GAD1 encodes a secreted peptide that controls critical transitions of rice domestication, such as from fewer, longer grains with long awns to more, shorter grains with no or short awns.
Abstract
Cultivated rice (Oryza sativa) was domesticated from wild rice (Oryza rufipogon), which typically displays fewer grains per panicle and longer grains than cultivated rice. In addition, wild rice has long awns, whereas cultivated rice has short awns or lacks them altogether. These changes represent critical events in rice domestication. Here, we identified a major gene, GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1), that regulates those critical changes during rice domestication. GAD1 is located on chromosome 8 and is predicted to encode a small secretary signal peptide belonging to the EPIDERMAL PATTERNING FACTOR-LIKE family. A frame-shift insertion in gad1 destroyed the conserved cysteine residues of the peptide, resulting in a loss of function, and causing the increased number of grains per panicle, shorter grains, and awnless phenotype characteristic of cultivated rice. Our findings provide a useful paradigm for revealing functions of peptide signal molecules in plant development and helps elucidate the molecular basis of rice domestication.
INTRODUCTION
Cell-to-cell communication helps coordinate developmental and environmental responses. Secreted peptides mediate cell-to-cell signaling during development and pattern formation in multicellular organisms. Plants contain several signal peptides, such as systemin, which functions in the wounding response (Pearce et al., 1991); phytosulfokine, which induces the proliferation of single mesophyll cells (Matsubayashi and Sakagami, 1996); S-locus cysteine-rich protein/S-locus protein 11, which determines self-incompatibility (Schopfer et al., 1999); LUREs, which direct pollen tube elongation (Okuda et al., 2009); and CLAVATA3, which mediates the signaling of cell fate decisions in the Arabidopsis thaliana shoot meristem (Fletcher et al., 1999). EPIDERMAL PATTERING FACTORs (EPFs), such as EPF1, EPF2, and EPF-LIKE9 (EPFL9)/Stomagen, act as key regulators of stomatal development in Arabidopsis (Engineer et al., 2014; Hara et al., 2007, 2009; Hunt and Gray, 2009; Sugano et al., 2010).
Cultivated rice (Oryza sativa), a staple food for more than half of the global population (Khush, 1997), was domesticated from the wild rice (Oryza rufipogon) ∼8000 years ago (Fuller et al., 2010; Zong et al., 2007). Both morphological traits and physiological characteristics changed remarkably during domestication. Thorough understanding of rice domestication processes and molecular mechanisms active in cultivated rice will be beneficial for rice improvement. During the past 10 years, the rice evolutionary mechanisms and several domestication-related genes, including Sh4/SHA1 (Konishi et al., 2006; Li et al., 2006; Lin et al., 2007), PROG1 (Jin et al., 2008; Tan et al., 2008), Bh4 (Zhu et al., 2011), Rc (Sweeney et al., 2006), OsLG1 (Ishii et al., 2013; Zhu et al., 2013), An-1 (Luo et al., 2013), and LABA1/An-2 (Gu et al., 2015; Hua et al., 2015), were characterized.
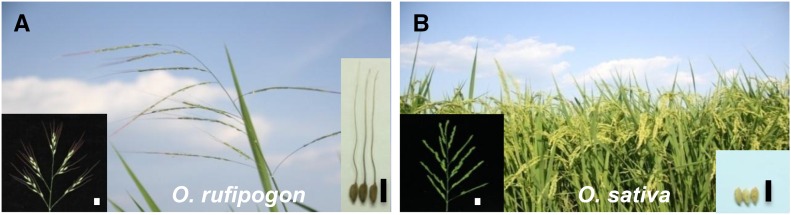
Wild rice typically produces a few, long grains per panicle, with long awns (Figure 1A) that are crucial for seed dispersal and deterring granivore predation. However, most current rice cultivars generate many grains per panicle and shorter grains and show no or only short awns (Figure 1B). These characteristics facilitate rice storage and processing (Hua et al., 2015), and the differences between wild and cultivated rice represent critical events in rice domestication. However, the molecular mechanisms responsible for these changes are still largely unknown.
Figure 1.
Phenotypes of Wild and Cultivated Rice.
(A) O. rufipogon. A panicle of wild rice is shown in the lower left corner, and the grains of wild rice are illustrated in the lower right corner. Bar = 10 mm.
(B) O. sativa. A panicle of cultivated rice is shown in the lower left corner, and the grains of cultivated rice are illustrated in the lower right corner. Bar = 10 mm.
Here, we found that an EPFL peptide, encoded by GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1), regulates grain number, grain length, and awn development, all of which were altered during rice domestication. A frame-shift insertion in gad1 destroyed the conserved cysteine residues of the EPFL peptide, causing the increased number of grains per panicle, the shorter grains, and the awnless phenotype found in cultivated rice. Our results thus establish the function of a peptide signal molecule in plant development and help uncover the genetic basis of rice domestication.
RESULTS
Characterization and Cloning of GAD1
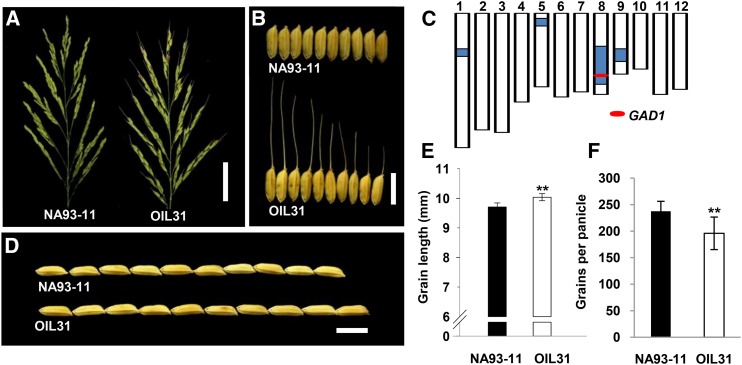
We obtained one introgression line (OIL31) from a set of introgression lines derived from a cross between indica variety NA93-11 and an accession of common wild rice (W2014, O. rufipogon). OIL31, which harbored four W2014 chromosomal segments, produced fewer grains per panicle and longer grains as well as a high percentage of awned seeds (∼80%), with long awns (18 ± 1.35 mm) in comparison with NA93-11 (Figure 2).
Figure 2.
Phenotypes of NA93-11 and OIL31.
(A) Panicle comparison between NA93-11 and OIL31. Bar = 50 mm.
(B) Grains of NA93-11 and OIL31. Bar = 10 mm.
(C) Genotype of OIL31. The white regions indicate the NA93-11 genetic background. The blue regions indicate wild rice introgressions. The red ellipse indicates the GAD1 locus.
(D) and (E) Grain length comparison between NA93-11 and OIL31. Bar = 10 mm.
(F) Comparison of grain number per panicle between NA93-11 and OIL31.
Sample sizes were n = 20. The statistical significance was set at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd.
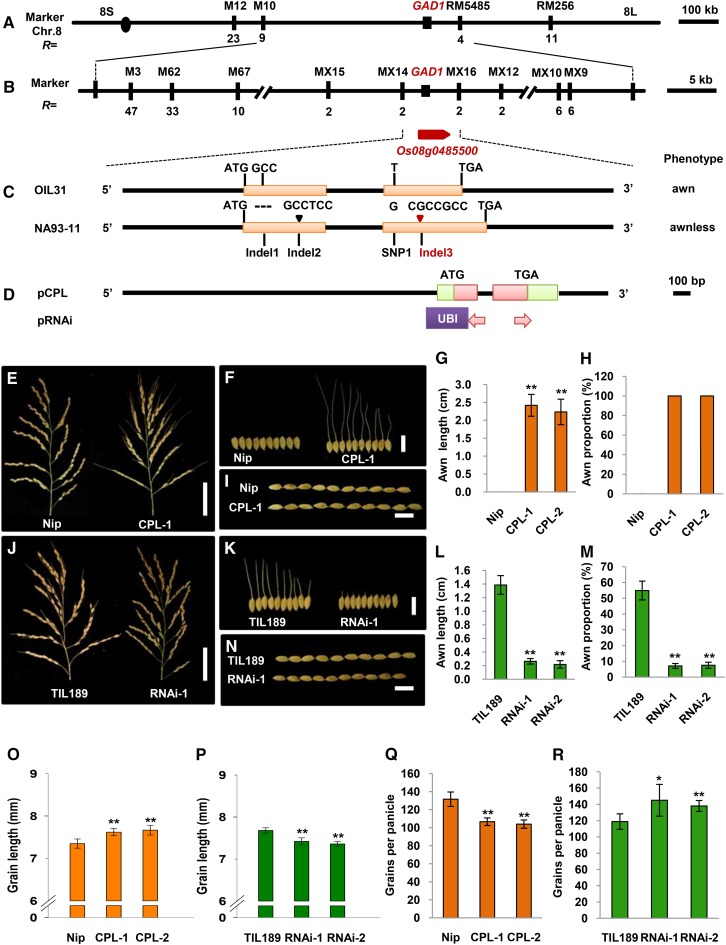
A genetic linkage analysis of 236 F2 individuals derived from a cross between OIL31 and NA93-11 indicated that the grain number per panicle, grain length, and awn phenotype were controlled by a single dominant gene, GAD1. We used the awn phenotype as a representative morphological marker to locate the GAD1 gene between simple sequence repeat markers M10 and RM5485 on the long arm of chromosome 8 (Figure 3A). Using 4250 F2 individuals, we further delimited GAD1 to within an ∼6-kb region between the markers MX14 and MX16, in which there was only one predicted gene (Os08g0485500) in the Nipponbare genome (The Rice Annotation Project Database) (Figure 3B). A comparison of the Os08g0485500 coding region between OIL31 and NA93-11 revealed one single nucleotide polymorphism (SNP1) and three insertion/deletions (Indel1, Indel2, and Indel3). SNP1 was a synonymous mutation, while Indel1 (a 3-bp deletion in NA93-11), Indel2 (a 6-bp insertion in NA93-11), and Indel3 (a 7-bp insertion in NA93-11) led to a single amino acid residue deletion, a two amino acid residue insertion, and a frame shift, respectively (Figure 3C; Supplemental Figure 1).
Figure 3.
Map-Based Cloning of GAD1.
(A) The target gene was mapped between the markers M10 and RM5485 on the long arm of chromosome 8. The numbers underneath the bars indicate the number of recombinants between GAD1 and the molecular markers.
(B) GAD1 was fine mapped to an ∼6-kb region between the markers MX14 and MX16. The numbers underneath the bars indicate the numbers of recombinants between GAD1 and molecular markers. Os08g0485500 was the candidate gene. The arrow indicates gene direction.
(C) Gene structure of GAD1 and the different sites between OIL31 and NA93-11. The black bars represent 5′ upstream and 3′ downstream regions and introns; rectangles represent exons; and triangles represent insertions. There were three Indels (Indel1, Indel2, and Indel3) and an SNP (SNP1) between OIL31 and NA93-11.
(D) The constructs used in the GAD1 functional investigation. pCPL represents the ∼4.0-kb W2014 genomic fragment covering the entire GAD1 gene region and the 2.5-kb 5′ upstream and 350-bp 3′ downstream sequences used for the complementation test. pRNAi denotes the RNAi construct that harbors a 350-bp specific region of the GAD1 mRNA. UBI is a maize Ubiquitin promoter. Green areas represent the 5′ and 3′ untranslated regions; red areas represent gene coding regions.
(E) Panicle comparison between Nipponbare (Nip) and the transgenic line (CPL-1). Bar = 50 mm.
(F) Grain comparison between Nip and the transgenic line (CPL-1). Bar = 10 mm.
(G) Awn length comparison between Nip and two independent transgenic lines, CPL-1 and CPL-2.
(H) The comparison of percentages of awned seeds between Nip and two independent transgenic lines, CPL-1 and CPL-2.
(I) and (O) Grain length comparison between Nip and transgenic lines, CPL-1 and CPL-2. Bar = 10 mm.
(J) Panicle comparison between a control plant (TIL189) and a transgenic line (RNAi-1). Bar = 50 mm.
(K) Grain comparison between TIL189 and a transgenic line (RNAi-1). Bar = 10 mm.
(L) Awn length comparison between TIL189 and two independent transgenic lines, RNAi-1 and RNAi-2.
(M) The comparison of percentages of awned seeds between TIL189 and two independent transgenic lines, RNAi-1 and RNAi-2.
(N) and (P) Grain length comparison between TIL189 and RNAi transgenic lines, RNAi-1 and RNAi-2. Bar = 10 mm.
(Q) Grain number per panicle comparison between Nip and two independent transgenic lines, CPL-1 and CPL-2.
(R) Grain number per panicle comparison between TIL189 and two independent transgenic lines, RNAi-1 and RNAi-2.
Sample sizes were n = 10. The statistical significance was set at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd.
To verify the function of Os08g0485500, we generated a construct (pCPL) that contained an ∼4.0-kb genomic fragment including the entire Os08g0485500 gene from wild rice W2014 (Figure 3D) and introduced the pCPL construct into the domesticated japonica variety Nipponbare. Compared with Nipponbare (controls), 32 independent transgenic lines of the wild rice Os08g0485500 (pCPL) showed fewer grains per panicle, longer grains and long awns (Figures 3E to 3I, 3O, and 3Q). These results demonstrated that Os08g0485500 in the O. rufipogon genome was a key gene (GAD1) regulating grain number, grain length, and awn development.
We further transformed a GAD1 interfering RNA (RNAi) construct (Figure 3D) into an introgression line harboring the GAD1 locus. The downregulation of the GAD1 transcript level in the RNAi transgenic plants led to more grains per panicle, shorter grains, and the short awns (or awnless) phenotype, compared with control plants (Figures 3J to 3N, 3P, and 3R). Thus, GAD1 is the key gene controlling the change in grain number, grain length, and awn development that occurred during rice domestication.
Expression of GAD1 Promotes Grain Elongation and Awn Development
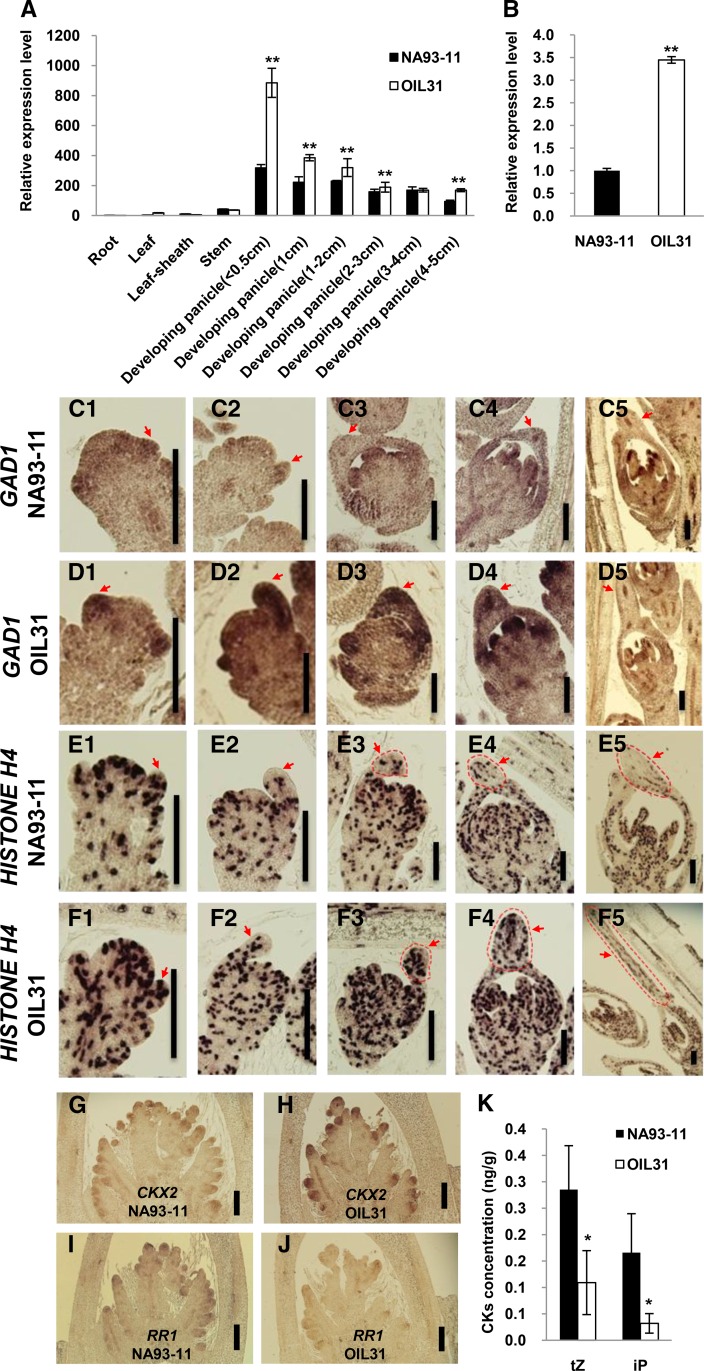
Using qRT-PCR, we analyzed the GAD1 expression pattern. GAD1 was preferentially expressed in the developing panicles of OIL31, especially in panicles <0.5 cm in length. Lower levels of transcript accumulation were detected in roots, leaves, leaf sheaths, and stems. With the development of panicles, GAD1 expression levels dropped dramatically in both NA93-11 and OIL31 (Figure 4A). Using RNA in situ hybridization, we detected GAD1 transcripts in the primary and secondary branch meristem in OIL31 and NA93-11 (Supplemental Figures 2 and 3). Because awns are organs at the tips of lemmas, we also compared the GAD1 expression in NA93-11 and OIL31 at the apices of glumes, which are composed of lemmas and paleae. GAD1 expression was higher in OIL31 than in NA93-11 at the glume apices (Figure 4B).
Figure 4.
The Expression Patterns and Effects of GAD1 on Spikelet Development.
(A) Comparison of GAD1 expression patterns between NA93-11 and OIL31.
(B) Comparison of GAD1 expression levels of at the apices of glumes between NA93-11 and OIL31.
(C1) to (C5) RNA in situ hybridization of GAD1 during spikelet development in NA93-11. Bar = 100 μm.
(D1) to (D5) RNA in situ hybridization of GAD1 during spikelet development in OIL31. Bar = 100 μm.
(E1) to (E5) RNA in situ hybridization of HISTONE H4 during spikelet development in NA93-11. Bar = 100 μm.
(F1) to (F5) RNA in situ hybridization of HISTONE H4 during spikelet development in OIL31. Bar = 100 μm.
(G) RNA in situ hybridization of CKX2 in a young panicle of NA93-11. Bar = 100 μm.
(H) RNA in situ hybridization of CKX2 in a young panicle of OIL31. Bar = 100 μm.
(I) RNA in situ hybridization of RR1 in a young panicle of NA93-11. Bar = 100 μm.
(J) RNA in situ hybridization of RR1 in a young panicle of OIL31. Bar = 100 μm.
(K) Comparison of cytokinin concentrations in 5- to 10-mm-long young panicles between NA93-11 and OIL31.
Red arrows (and red dashed lines in [E3] to [E5] and [F3] to [F5]) indicate the tips of lemmas. The statistical significance was set at P < 0.05 based on a two-tailed Student’s t test. Error bars represent the sd. In (A), (B), and (K), the data represent the average of three independent biological replicates.
To reveal the effects of GAD1, we compared awn development between OIL31 and NA93-11 using scanning electron microscopy. Based on the rice spikelet development (Sp) stages defined by Itoh et al. (2005), we detected no difference between OIL31 and NA93-11 until the Sp6 stage. At this stage, awn primordia appeared at the apices of the lemmas in OIL31 and extended from Sp6 to Sp8. However, during the same stages, no awns formed in NA93-11 (Supplemental Figure 4).
The detection of GAD1 expression during spikelet development using RNA in situ hybridization consistently showed no differences in the GAD1 expression patterns between OIL31 and NA93-11 until the Sp6 stage (Figures 4C1 and 4D1). At this stage, the transcripts of GAD1 were more abundant at the apices of lemmas in OIL31 than in NA93-11 (Figures 4C2 and 4D2). At the Sp7 stage and early Sp8 stage (Sp8e), the abundant expression of GAD1 was maintained at the apices of lemmas that formed the awn primordia in OIL31, and GAD1 expression was still low in NA93-11 (Figures 4C3, 4C4, 4D3, and 4D4). Thus, the GAD1 expression pattern was consistent with its role in the promotion of awn development. Additionally, at the late Sp8 stage (Sp8I), when spikelet development is almost complete, the GAD1 transcript was weakly detected in lemmas of both OIL31 and NA93-11 (Figures 4C5 and 4D5).
Previous studies indicated that cell division plays a crucial role in promoting awn formation and elongation (Hua et al., 2015; Luo et al., 2013). Therefore, we examined the expression of HISTONE H4, a marker gene of active cell division (Marzluff and Duronio, 2002), using RNA in situ hybridization. The HISTONE H4 expression patterns were similar between OIL31 and NA93-11 until the Sp7 stage (Figures 4E1, 4E2, 4F1, and 4F2). However, at the Sp7 and Sp8 stages, HISTONE H4 expression was higher in the awn primordia of OIL31 than in those of NA93-11 (Figures 4E3 to 4E5 and 4F3 to 4F5). Thus, there was more active cell division in the awn primordia of OIL31 than of NA93-11. A qRT-PCR analysis of HISTONE H1, another marker gene of active cell division (Luo et al., 2013; Marzluff and Duronio, 2002), consistently showed that the expression of HISTONE H1 was higher in the young panicles of OIL31 than those of NA93-11 (Supplemental Figure 5).
An analysis of cell cycle-related gene expression at the apices of glumes showed that 22 genes involved in the G1/S and G2/M phases regulating mitotic division (Li et al., 2011) were more highly expressed at the apices of glumes in OIL31 than in NA93-11 (Supplemental Figure 6). Thus, cell division was more active at the apices of glumes in OIL31 than in NA93-11, indicating that GAD1 promoted continuous cell division at the apices of glumes, which likely led to the promotion of grain elongation and awn development in wild rice.
Cytokinin Is Decreased in the Introgression Line OIL31
The plant hormone cytokinin is one of the factors controlling shoot apical meristem activity, which is a central determinant of grain number. Previous studies revealed that downregulation of CKX2 or DST causes cytokinin accumulation and increases grain number in rice (Ashikari et al., 2005; Li et al., 2013; Zhang and Yuan, 2014). Our RNA in situ hybridization and real-time PCR analysis showed that expression of CKX2 and DST were increased in the introgression line OIL31 compared with cultivar NA93-11 (Figures 4G and 4H; Supplemental Figure 7A). Consistent with the high expression of CKX2 and DST, the cytokinin (iP and tZ) contents were substantially decreased in the inflorescence meristem of OIL31, and expression of the cytokinin response marker genes RR1 and RR2 (Jain et al., 2006) was significantly decreased in the young panicles of OIL31 (Figures 4I to 4K; Supplemental Figure 7B). Together with our observations of higher GAD1 expression and fewer grains per panicle in OIL31 (Figures 2F and 4A) and previous genetic evidence (Ashikari et al., 2005; Li et al., 2013), our results suggest a hypothesis that GAD1 activates DST and CKX2 to reduce the cytokinin level and cause less grain number per panicle in wild rice.
GAD1 Encodes a Predicted Small Secreted Cysteine-Rich Peptide with a Conserved Cysteine Residue
The GAD1 cDNA in wild rice W2014 encodes a 127-amino acid protein that is a homolog of a small secreted cysteine-rich EPF/EPFL peptide in Arabidopsis (Engineer et al., 2014; Hara et al., 2007, 2009; Hunt and Gray, 2009; Sugano et al., 2010; Takata et al., 2013) (Supplemental Figures 8 and 9 and Supplemental Data Set 1). Similar to the Arabidopsis EPF/EPFL peptides, the GAD1 prepropeptide contained an N-terminal secretory signal peptide and a mature peptide with a conserved region of six cysteine residues (Supplemental Figures 10 and 11 and Supplemental Table 1). The six conserved cysteine residues are essential for proper formation of intramolecular disulfide bonds that are critical for peptide function (Abrash and Bergmann, 2010; Jewaria et al., 2013; Katsir et al., 2011; Lee et al., 2012; Ohki et al., 2011; Takata et al., 2013). A comparison of the predicted mature peptides between wild rice W2014 and cultivar NA93-11 indicated that the 7-bp (Indel3) insertion in gad1 caused a frame-shift mutation that led to the loss of two (5th and 6th) essential cysteine residues and, accordingly, likely prevented the formation of the proper mature peptide, resulting in the loss of the gad1 function in NA93-11 (Figure 5A).
Figure 5.
GAD1 Protein Variants in Wild and Cultivated Rice.
(A) Differences between GAD1 in W2014 and gad1 in NA93-11. The green rectangles represent the signal peptide. Reddish rectangles represent the pro-peptide. Purple rectangles represent the mature peptide. Red vertical lines represent cysteine residues. Blue rectangles represent the frame-shift region.
(B) The distribution of GAD1 protein variants in wild rice.
(C) The distribution of gad1 protein variants in cultivated rice.
To determine the amount of GAD1 variation among wild and cultivated rice, we sequenced the GAD1 coding region in 53 accessions of wild rice originating from 12 countries and 203 accessions of cultivated rice from 15 countries. We found 10 kinds of GAD1/gad1 haplotypes in wild and cultivated rice (Supplemental Figure 12). The GAD1 gene of most wild rice contained a 9-bp insertion or was similar to W2014 and encoded a mature protein containing six cysteine residues (6C). This gene was associated with the long-awn phenotype, suggesting that GAD1 with six cysteine residues is functional. In the cultivated rice, there were 180 varieties that, like NA93-11, had frame shifts that destroyed the classic 5th and 6th cysteine residues in GAD1. Among those, 137 varieties contained a 7-bp insertion in the Indel3 of gad1 to generate a dysfunctional mutated mature peptide containing seven cysteine residues (7C), while the remaining 43 varieties contained a 5-bp insertion in Indel3 to encode the dysfunctional mutated protein with four cysteine residues (4C). Among the 180 varieties with the dysfunctional gad1 peptide, 161 (∼90%) of the varieties showed the awnless phenotype (Figures 5B and 5C; Supplemental Figure 12), while 19 varieties displayed an awn phenotype, which may be caused by other awn-regulatory gene(s).
Neutrality Test and Positive Selection on gad1 in O. sativa
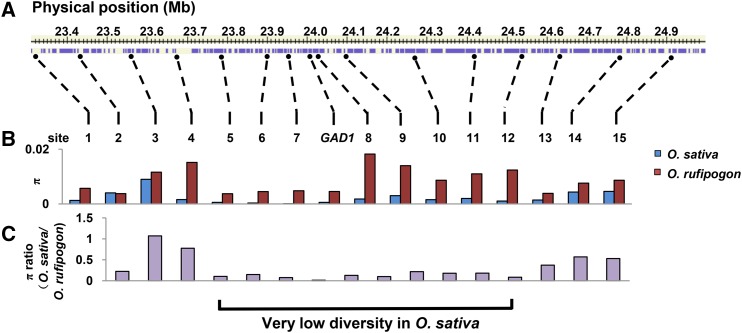
The nucleotide diversity of gad1 in cultivated rice was remarkably lower than that of GAD1 in wild rice. Tajima’s D test suggested that gad1 in cultivated rice deviated significantly from the neutral expectation (P < 0.01), while GAD1 in wild rice did not deviate from the neutral expectation (P > 0.10) (Supplemental Table 2). We further examined the nucleotide diversity of 16 loci in the GAD1-containing 1.9-Mb genomic region and identified an interval of ∼900 kb surrounding the GAD1 locus that exhibited a significantly decreased level of nucleotide diversity in cultivated rice relative to wild rice (Figure 6). Our results are consistent with recent whole-genome investigations (Huang et al., 2012; Xu et al., 2011). These data indicated that the gad1 gene regulating grain number, grain length, and awn development in O. sativa was strongly selected by humans during rice domestication.
Figure 6.
Nucleotide Diversity across the GAD1 Genomic Region on Chromosome 8.
(A) The 16 sampled loci (including GAD1) located in the ∼1.9-Mb genomic region around GAD1 gene on chromosome 8.
(B) Nucleotide diversity in cultivated rice and wild rice at the 16 sampled loci.
(C) Ratio of nucleotide diversity in O. sativa relative to that in O. rufipogon, including the ∼900-kb selective sweep region surrounding gad1 in cultivated rice.
To further investigate the selection of GAD1 during rice domestication, we used 159 accessions of cultivated rice from 15 countries to compare the appearance of GAD1 with two recently identified genes involved in awn development, An-1, encoding a basic helix-loop-helix transcription factor (Luo et al., 2013), and LABA1/An-2, encoding a cytokinin-activating enzyme (Gu et al., 2015; Hua et al., 2015). We found that the proportions of cultivars harboring the GAD1, LABA1, and An-1 alleles were 14, 30, and 30% respectively (Supplemental Figure 13), suggesting that the gad1 selection might be stronger than that for either laba1 or an-1 during rice domestication.
GAD1 Might Have Additive Effects with An-1 and LABA1/An-2
Two key genes, An-1 and LABA1/An-2, that affect awn development during rice domestication have been characterized previously. An-1 encodes a basic helix-loop-helix protein and regulates awn development (Luo et al., 2013). LABA1/An-2 encodes a cytokinin-activating enzyme and controls awn barb formation and awn elongation (Gu et al., 2015; Hua et al., 2015). To determine the relationships among An-1, LABA1/An-2, and GAD1, we generated a set of near-isogenic lines and pyramiding lines. The awn length of the pyramiding line harboring GAD1 and An-1 (6.27 ± 0.12 cm) was much longer than those of near-isogenic lines containing only GAD1 (1.89 ± 0.16 cm) or An-1 (4.6 ± 0.26 cm). Additionally, the awn length of a pyramiding line harboring GAD1 and LABA1/An-2 (2.68 ± 0.16 cm) was longer than that of a near-isogenic line containing LABA1/An-2 (no awn) and GAD1 (Supplemental Figure 14). Thus, GAD1 might have an additive effect with An-1 and LABA1/An-2.
DISCUSSION
In this article, we found that the GAD1-encoded EPFL peptide regulates grain number, grain length, and awn development in rice. A frame-shift insertion in coding region of the GAD1 gene was responsible for the transition from a few, long grains per panicle with long awns to many grains per panicle, shorter grains, and no or only short awns during rice domestication.
Crop domestication is the genetic modification of a wild species to generate a new form to meet human needs through cultivation (Doebley et al., 2006). Wild rice shows prostrate growth, severe shattering, spread panicles, and long awns, while cultivated rice exhibits erect growth, no seed-shattering, and compact panicles, which may be more amenable to cultivation and allow harvest of more seeds (Jin et al., 2008; Ishii et al., 2013; Konishi et al., 2006; Li et al., 2006; Lin et al., 2007; Tan et al., 2008; Zhu et al., 2013). Recently, several genes controlling rice domestication-related traits have been characterized. These genes encode transcription factors or enzymes (Gu et al., 2015; Hua et al., 2015; Ishii et al., 2013; Jin et al., 2008; Konishi et al., 2006; Li et al., 2006; Lin et al., 2007; Luo et al., 2013; Sweeney et al., 2006; Tan et al., 2008; Zhu et al., 2013). By contrast, our study revealed that key rice domestication-related traits, including awn development, and yield-related traits were regulated by a predicted small secretory signal peptide GAD1, related to the Arabidopsis EPFL family. Our findings not only provide evidence revealing novel functions of peptide signal molecules during plant development, but also offer intriguing insights on the role of peptide signal molecules in rice domestication.
The EPF/EPFL Peptides Have Multiple Biological Functions
Multiple biological functions of EPF/EPFL family members have been characterized in plants. In Arabidopsis, EPF1, EPF2 and STOMAGEN/EPFL9 regulate stomatal development (Engineer et al., 2014; Hara et al., 2007, 2009; Hunt and Gray, 2009; Sugano et al., 2010); EPFL4 and EPFL6/CHALLAH, two EPFL family members, promote inflorescence growth (Uchida et al., 2012); and EPFL5 promotes fertility and fruit growth (Abrash et al., 2011). In Brassica napus, EFFL6 regulates filament elongation and may also play affect organ morphology and floral organ specification (Huang et al., 2014). In this study, we verified that the EPFL peptide encoded by GAD1 regulates grain number, grain length, and awn development in rice.
EPF1, EPF2, and STOMAGEN/EPFL9 regulate plant development by binding ERECTA family members (Lee et al., 2012, 2015). Two genes (LOC_Os06g03970 and LOC_Os06g10230) in the rice genome show high homology to Arabidopsis ERECTA. These genes therefore appear to be good initial candidates to encode proteins that serve as GAD1 receptors.
GAD1 Has Pleiotropic Effects on Awn and Grain Development
Previous studies showed that some key genes controlling the domestication processes of crops have pleiotropic effects. For example, the selection of prog1 led to erect growth, greater grain number, and higher grain yields in cultivated rice (Tan et al., 2008). LG1 controls rice ligule development and the transition of panicle architecture in cultivars during rice domestication (Ishii et al., 2013; Lee et al., 2007; Zhu et al., 2013). Q is a major gene involved in wheat (Triticum aestivum) domestication-related traits, such as free-threshing character, glume shape and tenacity, and rachis fragility (Simons et al., 2006). In our study, we found that GAD1 also had pleiotropic effects—on grain length, grain number, and awn development—during rice domestication. These findings indicate that targeting a gene with pleiotropic effects might be an efficient strategy for crop domestication.
Crop domestication represents the genetic modification of a wild species to create a new form to meet human needs. Our findings provided insights into the puzzles surrounding rice domestication and revealed a previously unrecognized function of peptide signal molecules in plant development.
METHODS
Plant Materials and Growth Conditions
For the fine mapping, expression pattern analysis, and panicle development analysis, we used introgression line OIL31 and indica cultivar NA93-11, which is a sibling of 93-11 with no awn. OIL31 was derived from a cross between W2014 (common wild rice [Oryza rufipogon] from India) and NA93-11. An F2 segregation population, containing 4250 individuals, derived from a backcross between OIL31 and NA93-11 was used for the fine mapping of GAD1. The introgression line TIL189 was derived from a cross between the indica cultivar Teqing and common wild rice W2014. The japonica variety Nipponbare and the introgression line TIL189 were used as transgenic recipients. We generated two pyramiding lines (An1/GAD1-NIL and GAD1/LABA1-NIL) that harbored GAD1 and An-1 (Luo et al., 2013), and GAD1 and LABA1/An-2 (Gu et al., 2015; Hua et al., 2015), respectively, in the NA93-11 genetic background. The introgression and pyramiding lines were generated as described previously (Tan et al., 2007). The plant materials were growth in the field at the Experimental Stations of China Agriculture University, Beijing and Hainan, China. The wild rice and cultivated rice (Oryza sativa) accessions used in the haplotype analysis, neutrality test, and selective sweep analysis are listed in Supplemental Data Sets 2 to 4, respectively.
Primers
The primers used in this study are listed in Supplemental Table 3.
Generation of Constructs and Transformations
The functional complementation construct (pCPL), which harbored the ∼4.0-kb W2014 genomic fragment covering the entire GAD1 gene, was inserted into the binary vector pCAMBIA1300 (AF234296). The GAD1 DNA fragments were PCR amplified by 1300-3 (Supplemental Table 3) and cloned into the pCAMBIA1300 vector KpnI and SalI sites. The RNAi construct contained an inverted repeat harboring the 350-bp GAD1 cDNA fragment in the vector pTCK303. The two plasmid constructs were introduced to Agrobacterium tumefaciens strain EHA105 and then transformed into the japonica variety Nipponbare and the introgression line TIL189, respectively.
Phenotypic Evaluation
We used 20 plants to measure the grain number of main panicles, grain length, awn length, and the percentage of awned seeds in the introgression line OIL31 and NA93-11. The average awn length was measured on the apical spikelet of each primary branch on the main stem panicle. The percentages of awned seeds were determined from awned spikelets on the main stem panicle, with the awns >1 mm on mature grains considered to represent an awned phenotype. Two independent complemented (with pCPL) and RNAi T2 homozygous lines were used for the measurements. The main panicles of 10 plants were measured for each line. Ten Nipponbare and TIL189 plants harboring empty vectors were used as the controls.
5′- and 3′-RACE and qRT-PCR
We extracted total RNA using TRIzol reagent (Invitrogen Life Technologies) and purified RNA with a purification kit (Tiangen). Total RNA was reverse transcribed with an oligo(dT20) primer using SuperScript III reverse transcriptase (Life Technologies). The 5′ and 3′ RACE were conducted with 5′- and 3′-Full RACE Kits (TaKaRa) following the manufacturer’s instructions. qRT-PCR was performed on a CFX96 real-time system (Bio-Rad). Diluted cDNA was amplified using the SsoFast Evagreen Supermix (Bio-Rad). The transcript levels were normalized to endogenous ACTIN transcripts amplified with primers ActinF and ActinR. Each set of experiments was repeated three times, and the relative quantification method (2−ΔΔCT) was used to evaluate quantitative variation (Livak and Schmittgen, 2001).
Scanning Electron Microscopy
We fixed the young panicles in 2.5% glutaraldehyde-PBS fixative solution, dehydrated the samples using an ethanol series, and dried them with a carbon dioxide critical-point dryer. The dried panicles were gold plated and observed using a Hitachi S-2460 scanning electron microscope at 15 kV. Scanning electron microscopy was performed as described previously (Hua et al., 2015).
RNA in Situ Hybridization
RNA in situ hybridization was performed as described previously (De Block and Debrouwer, 1993) with minor modifications. The young panicles of all stages from OIL31 and NA93-11 were fixed in 4% (w/v) paraformaldehyde, subjected to a dehydration series and infiltration, and embedded in Paraplast (Paraplast Plus; Sigma-Aldrich). The tissues were sliced into 8- to 10-μm sections with a microtome (Leica RM2145). An ∼300-bp fragment of GAD1 cDNA was amplified and used as the template to generate sense and antisense RNA probes that were labeled by digoxigenin using a DIG Northern Starter Kit (catalog no. 2039672; Roche) following the manufacturer’s instructions. Five independent experiments have been performed for RNA in situ hybridization.
Sequencing and Data Analysis
Multiple sequences were aligned with ClustalX (Thompson et al., 1997). The program DnaSP, version 5.0, was used to calculate nucleotide diversity and Tajima’s D test (Rozas et al., 2003). The phylogenetic tree was constructed by the MEGA 5 program. The evolutionary history was inferred using the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Tamura et al., 2011).
Signal Peptide Analysis
The cleavage sites and a signal peptide/non-signal peptide in the GAD1 protein were predicted using SignalP4.1 (http://www.cbs.dtu.dk/services/SignalP) (Petersen et al., 2011).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: GAD1 cDNA from W2014 (KX827806) and gad1 cDNA from NA93-11 (KX827807).
Supplemental Data
Supplemental Figure 1. GAD1 sequence comparison between OIL31 and NA93-11.
Supplemental Figure 2. RNA in situ hybridization of GAD1 in inflorescence meristem.
Supplemental Figure 3. RNA in situ hybridization of the GAD1 sense probe at different developmental stages in OIL31.
Supplemental Figure 4. Scanning electron microscopy images of spikelets at different developmental stages.
Supplemental Figure 5. qRT-PCR analysis of HISTONE H1 in young panicles of NA93-11 and OIL31.
Supplemental Figure 6. Transcript level comparison of 22 cell cycle-associated genes between NA93-11 and OIL31.
Supplemental Figure 7. Comparison of CKX2, DST, and RR transcript levels between NA93-11 and OIL31.
Supplemental Figure 8. Full-length cDNA of GAD1 and its predicted amino acid sequence.
Supplemental Figure 9. Phylogenetic tree of EPF/EPFL family genes from rice and Arabidopsis.
Supplemental Figure 10. Protein structure and maturation process predictions of GAD1.
Supplemental Figure 11. Analysis of the consensus sequence from the GAD1 protein.
Supplemental Figure 12. Haplotypes of GAD1 in diverse wild and cultivated rice accessions.
Supplemental Figure 13. Distribution of GAD1/gad1, LABA1/laba1, and An-1/an-1 in 159 varieties of cultivated rice.
Supplemental Figure 14. Phenotypes of near-isogenic and pyramiding lines containing GAD1, An-1, and LABA1.
Supplemental Table 1. The signal peptide and cleavage site predictions in the GAD1 protein.
Supplemental Table 2. GAD1 nucleotide diversity and Tajima’s D test.
Supplemental Table 3. Primers used in this study.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 9.
Supplemental Data Set 2A. Wild rice accessions used in the haplotype and nucleotide diversity analyses.
Supplemental Data Set 2B. Rice cultivars used in the haplotype and nucleotide diversity analyses.
Supplemental Data Set 3A. Wild rice accessions used in the selective sweep analysis.
Supplemental Data Set 3B. Rice cultivars used in the selective sweep analysis.
Supplemental Data Set 4. Rice cultivars used to analyze the distribution of GAD1/gad1 and two other awn-related genes.
Supplementary Material
Acknowledgments
We thank the International Rice Research Institute, Chinese Rice Research Institute, Institute of Crop Sciences of Chinese Academy of Agricultural Sciences, Guangxi Academy of Agricultural Sciences, and Guangdong Academy of Agricultural Sciences for providing the wild and cultivated rice samples. This research was supported by the National Key R&D Program for Crop Breeding (2016YFD0100301) and National Natural Science Foundation of China (Grant 91535301).
AUTHOR CONTRIBUTIONS
C.S. designed and supervised this study. J.J. conducted the characterization of the introgression line, map-based cloning, genetic transformations, gene expression analysis, and the evolutionary analysis. X.Z. constructed the introgression lines. X.S. and Y.F. maintained plant materials and performed the field management. L.H., W.Z., L.T., F.L., Z.Z., H.C., and P.G. collected the rice germplasm and phenotypic data. C.S., J.J., and D.X. analyzed the data and wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abrash E.B., Bergmann D.C. (2010). Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455. [DOI] [PubMed] [Google Scholar]
- Abrash E.B., Davies K.A., Bergmann D.C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., Angeles E.R., Qian Q., Kitano H., Matsuoka M. (2005). Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- De Block M., Debrouwer D. (1993). RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal. Biochem. 215: 86–89. [DOI] [PubMed] [Google Scholar]
- Doebley J.F., Gaut B.S., Smith B.D. (2006). The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Engineer C.B., Ghassemian M., Anderson J.C., Peck S.C., Hu H., Schroeder J.I. (2014). Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fuller D.Q., Sato Y.I., Castillo C., Qin L., Weisskopf A.R., Kingwell-Banham E.J., Song J., Ahn S.M., Van Etten J. (2010). Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthropol. Sci. 2: 115–131. [Google Scholar]
- Gu B., Zhou T., Luo J., Liu H., Wang Y., Shangguan Y., Zhu J., Li Y., Sang T., Wang Z., Han B. (2015). An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol. Plant 8: 1635–1650. [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K.U., Bergmann D.C., Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Yokoo T., Kajita R., Onishi T., Yahata S., Peterson K.M., Torii K.U., Kakimoto T. (2009). Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50: 1019–1031. [DOI] [PubMed] [Google Scholar]
- Hua L., et al. (2015). LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27: 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Tao Z., Liu Q., Wang X., Yu J., Liu G., Wang H. (2014). BnEPFL6, an EPIDERMAL PATTERNING FACTOR-LIKE (EPFL) secreted peptide gene, is required for filament elongation in Brassica napus. Plant Mol. Biol. 85: 505–517. [DOI] [PubMed] [Google Scholar]
- Hunt L., Gray J.E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19: 864–869. [DOI] [PubMed] [Google Scholar]
- Ishii T., Numaguchi K., Miura K., Yoshida K., Thanh P.T., Htun T.M., Yamasaki M., Komeda N., Matsumoto T., Terauchi R., Ishikawa R., Ashikari M. (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45: 462–465, e1–e2. [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47. [DOI] [PubMed] [Google Scholar]
- Jain M., Tyagi A.K., Khurana J.P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewaria P.K., Hara T., Tanaka H., Kondo T., Betsuyaku S., Sawa S., Sakagami Y., Aimoto S., Kakimoto T. (2013). Differential effects of the peptides Stomagen, EPF1 and EPF2 on activation of MAP kinase MPK6 and the SPCH protein level. Plant Cell Physiol. 54: 1253–1262. [DOI] [PubMed] [Google Scholar]
- Jin J., Huang W., Gao J.P., Yang J., Shi M., Zhu M.Z., Luo D., Lin H.X. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40: 1365–1369. [DOI] [PubMed] [Google Scholar]
- Katsir L., Davies K.A., Bergmann D.C., Laux T. (2011). Peptide signaling in plant development. Curr. Biol. 21: R356–R364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush G.S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S.Y., Ebana K., Fukuta Y., Sasaki T., Yano M. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312: 1392–1396. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Hnilova M., Maes M., Lin Y.C., Putarjunan A., Han S.K., Avila J., Torii K.U. (2015). Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Park J.J., Kim S.L., Yim J., An G. (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 65: 487–499. [DOI] [PubMed] [Google Scholar]
- Li C., Zhou A., Sang T. (2006). Rice domestication by reducing shattering. Science 311: 1936–1939. [DOI] [PubMed] [Google Scholar]
- Li S., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 110: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., Shao D., Xu C., Li X., Xiao J., He Y., Zhang Q. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43: 1266–1269. [DOI] [PubMed] [Google Scholar]
- Lin Z., Griffith M.E., Li X., Zhu Z., Tan L., Fu Y., Zhang W., Wang X., Xie D., Sun C. (2007). Origin of seed shattering in rice (Oryza sativa L.). Planta 226: 11–20. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luo J., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Duronio R.J. (2002). Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14: 692–699. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki S., Takeuchi M., Mori M. (2011). The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2: 512. [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897. [DOI] [PubMed] [Google Scholar]
- Petersen T.N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J.C., Messeguer X., Rozas R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schopfer C.R., Nasrallah M.E., Nasrallah J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Simons K.J., Fellers J.P., Trick H.N., Zhang Z., Tai Y.S., Gill B.S., Faris J.D. (2006). Molecular characterization of the major wheat domestication gene Q. Genetics 172: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244. [DOI] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Pfeil B.E., McCouch S. (2006). Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata N., Yokota K., Ohki S., Mori M., Taniguchi T., Kurita M. (2013). Evolutionary relationship and structural characterization of the EPF/EPFL gene family. PLoS One 8: e65183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Liu F., Xue W., Wang G., Ye S., Zhu Z., Fu Y., Wang X., Sun C. (2007). Development of Oryza rufipogon and O. sativa introgression lines and assessment for yield-related quantitative trait loci. J. Integr. Plant Biol. 49: 871–884. [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., Zhu Z., Fu Y., Cai H., Wang X., Xie D., Sun C. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40: 1360–1364. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Lee J.S., Horst R.J., Lai H.-H., Kajita R., Kakimoto T., Tasaka M., Torii K.U. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109: 6337–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. (2011). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. [DOI] [PubMed] [Google Scholar]
- Zhang D., Yuan Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65: 553–578. [DOI] [PubMed] [Google Scholar]
- Zhu B.F., et al. (2011). Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 155: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Tan L., Fu Y., Liu F., Cai H., Xie D., Wu F., Wu J., Matsumoto T., Sun C. (2013). Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 4: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y., Chen Z., Innes J.B., Chen C., Wang Z., Wang H. (2007). Fire and flood management of coastal swamp enabled first rice paddy cultivation in east China. Nature 449: 459–462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.