Abstract
The ability to distinguish between self and nonself is the fundamental basis of the immune system in all organisms. The conceptual distinction between self and nonself, however, breaks down when it comes to endogenous retroviruses and other retroelements. While some retroelements retain the virus-like features including the capacity to replicate and reinvade the host genome, most have become inactive through mutations or host epigenetic silencing. And yet, accumulating evidence suggests that endogenous retroelements, both active and inactive, play important roles not only in pathogenesis of immune disorders, but also in proper functioning of the immune system. This review discusses the recent development in our understanding of the interaction between retroelements and the host innate immune system. In particular, it focuses on the impact of retroelement transcripts on the viral RNA sensors such as Toll-like receptors, RIG-I-like receptors, protein kinase R, and the inflammasomes.
1. INTRODUCTION
Endogenous retroelements are mobile genetic elements that constitute nearly 50% of the human genome. These elements are present in almost all organisms, and it is thought to have arisen from integration of retroviruses into the host genome. Due to their ability to rearrange genetic elements and to alter the global transcriptional patterns, endogenous retroelements have been frequently implicated in a variety of genetic disorders, including breast cancer, multiple sclerosis, and amyotrophic lateral sclerosis (Downey et al., 2015; Konkel & Batzer, 2010; Li et al., 2015; Nissen et al., 2013; Suntsova et al., 2015). While some of the retroelements can generate replication-competent viruses or can be retro-transposed into the genome, most are highly mutated and thus rendered inactive. Even the ones retaining the capacity to replicate are often transcriptionally silenced through a multitude of epigenetic regulatory mechanisms (Elsasser et al., 2015; Molaro & Malik, 2016; Rowe et al., 2010). And yet, accumulating evidence suggests that endogenous retroelements play important roles in both pathogenesis of immune disorders and normal physiological functioning of the immune system (Volkman & Stetson, 2014).
Retroelements can be divided into two groups using two different criteria (see Deininger & Batzer, 2002; Kassiotis & Stoye, 2016; Mita & Boeke, 2016 for more detailed reviews). First, they can be grouped into those with their genes flanked by long terminal repeats (LTRs) and those without LTRs. The LTR retroelements utilize the LTRs for transcription initiation and termination. Their transcripts often encode essential nucleic acid processing enzymes, such as the reverse transcriptase (RT) that copies RNA to DNA and the endonuclease that cleaves genomic sites for insertion. Endogenous retroviruses (ERVs) also belong to this category, but they additionally encode viral envelope proteins for the generation of infectious virus particles. Unlike the LTR type, the non-LTR retroelements utilize promoters and enhancers within their own 5′-untranslated region (UTR) or in the host genes nearby. They include long interspersed elements (LINEs) and short interspersed elements (SINEs).
Retroelements can be also divided into autonomous and nonautonomous retroelements. While no retroelement is truly autonomous in a sense that they all rely on cellular machineries (such as the ribosome), nonautonomous retroelements additionally rely on the proteins encoded by autonomous retroelements for retrotransposition. Both the LTR and non-LTR types have autonomous and nonautonomous kinds. Within the LTR type, those with intact ORFs that encode a functional RT and an appropriate endonuclease would be autonomous, while those with mutations that compromise the activities of these enzymes would be nonautonomous. Within the non-LTR types, the best-studied autonomous retroelement is LINE-1, which is transcribed by RNA polymerase II (pol II) and encodes two proteins: the RNA-binding protein (ORF1) and the RT and endonuclease (ORF2). In contrast to LINE-1, SINEs have no coding capacity and thus rely on enzymes produced by LINEs for replication and retrotransposition. For example, a primate-specific SINE, Alu, relies on ORF2 of LINE-1 for its own retrotransposition. This co-option is possible because SINEs have adopted 3′-end sequences from LINEs, which are required for recognition by RTs and endonucleases. These replication-competent SINEs are typically generated by pol III using its internal pol III promoter. Most SINEs, however, are inactive retroelements and synthesized within pol II transcripts in the form of introns or 3′/5′-UTRs.
In this review, we will discuss how each of these classes of retroelements is involved in regulation and activation of the innate immune functions. We will here focus on the impact of the retroelement transcripts on the innate immune receptors that are known to detect viral RNAs during infection. These include Toll-like receptors (TLRs) 3, 7–8, RIG-I-like receptors (RLRs), protein kinase R (PKR), and the NLRP3 inflammasome. Although some retroelements, especially the ones transit through DNA intermediates, were also shown to impact innate immune sensors that detect viral DNAs, we will not discuss this topic as excellent reviews are available elsewhere (Kassiotis & Stoye, 2016; Volkman & Stetson, 2014).
2. TOLL-LIKE RECEPTORS
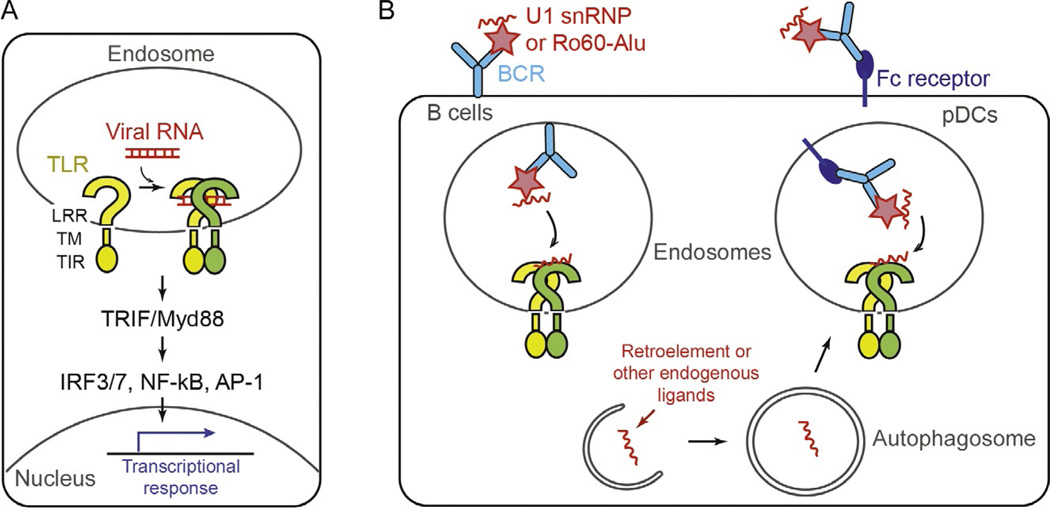
TLRs are membrane-bound receptors commonly characterized by the shared domain architectures. At the N-terminus, they have leucine-rich repeat (LRR) motifs that form the horseshoe-shaped ligand-binding domain (Fig. 1A). The LRR domain is located in the extracellular space or the lumen of endosomes for sensing microbial molecules (also known as pathogen-associated molecular patterns, PAMPs). Following the LRR domain are the central transmembrane domain and the C-terminal cytosolic signaling domain, namely Toll/IL-1 receptor homology (TIR; Fig. 1A). There are currently 10 and 12 TLRs characterized in human and mouse, respectively. Each TLR recognizes different types of PAMPs, thereby playing nonredundant functions in innate immune defense against pathogens. Among these, TLR3, TLR7, and TLR8 are the three TLRs in human shown to sense foreign RNA molecules, while additional TLRs (eg, TLR13) exist in mouse for RNA sensing (Ewald & Barton, 2011; Pelka et al., 2016). Upon ligand binding, TLRs undergo either dimerization of the LRR domain or reorganization of the preformed dimer (Ewald & Barton, 2011; Pelka et al., 2016). This conformational change in the LRR then propagates to the cytosolic TIR domain, leading to the recruitment of the adaptor molecules, TRIF and Myd88, and the activation of the transcription factors, IRF3, IRF7, NF-κB, and AP-1 (Fig. 1A). These transcription factors in turn upregulate a set of genes for inflammatory mediators and other restriction factors that limit pathogen spreading.
Fig. 1.
(A) Schematic of the TLR domain architecture and activation mechanism. The indicated mechanism was adopted from the TLR3 activation model. (B) Proposed mechanisms by which TLR7 recognizes self-RNAs. TLR7 was shown to be activated by the U1 snRNA or Alu elements upon entry into the endosome, which is mediated by autoantibodies against U1 snRNP and Ro60, or Fc receptors.
How do RNA-sensing TLRs distinguish between self- and nonself-RNAs? TLR3 detects double-stranded RNA structure in a sequence-independent, but length-dependent manner (Leonard et al., 2008; Liu et al., 2008). The longer RNA stimulates TLR3 better when compared among ~40–500 bp (Leonard et al., 2008). This observation led to the model that TLR3 may oligomerize on dsRNA to distinguish between long viral dsRNA and short cellular dsRNA. In contrast to TLR3, TLR7 and 8 recognize single-stranded RNAs (ssRNAs) with a preference for U-rich sequences (Diebold et al., 2006; Forsbach et al., 2008). A recent structure revealed that TLR8 binds to U-containing degradation products of ssRNA, instead of an intact RNA molecule, which could explain its dependence on the sequence content, not a specific sequence motif (Tanji et al., 2015).
The intrinsic biochemical specificity mentioned earlier, however, appears insufficient to allow robust discrimination of self- and nonself-RNAs by TLR3, 7–8. Two additional mechanisms have been proposed to further increase the fidelity of these receptors. First, posttranscriptional modification often occurs in cellular RNAs, and these modifications were shown to restrict activation of TLR3, 7–8 (Kariko et al., 2005). Second, TLR3, 7–8 are located in the endosome with the LRR domain exposed to the endosomal lumen. This localization would not only restrict their access to cytosolic self-RNAs but also to self-RNAs secreted from dying cells (Ewald & Barton, 2011). The protective nature of the endosomal localization was particularly well demonstrated with TLR9, a DNA-sensing TLR, which is also localized on endosomes. Engineered TLR9 targeted to the cell surface instead of the endosome responded to both self- (mammalian) and nonself- (bacterial mimic) DNA, while wild-type TLR9 in the endosome showed greater specificity for nonself-DNA (Barton, Kagan, & Medzhitov, 2006).
Despite the multitude of mechanisms to restrict self-recognition, multiple lines of evidence suggest that TLRs can be stimulated by self-ligands during both normal physiological and pathological processes. One of the first pieces of evidence supporting this notion came from the study of a mouse strain, in which the telomeric end of the X chromosome harboring the TLR7 gene is translocated to the Y chromosome, thereby duplicating the TLR7 gene. In the autoimmune-prone mouse background, this translocation event significantly enhanced the autoimmune pathology and was thus termed the Y-linked autoimmune accelerator (Yaa) locus mutation (Murphy & Roths, 1979; Pisitkun et al., 2006; Subramanian et al., 2006). This attribute was ascribed to TLR7 duplication as partial or complete deletion of TLR7 ablated the autoimmune phenotype (Christensen et al., 2006; Deane et al., 2007a). In further support of the role of TLR7 in immune disorders, overexpression of TLR7 alone caused a fatal acute inflammatory pathology in mice (Deane et al., 2007b). Finally, in human, the copy number variation of TLR7 was reported to correlate with the level of type I interferons and to be linked to childhood-onset of systemic lupus erythematosus (SLE; Garcia-Ortiz et al., 2010).
Studies suggest that the spontaneous activation of TLR7 in these disease models and SLE patients is due to the aberrant recognition of self- RNAs by TLR7 (Lau et al., 2005; Savarese et al., 2006). According to these studies, self-RNAs enter endosomes in the form of RNA– autoantigen complexes via B cell receptors (BCRs) or in the form of RNA-immune complexes via Fc receptors (Fig. 1B) (Lau et al., 2005; Savarese et al., 2006). This model is attractive as it explains why autoactivation of TLR7 (in both mouse models and human SLE patients) leads to selective proliferation of anti-ribonucleoprotein (RNP) B cells (Pisitkun et al., 2006). This is also highly analogous to the mechanisms by which TLR9 senses self-DNA and induces production of anti-DNA autoantibodies (Leadbetter et al., 2002).
What are the identities of the endogenous RNAs activating TLR7? Earlier studies showed that one of the major anti-RNP antibodies in SLE patients target the protein components of small nuclear ribonucleoproteins (snRNPs). These proteins, known as Smith or Sm proteins, are constituents of the spliceosome that interact with the spliceosomal U snRNAs (U1, 2, 4, 5; Migliorini et al., 2005). When complexed with the anti-Sm antibodies or the SLE patient sera containing the anti-RNP antibodies, purified U1 snRNP was shown to enter the endosome and activate TLR7, thereby inducing type I interferon production and inflammation (Savarese et al., 2006; Vollmer et al., 2005).
In a more recent report, Alu RNAs and another RNA-binding protein, Ro60, were also proposed to play a role in the pathogenesis of SLE. Ro60 is an abundant cellular protein and another common auto-antigen in patients with SLE and other immune disorders. Analysis of the RNA molecules copurified with Ro60 revealed that Alu transcripts (in particular, the intronic Alu’s) is the major RNA species bound by Ro60 (Hung et al., 2015). The Alu transcripts were also present in the anti-Ro60 immune complexes isolated from SLE patients, and it alone can directly activate TLR7 when delivered into the endosome (Hung et al., 2015). These observations fit the model mentioned earlier—that is, the autoantigen (in this case Ro60) acts as a mediator for endosomal delivery of self-RNAs, which then activate TLR7 (Fig. 1B). However, the role of Ro60 appears more complicated. The Ro60 knock-out mouse displayed the enhanced level of interferons, which in turn upregulated intronic Alu’s and further activated TLR7 (Hung et al., 2015). Although the exact role of Ro60 is still unclear, it was proposed that Ro60 has two seemingly opposing functions: first as an endosomal RNA delivery vehicle in the presence of autoantibody (extracellular role) and second as a suppressor of Alu production or release, which acts to block their access to TLR7 (intracellular role).
The activation of TLR7 in the absence of Ro60 begs the question of how intracellular Alu RNAs access the endosomal TLR7. Previous studies showed that TLR7 not only accesses ligands through endocytosis, but also through autophagy, a catabolic process by which cytosolic components are enclosed within the double-membrane structure and delivered to lysosomes for degradation (Lee et al., 2007). Autophagy was also shown to be important for TLR7-mediated sterile inflammation in a transgenic mouse model overexpressing TLR7 (Weindel et al., 2015). It remains to be investigated whether autophagy occurs selectively for Alu RNAs or other endogenous ligands, and if so, how.
TLR8 is closely related to TLR7 in terms of both protein sequence and RNA specificity. Compared to TLR7, however, TLR8 is relatively poorly understood because mouse TLR8 does not bind and respond to RNA, while human TLR8 does. Nevertheless, accumulating evidence suggests that human TLR8 also contributes to pathogenesis of a spectrum of autoimmune and autoinflammatory diseases likely through recognition of self-RNAs. As with TLR7, human TLR8 can be also stimulated by snRNPs in plasmacytoid dendritic cells (pDCs) in a manner dependent on anti- RNP autoantibodies or Fc receptors (Vollmer et al., 2005). A transgenic mouse harboring multiple copies of human TLR8 displayed a high level of autoinflammation in a TLR8-dose dependent manner (Guiducci et al., 2013). In human, a correlation between the level of TLR8 and the disease state has been observed in patients with SLE (Guiducci et al., 2013) and antiphospholipid syndrome (Doring et al., 2010), although the causal relation remains to be further investigated.
TLR3 was also shown to be responsive to endogenous RNAs under various experimental conditions (Bernard et al., 2012; Biswas et al., 2015; Brentano et al., 2005; Cavassani et al., 2008; Kariko et al., 2004; Mori et al., 2015). Green et al. (2012) found that a variety of cellular RNAs, such as a subset of tRNAs, can activate TLR3 and thus, activate B cells. TLR3 (together with TLR7 and 9) was also shown to induce ERV suppression, presumably by sensing ERVs and by eliciting immune responses against ERVs (Yu et al., 2012). Recently, treatment of cancer cells with DNA-methylation inhibitor was shown to induce ERV upregulation and activation of TLR3 and MDA5, which will be discussed in more details in the next section.
3. RIG-I AND MDA5
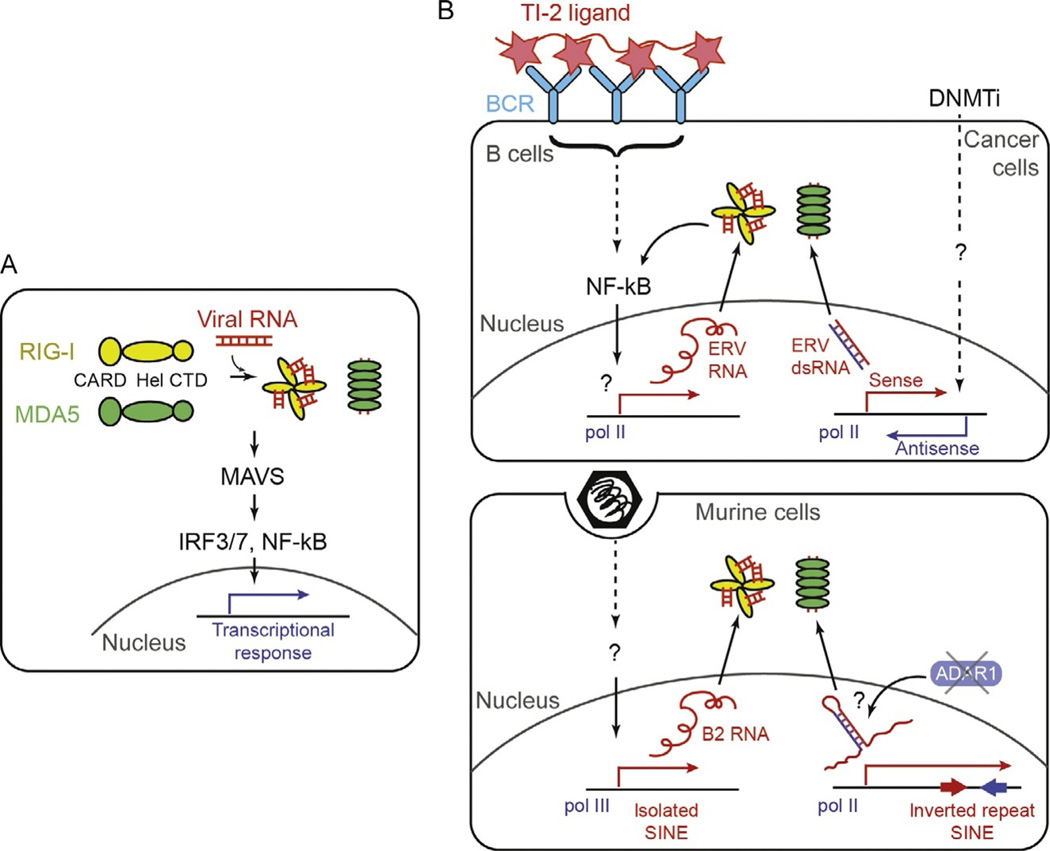
Unlike TLRs, RIG-I and MDA5 are soluble receptors that function in the cytoplasm. They share the same domain architecture, consisting of the N-terminal caspase activation recruitment domain (CARD), central helicase domain (Hel), and C-terminal zinc-coordinating domain (CTD; Fig. 2A). The helicase domain and CTD together function as an integrated RNA-binding unit, while the CARD activates the downstream signaling pathway by interacting with the signaling adaptor, MAVS (Kato, Takahasi, & Fujita, 2011; Wu & Hur, 2015). The interaction requires oligomerization of RIG-I and MDA5 through both RNA- and polyubiquitin-dependent mechanisms (Fig. 1A; Sohn & Hur, 2016). Upon its interaction with RIG-I or MDA5, MAVS polymerizes and recruits downstream signaling molecules, such as TRAF2, 3, 5, and 6, leading to the activation of the transcription factors, IRF3/7 and NF-κB (Saha, 2006).
Fig. 2.
(A) Schematic of the domain architecture and activation mechanism of RIG-I and MDA5. (B) Proposed mechanisms by which RIG-I and MDA5 recognize retroelements. In the upper panel, RLRs were proposed to be activated by ERVs upon BCR engagement of TI-2 type ligands (left) or DNMTi-mediated epigenetic remodeling of the genome of the cancer cells. In the lower panel, RLRs were proposed to be activated by SINEs upon viral infection (left) or in the absence of ADAR1-mediated RNA modification.
RIG-I and MDA5 play nonredundant roles by recognizing largely distinct groups of viral RNAs. RIG-I senses relatively short duplexed regions of RNA with 5′-triphosphate (5′-ppp) or diphosphate (5′-pp) groups, which are often present at the end of many viral genomic RNAs (Baum, Sachidanandam, & Garcia-Sastre, 2010; Goubau et al., 2014; Martinez-Gil et al., 2013; Pichlmair et al., 2006; Schlee et al., 2009; Xu et al., 2015). While all nascent transcripts (both cellular and viral) contain 5′-ppp, cellular RNAs normally undergo 5′-processing and therefore a removal of 5′-ppp prior to their nuclear export, which enables them to escape detection by RIG-I. In contrast to RIG-I, viral RNA recognition by MDA5 does not depend on 5′-ppp, but instead on the length of the RNA duplex region. MDA5 recognizes several kilobase-long duplex RNAs formed during replication of many positive strand RNA viruses (eg, picornaviruses; Feng et al., 2012; Kato et al., 2008; Triantafilou et al., 2012), although other types of viral RNAs were also reported to activate MDA5 (Deddouche et al., 2014; Runge et al., 2014). Cellular dsRNAs or hairpin structures are thought to be significantly shorter than a kilobase, which presumably helps escaping detection by MDA5. This distinction between self- and nonself-RNAs, however, is not absolute for either RIG-I or MDA5. Accumulating evidence suggests that RIG-I and MDA5 can also sense cellular RNAs, in particular retroelements, in various physiological contexts.
One of the first examples supporting this notion came fromthe investigation of the B cell activation mechanism (Zeng et al., 2014). B cell-dependent antibody response can occur in either the T helper cell-dependent or -independent manner. The T cell-independent B cell activation often requires TLR activation (termed TI-1 type activation). However, when B cell is stimulated with multivariate ligands such as bacterial polysaccharides and viral capsids, TLR is not required (TI-2 type activation). A recent study showed that the TI-2 type activation depends on MAVS, suggesting the involvement of RIG-I and/or MDA5 (Zeng et al., 2014). A protein, STING, was also shown to be required, which is the downstream adaptor of the cytosolic DNA sensor, cGAS. Detailed analysis revealed that the BCR engagement of TI-2 type ligands and the subsequent activation of NF-κB leads to transcriptional activation of ERVs, and that ERVs in turn activate RIG-I/MDA5/MAVS and cGAS/STING pathways (Fig. 2B) (Zeng et al., 2014). While activation of cGAS/STING could be inhibited by RT inhibitors, as expected from the DNA specificity of cGAS, the activity of RIG-I/MDA5/MAVS was independent of the RT inhibitors, suggesting that the active replication of ERV is not required, but its transcription likely suffices. Although ERV RNAs were copurified with RIG-I, it is unclear whether they in fact can directly activate RIG-I (or MDA5) and what features of ERVs allow recognition by RIG-I (or MDA5).
The role of ERVs in the RIG-I/MDA5 activation was also proposed in the case of cancer cells treated with DNA methyltransferase (DNMT) inhibitors, such as 5-azacytidine (Aza) and 5-aza-20-deoxycytidine (Chiappinelli et al., 2016; Roulois et al., 2015). DNMT inhibitors are common chemotherapeutic agents, but the exact mode of their action had been poorly understood. Recent mechanistic investigations led to two independent reports, which showed that the Aza treatment upregulates ERVs and this in turn activates MDA5 and TLR3 (Fig. 2B) (Chiappinelli et al., 2016; Roulois et al., 2015). The degree at which Aza induced the antiviral immune response correlated with the responsiveness to the immune checkpoint therapy, suggesting that harnessing the MDA5 and TLR3 pathways could have a therapeutic benefit. Although the exact mechanism by which ERVs are sensed byMDA5 and TLR3 is yet unclear, the observation that Aza treatment induces bidirectional transcription of ERVs (Chiappinelli et al., 2016) led to the speculation that the duplex formed between the sense and antisense transcripts activate MDA5 and TLR3 (Fig. 2B). It remains to be investigated, however, whether the sense:antisense duplex is in fact formed in the cells, and whether the duplex is long enough to activate MDA5.
Could other retroelements beside ERVs act as additional endogenous ligands for RIG-I/MDA5? Multiple lines of evidence suggest that SINEs, constituting more than 10% of the human and mouse genome, could serve this function. As mentioned in Section 1, SINEs can be transcribed either by pol II in the form of introns or UTRs, or by pol III as distinct independent transcripts. It is the latter that can be integrated into new genomic loci as the former lacks features recognized by RT and endonuclease encoded by LINEs. Accordingly, transcription of SINEs, in particular the pol III-derived SINEs, is epigenetically silenced in most tissues unless reactivated by a variety of environmental stresses (Varshney et al., 2015). These stimuli include heat shock, viral infection, and treatment with DNA-damaging agents, including Aza (Fornace &Mitchell, 1986; Hagan & Rudin, 2007; Leonova et al., 2013; Li et al., 1999; Liu et al., 1995). In fact, Aza-mediated IFN induction has been also ascribed to pol III-derived SINEs (Leonova et al., 2013) as well as bidirectional ERV transcription (Chiappinelli et al., 2016).
Evidence linking SINEs to RIG-I/MDA5 came from studies of cellular responses to viral infection. A herpes simplex virus immediate-early protein, ICP27, was shown to stimulate pol III-mediated transcription of Alu elements through the activation of the pol III general transcription factor, TFIIIC (Jang & Latchman, 1992). Similar induction of pol III-mediated SINEs was observed upon infection with murine gammaherpesvirus 68 (MHV68; Karijolich, Abernathy, & Glaunsinger, 2015). In this case, upregulation of SINEs, in particular mouse B2 elements, led to the activation of NF-κB in a MAVS-dependent and -independent manners (Fig. 2B). Activated NF-κB is in turn co-opted for the transcription of the MHV68 genes, exemplifying a case where virus exploits an aspect of inflammation to its own advantage. Given the wide range of environmental stresses that upregulate SINEs, similar inflammatory reactions may underlie a variety of cellular stress responses. The question remains, however, as to how SINEs transcribed by pol III, not pol II, activate RIG-I and/or MDA5. It is worth noting that pol III is functional in both the cytosol and the nucleus and that cytosolic pol III can generate RIG-I ligands when provided with the appropriate DNA template (Ablasser et al., 2009; Chiu, Macmillan, & Chen, 2009). It remains to be seen whether there is any mechanistic commonality shared between pol III-mediated cytosolic sensing by RIG-I and pol III-transcribed SINE sensing by RIG-I/MDA5.
The above studies illustrate the role of self-recognition by RIG-I and MDA5 in normal physiological response to cellular and environmental stresses. Could such self-recognition be involved in pathogenesis of immune disorders? While there are clear links between RIG-I, MDA5, and various autoinflammatory and autoimmune diseases, the mechanism by which RIG-I/MDA5 are activated in the disease process is still debated. In mouse, a chemically induced single point mutation or overexpression of MDA5 led to de novo development of autoinflammation or acceleration of the existing autoimmune pathology (Crampton et al., 2012; Funabiki et al., 2014). In human, multiple genome wide association studies have identified the links between MDA5 (encoded by the gene IFIH1) and type 1 diabetes, SLE, and various forms of arthritis (Cunninghame Graham et al., 2011; Nejentsev et al., 2009; Stuart et al., 2015). A stronger causal relationship was established from studies of Mendelian genetics of Aicardi– Goutieres Syndromes, Singleton–Merten syndrome, and SLE (Jang et al., 2015; Oda et al., 2014; Rice et al., 2014; Rutsch et al., 2015; Van Eyck et al., 2015). In these cases, several rare gain-of-function mutations in MDA5 and RIG-I strongly correlated with the high level of type I interferon in peripheral blood, although not all individuals with the interferon signature displayed the disease symptoms (Rice et al., 2014). It is currently debated whether hyperactivation of MDA5 and RIG-I in these patients are due to loss of autorepression or errorneous recognition of endogenous RNA (Funabiki et al., 2014; Lassig et al., 2015; Oda et al., 2014; Rice et al., 2014).
The potential role of retroelements in RIG-I/MDA5 activation was proposed from a study of the ADAR1 deficiency. ADAR1 is a dsRNA-dependent adenosine deaminase that modifies adenosines into inosines embedded within the duplex RNA. The modification occurs in a stochastic and nondiscriminatory fashion, although some sequence preference was observed especially in the context of imperfect duplexes (Bass, 2002). The best-known targets of ADAR1 in human and mouse are SINE RNAs—more specifically, those that occur in the inverted repeat configuration within the same RNA molecule (Fig. 2B) (Carmi, Borukhov, & Levanon, 2011; Chen, DeCerbo, & Carmichael, 2008). In human, the inverted repeat Alu RNAs (IR-Alu’s) form ~300 bp long fold-back hairpin structures and are often found in the 3′-UTR of mRNAs. The lack of ADAR1 or its enzymatic activity in mouse leads to embryonic lethality due to high level of inflammation and failure in hematopoietic stem cells (Hartner et al., 2008; Wang et al., 2000). In human, loss-of-function mutations of ADAR1 cause AGS (Rice et al., 2010), further supporting the role of ADAR1 in suppressing the type I interferon immunity. Three recent publications reported that the inflammatory signatures of ADAR1 knockout can be reversed (or at least partially relieved) by double deletion of MDA5 or MAVS (Liddicoat et al., 2015; Mannion et al., 2014; Pestal et al., 2015). Although the exact mechanism of how MDA5 is activated in deamination-defective cells is yet unclear, the deamination activity of ADAR1 against a subset of pol II-transcribed IR-SINEs led to the hypothesis that IR-SINEs activate MDA5 in the absence of ADAR1-editing (Fig. 2B) (George et al., 2016; Liddicoat et al., 2015). This, however, remains to be tested.
4. PROTEIN KINASE R
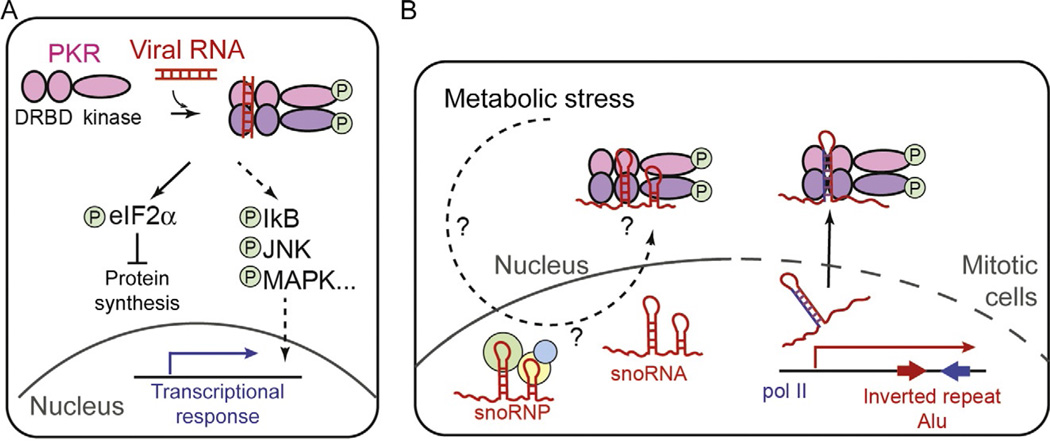
The PKR is a dsRNA-dependent kinase with N-terminal dsRNA-binding domains and a C-terminal kinase domain. It is often categorized as both a viral RNA receptor and an antiviral effector molecule due to its ability to sense the presence of viral RNAs and to directly suppress viral replication. In the absence of viral infection, PKR normally exists in the inactive monomeric form. Upon dsRNA binding, PKR phosphorylates itself and dimerizes to form an active kinase (Fig. 3A) (Lemaire & Cole, 2008; McKenna et al., 2007a, 2007b). Activated PKR then phosphorylates several cellular proteins, of which the best-known target is the alpha subunit of the translational initiation factor, eIF2 (Fig. 3A). The unphosphorylated eIF2α allows the eIF2 complex to deliver the initiator tRNA to the ribosome, whereas the phosphorylated eIF2α does not. Thus, activation of PKR leads to the global shutdown of protein synthesis, both host and virus, and in some cases triggers cell death.
Fig. 3.
(A) Schematic of the domain architecture and activation mechanism of PKR. Circled letter “P” indicates phosphorylation. (B) Proposed mechanisms by which PKR senses self-RNAs. Two examples are shown: PKR was proposed to sense snoRNAs in response to metabolic stress (left) and inverted repeat Alu elements (IR-Alu’s) during mitosis (right). It was proposed that mitotic access to IR-Alu's is possible due to mitotic breakdown of the nuclear envelope (indicated by the dotted line). It is unclear how PKR gains access to snoRNAs that are primarily nuclear, and whether this interaction is also dependent on the cell cycle.
In addition to eIF2α, PKR has been reported to phosphorylate a number of other proteins, thereby regulating multiple cellular processes beyond protein synthesis. The proposed PKR substrates include the inhibitor of NF-κB (IκB), p38, c-Jun N-terminal kinase (JNK), and mitogen-activated protein kinases (MAPKs) (Fig. 3B), although some of these may not be the direct targets of PKR (Marchal et al., 2014; McAllister et al., 2010; Zhang et al., 2009). PKR is also one of the major factors that drive formation of stress granules upon viral infection (Lloyd, 2013), which are thought to be the storage sites for translationally stalled mRNAs. Finally, a recent report suggested that PKR is an important requirement for the functioning of MDA5 (Pham et al., 2016), which adds another layer of complexity to the role of PKR in antiviral defense.
Early studies of PKR have shown that activation of PKR requires dsRNA of at least ~30 bp in length (Lemaire & Cole, 2008; Manche et al., 1992; Minks et al., 1979). This is consistent with the idea that RNA-mediated juxtaposition of at least two PKR molecules are required for its autophosphorylation and dimerization, the prerequisite for the activation (Lemaire & Cole, 2008). This notion is further supported by the observation that an excess amount of dsRNA suppresses PKR, presumably by breaking apart PKR molecules and preventing the dimer formation (Lemaire & Cole, 2008). Interestingly, this length requirement no longer applies when RNA contains a 5′-ppp group (Nallagatla et al., 2007). That is, a short stem loop (as short as ~15 bp) can stimulate PKR as long as it contains 5′-ppp and single-stranded overhangs at the 5′- and 3′-ends. While the dependence on the 5′-ppp is shared with RIG-I, the importance of the end overhangs makes the substrate specificity of PKR distinct from that of RIG-I.
Studies showed that PKR can not only respond to viral infection but also to a variety of cellular and environmental stresses, such as excess nutrient, metabolic abnormalities, and ER stress (Marchal et al., 2014). In both human and mouse, there is a correlation between obesity and hyperactivation of PKR (Cavalho et al., 2013; Nakamura et al., 2010). Activated PKR in the obesity mouse model induces phosphorylation of JNK and insulin receptor substrate 1 (IRS-1), leading to the diminished insulin sensitivity and glucose tolerance (Cavalho et al., 2013; Nakamura et al., 2010). Interestingly, stimulation of PKR by high-fat diet or its metabolic mimetic, free palmitic acid, is dependent on the ability of PKR to bind RNA, which led to the notion that PKR is activated by endogenous RNA (Nakamura et al., 2010). Analysis of RNA copurified with PKR identified small nucleolar RNAs (snoRNAs) as the major constituent of the PKR-bound species (Youssef et al., 2015). The snoRNAs are noncoding RNAs involved in posttranscriptional modification of ribosomal and spliceosomal RNAs. Although the levels of snoRNAs are unaffected by palmitic acid, the association between PKR and snoRNAs was found to be dependent on palmitic acid (Fig. 3B). It is yet unclear how nuclear snoRNAs (or snoRNPs) gain access to cytosolic PKR and how this interaction is regulated by the metabolic stress (Fig. 3B).
The role of endogenous RNA in virus-independent activation of PKR was also proposed in the context of mitotic translational control. Examination of the PKR activity during cell cycle revealed that PKR is transiently activated during mitosis and suppresses global protein synthesis (Kim et al., 2014). The mitotic activation of PKR was found to be sensitive to actinomycin D and ribonucleases, which led to the model that RNA transcribed during mitosis activates PKR. Interestingly, IR-Alu’s, which were previously implicated in PKR activation (Elbarbary et al., 2013), were found to associate with PKR only during mitosis, but not in the interphase (Kim et al., 2014). Because IR-Alu’s are known to be preferentially localized in the nucleus (Chen et al., 2008), it was proposed that the mitotic breakdown of the nuclear envelope allows their access to the cytosolic PKR (Fig. 3B). Interestingly, Kim et al. (2014) found that activated PKR (ie, phospho-PKR) co-localizes with the chromosomes on the metaphase plate during mitosis, the distribution distinct from those of IR-Alu’s. It remains to be investigated whether this chromosomal localization of PKR is involved in its activation mechanism and if so, whether additional RNAs beside IR-Alu’s are also involved in PKR activation.
5. INFLAMMASOME
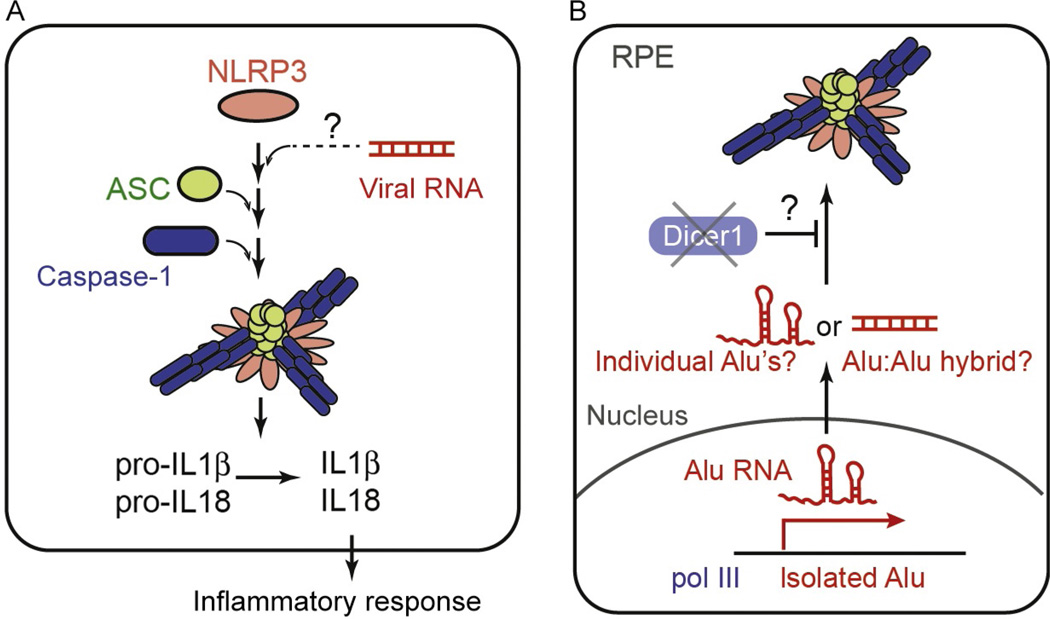
Inflammasome refers to a class of cytosolic molecular assemblies that form in response to a variety of microbial infection. These assemblies typically include nucleotide-binding domain/leucine-rich-repeat-containing receptor (NLR) or an AIM2-like receptor; the adaptor protein, ASC; and the effector protein, caspase-1 (Fig. 4A). Upon their assemblies, inflammasomes activate caspase-1, which in turn cleaves pro-IL-1β or pro-IL-18 to produce the mature proinflammatory cytokines (Fig. 4A). While most inflammasomes share the same effector molecules and perform similar downstream functions, individual inflammasomes play distinctive roles by recognizing different ligands using different receptor proteins. For example, the receptor AIM2 induces the inflammasome assembly upon binding to cytosolic DNAs, while the receptor NAIP2 does upon recognition of bacterial type III secretion systems (Hu et al., 2015; Zhang et al., 2015). Among the inflammasomes characterized to date, only a small number of them, most notably the one with NLRP3, have been implicated in viral RNA detection (Franchi et al., 2014; Li et al., 2015; Mitoma et al., 2013; Wang et al., 2015). Stimulants of the NLRP3 inflammasome, however, are not restricted to viral RNAs but include a variety of molecules or cellular conditions resulting from the loss of cellular homeostasis (Rathinam & Fitzgerald, 2016). These stimuli, so-called danger signals, can be derived directly or indirectly from lysosomal disruption, extracellular ATP, potassium ion efflux, etc. While the mechanism by which the NLRP3 inflammasome responds to the diverse stimuli with vastly distinct physicochemical properties is yet unclear, recent reports suggested that many of these stimuli converge on the potassium ion efflux (Munoz-Planillo et al., 2013), which are then sensed by another molecule upstream to NLRP3 (He et al., 2016; Schmid-Burgk et al., 2016; Shi et al., 2016).
Fig. 4.
(A) Schematic of the components and activation mechanism of the NLRP3 inflammasome. (B) A proposed mechanism by which the NLRP3 inflammasome is activated by self-RNAs. The Dicer1 deficiency was shown to cause accumulation of pol III-transcribed Alu elements, and this in turn was proposed to activate NLRP3 in RPE cells. It is unclear exactly how Alu can stimulate NLRP3; whether the stimulation involves the secondary structure within the individual Alu elements or intermolecular hybridization of distinct Alu elements.
Can endogenous RNA be another form of the danger signal that activates NLRP3? This possibility was examined in the context of geographic atrophy (GA), an advanced form of age-related macular degeneration, characterized by degeneration of the retinal pigmented epithelium (RPE). It was reported that RPEs from GA patients have a low level of Dicer1, a dsRNA-specific ribonuclease involved in miRNA biogenesis (Kaneko et al., 2011). Kaneko et al. further showed that the Dicer1 deficiency causes accumulation of dsRNA made of SINEs (Alu RNAs in human and B1 and B2 RNAs in mouse). These SINE RNAs in turn induce the activation of the NLRP3 inflammasome (Tarallo et al., 2012) and consequently, inflammation and cytotoxicity in RPE (Fig. 4B). Interestingly, pol III-transcribed Alu RNAs, instead of pol II-transcribed Alu’s (which is the more abundant form of Alu), were proposed to be the source of the cytotoxicity (Kaneko et al., 2011). Considering that pol III-transcribed Alu transcripts normally contain a single Alu element, not IR-Alu’s, it is unclear exactly how Alu can stimulate NLRP3; whether the stimulation involves the secondary structure within the individual Alu elements or intermolecular hybridization of distinct Alu transcripts (Fig. 4B; Gong & Maquat, 2011). It also remains to be investigated as to how Dicer1 regulates the level of Alu RNAs and whether it involves a direct cleavage by Dicer1.
6. CONCLUSION AND PERSPECTIVES
We here described the interactions between endogenous retroelements and the innate immune system, including TLRs, RLRs, PKR, and the NLRP3 inflammasome. While these receptors were originally characterized as innate immune sensors for viral RNAs, accumulating evidence suggests that they also react to endogenous RNAs during both pathogenesis of immune disorders and proper functioning of the immune system. Considering that these sensors are known to detect viral RNAs, it is not surprising to see ERVs and other retroelements having been the prime suspects as the endogenous ligands. The notion is especially attractive because some of these retroelements, in particular the active ERVs and SINE/LINEs, are normally transcriptionally silenced, but are upregulated in response to a variety of cellular stresses, the phenomenon that fits into the concept of “danger signals.” This notion has been applied not only to those active retroelements with the capability to replicate inside the host but has also been extended to those that are inactive, ubiquitously expressed, and bear little similarity to viruses.
The effort to define the causality between retroelements and immune activation, however, has been challenging. This reflects in large part the lack of feasibility to genetically deplete these elements and test their impact. The shear copy number of any one class of these elements in the genome not only exceeds far beyond the current technical limit, but such massive genetic manipulation, even if possible, would have a profound biological impact that would be difficult to dissect. For this reason, biochemical assays such as coimmunoprecipitation (co-IP) have been the primary method of choice. However, it is important to note that physical association is hardly a proof of their activity against these innate immune receptors. Many of these receptors have high-affinity binding to nonagonist RNAs as well as agonist RNAs, largely due to the electrostatic nature of their interactions. Thus, additional discrimination steps (such as conformational and kinetic discrimination) upon initial binding likely play critical roles, and this view is supported by multiple structural and biochemical studies of these receptors.
Then, how can one identify the endogenous RNA ligands for the innate immune sensors? Can one specifically pull-down the active form of the receptor to distinguish between the agonist and nonagonist RNAs? While it is theoretically possible, transient, or dynamic nature of many of these interactions (del Toro Duany, Wu, & Hur, 2015; McKenna et al., 2007b) could make implementation of this idea technically challenging. Proving the activity of a bound RNA in physiological condition is also nontrivial as a wide range of RNAs can artificially activate these receptors when introduced in isolation or at high concentration. Thus, it is necessary to examine the functional consequence of depleting the RNA of interest in order to examine its physiological importance. In the case of retroelements, one way to achieve this is to use cellular extract or permeabilized cells, where rapid depletion of a large population of RNA is feasible without eliciting confounding side effects. We await for additional technological and conceptual advances that could further advance our understanding of the relationship between retroelements and the innate immune system.
REFERENCES
- Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunology. 2009;10(10):1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nature Immunology. 2006;7(1):49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annual Review of Biochemistry. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2010;108:3092. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard JJ, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nature Medicine. 2012;18:1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, et al. Extracellular RNA facilitates hypoxia-induced leukocyte adhesion and infiltration in the lung through TLR3-IFN-γ-STAT1 signaling pathway. European Journal of Immunology. 2015;45:3158–3173. doi: 10.1002/eji.201545597. [DOI] [PubMed] [Google Scholar]
- Brentano F, et al. RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via Toll-like receptor 3. Arthritis and Rheumatism. 2005;52:2656–2665. doi: 10.1002/art.21273. [DOI] [PubMed] [Google Scholar]
- Carmi S, Borukhov I, Levanon EY. Identification of widespread ultra-edited human RNAs. PLoS Genetics. 2011;7(10):e1002317. doi: 10.1371/journal.pgen.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalho BM, et al. Modulation of double-stranded RNA-activated protein kinase in insulin sensitive tissues of obese humans. Obesity (Silver Spring) 2013;21:2452–2457. doi: 10.1002/oby.20410. [DOI] [PubMed] [Google Scholar]
- Cavassani KA, et al. TLR3 is an endogenous sensor of tissue necrosis during acute inflammatory events. The Journal of Experimental Medicine. 2008;205:2609–2621. doi: 10.1084/jem.20081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. The EMBO Journal. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2016;164(5):1073. doi: 10.1016/j.cell.2015.10.020. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Crampton SP, et al. Ifih1 gene dose effect reveals MDA5-mediated chronic type I IFN gene signature, viral resistance, and accelerated autoimmunity. The Journal of Immunology. 2012;188:1451–1459. doi: 10.4049/jimmunol.1102705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunninghame Graham DS, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genetics. 2011;7(10):e1002341. doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007a;27:801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007b;27(5):801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deddouche S, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Research. 2002;12(10):1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- del Toro Duany Y, Wu B, Hur S. MDA5-filament, dynamics and disease. Current Opinion in Virology. 2015;12:20–25. doi: 10.1016/j.coviro.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, et al. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. European Journal of Immunology. 2006;36(12):3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- Doring Y, et al. Human antiphospholipid antibodies induce TNFalpha in monocytes via Toll-like receptor 8. Immunobiology. 2010;215(3):230–241. doi: 10.1016/j.imbio.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Downey RF, et al. Human endogenous retrovirus K and cancer: Innocent bystander or tumorigenic accomplice? International Journal of Cancer. 2015;137(6):1249–1257. doi: 10.1002/ijc.29003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, et al. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes and Development. 2013;27(13):1495–1510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser SJ, et al. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522(7555):240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE, Barton GM. Nucleic acid sensing Toll-like receptors in autoimmunity. Current Opinion in Immunology. 2011;23(1):3–9. doi: 10.1016/j.coi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Reports. 2012;29:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace AJ, Jr, Mitchell JB. Induction of B2 RNA polymerase III transcription by heat shock: Enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Research. 1986;14(14):5793–5811. doi: 10.1093/nar/14.14.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbach A, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. The Journal of Immunology. 2008;180(6):3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- Franchi L, et al. Cytosolic double-stranded RNA activates the NLRP3 inflammasome via MAVS-induced membrane permeabilization and K+ efflux. The Journal of Immunology. 2014;193(8):4214–4222. doi: 10.4049/jimmunol.1400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki M, et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Garcia-Ortiz H, et al. Association of TLR7 copy number variation with susceptibility to childhood-onset systemic lupus erythematosus in Mexican population. Annals of the Rheumatic Diseases. 2010;69:1861–1865. doi: 10.1136/ard.2009.124313. [DOI] [PubMed] [Google Scholar]
- George CX, et al. Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. The Journal of Biological Chemistry. 2016;291(12):6158–6168. doi: 10.1074/jbc.M115.709014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, et al. Activation of autoreactive B cells by endogenous TLR7 and TLR3 RNA ligands. The Journal of Biological Chemistry. 2012;287:39789–39799. doi: 10.1074/jbc.M112.383000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C, et al. RNA recognition by human TLR8 can lead to autoimmune inflammation. The Journal of Experimental Medicine. 2013;210(13):2903–2919. doi: 10.1084/jem.20131044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CR, Rudin CM. DNA cleavage and Trp53 differentially affect SINE transcription. Genes, Chromosomes & Cancer. 2007;46(3):248–260. doi: 10.1002/gcc.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner JC, et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nature Immunology. 2008;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, et al. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530(7590):354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350(6259):399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- Hung T, et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350:455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KL, Latchman DS. The herpes simplex virus immediate-early protein ICP27 stimulates the transcription of cellular Alu repeated sequences by increasing the activity of transcription factor TFIIIC. The Biochemical Journal. 1992;284(Pt. 3):667–673. doi: 10.1042/bj2840667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M-A, et al. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. The American Journal of Human Genetics. 2015;96:266–274. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Abernathy E, Glaunsinger BA. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-kappaB pathway. PLoS Pathogens. 2015;11(11):e1005260. doi: 10.1371/journal.ppat.1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, et al. mRNA is an endogenous ligand for Toll-like receptor 3. The Journal of Biological Chemistry. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- Kariko K, et al. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kassiotis G, Stoye JP. Immune responses to endogenous retroelements: Taking the bad with the good. Nature Reviews. Immunology. 2016;16(4):207–219. doi: 10.1038/nri.2016.27. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: Cytoplasmic sensors for non-self RNA. Immunological Reviews. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. The Journal of Experimental Medicine. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, et al. PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes and Development. 2014;28(12):1310–1322. doi: 10.1101/gad.242644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Seminars in Cancer Biology. 2010;20(4):211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassig C, et al. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife. 2015;4:e10859. doi: 10.7554/eLife.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CM, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. The Journal of Experimental Medicine. 2005;202(9):1171–1177. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416(6881):603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Lee HK, et al. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lemaire PA, Cole JL. Mechanism of PKR activation by dsRNA. Journal of Molecular Biology. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JN, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(1):258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(1):E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, et al. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239(2):367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- Li J, et al. DDX19A senses viral RNA and mediates NLRP3-dependent inflammasome activation. The Journal of Immunology. 2015a;195(12):5732–5749. doi: 10.4049/jimmunol.1501606. [DOI] [PubMed] [Google Scholar]
- Li W, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Science Translational Medicine. 2015b;7(307):307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349(6252):1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WM, et al. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Research. 1995;23(10):1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320(5874):379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RE. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdisciplinary Reviews. RNA. 2013;4(3):317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, et al. Interactions between double-stranded RNA regulators and the protein kinase DAI. Molecular and Cellular Biology. 1992;12(11):5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion NM, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Reports. 2014;9(4):1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal JA, et al. The impact of PKR activation: From neurodegeneration to cancer. The FASEB Journal. 2014;28:1965–1974. doi: 10.1096/fj.13-248294. [DOI] [PubMed] [Google Scholar]
- Martinez-Gil L, et al. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. Journal of Virology. 2013;87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, et al. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. Journal of Virology. 2010;84(1):380–386. doi: 10.1128/JVI.02630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna SA, et al. Biophysical and biochemical investigations of dsRNA-activated kinase PKR. Methods in Enzymology. 2007a;430:373–396. doi: 10.1016/S0076-6879(07)30014-1. [DOI] [PubMed] [Google Scholar]
- McKenna SA, et al. Molecular framework for the activation of RNA-dependent protein kinase. The Journal of Biological Chemistry. 2007b;282(15):11474–11486. doi: 10.1074/jbc.M700301200. [DOI] [PubMed] [Google Scholar]
- Migliorini P, et al. Anti-Sm and anti-RNP antibodies. Autoimmunity. 2005;38(1):47–54. doi: 10.1080/08916930400022715. [DOI] [PubMed] [Google Scholar]
- Minks MA, et al. Structural requirements of double-stranded RNA for the activation of 2′-5′-Oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. The Journal of Biological Chemistry. 1979;254:10180–10183. [PubMed] [Google Scholar]
- Mita P, Boeke JD. How retrotransposons shape genome regulation. Current Opinion in Genetics and Development. 2016;37:90–100. doi: 10.1016/j.gde.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H, et al. The DHX33RNAhelicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39(1):123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Malik HS. Hide and seek: How chromatin-based pathways silence retroelements in the mammalian germline. Current Opinion in Genetics and Development. 2016;37:51–58. doi: 10.1016/j.gde.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, et al. Necrosis-induced TLR3 activation promotes TLR2 expression in gingival cells. Journal of Dental Research. 2015;94:1149–1157. doi: 10.1177/0022034515589289. [DOI] [PubMed] [Google Scholar]
- Munoz-Planillo R, et al. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38(6):1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ED, Roths JB. A Y chromosome associated factor in strain BXSB producing accelerated autoimmunity and lymphoproliferation. Arthritis and Rheumatism. 1979;22:1188–1194. doi: 10.1002/art.1780221105. [DOI] [PubMed] [Google Scholar]
- Nakamura T, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140(3):338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallagatla SR, et al. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- Nejentsev S, et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen KK, et al. Endogenous retroviruses and multiple sclerosis-new pieces to the puzzle. BMC Neurology. 2013;13:111. doi: 10.1186/1471-2377-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, et al. Aicardi-Goutières syndrome is caused by IFIH1 mutations. The American Journal of Human Genetics. 2014;95:121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K, et al. Nucleic acid-sensing TLRs and autoimmunity: Novel insights from structural and cell biology. Immunological Reviews. 2016;269(1):60–75. doi: 10.1111/imr.12375. [DOI] [PubMed] [Google Scholar]
- Pestal K, et al. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43(5):933–944. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AM, et al. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathogens. 2016;12(3):e1005489. doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Fitzgerald KA. inflammasome complexes: Emerging mechanisms and effector functions. Cell. 2016;165(4):792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nature Genetics. 2010;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature Genetics. 2014;46(5):503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Runge S, et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathogens. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsch F, et al. A specific IFIH1 gain-of-function mutation causes Singleton-Merten syndrome. The American Journal of Human Genetics. 2015;96:275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, 2006 #166 (PMID: 16858409); Paz, 2011 #167 (PMID: 21200404); Liu, 2013 #558 (PMID: 23951545); Wu, 2015 #s1081 (PMID: 25942693) [Google Scholar]
- Savarese E, et al. U1 small nuclear ribonucleoprotein immune complexes induce type I interferon in plasmacytoid dendritic cells through TLR7. Blood. 2006;107(8):3229–3234. doi: 10.1182/blood-2005-07-2650. [DOI] [PubMed] [Google Scholar]
- Schlee M, et al. Recognition of 5′triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, et al. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. The Journal of Biological Chemistry. 2016;291(1):103–109. doi: 10.1074/jbc.C115.700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nature Immunology. 2016;17(3):250–258. doi: 10.1038/ni.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J, Hur S. Filament assemblies in foreign nucleic acid sensors. Current Opinion in Structural Biology. 2016;37:134–144. doi: 10.1016/j.sbi.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart PE, et al. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. The American Journal of Human Genetics. 2015;97(6):816–836. doi: 10.1016/j.ajhg.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(26):9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntsova M, et al. Molecular functions of human endogenous retroviruses in health and disease. Cellular and Molecular Life Sciences. 2015;72(19):3653–3675. doi: 10.1007/s00018-015-1947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji H, et al. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nature Structural and Molecular Biology. 2015;22(2):109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- Tarallo V, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149(4):847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K, et al. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. Journal of Cell Science. 2012;125(Pt. 20):4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- Van Eyck L, et al. IFIH1 mutation causes systemic lupus erythematosus with selective IgA deficiency. Arthritis & Rheumatology (Hoboken, N.J.) 2015;67:1592–1597. doi: 10.1002/art.39110. [DOI] [PubMed] [Google Scholar]
- Varshney D, et al. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nature Communications. 2015;6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Stetson DB. The enemy within: Endogenous retroelements and autoimmune disease. Nature Immunology. 2014;15(5):415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer J, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. The Journal of Experimental Medicine. 2005;202(11):1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Wang P, et al. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350(6262):826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindel CG, et al. B cell autophagy mediates TLR7-dependent autoimmunity and inflammation. Autophagy. 2015;11:1010–1024. doi: 10.1080/15548627.2015.1052206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Hur S. How RIG-I like receptors activate MAVS. Current Opinion in Virology. 2015;12:91–98. doi: 10.1016/j.coviro.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. Identification of a natural viral RNA motif that optimizes sensing of viral RNA by RIG-I. MBio. 2015;6:e01265–e01215. doi: 10.1128/mBio.01265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef OA, et al. Potential role for snoRNAs in PKR activation during metabolic stress. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(16):5023–5028. doi: 10.1073/pnas.1424044112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37(5):867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Zeng M, et al. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 2014;346:1486–1492. doi: 10.1126/science.346.6216.1486. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang P, et al. Protein kinase PKR-dependent activation of mitogen-activated protein kinases occurs through mitochondrial adapter IPS-1 and is antagonized by vaccinia virus E3L. Journal of Virology. 2009;83(11):5718–5725. doi: 10.1128/JVI.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350(6259):404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]