2. With the exception of Rubisco, which remained enriched in band 1, the most abundant host protein precipitating with anti-P20 IgG was a nucleolar protein, fibrillarin, which was detected in band 2 in three independent experiments (Table 1). Fibrillarin is known to interact with viral MPs and is important for long-distance trafficking of several RNA viruses (Kim et al., 2007; Taliansky et al., 2010; Semashko et al., 2012; Zheng et al., 2015). The LC-MS/MS results were confirmed by immunoblot analysis, revealing the presence of P20 and fibrillarin in the P20 co-IP complex in the HV coinfected tissues (Figures 4C and 4D).
Figure 4.
Identification of P20-Interacting Protein Complex from Grafting N. benthamiana Leaves with HV-Dependent or -Independent SatBaMV Infection by Co-IP.
(A) Coomassie blue staining of total protein extracted from leaves of healthy or BaMV (pKB) + satBaMV (pKF4) (Liou et al., 2013) coinfected N. benthamiana at 7 DPI.
(B) Co-IP protein complexes by preimmune IgG (PIS) or anti-P20 IgG (P20) were separated by SDS-PAGE. Protein bands were visualized by silver staining; frames indicate protein bands excised for LC-MS/MS protein identification. The gel is representative of three independent experiments.
(C) and (D) Detection of P20 and fibrillarin in co-IP complex from anti-P20 IgG or PIS antibody. Input (−) and complex were separated by SDS-PAGE followed by immunoblot analyses with anti-P20 (C) or anti-FIB IgG (D).
(E) HV-independent grafting experiments are illustrated in the left as in Figure 2B. Total protein was extracted from wild-type scion leaves after grafting at 15 DAG. Co-IP and protein analysis were performed as in (B).
Table 1. Proteins Identified by LC-MS/MS after Immunoprecipitation of P20 IgG from BaMV and satBaMV Coinfected N. benthamiana.
| Band | Accession | Protein Name | Mass (kD) | Score |
|---|---|---|---|---|
| 1 | ATCG00490.1 | RBCL ribulose-bisphosphate carboxylase | 53.4 | 59 |
| 2 | AT4G25630.1 | FIB2, ATFIB2 fibrillarin 2 | 33.8 | 42 |
| 2 | *gi|345134903|dbj|BAK64672.1| | *BaMV TGBp1 | 27.7 | 43 |
| 3 | AT5G01530.1 | Light-harvesting complex PSII | 25.2 | 130 |
| 3 | *gi|2407623|gb|AAB70566.1| | *BaMV CP | 25.5 | 818 |
| 4 | AT3G16640.1 | TCTP translationally controlled tumor protein | 21.3 | 55 |
| 4 | *gi|37782235|gb|AAP31339.1| | *SatBaMV P20 | 19.9 | 26 |
| 5 | AT1G77300.1 | Histone H3 | 17.0 | 25 |
| 6 | ATCG00580.1 | PSBE PSII reaction center protein | 10.0 | 36 |
Asterisks indicate BaMV- or satBaMV-encoded proteins. Score is parameter characterizing identification reliability of a certain protein. In general, at score value > 23, identification is considered as reliable (P < 0.05).
As shown in Figures 2B to 2D, satBaMV systemic trafficking could be HV independent; therefore, we also verified the satBaMV movement complex in the absence of HV. The wild-type scion N. benthamiana, grafted onto satBaMV transgenic line 2-6 (Figure 4E), was harvested at 15 DAG and proteins were precipitated with anti-P20 IgG. In three independent experiments, P20 was detected in band 8 and fibrillarin was also detected in band 7 independent of HV by LC-MS/MS (Table 2). In summary, with or without HV, P20 and fibrillarin may form a complex in vivo.
Table 2. Proteins Identified by 1D LC-MS/MS after Immunoprecipitation of P20 IgG from Wild-Type Scion Grafted onto satBaMV Transgenic Stock.
| Band | Accession | Protein Name | Mass (kD) | Score |
|---|---|---|---|---|
| 1 | AT5G04140.2 | FD-GOGAT glutamate synthase 1 | 181.2 | 76 |
| 2 | AT2G26080.1 | GLDP2 glycine decarboxylase P-protein 2 | 114.6 | 246 |
| 3 | AT3G09440.1 | Heat shock protein 70 (Hsp 70) family protein | 71.5 | 181 |
| 4 | AT1G55490.1 | LEN1 chaperonin 60 beta | 64.1 | 85 |
| 5 | AT4G38970.1 | FBA2 fructose-bisphosphate aldolase 2 | 43.1 | 486 |
| 6 | ATCG00020.1 | PSII reaction center protein A | 39.0 | 218 |
| 7 | AT4G25630.1 | FIB2, ATFIB2 fibrillarin 2 | 33.8 | 28 |
| 8 | *gi|37782235|gb|AAP31339.1| | *SatBaMV P20 | 19.9 | 27 |
| 8 | AT2G36160.1 | Ribosomal protein S11 family protein | 16.3 | 56 |
Asterisks indicate satBaMV-encoded proteins. Score is parameter characterizing identification reliability of a certain protein. In general, at score value > 23, identification is considered as reliable (P < 0.05).
Fibrillarin Silencing Suppresses Long-Distance Trafficking of SatBaMV
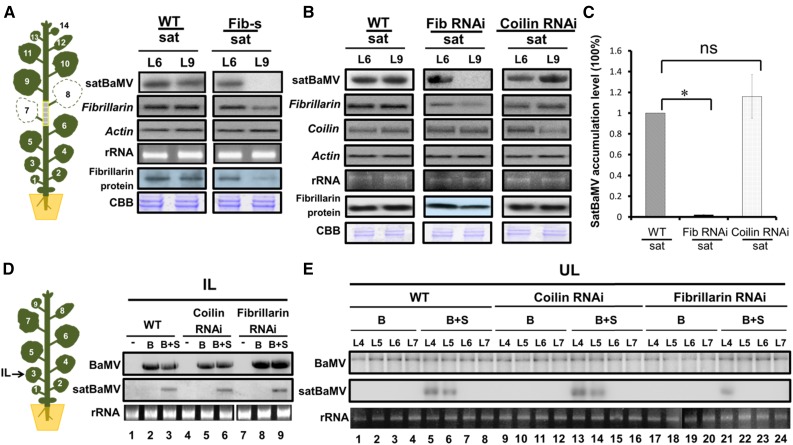
To examine whether fibrillarin has a role in satBaMV trafficking in the absence of HV, we used virus-induced gene silencing (VIGS) with a Tobacco rattle virus (TRV) vector (Ratcliff et al., 2001) to reduce fibrillarin expression in N. benthamiana plants. PHYTOENE DESATURASE, the silencing of which is reflected by photobleaching of leaves, was used as a control. Agroinfection with pTRV-NbFib carrying a fragment of the fibrillarin gene from N. benthamiana (Kim et al., 2007) resulted in 60% reduction in levels of both fibrillarin mRNA and protein in VIGS plants (Figure 5A). RNA gel blot analysis revealed that the satBaMV RNA was no longer detectable in fibrillarin-silenced scion plants after grafting onto the satBaMV transgenic line at 9 DAG (Figure 5A).
Figure 5.
Fibrillarin Silencing Suppresses SatBaMV Trafficking.
(A) Accumulation of satBaMV RNA and fibrillarin mRNA and protein in N. benthamiana leaves from satBaMV transgenic stock and grafted wild-type and TRV-induced fibrillarin silenced (Fib-s) scions. Plants were first infiltrated with Agrobacterium strain LBA4404 carrying binary vectors expressing pTRV1 and pTRV2-fibrillarin (Fib-s); 7 d later, a fib-s scion was grafted onto satBaMV transgenic line 2-6 (sat). At 9 DAG, stock and scion leaves were harvested for RNA gel blot and immunoblot analyses. CBB, Coomassie Brilliant Blue staining. Four independent biological samples, each involving four plants, generated similar results. Actin and CBB were used as loading controls.
(B) Accumulation of satBaMV RNA, coilin, and fibrillarin mRNA and fibrillarin protein in N. benthamiana leaves from satBaMV transgenic stock and grafted wild-type, fibrillarin, and coilin RNAi (Shaw et al., 2014) transgenic scions. RNA and protein analysis were performed at 9 DAG as described in (A). Four independent biological samples, each involving four plants, generated similar results.
(C) Statistical analysis of satBaMV accumulation in grafted coilin or fibrillarin RNAi transgenic scions. Values are normalized against the wild-type sample. Data are mean ± sd from four independent biological samples, each involving four plants and were analyzed by Student’s t test. The asterisk represents significant difference between wild-type and fibrillarin RNAi lines (*P < 0.001; ns, not significant).
(D) and (E) Accumulation of BaMV and satBaMV in wild-type, coilin, and fibrillarin RNAi transgenic plants. RNA gel blot analyses of BaMV and satBaMV RNAs in wild-type, coilin, and fibrillarin RNAi transgenic plants agroinfected with pKB (B) or pKB + pKF4 (B+S) in inoculated leaves (IL) harvested at 5 DPI (D) and uninoculated upper leaves (UL) at 15 DPI (E). Four independent biological samples, each involving four plants generated similar results. −, Uninoculated (healthy) plant control; rRNA, loading control.
To further confirm that satBaMV systemic movement without HV depends on fibrillarin, we grafted the N. benthamiana fibrillarin knockdown line (Shaw et al., 2014) onto satBaMV transgenic plants. RNAi knockdown of coilin, encoding one of the main components of Cajal bodies (Ogg and Lamond, 2002), served as a control. SatBaMV did not move into fibrillarin RNAi scions, but substantial movement was evident in the coilin RNAi scions at 9 DAG (Figure 5B); quantitative analysis revealed that satBaMV mRNA was greatly reduced in fibrillarin RNAi scions but not coilin RNAi scions (Figure 5C). Thus, this result supports the crucial role of fibrillarin in satBaMV long-distance transport without HV.
To examine the effect of fibrillarin on satBaMV coinfection with HV, we agroinfiltrated N. benthamiana plants with pKB (Liou et al., 2014) and pKF4 (Liou et al., 2014), carrying the infectious cDNA clones of BaMV and satBaMV, respectively. Although mRNA expression of coilin and fibrillarin was greatly suppressed in infiltrated and upper uninfiltrated leaves of RNAi N. benthamiana plants (Figure 5B; data not shown), the inoculated leaves of wild-type, coilin, or fibrillarin RNAi plants contained similar levels of BaMV or satBaMV RNA accumulation after infiltration with BaMV or coinfiltration with HV+satBaMV at 5 DPI (Figure 5D). Notably, satBaMV RNA was greatly decreased in the upper leaves of fibrillarin RNAi plants (Figure 5E, lanes 21 to 24 versus 5 to 8) but not coilin RNAi plants (Figure 5E, lanes 13 to 16 versus 5 to 8; Supplemental Figure 4B) coagroinfiltrated with pKB and pKF4. Quantitative analyses of three independent experiments revealed that satBaMV accumulation in the upper, noninfiltrated leaf L4 of fibrillarin RNAi plants was ∼20% of that of the wild-type or coilin RNAi plants but undetectable in L5 of fibrillarin RNAi plants (Supplemental Figure 4B). SatBaMV accumulation in L4 or L5 of wild-type and coilin RNAi plants did not differ (Supplemental Figure 4B). However, BaMV accumulation was not greatly affected in the upper noninfiltrated leaves (L4-L7) among the wild-type, fibrillarin RNAi, or coilin RNAi plants after BaMV or satBaMV coinfection at 15 DPI (Figure 5E; Supplemental Figure 4A). Therefore, fibrillarin silencing impaired only the long-distance trafficking of satBaMV in HV and satBaMV coinfected plants.
Fibrillarin and P20 Form Complexes with SatBaMV RNA
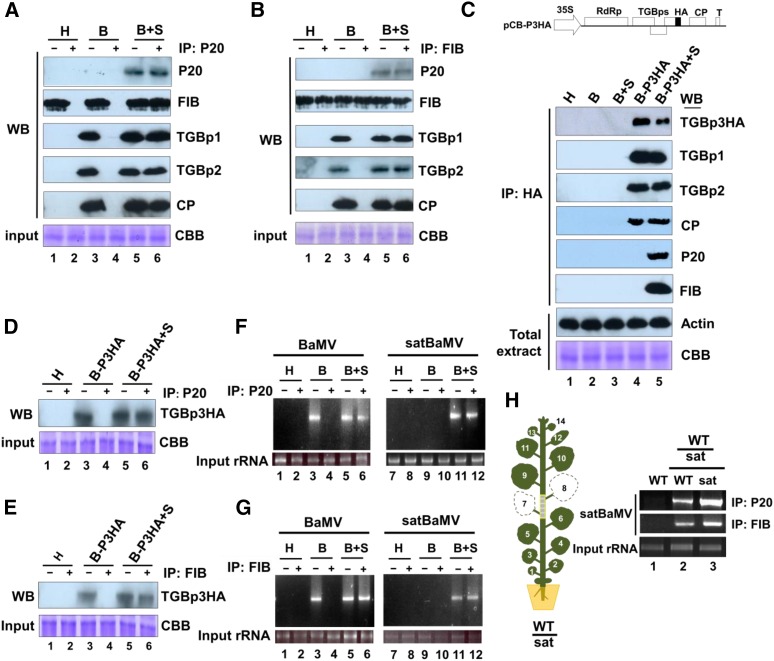
The previous results suggest that fibrillarin may be important for long-distance trafficking of satBaMV during BaMV coinfection. Therefore, we performed co-IP assays to determine the nature of the putative protein-protein interactions. Total proteins were extracted from the leaves of N. benthamiana plants infected with HV or coinfected with satBaMV, and co-IP assays were then performed with anti-P20 or antifibrillarin IgG. Input proteins and co-IP fractions were confirmed by immunoblot analysis (Figure 6).
Figure 6.
Mobile SatBaMV-P20 Complexes Contain FIB, TGBp1, TGBp2, TGBp3, CP, BaMV, and SatBaMV RNAs in N. benthamiana Agroinfected with pKB and pKF4.
(A) and (B) Wild-type N. benthamiana plants were agroinfected with pKB (B) or pKB + pKF4 (B+S). Healthy (H) leaves were used as a control. Total proteins were extracted from uninoculated upper leaves of healthy plants or agroinfected plants at 15 DPI, and co-IP was performed with anti-P20 (A) or anti-fibrillarin (FIB) (B) IgG followed by protein A agarose immunoprecipitation. Input (−) and eluted (+) proteins were separated by SDS-PAGE followed by immunoblot analyses with anti-P20, anti-FIB, anti-TGBp1, anti-TGBp2, or anti-CP IgG. Lane 6 was loaded in 20-fold dilution. CBB indicates the input protein before co-IP. Four independent biological samples generated similar results.
(C) Genomic map of BaMV infectious clone pCB-P3HA (Chou et al., 2013). N. benthamiana plants were inoculated with wild-type pCB (B), pCB + pCBSF4 (B+S), or pCB-P3HA (B-P3HA), or pCB-P3HA + pCBSF4 (B-P3HA+S). Co-IP and immunoblot analyses were as described in (A) and (B), except that the HA antibody was used for immunoprecipitation. Input proteins before co-IP were detected by immunoblot against actin and Coomassie blue staining. Four independent biological samples generated similar results.
(D) and (E) Immunoblot analysis of TGBp3HA in co-IP complex. Co-IP with anti-P20 (D) or anti-FIB (E) IgG followed by protein A agarose. Input (−) and eluted (+) proteins were separated by SDS-PAGE followed by immunoblot analyses with anti-HA antibody. Lane 6 was loaded in 20-fold dilution. Four independent biological samples were examined.
(F) and (G) RT-PCR detection of BaMV (left panels) and satBaMV RNA (right panels) in co-IP fractions. RNA was extracted from anti-P20 (F) or anti-FIB (G) IgG co-IP fractions from healthy (H) or agroinfected with pKB (B) or pKB + pKF4 (B+S) N. benthamiana leaves. RT-PCR was used to detect the presence of BaMV and satBaMV RNAs. Amplified products were separated with agarose gel and the expected sizes of the BaMV (0.8 kb) and satBaMV (0.8 kb) fragments were shown. Four independent biological samples generated similar results.
(H) RT-PCR detection of satBaMV RNA in co-IP fractions from nongrafted or grafted plants. The grafting experiment was illustrated in Figure 2B. Total protein from nongrafted wild-type (lane 1) or grafted L6 (lane 2) and L9 (lane 3) leaves was extracted and used for co-IP at 15 DAG. RNA was extracted and detected by RT-PCR from anti-P20 or anti-FIB IgG co-IP fractions. Four independent biological samples were examined.
With anti-P20 IgG, protein samples extracted from leaves coinfected with HV and satBaMV revealed signals for P20, fibrillarin, TGBp1, TGBp2, and CP (Figure 6A, lane 6). These proteins were not detected in samples from plants infected with HV alone or in healthy control leaves (Figure 6A, lanes 2 and 4). Likewise, co-IP assay with antifibrillarin IgG revealed equivalent signals for P20, fibrillarin, TGBp1, TGBp2, and CP in HV and satBaMV coinfected leaves (Figure 6B, lane 6), indicating that fibrillarin interacted directly or indirectly with P20, TGBp1, TGBp2, and CP. To determine whether fibrillarin interacts with P20 directly, we generated fusions of activation domain (AD) or binding domain with fibrillarin or P20 for yeast two-hybrid assays. We observed strong self-interactions for both P20 and fibrillarin and also a direct interaction between fibrillarin and P20, regardless of which protein was fused to AD (Supplemental Figure 6). This interaction is consistent with the results obtained by co-IP assay with anti-P20 or antifibrillarin IgG.
Like potexviruses, all three TGBps and CP are required for BaMV cell-to-cell and systemic movement (Lin et al., 2004, 2006; Lan et al., 2010; Lee et al., 2011; Wu et al., 2011; Chou et al., 2013). To examine the interaction of TGBp3 with P20 and/or fibrillarin, we inoculated N. benthamiana leaves with a 35S promoter-driven HV derivative (pCB-P3HA) carrying a TGBp3::HA fusion (Figure 6C; Chou et al., 2013) with or without the satBaMV pCBSF4 plasmid (Lin et al., 2004). Co-IP assays with an HA antibody precipitated TGBp3HA, TGBp1, TGBp2, and CP but not P20 or fibrillarin in fractions, from pCB-P3HA-infected plants (Figure 6C, lane 4). The HA antibody additionally precipitated fibrillarin and P20 from protein extracts of plants coinoculated with pCB-P3HA and pCBSF4 (Figure 6C, lane 5) but not from uninoculated wild-type, pCB-inoculated, or pCB and pCBSF4 coinoculated plants. However, co-IP assays with anti-P20 IgG (Figure 6D) or anti-fibrillarin IgG (Figure 6E) revealed positive signals for TGBp3HA in HV and satBaMV coinfected leaves (Figures 6D and 6E, lane 6). Hence, consistent with observations in other potexviruses (Park et al., 2013), BaMV TGBps and CP may interact with each other, and P20 may form protein complexes with fibrillarin, BaMV TGB proteins, and CP in plant tissues coinfected with HV and satBaMV.
To further confirm that the P20-fibrillarin protein complexes are RNP complexes, we extracted RNA from total sap and co-IP fractions after incubation with anti-P20 or antifibrillarin IgG. In plants coinfected with HV and satBaMV, both HV and satBaMV RNAs were present in the total sap and co-IP fractions, as shown using anti-P20 (Figure 6F, lanes 5, 6, 11, and 12) or antifibrillarin IgG (Figure 6G, lanes 5, 6, 11, and 12). In the negative control (immunoprecipitation of total sap of HV-infected plants using anti-P20 IgG), HV RNA was detected in only total sap from HV-infected plants but not in co-IP fractions with anti-P20 or anti-fibrillarin IgG (Figures 6F and 6G, lanes 3 and 4). These results support the conclusion that P20 can form RNP complexes with fibrillarin and also complexes with HV proteins as well as HV and satBaMV RNAs in satBaMV coinfected plants.
However, wild-type scions grafted onto the satBaMV transgenic stock, 2-6 line, without HV, showed satBaMV RNA in co-IP fractions with anti-P20 or anti-fibrillarin IgG for the formation of a satBaMV-P20-fibrillarin RNP complex (Figure 6H).
The Fibrillarin-SatBaMV-P20 RNP Complex Is Absent from the Fibrillarin RNAi Transgenic Line
To verify whether the composition of the satBaMV-P20 RNP complex with or without HV was changed by reduced fibrillarin level, the fibrillarin RNAi plants were grafted onto satBaMV transgenic lines 2 to 6 (Supplemental Figure 5A). Fibrillarin RNAi scion tissues were harvested and proteins were precipitated with anti-P20 IgG. In three independent experiments, P20 was detected in band 7; other detected proteins were chloroplast, ribosomal, and helicase proteins (Supplemental Table 1). Total proteins were extracted from the leaves of fibrillarin RNAi plants coinfected with HV and satBaMV, and co-IP assays were performed with anti-P20 IgG at 15 DPI (Supplemental Figure 5B). The most abundant P20 was detected in band 9, whereas viral CP and TGBp1 comigrated into band 8 (Supplemental Table 2), and ATP synthase, pyrophosphorylase 2, glycine decarboxylase P-protein 2, and ribosomal and helicase proteins were detected in three independent experiments. These data show that the composition of the satBaMV-P20 RNP complex with and without HV was distinct in plants with reduced fibrillarin levels. Furthermore, no fibrillarin was detected in the satBaMV-P20 RNP complex in fibrillarin RNAi transgenic line (Supplemental Tables 1 and 2), suggesting that the residual fibrillarin produced in the RNAi line is completely or nearly completely used for its primary function in ribosome biogenesis.
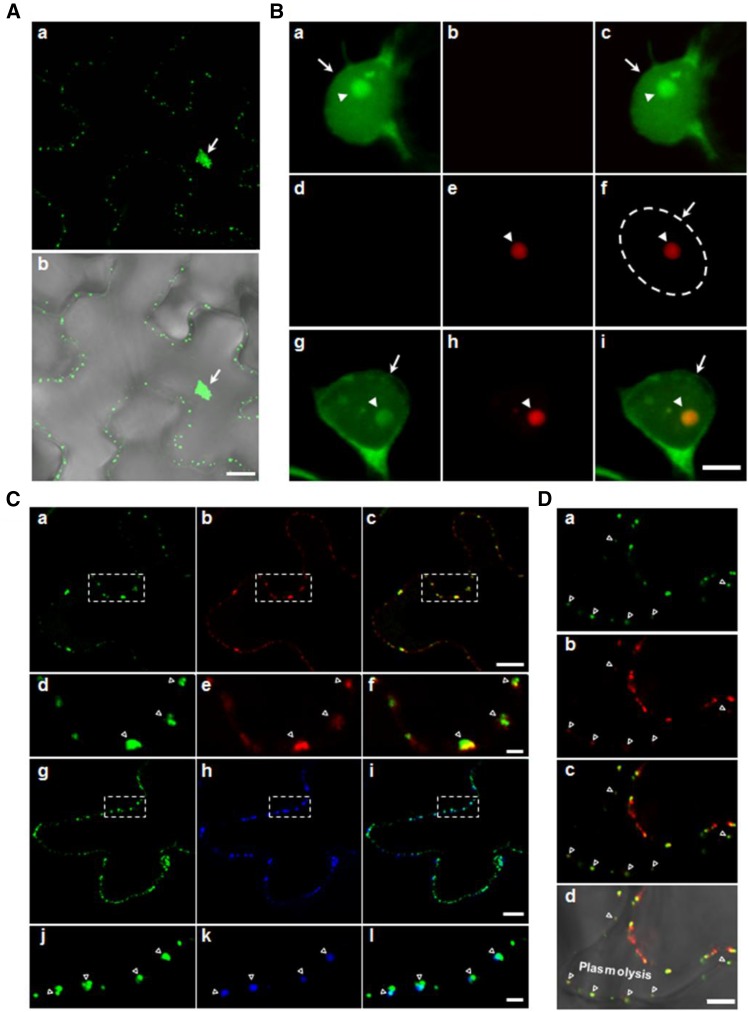
P20 Colocalizes with Fibrillarin and Forms Punctate Structures at the Cell Periphery
To reveal the subcellular localization of P20, we constructed an Agrobacterium tumefaciens-compatible plasmid, pBin-P20-eGFP, for transient expression of GFP-tagged P20. P20-eGFP was located in the nucleus and also at the cell periphery, where it formed punctate structures (Figure 7A, a), as described previously (Vijaya Palani et al., 2006; Palani et al., 2009). At higher magnification, P20-eGFP could be seen to form one or two foci in the nucleus (Figure 7B, a to c). To determine whether P20-eGFP colocalized with fibrillarin (FIB2), we agroinfiltrated leaves with pBin-mCherry-NbFIB2 alone or with pBin-P20-eGFP. Transiently expressed mCherry-NbFIB2 was mainly localized in the nucleolus (Figure 7B, d to f). When P20-eGFP was coexpressed with mCherry-NbFIB2, these two proteins exhibited perfect colocalization in the nucleolus (Figure 7B, g to i), indicating that P20 can be imported into the nucleus and become enriched in the nucleolus.
Figure 7.
Subcellular Localization of satBaMV-Encoded Protein P20 and Fibrillarin in N. benthamiana Epidermal Cells.
Proteins were fused to eGFP or DsRed/mCherry and expressed in N. benthamiana leaves by agroinfiltration. Two days after agroinfiltration, leaf sections were examined under a confocal laser scanning microscope.
(A) P20-eGFP localizes to the nucleus and cell periphery as punctate structures (a). The cell periphery is shown through the merging of the fluorescent signal with the bright field (b).
(B) Localization of P20-eGFP and mCherry-NbFIB2 in nucleus and nucleolus. Epidermal cells expressing P20-eGFP ([a] to [c]) or mCherry-NbFIB2 ([d] to [f]), or co-expressing both proteins ([g] to [i]) were examined. The nuclear area is indicated by the dashed circle.
(C) Localization of P20-eGFP peripheral punctate adjacent to plasmodesmata. P20-eGFP coexpressed with TMVMP-DsRed ([a] to [f]) or callose deposition (as stained with aniline blue) ([g] to [l]). The images in the dashed square areas of (a) to (c) and (g) to (i) are shown magnified in (d) to (f) and (j) to (l), respectively.
(D) Localization of P20-eGFP after plasmolysis. The leaf tissues were plasmolyzed with 1 M NaCl before confocal microscopy. Images show localization of P20-eGFP (a), TMVMP-DsRed (b), and merged images ([c] and [d]) in the plasmolyzed cell. The plasmolyzed region is labeled (Plasmolysis). Green, red, and blue represent eGFP, DsRed/mCherry, and aniline blue signals, respectively. Arrows and arrowheads indicate nucleus and nucleolus, respectively. Open triangles indicate the plasmodesmata.
Bars = 40 μm in (A), 5 μm in (B) and (D), 10 μm in (C) ([a] to [c] and [g] to [i]), and 2 μm in (C) ([d] to [f] and [j] to [l]). Individual and merged images were edited using Photoshop CS5.
P20 protein was previously found to move cell-to-cell autonomously (Vijaya Palani et al., 2006); accordingly, we examined whether the P20 peripheral punctae were associated with the PD. We used both DsRed-tagged TMV MP (Figure 7C, a to f) and aniline blue staining (Figure 7C, g to l) as PD markers. Regardless of which PD marker was used, most, if not all, of the P20-eGFP colocalized with labeled PD.
To further determine whether the P20-eGFP localized in the PD channel, we used leaves expressing P20-eGFP and TMVMP-DsRed plasmolyzed with 1 M NaCl. Nearly half of the leaf cells were plasmolyzed after NaCl treatment. A large amount of P20-eGFP localized along the shrunken cell membrane. However, substantial P20-eGFP still remained in the cell wall and colocalized with the PD marker TMVMP-DsRed after plasmolysis (Figure 7D). Taken together, these findings suggest that P20 can localize to the nucleolus with fibrillarin and form punctate structures at the cell periphery with integration into the PD channel.
DISCUSSION
Although satRNAs are subviral agents that require HVs for efficient replication and long-distance trafficking in coinfected plants, our data clearly show that transgenic satBaMV alone can move systemically across a graft union in N. benthamiana. We also demonstrated that expression of P20 in phloem (Figure 1B) can complement long-distance trafficking of P20-defective satBaMV in P20-transgenic N. benthamiana (Figures 1C and 1D; Supplemental Figure 1). In addition, autonomous satBaMV trafficking into wild-type scions is fibrillarin dependent (Figure 5). In the absence of HV, satBaMV RNA appears to exist as an RNP complex composed of P20 and fibrillarin (Figure 4E, Table 2), whereas in the presence of HV, viral MPs and CP are also recruited into the mobile RNP complex for efficient trafficking (Figure 6). Therefore, satBaMV trafficking appears not to require prior replication of satBaMV with HV or encapsidation by HV CP (Lin and Hsu, 1994), and the mobility of satBaMV can be independent of replication. This observation implies that satBaMV may have an advantage for survival in nature because its capacity for autonomous trafficking to the distal leaves may enhance its chances to encounter HV for further amplification and spread once satBaMV and HV initially infect different cells. Fibrillarin is required for only satBaMV, but not HV long-distance trafficking, which suggests that HV and satRNA may have evolved distinct methods for trafficking. A previous study also found a differential requirement for the host factor heat shock protein 90 in the replication of HV and satBaMV (Huang et al., 2012).
Viroids do not encode any translatable products and are not encapsidated. Viroid RNA alone can replicate and traffic efficiently without the need for an HV in host plants (Flores et al., 2009). However, differentiating between replication and movement of viroids through their interactions with cellular factors is problematic. Although replication-independent long-distance trafficking of Brome mosaic virus RNA3 can be recapitulated by agroinfection of individual cDNA components into different expression sites in N. benthamiana, the detection of the movement signal of RNA3 requires the subsequent replication of RNA1 and RNA2 (Gopinath and Kao, 2007). Thus, uncoupling of satBaMV replication from trafficking will provide an opportunity to gain greater molecular insights into satRNA systemic movement in planta.
Unlike most satRNA-encoded proteins required for satRNA replication (Hu et al., 2009), the satBaMV-encoded P20 nonstructural protein assists satBaMV long-distance trafficking in plants (Lin et al., 1996; Vijaya Palani et al., 2006; Vijayapalani et al., 2012) (Figure 1). Previously, we showed that P20 can preferentially bind to the 5′- and 3′-untranslated regions of satBaMV to form satBaMV-P20 RNP complexes (Tsai et al., 1999; Vijayapalani et al., 2012) and that formation of the complexes is negatively regulated by P20 phosphorylation (Vijayapalani et al., 2012). Our current results show that P20 can form punctate structures localized at PD (Figure 7) and the satBaMV-P20 RNP complexes can traffic autonomously through the phloem in satBaMV-transgenic stocks or scions (Figures 2B to 2D). In situ RT-PCR experiments also provided strong support that satBaMV moves within the functional phloem system. On grafting, the satBaMV is present in phloem of the stem, petiole and major vein of leaves in wild-type scions (Figure 3). The accumulation pattern of visualized satBaMV within vascular tissues in N. benthamiana phloem resembles that of SEO1 in N. tabacum (Ernst et al., 2012). Thus, evidence in support of the satBaMV phloem trafficking was provided by the combination of grafting, RNA gel blot analysis, in situ RT-PCR, and tissue blotting techniques. With these experimental approaches, we can conclude that the satBaMV is translocated through satBaMV transgenic stock into the wild-type scion via the phloem. This finding agrees with other recent studies of cellular or viral RNAs indicating that there are systemic recombination signals, that small RNAs alone can undergo systemic transport across a graft union (Turgeon and Wolf, 2009; Dunoyer et al., 2010), and that grafting is able to determine the specificity and efficiency of RNA trafficking (Kehr and Buhtz, 2008). We found that GFP RNA, BaMV CP RNA, and CMV satRNA in transgenic lines were all restricted to stocks (Figure 2E). Therefore, satBaMV may contain long-distance trafficking determinants that involve specific RNA sequences or structural elements, and the P20 protein may interact with host factors.
Using a combination of co-IP and LC-MS/MS along with VIGS assays or the fibrillarin RNAi transgenic line, we further determined that the HV-independent or -dependent systemic movement of the satBaMV RNP complexes depends on fibrillarin (Figure 5; Supplemental Figure 5 and Supplemental Tables 1 and 2). Fibrillarin is required for rRNA processing (Barneche et al., 2000; Sáez-Vasquez et al., 2004) and is a nucleolar-localized RBP required for systemic infection of GRV (Kim et al., 2007; Canetta et al., 2008), Potato leafroll virus (Haupt et al., 2005), and Rice stripe tenuivirus (Zheng et al., 2015). Nucleolar colocalization of fibrillarin and P20 protein (Figure 7), along with detection of fibrillarin, P20 protein, and satBaMV RNA in co-IP complexes (Figures 4 and 6) but not in the fibrillarin RNAi transgenic line (Supplemental Figure 5 and Supplemental Tables 1 and 2), is consistent with the evidence for interaction of fibrillarin with satBaMV-P20 RNP complexes (Figure 4E, Table 2). Experiments with transgenic RNAi lines reinforced our findings that fibrillarin is crucial for systemic movement of satBaMV but not BaMV trafficking (Figure 5E). Hence, fibrillarin is the identified host factor that is differentially required for satRNA and HV long-distance trafficking. Our results also suggest that satBaMV and BaMV may move independently by interacting with distinct cellular factors. Increasing evidence suggests that interactions between viral MPs and nuclear-localized proteins may represent an essential step for virus systemic trafficking (Solovyev and Savenkov, 2014), but determining where and how such interactions occur requires additional experimentation. Mobile satBaMV RNA complexes containing P20, fibrillarin, satBaMV RNA, TGBp1-3, and CP are also present with HV coinfection (Figure 6). In this way, BaMV TGBps may participate in PD gating (Howard et al., 2004) or by interaction with fibrillarin-satBaMV-P20 RNP complex to help with satBaMV movement. Taken together, our findings for the fibrillarin-satBaMV-P20 RNP complex suggest that the nuclear/nucleolar-localized fibrillarin activity may involve cytosolic interactions that require a high degree of coordination of the P20 protein of satBaMV during phloem-mediated trafficking.
Phloem-mobile RNP complexes can move in the translocation stream from source to sink tissues (Ursache et al., 2014). This RNP complex transport pathway can be regulated when plants respond to environmental cues or pathogen attack (Pallas and Gómez, 2013; Ursache et al., 2014). Dark treatment may alter the source-sink relationship and increase virus susceptibility (Lemoin et al., 2013; Helms and McIntyre, 1967). Our results demonstrate that dark treatment facilitates long-distance trafficking of fibrillarin-based satBaMV RNP complexes, presumably via a change in photoassimilate allocation. Several studies have also indicated that fibrillarin can exit the nucleoli and move to other cell compartments during exposure to biotic or abiotic stresses, such as actinomycin D (Chen and Jiang, 2004), mercury treatment (Chen et al., 2002), or GRV infection (Kim et al., 2007). Hence, fibrillarin may function in defense responses under stress conditions. However, whether P20 co-moves with fibrillarin or whether the nucleolar activity of P20 results in the relocalization of fibrillarin to the cytoplasm during satBaMV transport through the phloem remains to be determined.
In addition to identifying fibrillarin, we identified other host proteins, such as histone H3, which is crucial for trafficking of a geminivirus DNA through the nuclear pore complex and PD (Zhou et al., 2011), in our co-IP experiments (Table 1). These proteins may also interact with P20, directly or indirectly, to form fibrillarin-based satBaMV RNP complexes. Whether the phloem-mobile satBaMV RNP complexes contain any phloem proteins or RNA-specific chaperones that could modify the satBaMV RNA structure, thereby facilitating systemic spread, requires further investigation.
METHODS
Construction of Plasmids
The gene encoding the satBaMV P20 protein was placed under the control of the SUC2 promoter in companion cells (Haywood et al., 2005) by inserting the SUC2 promoter at the HindIII/XbaI sites of p1390 (Haywood et al., 2005) to generate p1390-SUC2pro. The P20 gene was then inserted at the XbaI/BamHI sites of p1390-SUC2pro vector to generate SUC2pro:P20 for transformation. To generate SUC2pro:P20-eGFP for transient expression P20, the P20-eGFP DNA fragment was amplified from pCass-P20-EGFP (Vijaya Palani et al., 2006) with the primers P20-eGFP-XbaI-F and P20-eGFP-BamHI-R (Supplemental Table 3) and then cloned in the p1390-SUC2:P20 plasmid at the XbaI/BamHI sites.
To express P20-eGFP in Nicotiana benthamiana, we constructed pBIN-P20-eGFP. The P20-eGFP DNA fragment was amplified from pCass-P20-EGFP (Vijaya Palani et al., 2006) using primers Tf-XmaI-F and Tf-XmaI-R (Supplemental Table 3) and then cloned into the pBIN61 plasmid at the XmaI site to generate pBIN-P20-eGFP. For transient expression of mCherry-NbFIB2 in N. benthamiana, we constructed pBIN-mCherry-NbFIB2. The mCherry DNA fragment was first amplified from pBA-mCh-p1 (Chou et al., 2013) using primers mCherry-SmaI-F and mCherry-KpnI-R (Supplemental Table 3) and then cloned into the pCass plasmid (Ding et al., 1995) at the SmaI and KpnI sites to generate pCass-mCherry. Then, the FIB2 coding region was amplified from N. benthamiana cDNA using primers NbFIB2-KpnI-F and NbFIB2-EcoRI-R (Supplemental Table 3). The amplified FIB2 fragment was then cloned into pCass-mCherry to generate pCass-mCherry-NbFIB2. Finally, pBIN-mCherry-NbFIB2 was generated by amplifying the mCherry-NbFIB2 fragment from pCass-mCherry-NbFIB2 using primers Tf-XmaI-F and Tf-XmaI-R (Supplemental Table 3) and then cloning it into the pBIN61 vector at the XmaI site.
To construct the pBIN-TMVMP-DsRed plasmid required as a PD marker, we first amplified the DsRed DNA fragment from pdNR (Addgene) using primers DsRed KpnI-F and DsRed EcoRI-R (Supplemental Table 3); the fragment was then cloned into the pCass plasmid at the KpnI and EcoRI sites to generate pCass-DsRed. The DNA of TMVMP was amplified from pTMV-ΔCP using primers TMV-MP-StuI-F and TMV-MP-KpnI-F (Supplemental Table 3) and then cloned into the pCass-DsRed plasmid at the StuI and KpnI sites to generate pCass-TMVMP-DsRed. Finally, the DNA of TMVMP-DsRed was amplified from pCass-TMVMP-DsRed using primers Tf-XmaI-F and Tf-XmaI-R and cloned into the pBIN61 plasmid at the XmaI site to generate pBIN-TMVMP-DsRed.
Generation of Transgenic Plants
The procedures for transformation and regeneration of transgenic SUC2pro:P20 N. benthamiana plants were previously described (Lin et al., 2013). The transgenic lines were selected based on their resistance to 50 mg/L hygromycin on Murashige and Skoog agar medium (Sigma-Aldrich). Seedlings exhibiting resistance to hygromycin were examined for P20 gene expression by RT-PCR using primers P20-EcoRI-F and P20-EcoRI-R (Supplemental Table 3). In total, we obtained 20 independent homozygous lines after segregation analysis. The homozygous lines 1-29 and 3-1 were used in this study. Transgenic N. benthamiana lines expressing satBaMV driven by the 35S promoter were a gift from Yau-Heiu Hsu (National Chung Hsing University, Taichung, Taiwan). Five homozygous lines were obtained and lines 2-6 and 9-2 were used for grafting experiments. The transgenic N. tabacum line expressing satCMV driven by the 35S promoter was a gift from Yau-Heiu Hsu (National Chung Hsing University, Taichung, Taiwan).
Fibrillarin RNAi transgenic N. benthamiana plants were generated by transformation with Agrobacterium tumefaciens LBA4404 carrying the plasmid pFGC5941.Fibrillarin3′, as described for coilin RNAi plants (Shaw et al., 2014). The plasmid consisted of a 321-bp fragment (region 501 to 821) of the N. benthamiana fibrillarin gene (AM269909) cloned in opposite orientations flanking the CHALCONE SYNTHASE intron. Three independent N. benthamiana fibrillarin RNAi lines were selected and were found to elicit an ∼60% reduction in fibrillarin expression. None of the lines exhibited obvious phenotype alterations, and line #1 was used in this study.
Coilin RNAi transgenic N. benthamiana plants were previously characterized (Shaw et al., 2014). Transformation and regeneration of fibrillarin and coilin RNAi transgenic plants were as previously described (Taliansky et al., 2004). To avoid the possibility of “off-target” silencing, we confirmed that the fibrillarin and coilin fragments used in the RNAi constructs did not contain any 21-nucleotide stretches showing similarity to other genes using the small interfering RNA scan website (http://bioinfo2.noble.org/RNAiScan.htm).
Protein Analysis
Leaves of SUC2pro:P20 transgenic plants, or those of BaMV- and satBaMV-infected plants, were ground in liquid nitrogen and resuspended in extraction buffer (50 mM Tris-HCl, pH 8, 10% glycerol, 1 mM EDTA, 100 mM NaCl, and 1 mM PMSF). For co-IP, 20-fold diluted protein samples were used for immunoblot analysis with anti-P20, anti-TGBp1, anti-TGBp2, anti-HA (Chou et al., 2013), rabbit anti-actin (1/5000 dilution, provided by Y.-Y. Hsu), or anti-CP antibodies (Lin et al., 1992; Chou et al., 2013) together with anti-fibrillarin IgG, H-140 (sc-25397) (Santa Cruz Biotech), followed by staining with goat anti-rabbit IgG HRP secondary antibody (Abcam).
Plant Growth, Inoculation, and Grafting
All wild-type and transgenic N. benthamiana or Nicotiana tabacum plants were grown at 28°C in a walk-in plant growth chamber under a 16-h-light/8-h-dark cycle with a white light (Philips TLD 36W/840 ns) intensity of 185 to ∼222 μmol m−2 s−1 at the leaf surface. For each set of experiments, we used 4-week-old plants for inoculation. The methods for inoculation by Agrobacterium expressing BaMV or satBaMV were described previously (Liou et al., 2014).
Wild-type or grafted N. benthamiana plants were agroinfected with Agrobacterium expressing full-length infectious cDNA clones of BaMV, pKB (Liou et al., 2014), or satBaMV, pKF4 (Liou et al., 2014) in pKYLX7 (Schardl et al., 1987) binary vector. For complementation assays, wild-type and SUC2pro:P20 transgenic plants were inoculated with 0.5 μg pCB alone or coinoculated with 0.5 μg pCBSF4 or pCBSGFP (Lin et al., 2004; Vijayapalani et al., 2012).
Grafting was performed as described (Turnbull et al., 2002) except that ∼40-d-old N. benthamiana or N. tabacum plants were used for cleft grafting. Each grafting experiment was repeated at four times, each including four plants.
Agrobacterium Culture, Infiltration, and VIGS
Plasmids (pKB or pKF4) for protein transient expression were transformed into Agrobacterium C58C1 by electroporation. Agrobacterium cultures were grown as described (Liou et al., 2014) and diluted to an optical density of 0.4 to 1.0 at 600 nm for infiltration into leaves of N. benthamiana plants. For coexpression of two plasmids, two solutions of Agrobacterium, each harboring a specific plasmid, were mixed in a 1:1 ratio prior to agroinfiltration. Agrobacterium was infiltrated into the intercellular space of N. benthamiana leaves.
For VIGS assays, 4-week-old plants of N. benthamiana were infiltrated with mixture of Agrobacterium LBA4404 harboring either the TRV1 or TRV2 RNA2 vector (p0704) containing the fibrillarin fragment (Kim et al., 2007). In parallel, silencing of the PHYTOENE DESATURASE leading to a photobleached phenotype was used as a marker for monitoring the effectiveness of VIGS. At 7 d after agroinfiltration, we harvested systemically infected leaves for RNA accumulation assays.
RNA Analysis
Total RNA was extracted from N. benthamiana or N. tabacum tissues using Tripure in accordance with the manufacturer’s instructions (Roche). RNA gel blot analysis was performed as described previously (Lin et al., 1996). BaMV and satBaMV accumulation was analyzed using 32P-labeled RNA probes specific for the BaMV 3′ end generated from HindIII-linearized pBaHB (Lin et al., 1993) and specific for full-length satBaMV generated from EcoRI-linearized pBSHE (Lin et al., 2013), respectively. The satCMV probe was transcribed from HindIII-linearized pGEM4 (provided by Yau-Heiu Hsu) using SP6 RNA polymerase. The GFP probe was transcribed from EcoRI-linearized pGEM-T Easy GFP using SP6 RNA polymerase (New England Biolabs) (Vijayapalani et al., 2012). Accumulation of fibrillarin, coilin, and actin mRNA was conducted as described by Kim et al. (2007). Four independent replicates were performed for each experiment.
For RT-qPCR, 2 μg total RNA extracted from plants was reverse-transcribed into poly d(T) cDNA using SuperScript III reverse transcriptase (Invitrogen) in triplicates with the GeneAmp 9700 sequence detection Real-Time PCR system (Life Technologies) and SYBR Green I core reagent (Life Technologies). Normalization of satBaMV accumulation was to the 18S gene. Primers for satBaMV (satBaMVrt-F and satBaMVrt-R) and 18S (18S-F and 18S-R) are in Supplemental Table 3.
To analyze BaMV or satBaMV RNA in the co-IP fractions, we extracted total RNA from the anti-P20 or antifibrillarin co-IP fractions followed by RT-PCR. Primer set BaMV-F and BaMV-R (Supplemental Table 3) was used to amplify a 729-nucleotide BaMV cDNA 3′ fragment (produced after 25 cycles of PCR). Similarly, primers satBaMV-F and satBaMV-R (Supplemental Table 3) were used to amplify full-length satBaMV cDNA.
Tissue Blotting
Sections were cut from fresh stem tissues by hand with a new razor blade. Tissue blots were made by pressing the newly cut surface onto a membrane; a 32P-labeled RNA probe specific for full-length satBaMV was generated from EcoRI-linearized pBSHE (Lin et al., 2013) and then used to detect the distribution of satBaMV in the tissues (Lin et al., 1990).
In Situ RT-PCR
The localization of satBaMV was determined using established protocols for in situ RT-PCR (Yoo et al., 2004). A reverse transcriptase cocktail (containing SuperScript III reverse transcriptase components and satBaMV-R primer [Supplemental Table 3] at 75 μM) was prepared immediately before use. Fresh 200-μm thick sections were obtained using a D.S.K. Microslicer DTK-1000. Sections were placed onto a glass slide, covered by a 25-μL aliquot of the above cocktail and sealed with amplicover discs and clips. The reverse transcription step was performed at 50°C for 60 min. For the PCR step, the satBaMV-F primer of 75 μM (Supplemental Table 3) and ChromaTide Alexa Fluor 488-5-dUTP (20 μM; Molecular Probes) were added; dTTP was reduced to 10 μM. The amplification protocol consisted of 10 cycles: 30 s at 94°C, 30 sat 55°C, and 60 s at 72°C. A MJ Research PTC-200 Peltier Thermal cycler was used for these experiments.
After this reaction series, sections were incubated (1 min) in absolute ethanol, followed by rinsing in 1 mM EDTA and then overnight washing (16 h), at 22°C, in this EDTA solution.
Co-IP and LC-MS/MS
Plant total proteins were extracted using Tris-HCl buffer (Dharmasiri et al., 2005) from systemically infected leaves of N. benthamiana agroinfiltrated with pKB at 7 DPI or together with pKF4. Plant Rubisco was removed using Seppro RuBisCO Spin Columns (catalog number SEP070; Sigma-Aldrich) in accordance with the manufacturer’s instructions.
In vivo co-IP experiments with anti-P20 (Palani et al., 2009) and anti-fibrillarin IgG, H-140 (sc-25397; Santa Cruz Biotech) were performed as described (Dharmasiri et al., 2005). Briefly, plant extracts containing 1 mg protein were incubated with anti-P20 IgG (1:150 v/v) for 1 h at 4°C on a rotary shaker. Then, 20 μL Protein A agarose beads (GE Healthcare Life Science) was added, and the mixture was incubated for 3 h at 4°C. After washing with Tris-HCl buffer (Dharmasiri et al., 2005), the immunoprecipitate resuspended in 2× sample buffer (4% SDS, 20% glycerol, 0.12 M Tris, pH 6.8, and 10% β-mercaptoethanol) and separated on a NuPAGE Novex 4 to 12% Bis-Tris protein gel (Invitrogen Life Science Technologies).
Gels were silver-stained, and protein bands were then excised and digested with trypsin for analysis by LC-MS/MS (Lo et al., 2011). LC-MS/MS fragmented ions were used with Mascot (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=MIS) to search against the most recent Arabidopsis thaliana, BaMV, and satBaMV databases in the National Center for Biotechnology Information.
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were performed as recommended by the manufacturers of the GAL4 Two-Hybrid Phagemid Vector kits (Agilent Technologies). The full-length coding sequence of fibrillarin was generated using NbFIB2-EcoRI-F and NbFIB2-EcoRI-R, and P20 was generated using P20-EcoRI-F and P20-EcoRI-R (Supplemental Table 3). Fibrillarin or P20 was cloned into downstream of the GAL4 AD at EcoRI site or downstream of the GAL4 DNA binding domain at the EcoRI site. The rich medium yeast extract, peptone, dextrose (YPD) is most commonly used for growing yeast under nonselective conditions. To test interactions between fibrillarin and P20, we coexpressed the constructs in YRG-2 yeast cells and selected by incubation in leucine-tryptophan-histidine medium at 28°C for 2 to 3 d until colonies appeared. Each experiment was repeated three times.
Confocal Microscopy
To visualize satBaMV accumulation from in situ RT-PCR (Figure 3), fresh tissues were examined under a Zeiss LSM880 laser scanning microscope with a 40×/1.2 W Korr UV-VIS-IR objective lens. Images were captured using the ZEN software with excitation/emission of 488 nm/505 to 550 nm.
To visualize PD in leaf epidermal cells, we infiltrated aniline blue fluorochrome (Biosupplies) (0.1 mg/mL in water) into agroinfected leaves of N. benthamiana and then immediately examined the leaves using a Zeiss LSM510 laser scanning microscope with a 40×/1.2 W Korr UV-VIS-IR objective lens (Figure 7). Images were captured using the LSM510 software with filters for aniline blue fluorochrome (excitation/emission: 405 nm/480 to 510 nm), GFP (excitation/emission: 488 nm/505 to 575 nm), and mCherry/DsRed (excitation/emission: 543 nm/560 to 615 nm). All images were processed and cropped using Zeiss LSM Image Browser and Photoshop CS5 (Adobe).
The agroinfiltrated leaves expressing P20-eGFP and TMVMP-DsRed at 2 d after inoculation were used for plasmolysis studies (Figure 7D). The leaf discs were treated with 1 M NaCl for 20 min before observation. Samples were scanned under a Zeiss LSM880 laser scanning microscope with a C-Apochromat 40×/1.2 W Korr FCS M27 objective lens. Images were captured using ZEN2 software with the filters of GFP (excitation/emission: 488 nm/500 to 550 nm) and DsRed (excitation/emission: 561 nm/570 to 619 nm).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under accession numbers shown in Tables 1 and 2 and Supplemental Tables 1 and 2.
Supplemental Data
Supplemental Figure 1. Trans-complementation of the systemic movement of P20-defective satellite RNA of Bamboo mosaic virus in P20 transgenic N. benthamiana.
Supplemental Figure 2. Systemic movement of SatBaMV with and without HV.
Supplemental Figure 3. RT-qPCR analysis of satBaMV mRNA accumulation in N. benthamiana grafting experiments.
Supplemental Figure 4. BaMV and SatBaMV accumulation in coilin or fibrillarin RNAi transgenic lines and fibrillarin accumulation in fibrillarin VIGS plants.
Supplemental Figure 5. Identification of P20-interacting protein complex from grafting N. benthamiana fibrillarin RNAi leaves by coimmunoprecipitation.
Supplemental Figure 6. Yeast two-hybrid analysis of interactions between fibrillarin and P20.
Supplemental Table 1. Proteins identified by LC-MS/MS after the immunoprecipitation of P20 IgG from fibrillarin RNAi scions grafted onto satBaMV transgenic stock.
Supplemental Table 2. Proteins identified by LC-MS/MS after the immunoprecipitation of P20 IgG from BaMV and satBaMV coinfected fibrillarin RNAi plants.
Supplemental Table 3. List of primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Andy Jackson (Plant and Microbial Biology Department, University of California, Berkeley, CA) for editing the manuscript, M.-Z. Fang for sequencing, and W.-N. Jane for in situ RT-PCR. We also thank the Proteomics Core Lab, Transgenic Plant Core Lab, and Plant Cell Biology Core Lab at the Institute of Plant and Microbial Biology, Academia Sinica, Taiwan for technical assistance. This research was supported by grants from the Ministry of Science and Technology (NSC 1002313B001002MY3) and the Academia Sinica Investigator Award, Taiwan, to N.-S.L. The work of J.S. and M.T. was funded by the Scottish Government Rural and Environmental Science and Analytical Services Division.
AUTHOR CONTRIBUTIONS
C.-H.C., N.-S.L., and Y.-H.H. designed the project. C.-H.C. and Y.-S.L. conducted most of the experiments. S.-C.L. and J.-D.W. obtained confocal microscope images. J.S., M.T., B.-Y.C., and Y.-H.H. designed the viral constructs and characterized the transgenic lines. C.-H.C., F.-C.H, Y.-H.H., M.T., and N.-S.L. analyzed data. C.-H.C., F.-C.H., M.T., and N.-S.L. wrote the article.
Glossary
- PD
plasmodesmata
- RNP
ribonucleoprotein
- RBP
RNA binding protein
- MP
movement protein
- TMV
Tobacco mosaic virus
- GRV
Groundnut rosette virus
- satRNA
satellite RNA
- HV
helper virus
- BaMV
Bamboo mosaic virus
- satBaMV
BaMV-associated satRNA
- CP
capsid protein
- co-IP
coimmunoprecipitation
- DPI
days postinoculation
- DAG
days after grafting
- SA
shoot apex
- satCMV
satRNA of Cucumber mosaic virus
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- VIGS
virus-induced gene silencing
- AD
activation domain
Footnotes
Articles can be viewed without a subscription.
References
- Amari K., Di Donato M., Dolja V.V., Heinlein M. (2014). Myosins VIII and XI play distinct roles in reproduction and transport of tobacco mosaic virus. PLoS Pathog. 10: e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Suzui N., Fujimaki S., Dohmae N., Yonekura-Sakakibara K., Fujiwara T., Hayashi H., Yamaya T., Sakakibara H. (2005). Destination-selective long-distance movement of phloem proteins. Plant Cell 17: 1801–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Sorenson R., Juntawong P. (2009). Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 14: 443–453. [DOI] [PubMed] [Google Scholar]
- Barneche F., Steinmetz F., Echeverría M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275: 27212–27220. [DOI] [PubMed] [Google Scholar]
- Canetta E., Kim S.H., Kalinina N.O., Shaw J., Adya A.K., Gillespie T., Brown J.W.S., Taliansky M. (2008). A plant virus movement protein forms ringlike complexes with the major nucleolar protein, fibrillarin, in vitro. J. Mol. Biol. 376: 932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Jiang P. (2004). Altered subcellular distribution of nucleolar protein fibrillarin by actinomycin D in HEp-2 cells. Acta Pharmacol. Sin. 25: 902–906. [PubMed] [Google Scholar]
- Chen M., Rockel T., Steinweger G., Hemmerich P., Risch J., von Mikecz A. (2002). Subcellular recruitment of fibrillarin to nucleoplasmic proteasomes: implications for processing of a nucleolar autoantigen. Mol. Biol. Cell 13: 3576–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.H., Sheng J., Hind G., Handa A.K., Citovsky V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.L., Hung Y.J., Tseng Y.H., Hsu H.T., Yang J.Y., Wung C.H., Lin N.S., Meng M.S., Hsu Y.H., Chang B.Y. (2013). The stable association of virion with the triple-geneblock protein 3-based complex of Bamboo mosaic virus. PLoS Pathog. 9: e1003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- Ding S.W., Rathjen J.P., Li W.X., Swanson R., Healy H., Symons R.H. (1995). Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J. Gen. Virol. 76: 459–464. [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916. [DOI] [PubMed] [Google Scholar]
- Ernst A.M., Jekat S.B., Zielonka S., Müller B., Neumann U., Rüping B., Twyman R. M., Krzyzanek V., Prüfer D., Noll G. A. (2012). Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc. Natl. Acad. Sci. USA 109: E1980–E1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R., Gas M.E., Molina-Serrano D., Nohales M.A., Carbonell A., Gago S., De la Peña M., Daròs J.A. (2009). Viroid replication: rolling-circles, enzymes and ribozymes. Viruses 1: 317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman M.H.S., Seurinck J., Marins M., Goldman G.H., Mariani C. (1998). A tobacco flower-specific gene encodes a polyphenol oxidase. Plant Mol. Biol. 36: 479–485. [DOI] [PubMed] [Google Scholar]
- Gopinath K., Kao C.C. (2007). Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 19: 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham B.K., Brandom J.L., Xoconostle-Cázares B., Ringgold V., Lough T.J., Lucas W.J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21: 197–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P.A., Park J.W., Sasaki N., Ballard K.D., Maule A.J., Nelson R.S. (2009). Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc. Natl. Acad. Sci. USA 106: 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S., Stroganova T., Ryabov E., Kim S.H., Fraser G., Duncan G., Mayo M.A., Barker H., Taliansky M. (2005). Nucleolar localization of potato leafroll virus capsid proteins. J. Gen. Virol. 86: 2891–2896. [DOI] [PubMed] [Google Scholar]
- Haywood V., Yu T.S., Huang N.C., Lucas W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42: 49–68. [DOI] [PubMed] [Google Scholar]
- Helms K., McIntyre G.A. (1967). Light-induced susceptibility of Phaseolus vulgaris L. to tobacco mosaic virus infection. I. Effects of light intensity, temperature, and the length of the preinoculation dark period. Virology 31: 191–196. [DOI] [PubMed] [Google Scholar]
- Hipper C., Brault V., Ziegler-Graff V., Revers F. (2013). Viral and cellular factors involved in Phloem transport of plant viruses. Front. Plant Sci. 4: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A.R., Heppler M.L., Ju H.J., Krishnamurthy K., Payton M.E., Verchot-Lubicz J. (2004). Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328: 185–197. [DOI] [PubMed] [Google Scholar]
- Hu C.C., Hsu Y.H., Lin N.S. (2009). Satellite RNAs and satellite viruses of plants. Viruses 1: 1325–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Hu C.C., Liou M.R., Chang B.Y., Tsai C.H., Meng M., Lin N.S., Hsu Y.H. (2012). Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 8: e1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J., Buhtz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59: 85–92. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Macfarlane S., Kalinina N.O., Rakitina D.V., Ryabov E.V., Gillespie T., Haupt S., Brown J.W.S., Taliansky M. (2007). Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc. Natl. Acad. Sci. USA 104: 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F. (2013). Plasmodesmata: intercellular tunnels facilitating transport of macromolecules in plants. Cell Tissue Res. 352: 49–58. [DOI] [PubMed] [Google Scholar]
- Lan P., Yeh W.B., Tsai C.W., Lin N.S. (2010). A unique glycine-rich motif at the N-terminal region of Bamboo mosaic virus coat protein is required for symptom expression. Mol. Plant Microbe Interact. 23: 903–914. [DOI] [PubMed] [Google Scholar]
- Lee C.C., Ho Y.N., Hu R.H., Yen Y.T., Wang Z.C., Lee Y.C., Hsu Y.H., Meng M. (2011). The interaction between bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85: 12022–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoin N., et al. (2013). Soure-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.Y., Hsu Y.H., Chen H.C., Lin N.S. (2013). Transgenic resistance to Bamboo mosaic virus by expression of interfering satellite RNA. Mol. Plant Pathol. 14: 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.K., Chang B.Y., Liao J.T., Lin N.S., Hsu Y.H. (2004). Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 85: 251–259. [DOI] [PubMed] [Google Scholar]
- Lin M.K., Hu C.C., Lin N.S., Chang B.Y., Hsu Y.H. (2006). Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the bamboo mosaic virus satellite RNA-mediated expression system. J. Gen. Virol. 87: 1357–1367. [DOI] [PubMed] [Google Scholar]
- Lin N.S., Hsu Y.H. (1994). A satellite RNA associated with bamboo mosaic potexvirus. Virology 202: 707–714. [DOI] [PubMed] [Google Scholar]
- Lin N.S., Hsu Y.H., Hsu H.T. (1990). Immunological detection of plant viruses and a mycoplasmalike organism by direct tissue blotting on nitrocellulose membranes. Phytopathology 80: 824–828. [Google Scholar]
- Lin N.S., Lin F.Z., Huang T.Y., Hsu Y.H. (1992). Genome properties of Bamboo mosaic virus. Phytopathology 82: 731–734. [Google Scholar]
- Lin N.S., Chai Y.J., Huang T.Y., Chang T.Y., Hsu Y.H. (1993). Incidence of Bamboo Mosaic Potexvirus in Taiwan. Plant Disease 77: 448–450. [Google Scholar]
- Lin N.S., Lee Y.S., Lin B.Y., Lee C.W., Hsu Y.H. (1996). The open reading frame of bamboo mosaic potexvirus satellite RNA is not essential for its replication and can be replaced with a bacterial gene. Proc. Natl. Acad. Sci. USA 93: 3138–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N.S., Lin B.Y., Lo N.W., Hu C.C., Chow T.Y., Hsu Y.H. (1994). Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75: 2513–2518. [DOI] [PubMed] [Google Scholar]
- Liou M.R., Huang Y.W., Hu C.C., Lin N.S., Hsu Y.H. (2014). A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 12: 330–343. [DOI] [PubMed] [Google Scholar]
- Lo Y.S., Cheng N., Hsiao L.J., Annamalai A., Jauh G.Y., Wen T.N., Dai H., Chiang K.S. (2011). Actin in mung bean mitochondria and implications for its function. Plant Cell 23: 3727–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J., Yoo B.C., Kragler F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2: 849–857. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Ham B.K., Kim J.Y. (2009). Plasmodesmata - bridging the gap between neighboring plant cells. Trends Cell Biol. 19: 495–503. [DOI] [PubMed] [Google Scholar]
- Middleton A.M., Farcot E., Owen M.R., Vernoux T. (2012). Modeling regulatory networks to understand plant development: small is beautiful. Plant Cell 24: 3876–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875. [DOI] [PubMed] [Google Scholar]
- Ogg S.C., Lamond A.I. (2002). Cajal bodies and coilin--moving towards function. J. Cell Biol. 159: 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani P.V., Chiu M., Chen W., Wang C.C., Lin C.C., Hsu C.C., Cheng C.P., Chen C.M., Hsu Y.H., Lin N.S. (2009). Subcellular localization and expression of bamboo mosaic virus satellite RNA-encoded protein. J. Gen. Virol. 90: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas V., Gómez G. (2013). Phloem RNA-binding proteins as potential components of the long-distance RNA transport system. Front. Plant Sci. 4: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.R., Seo J.K., Kim K.H. (2013). Viral and nonviral elements in potexvirus replication and movement and in antiviral responses. Adv. Virus Res. 87: 75–112. [DOI] [PubMed] [Google Scholar]
- Raffaele S., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez A.M., Baulcombe D.C. (2001). Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25: 237–245. [DOI] [PubMed] [Google Scholar]
- Ruiz M.T., Voinnet O., Baulcombe D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverría M. (2004). A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24: 7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C.L., Byrd A.D., Benzion G., Altschuler M.A., Hildebrand D.F., Hunt A.G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–11. [DOI] [PubMed] [Google Scholar]
- Semashko M.A., González I., Shaw J., Leonova O.G., Popenko V.I., Taliansky M.E., Canto T., Kalinina N.O. (2012). The extreme N-terminal domain of a hordeivirus TGB1 movement protein mediates its localization to the nucleolus and interaction with fibrillarin. Biochimie 94: 1180–1188. [DOI] [PubMed] [Google Scholar]
- Shaw J., Love A.J., Makarova S.S., Kalinina N.O., Harrison B.D., Taliansky M.E. (2014). Coilin, the signature protein of Cajal bodies, differentially modulates the interactions of plants with viruses in widely different taxa. Nucleus 5: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev A.G., Savenkov E.I. (2014). Factors involved in the systemic transport of plant RNA viruses: the emerging role of the nucleus. J. Exp. Bot. 65: 1689–1697. [DOI] [PubMed] [Google Scholar]
- Takeda R., Petrov A.I., Leontis N.B., Ding B. (2011). A three-dimensional RNA motif in Potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana. Plant Cell 23: 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliansky M., Kim S.H., Mayo M.A., Kalinina N.O., Fraser G., McGeachy K.D., Barker H. (2004). Escape of a plant virus from amplicon-mediated RNA silencing is associated with biotic or abiotic stress. Plant J. 39: 194–205. [DOI] [PubMed] [Google Scholar]
- Taliansky M.E., Brown J.W.S., Rajamäki M.L., Valkonen J.P.T., Kalinina N.O. (2010). Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems. Adv. Virus Res. 77: 119–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.S., Hsu Y.H., Lin N.S. (1999). Bamboo mosaic potexvirus satellite RNA (satBaMV RNA)-encoded P20 protein preferentially binds to satBaMV RNA. J. Virol. 73: 3032–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R., Wolf S. (2009). Phloem transport: cellular pathways and molecular trafficking. Annu. Rev. Plant Biol. 60: 207–221. [DOI] [PubMed] [Google Scholar]
- Turnbull C.G.N., Booker J.P., Leyser H.M.O. (2002). Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32: 255–262. [DOI] [PubMed] [Google Scholar]
- Ursache R., Heo J.O., Helariutta Y. (2014). Plant Vascular Biology 2013: vascular trafficking. J. Exp. Bot. 65: 1673–1680. [DOI] [PubMed] [Google Scholar]
- Vijayapalani P., Chen J.C.F., Liou M.R., Chen H.C., Hsu Y.H., Lin N.S. (2012). Phosphorylation of bamboo mosaic virus satellite RNA (satBaMV)-encoded protein P20 downregulates the formation of satBaMV-P20 ribonucleoprotein complex. Nucleic Acids Res. 40: 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaya Palani P., Kasiviswanathan V., Chen J.C.F., Chen W., Hsu Y.H., Lin N.S. (2006). The arginine-rich motif of Bamboo mosaic virus satellite RNA-encoded P20 mediates self-interaction, intracellular targeting, and cell-to-cell movement. Mol. Plant Microbe Interact. 19: 758–767. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Lee S.C., Wang C.W. (2011). Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 193: 521–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo B.C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y.M., Lough T.J., Lucas W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Du Z., Lin C., Mao Q., Wu K., Wu J., Wei T., Wu Z., Xie L. (2015). Rice stripe tenuivirus p2 may recruit or manipulate nucleolar functions through an interaction with fibrillarin to promote virus systemic movement. Mol. Plant Pathol. 16: 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Rojas M.R., Park M.R., Seo Y.S., Lucas W.J., Gilbertson R.L. (2011). Histone H3 interacts and colocalizes with the nuclear shuttle protein and the movement protein of a geminivirus. J. Virol. 85: 11821–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.