Summary
AGAMOUS, a key player in floral morphogenesis, specifies reproductive organ identities and regulates the timely termination of stem cell fates in the floral meristem. Here, we report that strains carrying mutations in three genes, HUA1, HUA2, and HUA ENHANCER4 (HEN4), exhibit floral defects similar to those in agamous mutants: reproductive-to-perianth organ transformation and loss of floral determinacy. HEN4 codes for a K homology (KH) domain-containing, putative RNA binding protein that interacts with HUA1, a CCCH zinc finger RNA binding protein in the nucleus. We show that HUA1 binds AGAMOUS pre-mRNA in vitro and that HEN4, HUA1, and HUA2 act in floral morphogenesis by specifically promoting the processing of AGAMOUS pre-mRNA. Our studies underscore the importance of RNA processing in modulating plant development.
Introduction
Flowers arise from groups of florally determined undifferentiated cells known as floral meristems. The fates of the four major types of floral organs (sepals, petals, stamens, and carpels), arranged in concentric whorls, are specified by the combinatorial activities of the A, B, and C classes of floral homeotic genes (reviewed in Coen and Meyerowitz, 1991; Lohmann and Weigel, 2002). As a class C gene, AGAMOUS (AG) specifies stamen and carpel identities. In addition, AG controls floral determinacy by repressing the expression of the stem cell fate maintenance gene WUSCHEL (WUS; Laux et al., 1996; Lenhard et al., 2001; Lohmann et al., 2001).
The region-specific accumulation of AG RNA in the floral meristem is primarily controlled by transcriptional regulation. Whereas positive regulators such as LEAFY and WUS activate AG expression (Busch et al., 1999; Lenhard et al., 2001; Lohmann et al., 2001), negative regulators, such as APETALA2 (AP2), LEUNIG, SEUSS, AINTEGUMENTA, and STERILE APETALA, ensure that AG is not expressed in the outer two floral whorls (Drews et al., 1991; Liu and Meyerowitz, 1995; Byzova et al., 1999; Krizek et al., 2000; Franks et al., 2002). Both positive and negative transcriptional regulation require DNA elements present in the second intron of AG (Bomblies et al., 1999; Busch et al., 1999; Deyholos and Sieburth, 2000; Lohmann et al., 2001). While these studies clearly document transcriptional regulation of AG, our studies have begun to reveal genes that specifically facilitate the expression of AG at the posttranscriptional level (see below).
In a genetic screen aimed at isolating extragenic mutations that enhanced the weak ag allele ag-4, HUA1 and HUA2 were identified as genes that act in the specification of stamen and carpel identities and in the control of floral determinacy (Chen and Meyerowitz, 1999). While HUA2 codes for a novel protein (Chen and Meyerowitz, 1999), HUA1 codes for an RNA binding protein with six tandem CCCH zinc fingers (Li et al., 2001). Due to the weak loss-of-C function phenotypes of hua1-1 hua2-1 flowers (Chen et al., 2002; Western et al., 2002), the exact regulatory relationship between HUA1, or HUA2, and AG was not known.
In order to identify additional components of the C pathway as well as explore the functions of HUA1 and HUA2, we performed another mutagenesis screen with the hua1-1 hua2-1 mutant. Two genes identified from this screen, HEN1 and HEN2, act in the C pathway in the flower while also acting in other developmental processes in the plant. HEN1 encodes a novel protein (Chen et al., 2002) that is required for the accumulation of microRNAs (Park et al., 2002), indicating a role of microRNAs in flower development. A mutation in the putative RNA helicase HEN2, in the hua1-1 hua2-1 background, results in the transformation of stamens to petal/sepal mosaic organs and carpels to organs with sepal character, suggesting that HEN2 acts in both B and C pathways in the flower (Western et al., 2002). These studies added more evidence of posttranscriptional regulation in flower development, but did not reveal the molecular relationship between HEN1, or HEN2, with AG.
In this study, we have identified another gene, named HUA ENHANCER4 (HEN4), mutations in which, in the hua1-1 hua2-1 background, cause severe floral phenotypes similar to those of strong ag alleles. We demonstrate that all three genes, HEN4, HUA1, and HUA2, function in floral reproductive organ identity and floral determinacy specification by facilitating the processing of AG pre-mRNA. In addition, we show that HEN2 also likely acts in AG pre-mRNA processing. HEN4 encodes a K homology (KH) domain-containing, putative RNA binding protein that colocalizes and interacts with HUA1, a CCCH zinc finger protein, in nuclear speckles. Our studies extend the significance of KH domain proteins in development from metazoans to plants by providing an example of a KH domain protein acting in plant development. Our studies also provide an intriguing parallel in the requirement of the CCCH zinc finger and KH domain proteins in cell fate specification in Arabidopsis and C. elegans, and suggest that the physical association between CCCH and KH domain proteins is evolutionarily conserved.
Results
HEN4 Acts in Stamen Identity and Floral Determinacy Specification
Two recessive mutations in HEN4, hen4-1 and hen4-2, caused stamen-to-petal transformation in the third whorl of hua1 hua2 flowers (for simplicity, we will be referring to hua1-1 and hua2-1 as hua1 and hua2, respectively). While wild-type and hua1 hua2 flowers had stamens in the third whorl (Figures 1A and 1B), early-arising flowers in hua1 hua2 hen4-1 plants had petaloid stamens in the third whorl (Figure 1C). Later-arising flowers in hua1 hua2 hen4-1 plants had six petals in the third whorl (Figures 1D and 1E). hen4-2 appeared to cause more severe floral homeotic phenotypes than hen4-1. Nearly all hua1 hua2 hen4-2 flowers had six petals in the third whorl (Figure 1F). Because both hen4-1 and hen4-2 were recessive and therefore likely reduce the gene function, we conclude that HEN4 acts in stamen identity specification in flower development.
Figure 1. Floral Phenotypes Caused by hua1, hua2, and hen4 Mutations.

(A–G) Floral phenotypes of various genotypes. Arrows indicate third whorl organs.
(A) An Ler flower.
(B) A hua1 hua2 flower.
(C–E) An early, an intermediate, and a late flower, respectively, from a hua1 hua2 hen4-1 plant. Note petaloid stamens or petals in the third whorl. The black and white arrows in (C) indicate lateral and medial third whorl organs, respectively.
(F) An early-arising flower from a hua1 hua2 hen4-2 plant. Note stamen-to-petal transformation in the third whorl.
(G) A hua1 hen4-1 flower.
(H) Gynoecia dissected out from stage 14 flowers. From left to right are gynoecia from a hua1 hua2 flower, an early-arising hua1 hua2 hen4-1 flower, a late-arising hua1 hua2 hen4-1 flower, and an early-arising hua1 hua2 hen4-2 flower. The latter two gynoecia are found on gynophores (arrowheads), indicating that the gynoecia have partial floral character.
(I and J) Scanning electron micrographs of stage 7 flowers of the hua1 hua2 and hua1 hua2 hen4-2 genotypes, respectively. Two sepals from each flower were dissected away to reveal the third whorl organs. The second whorl organs are small (purple arrows). The third whorl organs in the hua1 hua2 flower have begun to differentiate into anthers (arrowhead) and filaments (black arrow), whereas those (black arrows) in the hua1 hua2 hen4-2 flower show no sign of such differentiation.
(K–N) Scanning electron micrographs of ovary epidermal cells of various genotypes.
(K and L) The top and bottom regions of a hua1 hua2 ovary. Some cells in the top lateral region show epicuticular thickenings (K), whereas all cells in the bottom half of the ovary have a smooth surface (L).
(M and N) The bottom region of a hua1 hua2 hen4-1 and a hua1 hua2 hen4-2 ovary, respectively. All cells show epiculticular striation. The scale bars represent 50 μm in (I) and (J) and 10 μm in (K)–(N).
(O–V) Floral phenotypes of quadruple mutants and transgenic lines.
(O) hua1 hua2 hen4-1 ap1-1. Note stamens in the third whorl.
(P) hua1 hua2 hen4-1 ap2-2. Leaf-like organs are found in all four whorls.
(Q) hua1 hua2 hen4-1 ag-4, which resembles ag-1.
(R) hua1 hua2 hen4-1 ag-1, which is similar to ag-1.
(S and T) Flowers of hua1 hua2 hen4-1 plants transformed with the vector alone or HEN4 genomic DNA, respectively. Note the difference in the morphology of the third whorl organs (arrows).
(U) Gynoecia of stage 14 flowers from hua1 hua2 (left), hua1 hua2 hen4-1 transformed with HEN4 genomic DNA (middle), and hua1 hua2 hen4-1 (right).
(V) hua1 hua2 hen4-1 35S∷AG. Note stamens in the second and third whorls (arrow).
hen4 mutations (both hen4-1 and hen4-2 are referred to when the number is not specified) also caused floral determinacy defects. Although early-arising flowers in hua1 hua2 hen4 plants had gynoecia with two well-fused carpels topped by stigmatic papillae (Figures 1C, 1D, 1F, and 1H), later-arising flowers that constituted the majority of the flowers produced in these plants had enlarged, heart-shaped gynoecia in the fourth whorl (Figure 1H). Additional floral organs were found in the gynoecia upon dissection, indicating loss of floral determinacy. Occasionally, flowers that resembled those of severe ag alleles (such as ag-1 or ag-3) with internal flowers were observed (Figure 1E). Therefore, HEN4 acts in the proper termination of floral meristem activity during flower development.
The stamen-to-petal transformation in hua1 hua2 hen4 flowers was further confirmed by scanning electron microscopy (SEM). Unlike stamen epidermal cells that are shaped like jigsaw puzzle pieces, the epidermal cells of hua1 hua2 hen4 third whorl organs resembled petal cells (data not shown), which are uniform in size and cone shaped (Smyth et al., 1990). In addition, the stamen-to-petal transformation in hua1 hua2 hen4-2 flowers occurred early in development. By stage 7, stamen primordia in wild-type (Smyth et al., 1990) and hua1 hua2 flowers (Figure 1I) assumed the first sign of differentiation into stalked structures. In hua1 hua2 hen4-2 stage 7 flowers, however, the third whorl organ primordia assumed a flat shape, resembling perianth organs (Figure 1J).
The stamen identity and floral determinacy defects were only found in the hua1 hua2 hen4 triple mutants. hua1 hen4, hua2 hen4, and hen4 flowers appeared normal (Figure 1G and data not shown).
HEN4 Acts in Carpel Identity Specification
hua1 hua2 hen4 gynoecia exhibited more extensive sepal character than hua1 hua2 gynoecia. While some valve cells located in the top, lateral positions in hua1 hua2 gynoecia exhibited epicuticular striation patterns similar to those in sepal cells, most hua1 hua2 valve cells had a smooth surface (Chen et al., 2002; Western et al., 2002; Figures 1K and 1L). However, all valve cells in hua1 hua2 hen4 flowers (early-arising or late-arising) had epicuticular thickenings that resembled sepal cells (Figures 1M and 1N). Therefore, HEN4 also acts in carpel identity specification.
HEN4 Also Functions Outside the Flower
HUA1, HUA2, and HEN4 all appeared also to act in vegetative development. While hua1 and hen4 single mutants had no obvious vegetative defects, hua2 plants were slightly smaller than wild-type (see Supplemental Figures S1A–S1D at http://www.developmentalcell.com/cgi/content/full/4/1/53/DC1, and data not shown). All combinations of double mutants, hua1 hen4, hua2 hen4, and hua1 hua2, were smaller and shorter than any of the single mutants (Supplemental Figures S1E–S1G; Supplemental Table S1 and data not shown).
HEN4 Behaves Genetically as a C Function Gene in Flower Development
We introduced representative mutations in A and C genes into the hua1 hua2 hen4-1 background to investigate the genetic relationship between HEN4 and these genes.
hua1 hua2 hen4-1 ap1-1
APETALA1 (AP1) is a class A gene that specifies perianth identities. ap1-1, a severe loss-of-function mutation (Irish and Sussex, 1990), was essentially epistatic to hua1 hua2 hen4-1 in the third whorl—hua1 hua2 hen4-1 ap1-1 flowers had stamens in the third whorl (Figure 1O). This suggests that the third whorl stamen-to-petal transformation in hua1 hua2 hen4-1 is due to ectopic AP1 activity.
hua1 hua2 hen4-1 ap2-2
AP2 is the other A function gene that specifies perianth identities. hua1 hua2 hen4-1 ap2-2 flowers consisted primarily of leaf-like organs in all four whorls (Figure 1P), a phenotype that closely resembled that of ag-1 ap2-2 flowers (Bowman et al., 1991). This confirms that C function is greatly compromised in hua1 hua2 hen4-1.
hua1 hua2 hen4-1 ag-4 and hua1 hua2 hen4-1 ag-1
ag-4 flowers have stamens in the third whorl and internal flowers in the center of the flower (Sieburth et al., 1995). hua1 hua2 ag-4 flowers have primarily petals and occasional staminoid petals in the third whorl (Chen and Meyerowitz, 1999; Western et al., 2002). hua1 hua2 hen4-1 ag-4 flowers resembled ag-1 flowers (Figure 1Q), consistent with HEN4 acting in C function.
In order to address the question of whether HEN4 acts genetically in the AG pathway or in a parallel pathway, we introduced the severe loss-of-function mutation ag-1 into the hua1 hua2 hen4-1 background. The resulting quadruple mutant flowers were composed of many whorls of perianth organs as found in ag-1 flowers (Figure 1R), suggesting that ag-1 was essentially epistatic to hen4-1. Because stamen and carpel identities are completely lost in ag-1, the epistasis of ag-1 to hen4 does not necessarily imply that HEN4 acts in the AG pathway. However, since mutations in genes such as CLAVATA1 and SUPERMAN are known to enhance the floral determinacy defect of ag-1 (Schultz et al., 1991; Clark et al., 1993), the fact that hua1 hua2 hen4-1 ag-1 flowers resemble ag-1 in floral determinacy defects strongly suggests that HUA1, HUA2, and HEN4 act genetically in the AG pathway.
The Spatial Regulation of AP1 Expression and the Temporal Control of WUS Expression Are Defective in hua1 hua2 hen4 Flowers
We examined the expression of AP1 and WUS in hua1 hua2 hen4 and hua1 hua2 flowers to determine whether HEN4 acts similarly to AG in the regulation of these genes. In wild-type flowers, AP1 RNA is present throughout the floral meristem in stages 1 and 2 flowers but is restricted to the outer two floral whorls due to the negative regulation by AG starting from stage 3 (Gustafson-Brown et al., 1994). In hua1 hua2 hen4, AP1 RNA accumulation patterns differed from those in wild-type and hua1 hua2 flowers (Chen et al., 2002) as early as stage 3, when AP1 RNA was still detectable in the inner two whorls in the triple mutant at a low level (Figures 2A and 2B). During later stages of flower development, the ectopic AP1 expression was more extensive in hua1 hua2 hen4 as compared to hua1 hua2, with more cells in the inner two whorls expressing AP1 at higher levels (Figures 2C and 2D). In mature hua1 hua2 hen4 flowers, AP1 RNA was present throughout the carpel walls (Figure 2E and data not shown). WUS is expressed in a few cells underneath the stem cells in the shoot apical meristem and in stage 6 or younger floral meristems in wild-type (Mayer et al., 1998). WUS RNA disappears from the flower at around stage 7 due to negative regulation by AG (Lenhard et al., 2001; Lohmann et al., 2001). In hua1 hua2, WUS RNA was never observed in stage 7 or older flowers (Figure 2J and data not shown). However, in hua1 hua2 hen4-2, WUS RNA was found in some older flowers (Figure 2K), consistent with the floral determinacy defect observed in late-arising flowers.
Figure 2. RNA Accumulation Patterns of AP1, AG, WUS, and HEN4 as Determined by In Situ Hybridization.

(A–E) AP1 RNA accumulation patterns.
(A) A stage 3 hua1 hua2 flower, with no AP1 RNA in the center of the flower.
(B) A stage 3 hua1 hua2 hen4-2 flower, with a low level of AP1 RNA in the center of the flower.
(C and D) Stage 8 hua1 hua2 and hua1 hua2 hen4-2 flowers, respectively. The arrow indicates a patch of third whorl cells expressing AP1. Ectopic AP1 RNA accumulation in the inner two whorls is more extensive in the triple mutant than in the double mutant.
(E) A stage 14 hua1 hua2 hen4-2 flower. Note internal flower (arrowhead) and strong AP1 expression in the fourth whorl.
(F) AG RNA accumulation in hua1 hua2 hen4-2 stage 3 (s3) and stage 6 (s6) flowers. The onset and the domain of AG RNA accumulation are normal.
(G and H) AG RNA accumulation in stage 9 hua1 hua2 hen4-2 and hua1 hua2 flowers, respectively. In hua1 hua2 hen4-2, the domain of AG RNA accumulation is normal but the level is reduced.
(I) In situ hybridization using a probe specific for the larger AG RNAs on a stage 7 hua1 hua2 hen4-2 flower.
(J and K) WUS RNA accumulation in a hua1 hua2 (J) and a hua1 hua2 hen4-2 (K) flower. The arrow in (K) marks the WUS-expressing cells.
(L and M) HEN4 RNA accumulation patterns.
(L) Hybridization with a HEN4 sense probe.
(M) Hybridization with a HEN4 antisense probe. HEN4 RNA is found throughout the stage 4 flower. The scale bars represent 50 μm. Numbers in (A)–(E) and (G)–(I) indicate floral whorls.
AG Expression Domain Is Normal in hua1 hua2 hen4 Flowers
AG RNA is found in the inner two whorls starting at stage 3 in wild-type and hua1 hua2 plants (Drews et al., 1991; Chen and Meyerowitz, 1999; Figure 2H). The onset and the domain of AG expression were not affected in the hua1 hua2 hen4 triple mutants (Figure 2F). In stage 7 and older flowers, AG RNA continued to be found in the inner two whorls, but often in a patchy pattern or barely detectable in some organs (Figure 2G). We believe this is due to the reduced level of AG RNA in the triple mutant flowers (see below).
HEN4 Contains KH Domains
We first mapped HEN4 to a 48 kb region on chromosome V covered by a P1 clone MSJ1 and a BAC T12B11 (Figure 3A). Three candidate genes in this region were sequenced from hen4-1. One gene, At5g64390, was found to contain a G-to-A mutation that disrupts the splice acceptor site of the fourth intron. Sequencing of this gene from hen4-2 identified a C-to-T mutation in the fourth exon that results in a stop codon. To confirm that At5g64390 is HEN4, we cloned a genomic fragment containing only this gene into the plant transformation vector pPZP222 and transformed plants that were homozygous for hua1 and hen4-1 but segregating for hua2 with the construct. Seven independent T1 lines had flowers indistinguishable from those in hua1 hua2 (Figures 1T and 1U). Two other T1 lines showed floral phenotypes that were intermediate between hua1 hua2 and hua1 hua2 hen4-1. Molecular genotyping of the nineT1 plants for hua2-1 and phenotypic analyses of T2 plants grown with or without selection for the transgene confirmed that these T1 lines were hua1 hua2 hen4-1 plants containing the HEN4 transgene. Therefore, the HEN4 transgene rescued or partially rescued the floral homeotic defect caused by hen4-1. The pPZP222 vector alone did not rescue the floral homeotic phenotypes (Figure 1S). These data demonstrated that At5g64390 is indeed HEN4.
Figure 3. Map-Based Cloning of HEN4 and Features of the HEN4 Gene and Protein.

(A) HEN4 was mapped to a 48 kb region on chromosome V covered by the P1 clone MSJ1 and the BAC T12B11. The numbers under the BAC or P1 clones indicate the numbers of recombination breakpoints between markers in the clones and HEN4 in 684 chromosomes. The structure of the HEN4 gene is shown with the rectangles representing exons and the lines representing introns. The hatched rectangle represents a region that is part of an exon in transcript 2 but is spliced out of transcript 1. The position and nature of the two hen4 mutations are indicated.
(B) Diagrams of the two putative HEN4 protein forms with the KH domains and the potential nuclear localization signal (NLS) as indicated. Note that proteins 1 and 2 are from the short (1) and the long (2) HEN4 transcripts, respectively. The hen4 mutations would terminate the proteins at the indicated positions.
(C) A clustalW alignment of the five KH domains from HEN4 and the KH domains from three animal proteins, MEX-3 from C. elegans, and αCP1 and Nova-2 from humans. If six or more amino acids are identical at one position, the amino acids are in dark shade. Amino acids that are similar in nature at one position are lightly shaded.
Two HEN4 cDNA species (1 and 2, due to the exclusion or inclusion of the last intron) were obtained by RT-PCR (Figure 3A). The corresponding conceptual proteins contain five and four KH domains, respectively, and a putative nuclear localization signal (Figure 3B). KH domains are usually 70 amino acids in length with a characteristic pattern of hydrophobic residues, an invariant Gly-x-x-Gly segment, and a variable loop (Lewis et al., 2000; Figure 3C). X-ray crystallographic studies clearly demonstrated the affinity of KH domains for singlestranded RNA (Musco et al., 1996; Lewis et al., 2000).
Among the 27 reported KH domain proteins in the Arabidopsis genome (Chekanova et al., 2002; Lorkovic and Barta, 2002), HEN4 is the only one with a known developmental function so far. HEN4 is more closely related to ten other KH domain proteins from Arabidopsis and rice. The similarity between HEN4 and these proteins, however, is primarily limited to the KH domains. No clear HEN4 orthologs are present in metazoans, which implies that HEN4 is not part of a general machinery for RNA metabolism and instead may have specific developmental functions.
HEN4 Expression Patterns
In RNA filter hybridization, two HEN4 transcripts were detected in various organs, such as leaves, stems, roots, and inflorescences, in wild-type plants (Figure 4 and data not shown). Both transcripts were found in ag-2 and hua1 hua2 inflorescences (Figure 4). Only the smaller RNA species (transcript 1, which would give rise to the larger HEN4 protein) was found in hen4-1 and hen4-2 genotypes (Figure 4). Transcript 1 as an authentic HEN4 transcript was confirmed by RT-PCR and sequencing, as well as by expression studies using EYFP as the reporter gene (data not shown). The absence of HEN4 transcript 2 in hen4 genotypes was unlikely due to nonsense-mediated decay, because both transcripts in hen4 genotypes contained nonsense codons but yet transcript 1 was not affected. Instead, it is more likely that HEN4 acts to regulate the splicing of its own pre-mRNA.
Figure 4. HEN4 RNA Accumulation in Inflorescences from Various Genotypes.
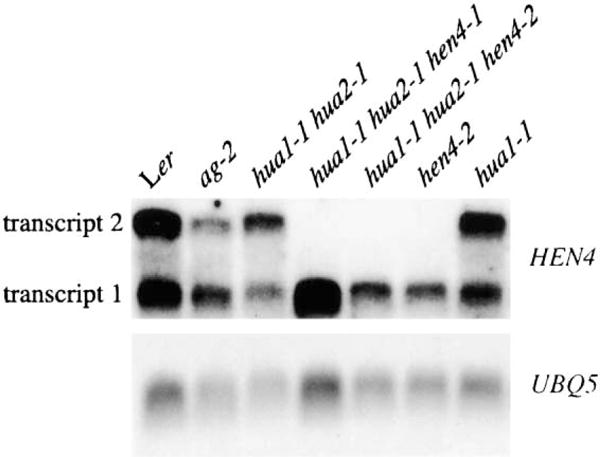
Both HEN4 transcripts are present in wild-type, ag-2, and hua1 hua2 inflorescences. HEN4 transcript 2 is absent in hen4 genotypes. Transcripts 1 and 2 correspond to HEN4 cDNAs 1 and 2, respectively. UBQ5 was the loading control.
Using a probe that can hybridize to both HEN4 RNA forms, we detected signals throughout the inflorescence meristem and throughout the flower in stage 6 and younger flowers (Figures 2L and 2M, and data not shown) by in situ hybridization. A probe specific to HEN4 cDNA2 failed to detect any signals in inflorescences or flowers, although RNA filter hybridization showed that this RNA form was found in inflorescences (Figure 4).
HEN4 Interacts with HUA1 in Nuclear Speckles
Because hua1 and hen4 mutations behave similarly and both HUA1 and HEN4 are probably RNA binding proteins, we speculated that the two proteins act together in the cell. First, we demonstrated that HEN4, like HUA1 (Li et al., 2001), was localized in the nucleus by visualizing YFP signals in the roots of the transgenic plants carrying a functional translational fusion of YFP to the C terminus of HEN4 protein 1 under the control of the HEN4 promoter (HEN4p∷HEN4-EYFP). Next, we determined whether HUA1 and HEN4 colocalize within the nucleus. The HEN4p∷HEN4-EYFP Agrobacteria were infiltrated into tobacco leaves alone or in combination with those containing 35S∷HUA1-ECFP. No YFP fluorescence was detected when HEN4-EYFP was introduced alone, while nuclear CFP fluorescence was detected in cells from the leaf area infiltrated with HUA1-CFP alone (Figures 5A, 5B, 5G, and 5H). When the two types of Agrobacteria were coinfiltrated, however, nuclear signals through both YFP and CFP filter sets were detected (Figures 5C and 5I). Coinfiltration of HEN4p∷HEN4-EYFP together with 35S∷GAL4-ECFP did not result in any YFP signal (Figures 5D and 5J), suggesting that HUA1 was responsible for the accumulation or localization of HEN4-EYFP.
Figure 5. Interaction of HEN4 and HUA1 in Tobacco Nuclei.

(A–D and G–J) Low-magnification fluorescence images collected with the CFP or YFP filter sets as indicated from tobacco leaves infiltrated with Agrobacteria containing 35S∷HUA1-ECFP (A and G), HEN4p∷HEN4-EYFP (B and H), 35S∷HUA1-ECFP and HEN4p∷HEN4-EYFP (C and I), or 35S∷GAL4-ECFP and HEN4p∷HEN4-EYFP (D and J). The bright dots represent multiple nuclei showing CFP orYFP fluorescence.
(E, F, K, and L) High-magnification images collected with the CFP or YFP filter sets as indicated showing single nuclei from tobacco cells infiltrated with Agrobacteria containing 35S∷HUA1-ECFP and HEN4p∷HEN4-EYFP (E and K) or 35S∷HUA1-ECFP and 35S∷EYFP-TGA5 (F and L). The dots represent CFP or YFP fluorescence in nuclear speckles. HUA1 and HEN4 are concentrated in the same speckles (arrows and arrowheads in [E] and [K]). TGA5 appears to be more uniform throughout the nucleus.
(M and N) cFRET images as displayed using quantitative pseudocolor to show the interaction between HEN4 and HUA1. cFRET values, in arbitrary linear units of fluorescence intensity (a.l.u.f.i.), were calculated by subtracting the crossover fluorescence from the fluorescence collected with the FRET filter set (see Supplemental Data for details). The reason why only some nuclear speckles appear to be cFRET positive is that cFRET values are proportional to fluorescence intensity. In fact, the most accurate measure of FRET is cFRETN/Y/C (see Table 1), which is normalized against EYFP and ECFP fluorescence intensity. The scale bar represents 10 μm. Magnification in (E), (F), and (K)-(N) is the same.
Examination of HEN4-EYFP and HUA1-ECFP signals at higher magnification showed that the two proteins were present in speckled patterns in the nuclei. Furthermore, HUA1, but not another nuclear protein, TGA5 (Kato et al., 2002), appeared to be concentrated in the same speckles with HEN4 (Figures 5E, 5F, 5K, and 5L). To determine whether HUA1 and HEN4 physically interacted with each other, we measured fluorescence resonance energy transfer (FRET) between the two proteins and between HUA1 and TGA5 as the negative control. Note that the images in Figures 5F and 5L were taken at the same Z section (depth into the nucleus). Therefore, EYFP-TGA5, although not concentrated in the nuclear speckles containing HUA1, was also present in the same speckles, which allowed us to use TGA5 as a negative control for quantitative FRET measurements. We performed the FRET measurements on a central area approximately 5–6 μm in diameter in each nucleus, such that regions without HUA1/HEN4 speckles were also included in the measurements. Indeed, FRET was observed between HUA1 and HEN4, but not between HUA1 and TGA5 (Figures 5M and 5N). The normalized FRET value between HUA1 and HEN4 was approximately six times that between HUA1 and TGA5 (Table 1), and comparable to that between ECFP-TGA5 and EYFP-TGA5, which are known to dimerize (Kato et al., 2002), suggesting that HEN4 and HUA1 interacted with each other in vivo. To further test the potential interaction between HEN4 and HUA1, we performed yeast two-hybrid assays on the two proteins. Indeed, HUA1 and HEN4 were found to interact in the two-hybrid assays (Supplemental Figure S2).
Table 1.
Collected and Normalized FRET Values in Nuclear Regions of Tobacco Cells
| Accumulated proteins (n, sample number) | cFRETa × 105 | cFRET/FRETb | cFRETN/Yc | cFRETN/Cc | cFRETN/Y/Cc |
|---|---|---|---|---|---|
| HUA1-ECFP/HEN4-EYFP (n = 9) | 1.08 ± 2.57 | 0.22 ± 0.04 | 0.53 ± 0.12 | 0.41 ± 0.09 | 1.5E-06 ± 2.1E-07 |
| HUA1-ECFP/EYFP-TGA5 (n = 9) | 1.71 ± 1.70 | 0.05 ± 0.05 | 0.08 ± 0.09 | 0.09 ± 0.08 | 2.5E-07 ± 2.5E-07 |
| ECFP-TGA5/EYFP-TGA5 (n = 6) | 3.50 ± 1.39 | 0.16 ± 0.04 | 0.30 ± 0.06 | 0.27 ± 0.10 | 1.1E-06 ± 1.7E-07 |
FRET values are shown as average ± standard deviation.
cFRET values are presented in arbitrary linear units of fluorescence intensity, which was calculated by subtracting the crossover fluorescence from the fluorescence collected with the FRET filter set (CFP excitation/YFP emission). See Experimental Procedures for details.
cFRET/FRET values were calculated by dividing cFRET by the intensity that was collected with the FRET filter set.
Normalized FRET values were calculated by dividing cFRET by the intensity of EYFP (Y), ECFP (C), or both EYFP and ECFP (Y/C) in the nuclear region to yield cFRETN/YFP, cFRETN/CFP, and cFRETN/YFP/CFP, respectively.
hua1, hua2, and hen4 Mutations Compromise AG Pre-mRNA Processing
The nuclear localization of the two RNA binding proteins implies a function in nuclear RNA metabolism. To test whether these genes act in the processing of AG pre-mRNA, we performed RNA filter hybridization with a probe that encompassed the entire AG genomic region on total RNA isolated from various genotypes (Figure 6A). We included hua1, hua2, and hen4 single mutants and all combinations of double and triple mutants in order to determine the contribution, if any, of each gene to AG pre-mRNA processing. In addition to the mature AG RNA, three larger RNA bands (named 1, 2, and 3) of approximately 2900, 2600, and 1400 nucleotides, respectively, were also detected in all single, double, and triple mutant flowers (Figure 6A). RNA bands 1 and 3 were also present in wild-type (Ler). Bands 1 and 2 hybridized with a probe from the AG second intron (Figure 6A), suggesting the presence of intron 2 sequences in the RNA species. While the abundance of RNA band 3 was not affected by the hua1, hua2, or hen4 mutations, the abundance of the two larger bands increased with the number of mutations (Figure 6A). hua1, hua2, and hen4 mutations all contributed to the increased abundance of RNA bands 1 and 2 in the hua1 hua2 hen4 triple mutants. The level of the mature AG RNA was concomitantly reduced (Figure 6A). Therefore, all three genes, HEN4, HUA1, and HUA2, act in AG pre-mRNA processing.
Figure 6. AG Pre-mRNA Processing Defects in Various Genotypes.
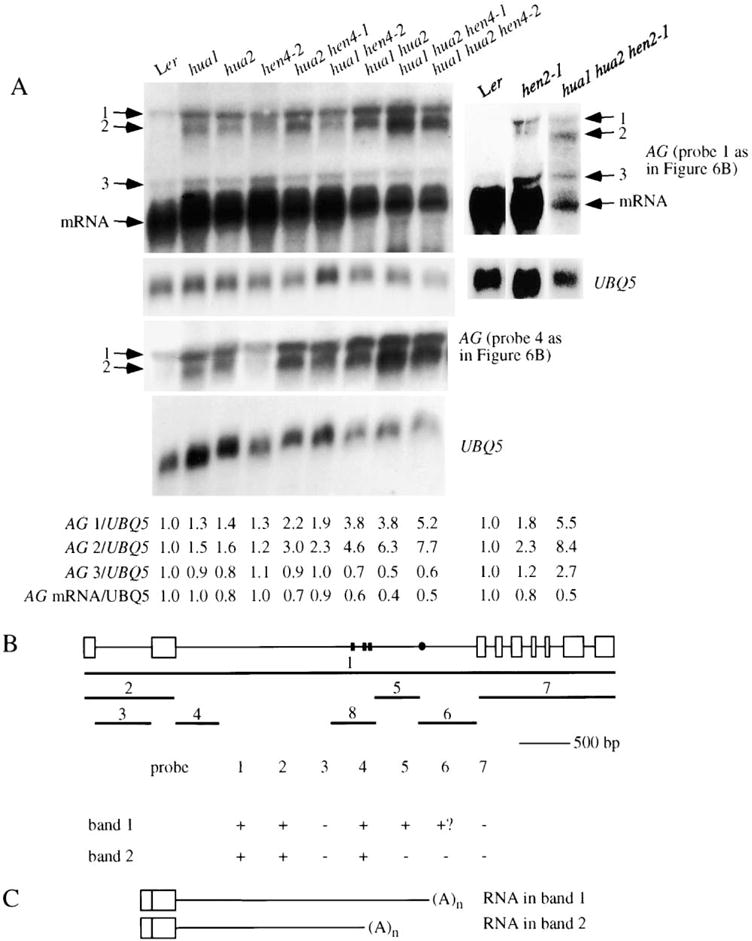
(A) RNA filter hybridization using probes corresponding to the entire AG genomic region or part of the second intron (probes 1 and 4, respectively; see [B]) on 50 μg of total RNA isolated from various genotypes. The corresponding UBQ5 control hybridization is shown below the AG hybridization. The abundance of the three larger bands (1–3) and AG mRNA in various genotypes is indicated by the numbers below the gel images, with wild-type levels arbitrarily set to 1.0.
(B) A summary of various RNA filter hybridization experiments aimed to determine the nature of the large transcripts in bands 1 and 2. The AG genomic region is diagrammed on top, with rectangles and lines representing exons and introns, respectively. The lines numbered 1 to 7 represent various probes used for RNA filter hybridization. Line 8 represents the region corresponding to AG RNA III used for in vitro binding assays. Results from various hybridization experiments are shown in the table below. Positive and negative hybridization signals are represented by “+” and “−,” respectively. “+?” indicates a weak signal that may or may not be real.
(C) Diagrams of the nucleotide composition of large transcripts represented by bands 1 and 2. The ends of the transcripts are represented by small, black rectangles or the dot in (B). The ends represented by the rectangles may belong to transcripts in band 2, whereas the one represented by the dot may belong to a transcript in band 1. All large transcripts contain the first two exons and are polyadenylated.
To determine whether HEN2, a putative nuclear RNA helicase, was also involved in AG pre-mRNA processing, we analyzed AG RNA accumulation in wild-type, hen2-1, and hua1 hua2 hen2-1 plants (Figure 6A). In hen2-1 and hua1 hua2 hen2-1, the abundance of RNA bands 1 and 2 was increased relative to Ler and hua1 hua2, respectively. AG mRNA level was greatly reduced in hua1 hua2 hen2-1 compared to Ler, which is consistent with the loss-of-C function phenotypes associated with hua1 hua2 hen2-1 flowers.
In order to determine the nature of RNA bands 1 and 2, we tested probes from different regions of the gene for their ability to hybridize to the large RNAs (Figure 6B). We found that probes covering the first two exons or the 5′ portion of the second intron were able to detect RNA bands 1 and 2, whereas probes corresponding to the first intron, the 3′ end of the second intron, or the 3′ portion of the AG genomic region (exon 3 and beyond) did not hybridize to the two RNA bands. As summarized in Figure 6C, results from the hybridization experiments suggested that the RNAs in bands 1 and 2 both contained the first two exons, and part or most of intron 2 for bands 1 and 2, respectively. In addition, the two transcripts were present in polyA+ RNA fractions, suggesting that they were polyadenylated. 3′ RACE was carried out to determine the exact 3′ end(s) of the RNA(s) in bands 1 and 2. Four 3′ ends were found at nucleotides 1775, 1859, 1864, and 2362 from the beginning of the second intron (Figure 6B). The first three may correspond to band 2, whereas the last one may correspond to band 1. Sequences immediately downstream of the four positions were not rich in A’s. Therefore, the RNAs were unlikely to be in the polyA+ fraction due to fortuitous pairing with oligo dT.
It is worth noting that the AG in situ probe as used in Figure 2 contained 90 nucleotides that are complementary to both the AG mRNA and the larger RNAs in bands 1 and 2. We have since repeated the experiments with a new AG mRNA-specific probe and obtained similar results (data not shown). A probe that was specific for the larger RNAs detected signals in hua1 hua2 hen4 flowers in the inner two whorls starting at stage 7 (Figure 2I). Because ectopic AP1 RNA in the inner two whorls was detected as early as stage 3, we suspect that AG RNA processing defects also started from this early stage but the level of the larger RNAs was too low to be detected with confidence.
HUA1, HUA2, and HEN4 appear to act specifically in the processing of AG pre-mRNA rather than in a general RNA processing machinery, because mutations in these genes did not lead to a higher abundance of larger RNAs from other floral homeotic genes, such as AP1, APET-ALA3, or PISTILLATA (Supplemental Figure S3).
Two lines of evidence indicate that the reduction of AG mRNA and protein (Supplemental Figure S4) in hua1 hua2 hen4 is likely the cause of the floral homeotic phenotype in the triple mutants. First, studies of transgenic plants containing antisense AG constructs demonstrated that reduction of AG RNA to 31%–58% of wild-type levels resulted in flowers with defects in floral determinacy, and partial stamen-to-petal and carpel-to-sepal transformation (Mizukami and Ma, 1995). hua1 hua2 hen4 flowers, in which AG mRNA is reduced to 40%– 50% of wild-type levels, largely resemble those of the AG antisense plants. Second, AG cDNA under the control of the 35S promoter (35S∷AG) rescued the homeotic phenotype of the hua1 hua2 hen4-1 triple mutant (Figure 1V). Whereas two hua1 hua2 hen4-1 plants containing the vector alone were indistinguishable from hua1 hua2 hen4-1 in floral phenotypes, three containing 35S∷AG had stamens in whorls 2 and 3 (Figure 1V).
HUA1 Binds AG RNA In Vitro
We performed a GST pulldown assay to test whether HUA1 could bind three AG RNA probes (I, II, and III, which correspond to regions 4, 5, and 8 in Figure 6B, respectively) from the second intron. The AG RNA probes were transcribed in the presence of [α-32P]UTP and incubated with GST-HUA1 or the control GST-αCP1, an unrelated RNA binding protein. The bound RNAs were eluted and resolved on a polyacrylamide gel. Efficient binding of GST-HUA1, but not GST-αCP1, to all three RNA segments was detected (Figure 7 and data not shown). To determine whether HUA1 was specific for these regions in the AG pre-mRNA, we tested a probe that corresponded to the first intron. GST-HUA1 was also able to bind this probe, which argues against region-specific affinity of HUA1 to AG pre-mRNA. While these observations cannot explain why AG intron 2 but not intron 1 was retained in the hua1, hua2, and hen4 mutants, the binding of HUA1 to AG pre-mRNA suggests that AG can be a direct in vivo target of HUA1 and HEN4, which act together.
Figure 7. Binding of HUA1 to AG RNA In Vitro.
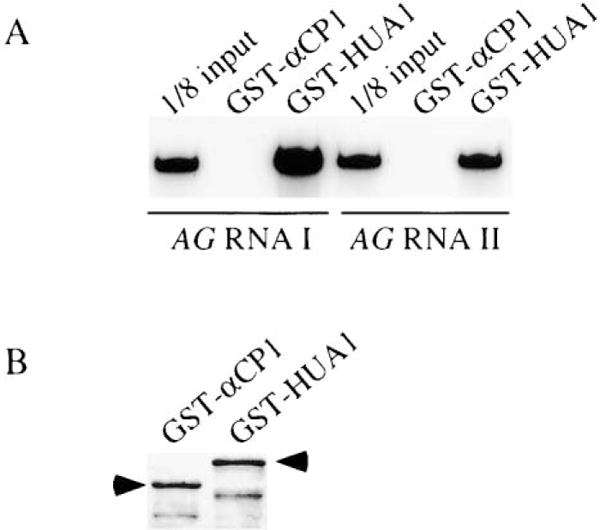
(A) In vitro-synthesized and 32P-labeled AG RNA I or II, which corresponds to fragments 4 or 5 in Figure 6B, respectively, was incubated with purified GST-αCP1 or GST-HUA1. After washing, the bound transcripts were eluted and resolved on a polyacrylamide gel.
(B) One sixtieth of the two proteins used for the binding assay was run on an SDS-polyacrylamide gel to show that near equal amounts of the two proteins were used in the binding assay. The arrowheads indicate the full-length GST fusion proteins. The lower bands were likely truncated proteins.
Discussion
HUA1, HUA2, HEN4, and HEN2 Facilitate AG Pre-mRNA Processing
Our studies demonstrated a role in AG pre-mRNA processing for not only HEN4, but also HUA1, HUA2, and HEN2. hen1-2, a mutation in HEN1, however, did not result in increased abundance of the large AG RNAs (data not shown). We showed that HEN1 is required for the accumulation of microRNAs and hypothesized that the loss-of-C function phenotypes in hua1 hua2 hen1 may result from increased expression of AP2 (Park et al., 2002), which is known to repress AG expression. Although not proven, HEN1 may act differently at the molecular level from HUA1, HUA2, HEN2, and HEN4.
We envision two molecular mechanisms by which HUA1, HUA2, and HEN4 facilitate AG pre-mRNA processing. HEN2 may act by similar or different mechanisms (see below). In one scenario, these proteins inhibit the recognition or usage of polyadenylation signals present in the AG second intron. Compromised activities of these proteins result in premature transcription termination and polyadenylation at these sites. In a second scenario, these proteins promote AG pre-mRNA splicing. Defects in AG pre-mRNA splicing caused cryptic polyadenylation signals in the second intron to linger long enough to be recognized, resulting in transcript cleavage followed by polyadenylation. The remaining 3′ portion of the AG pre-mRNA is then degraded. Upon examination of the sequences surrounding the four identified ends of the large transcripts in RNA bands 1 and 2, we found two potential polyadenylation signals that can theoretically be used to generate three of the four transcript ends (assuming that the two ends that are six nucleotides apart were generated with the same polyadenylation signal). The presence of the potential polyadenylation signals in the second intron of AG provides an explanation for the observed polyadenylated large transcripts in hua1 hua2 hen4 mutants, but does not favor a role of the proteins either in the inhibition of premature polyadenylation or in the promotion of splicing. However, if HEN4 acts in the alternative splicing of its own RNA, a role for HEN4 in splicing of AG pre-mRNA maybe more likely.
A similar example of alternative transcript processing playing a role in plant development involves FCA, which encodes an RNA binding protein required for the promotion of flowering in Arabidopsis (Macknight et al., 1997). Results from a recent study suggest that polyadenylation within intron 3 of FCA, which results in the production of nonfunctional transcripts, acts to limit the production of the functional transcript (Macknight et al., 2002). We do not know whether AG expression level is similarly controlled by splicing/polyadenylation. Although two large AG RNAs can be detected in the wild-type background, their abundance is much lower than that of the mature mRNA. However, it is possible that these large RNAs are generated at a higher level (thus reducing the amount of mRNA) but rapidly degraded. In this scenario, splicing of, or polyadenylation within, intron 2 can play an important role in controlling the level of AG mRNA. The functions of the HUA and HEN proteins would be to promote the splicing of, or counteract premature polyadenylation within, the second intron, and thus positively regulate AG expression.
While our studies in floral organ identity have uncovered several proteins with RNA-related roles acting to regulate AG expression, studies on flowering time control have also revealed RNA binding proteins, such as FCA and FPA, that control the expression of FLOWERING LOCUS C (FLC), a repressor of flowering (Michaels and Amasino, 1999; Sheldon et al., 1999; Schomburg et al., 2001). Interestingly, FLC also has a large intron (about 3.4 kb in the Columbia accession). A recent study showed that the FLC genomic region encompassing exon 1, the large intron 1, and exon 2 was able to confer FCA-dependent expression to a reporter gene (Sheldon et al., 2002).
HUA1, HUA2, and HEN4 appear to be specifically required for the processing of, or for preventing transcription termination within, the AG second intron, which is relatively large (2999 base pairs) and contains key transcriptional regulatory elements. Interestingly, the ends of the polyadenylated large transcripts are in regions known to mediate key regulatory functions, such as domain-specific expression (Deyholos and Sieburth, 2000) or responsiveness to upstream regulators such as LFY and WUS (Busch et al., 1999; Lohmann et al., 2001). It is possible that it is the size of the intron or the presence of certain sequences in the intron that necessitates the requirement for HUA1, HUA2, and HEN4 for the processing of the intron. Our preliminary studies on FLC, another gene with a large intron, suggest that intron size alone may not be the answer. With the entire, large intron 1 of FLC as a probe, we were unable to detect any aberrant FLC transcripts in wild-type, hua1 hua2, or hua1 hua2 hen4-1 plants (Supplemental Figure S3). However, the level of FLC mRNA appears to be reduced in hua1 hua2 and hua1 hua2 hen4-1 as compared to wild-type. Therefore, posttranscriptional regulation of FLC by HUA1, HUA2, or HEN4 cannot be totally ruled out.
Potential Molecular Functions of the HUA and HEN Proteins
We hypothesize that HUA1 and HEN4 bind to AG pre-mRNA in vivo to either block the potential polyadenylation sites or to promote splicing. In vitro, HUA1 binds three tested AG RNA fragments from the second intron, one including the 5′ splice site and the other two including the putative polyadenylation sites for the large AG RNAs. While this does not help distinguish the two possibilities, it demonstrates that HUA1 is capable of directly binding AG RNA. It remains to be tested whether the HUA1/HEN4 complex binds AG pre-mRNA at a specific site in vivo, with the specificity conferred either by HEN4 or by other proteins.
The HUA2 protein is 25% identical (37% similar) to human transcriptional coactivator proteins p52 and p75 in the N-terminal 200 amino acids of these proteins. p52 and p75 are known to interact with several components of the general transcriptional machinery (Ge et al., 1998). HUA2 also has an RPR domain (amino acids 778–909) found in many proteins, some of which are known to act in nuclear RNA metabolism (Steinmetz and Brow, 1998; Steinmetz et al., 2001). In particular, the RPR domain in the rat SR protein rA4 binds the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Yuryev et al., 1996). In addition, we found that HUA2 interacts with a homolog of a yeast splicing factor, Prp40p, both in a yeast two-hybrid assay and in vitro (Y.C. and X.C., unpublished results). Therefore, we hypothesize that HUA2 acts in coupled transcription and RNA processing, perhaps by recruiting splicing factors to sites of active transcription.
HEN2 is highly homologous to yeast Dob1p (Mtr4p), an RNA helicase required for the activity of the nuclear exosome, which acts in rRNA maturation and in the degradation of inefficiently spliced pre-mRNAs (reviewed by Butler, 2002). Studies of the two Arabidopsis homologs of core yeast exosome components AtRrp41p and AtRrp4p indicate that the Arabidopsis exosome may be functionally similar to its yeast counterpart (Chekanova et al., 2000, 2002). Assuming that HEN2 functions similarly to Dob1p, the increased abundance of the larger AG RNAs in hen2-1 and hua1 hua2 hen2-1 may be explained by a role of HEN2 in nuclear RNA degradation—in hen2-1, the larger AG RNAs are not degraded efficiently and thus accumulate to a higher level. However, if HEN2 acts solely in the degradation of unspliced RNAs, one has to evoke other mechanisms to explain why AG mRNA level is also reduced in hen2-1 or hua1 hua2 hen2-1. One possibility is that aberrant transcripts with introns from many genes accumulate in hen2-1, and they sequester the splicing machinery, resulting in inefficient splicing of AG and other RNAs. Another possibility is that HEN2 also plays a direct role in AG pre-mRNA processing in addition to its role in the exosome.
KH Domain and CCCH Zinc Finger Proteins in the Development of Multicellular Organisms
The yeast two-hybrid and the in planta FRET analyses clearly demonstrated that HUA1 and HEN4 interact with each other. In addition, the lack of YFP signals in tobacco infiltrated with Agrobacteria containing HEN4-EYFP alone and the presence of YFP signals in tobacco infiltrated with Agrobacteria harboring both HUA1-ECFP and HEN4-EYFP suggest that HEN4 expression (at the transcriptional or posttranscriptional levels) or nuclear localization requires HUA1. The physical interaction between HUA1 and HEN4 would be more consistent with a role of HUA1 in either the stabilization or the nuclear localization of HEN4.
Although HEN4 is, at present, the only KH domain protein in the plant kingdom with a known developmental function, many KH domain proteins have been found to perform key developmental functions by regulating different aspects of RNA metabolism in metazoans. In humans, the fragile X mental retardation protein, FMRP, is thought to regulate mRNA translation in the brain (Feng et al., 1997). Nova-1, which is implicated in paraneoplastic opsoclonus-myoclonus ataxia (POMA), a neurodegenerative syndrome (Buckanovich et al., 1993), regulates neuron-specific alternative splicing (Jensen et al., 2000). Two other KH domain proteins, αCP1 and αCP2, stabilize globin mRNAs during erythroid differentiation (Kiledjian et al., 1995). In Drosophila, Bicaudal-C, a protein with five KH domains, acts in embryo patterning by mediating the posterior movement of RNAs (Mahone et al., 1995).
CCCH zinc finger proteins also play critical posttranscriptional roles in animal development. The murine TTP protein regulates the stability of tumor necrosis factor α (TNF-α) RNA (Carballo et al., 1998). In C. elegans, several CCCH zinc finger proteins, Pie-1, Mex-1, and Pos-1, specify the identity of germline blastomeres during early embryonic development, probably by regulating the translation of maternal RNAs (Mello et al., 1996; Guedes and Priess, 1997; Tabara et al., 1999; Tenenhaus et al., 2001).
CCCH zinc finger proteins and KH domain proteins have also been found to act in similar developmental processes. In C. elegans, MEX3, a KH domain protein, functions in the establishment of soma/germline asymmetry in the early embryo by inhibiting the translation of pal-1 RNA (Draper et al., 1996; Hunter and Kenyon, 1996). In a yeast two-hybrid assay, MEX-3 interacts with MEX-6, a CCCH zinc finger protein (Huang et al., 2002). MEX-6, together with MEX-5, another CCCH zinc finger protein that shares high sequence similarity with MEX-6, also acts in soma/germline asymmetry specification in the early embryo (Schubert et al., 2000). In fact, MEX-5 and MEX-6 appear to stabilize MEX-3 in the anterior of the early embryo (Huang et al., 2002). This, and our finding that HUA1 interacts with HEN4, suggest that the interaction between CCCH and KH domain proteins is an ancient evolutionary phenomenon.
Experimental Procedures
Map-Based Cloning of HEN4
Wild-type plants of the Columbia ecotype were used to pollinate the stigma of early-arising flowers from hua1-1 hua2-1 hen4-1 plants (in the Ler background). A small number of seeds were obtained from these crosses. In the F2 population, plants showing the hua1-1 hua2-1 hen4-1 or the hua1-1 hua2-1 floral phenotypes were selected as the mapping population. The genotypes of those in the latter category were determined by the segregation of the hua1-1 hua2-1 hen4-1 floral phenotype in the F3 generation. Initial mapping with 52 hua1-1 hua2-1 hen4-1 plants showed that HEN4 was linked to the marker AthS0191 on chromosome V. New markers in this region were developed according to polymorphisms between Ler and Col, as reported by Cereon (http://www.Arab/dops/s.org/Cereon/index.html). Using these markers and 342 plants of known HEN4 genotypes, we mapped HEN4 to a 48 kb region covered by a P1 clone MSJ1 and a BAC T12B11 (see Figure 3).
A 5076 bp DNA fragment containing 583 bp of sequences upstream of the start codon and 554 bp of sequences downstream of the stop codon of At5g64390 was amplified by PCR from MSJ1 and cloned into the plant transformation vector pPZP222 (Hajdukiewicz et al., 1994). The resulting plasmid, pCC214, was used to transform hua1 hen4-1 plants that segregated for hua2 by vacuum infiltration.
Isolation of HEN4 cDNAs
The 5′ and 3′ ends of HEN4 transcripts were determined by 5′ and 3′ RACE, respectively. Full-length cDNAs corresponding to the two HEN4 transcripts were then cloned by RT-PCR using Pfu polymerase with RBp17 (5′-agcaacttagcgttaaagaccaccaaag-3′) and RBp18 (5′-agacatcaaactgattccttcaaagtacttaaa-3′). The two products, 2876 bp and 3410 bp in length, were cloned into pCR-BluntII-TOPO and pCRII (Invitrogen) to result in plasmids pCC055 and pCC033, respectively. The plasmids were sequenced to ensure the absence of mutations introduced by PCR.
Construction of Genotypic Combinations
Molecular genotyping for hua1-1 (Western et al., 2002), hua2-1 (Western et al., 2002), hen4-1, and hen4-2 greatly facilitated the identification of various genotypes in genetic crosses. hen4-1 resulted in adesigned Mwol polymorphism: the RBp11 (5′-ctcccattcttcctctacacattcag-3′)/RBp12 (5′-agataagcctcttggaggcgctggagg-3′) PCR product from wild-type could be digested with Mwol, whereas that from hen4-1 could not. For hen4-2 genotyping, the RBp5 (5′-cattgttccttaagttgtctggcata-3′)/HEN4p40 (5′-gctcttggctaaggaattttt cat-3′) PCR product was digested with Fnu4HI. hen4-2 resulted in the loss of an Fnu4HI site.
The construction of various genotypic combinations, such as hua1 hen4, hua2 hen4, hua1 hua2 hen4-1 ap1-1, hua1 hua2 hen4-1 ap2-2, hua1 hua2 hen4-1 ag-4, hua1 hua2 hen4-1 ag-1, and hua1 hua2 hen4-1 35S∷AG, is described in Supplemental Data.
In Situ Hybridization, RNA Filter Hybridization, and SEM
In situ hybridization and RNA filter hybridization were carried out as described (Li et al., 2001). Details for probes used can be found in Supplemental Data. For the detection of AG, AP1, AP3, PI, and FLC RNAs by RNA filter hybridization, 50 μg of total RNA or polyA+ RNA isolated from 50 μg of total RNA was used. For the detection of HEN4 RNA, polyA+ RNAwas isolated from approximately100 μg of total RNA. SEM was performed as described by Western et al. (2002).
Transient Expression Assay and Image Acquisition
Transient expression assays and image acquisition were performed according to Kato et al. (2002).
The construction of plasmids, HEN4p∷HEN4-EYFP, 35S∷HUA1-ECFP, 35S∷GAL4-ECFP, 35S∷EYFP-TGA5, and 35S∷ECFP-TGA5, is described in Supplemental Data.
FRET Analysis
Images were acquired at 0.2 μm Z steps and with a 0.3 s exposure time. Three images in each step were acquired in the same order through (1) a YFP filter set (excitation 500/20 nm, emission 535/30 nm), (2) a CFP filter set (excitation 436/10 nm, emission 470/30 nm), and (3) a FRET filter set (excitation 436/10 nm, emission 535/30 nm). Three stacked images were then deconvolved, and the image in the middle layer was used to measure the fluorescence intensity.
FRET calculation was performed based on the “microFRET” method (Sorkin et al., 2000). It was shown that the best way to obtain a precise background fluorescence value for nuclei in tobacco leaves is to measure the fluorescence intensity value from the cytosolic region in the same cell (Kato et al., 2002). Therefore, the mean fluorescence intensity values in the cytosolic area were subtracted from the nuclear images before carrying out FRET calculations.
Fluorescence through the FRET filter set consisted of a true FRET component (“corrected” FRET, cFRET) and a non-FRET component (both donor and acceptor fluorescence through the FRET filter set). See Supplemental Data for how cFRET values were derived.
RNA Binding Assays
The full-length HUA1 coding region was amplified and cloned into pGEX-2TKto generate the GST-HUA1 plasmid. The GST-αCP1 expression plasmid was a gift from Dr. Kiledjian at Rutgers University. RNA binding assays were performed as described by Trifillis et al. (1999) and Jiao et al. (2002).
Acknowledgments
We are grateful to Drs. Hong Ma and Wonkeun Park for valuable comments on the manuscript. We thank Drs. Hugo Dooner, Kenneth Irvine, Megerditch Kiledjian, and Samuel Gunderson for helpful discussions and Dr. Eric Lam for providing equipment and expertise for FRET analysis. We are grateful to Dr. Megerditch Kiledjian for the GST-αCP1 strain and to Drs. Kiledjian and Xinfu Jiao for help with the RNA binding assays. We thank Jiqun Zhao and Junhong Sun for technical assistance and Dr. Eric Lam and Yuda Fang for helping to construct the pGAL4-ECFP-EL103 plasmid. We are grateful to Dr. Philip James for yeast strain PJ69-4A and Dr. Tamara Western for hen2-1 seeds. We acknowledge Cereon for making its SNP data available to the public. This work was supported by a Charles and Johanna Busch predoctoral fellowship to Y.C. and by a National Institutes of Health grant (GM61146-01) to X.C.
Footnotes
References
- Bomblies K, Dagenais N, Weigel D. Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev Biol. 1999;216:260–264. doi: 10.1006/dbio.1999.9504. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Butler JS. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- Byzova MV, Franken J, Aarts MGM, de Almeida-Engler J, Engler G, Mariani C, Van Lookeren Campagne MM, Angenent GC. Arabidopsis STERILE APETALA, a multifunctional gene regulating inflorescence, flower, and ovule development. Genes Dev. 1999;13:1002–1014. doi: 10.1101/gad.13.8.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA. Poly (A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J Biol Chem. 2000;275:33158–33166. doi: 10.1074/jbc.M005493200. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Dutko JA, Mian IS, Belostotsky DA. Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3′ to 5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res. 2002;30:695–700. doi: 10.1093/nar/30.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Meyerowitz EM. HUA1 and HUA2 are two members of the floral homeotic AGAMOUS pathway. Mol Cell. 1999;3:349–360. doi: 10.1016/s1097-2765(00)80462-1. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Deyholos MK, Sieburth LE. Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell. 2000;12:1799–1810. doi: 10.1105/tpc.12.10.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Roeder RG. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes S, Priess JR. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. 1997;124:731–739. doi: 10.1242/dev.124.3.731. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF. Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell. 1994;76:131–143. doi: 10.1016/0092-8674(94)90178-3. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Huang NN, Mootz DE, Walhout AJM, Vidal M, Hunter CP. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development. 2002;129:747–759. doi: 10.1242/dev.129.3.747. [DOI] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the apetala1 gene during Arabidopsis floral development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YYL, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermialike RNA-binding protein. Biol Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- Kato N, Pontier D, Lam E. Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer (FRET) in tobacco leaf nuclei. Plant Physiol. 2002;129:931–942. doi: 10.1104/pp.005496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Wang X, Liebhaber SA. Identification of two KH domain proteins in the α-globin mRNP complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Prost V, Macias A. AINTEGUMENTA promotes petal identity and acts as a negative regulator of AGAMOUS. Plant Cell. 2000;12:1357–1366. doi: 10.1105/tpc.12.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KFX, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jurgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Li J, Jia D, Chen X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell. 2001;13:2269–2281. doi: 10.1105/tpc.010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development. 1995;121:975–991. doi: 10.1242/dev.121.4.975. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Weigel D. Building beauty: the genetic control of floral patterning. Dev Cell. 2002;2:135–142. doi: 10.1016/s1534-5807(02)00122-3. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30:623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt L, Murphy G, Sherson S, Cobbett C, Dean C. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Macknight R, Duroux M, Laurie R, Dijkwel P, Simpson G, Dean C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell. 2002;14:877–888. doi: 10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahone M, Saffman EE, Lasko PF. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382:710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- Musco G, Stier G, Joseph C, Morelli MAC, Nilges M, Gibson TJ, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CM, Lin R, de Bries CJ, Plasterk RHA, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Pickett FB, Haughn GW. The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. Plant Cell. 1991;3:1221–1237. doi: 10.1105/tpc.3.11.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell. 2002;14:1–11. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM. Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell. 1995;7:1249–1258. doi: 10.1105/tpc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and Grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Tabara H, Hill RJ, Mello CC, Priess JR, Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. 1999;126:1–11. doi: 10.1242/dev.126.1.1. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Subramaniam K, Dunn MA, Seydoux G. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 2001;15:1031–1040. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifillis P, Day N, Kiledjian M. Finding the right RNA: identification of cellular mRNA substrates for RNA-binding proteins. RNA. 1999;5:1071–1082. doi: 10.1017/s1355838299981803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Cheng Y, Jun L, Chen X. HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis. Development. 2002;129:1569–1581. doi: 10.1242/dev.129.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi R, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]


