Summary
The oral anaerobe Porphyromonas gingivalis is associated with the development of cancers including oral squamous cell carcinoma (OSCC). Here we show that infection of gingival epithelial cells with P. gingivalis induces expression and nuclear localization of the ZEB1 transcription factor which controls epithelial-mesenchymal transition (EMT). P. gingivalis also caused an increase in ZEB1 expression as a dual species community with Fusobacterium nucleatum or Streptococcus gordonii. Increased ZEB1 expression was associated with elevated ZEB1 promoter activity and did not require suppression of the miR-200 family of micro RNAs. P. gingivalis strains lacking the FimA fimbrial protein were attenuated in their ability to induce ZEB1 expression. ZEB1 levels correlated with an increase in expression of mesenchymal markers, including vimentin and MMP-9, and with enhanced migration of epithelial cells into matrigel. Knockdown of ZEB1 with siRNA prevented the P. gingivalis-induced increase in mesenchymal markers and epithelial cell migration. Oral infection of mice by P. gingivalis increased ZEB1 levels in gingival tissues, and intracellular P. gingivalis were detected by antibody staining in biopsy samples from OSCC. These findings indicate that FimA-driven ZEB1 expression could provide a mechanistic basis for a P. gingivalis contribution to OSCC.
Introduction
Once considered implausible, the concept that bacteria can be associated with cancer development is now well established. Indeed, a causal relationship between Helicobacter pylori and gastric cancer has been demonstrated (Kim et al., 2011), and a growing body of evidence supports the relationship between specific bacteria and various types of cancer (Garrett, 2015, Sahingur and Yeudall, 2015). For example, Fusobacterium nucleatum, a common inhabitant of the oral cavity, is over-represented in colorectal carcinoma (Castellarin et al., 2012, Kostic et al., 2012) and can induce colorectal carcinogenesis by activating E-cadherin/β-catenin signaling (Rubinstein et al., 2013). F. nucleatum can also inhibit natural killer (NK) cell cytotoxicity and killing of various tumors (Gur et al., 2015). High levels of antibodies to Porphyromonas gingivalis, a keystone pathogen in periodontal diseases, correlate with a greater than 2-fold increased risk of pancreatic cancer (Michaud, 2013). P. gingivalis is also associated with oral squamous cell carcinoma (OSCC). The surfaces of OSCCs harbor higher levels of Porphyromonas compared to contiguous healthy mucosa (Nagy et al., 1998), and P. gingivalis can be detected within gingival carcinomas by immunohistochemistry (Katz et al., 2011). Moreover, recent studies have established that combined infection with P. gingivalis and F. nucleatum promotes tumor progression in an oral-specific chemical carcinogenesis mouse model (Gallimidi et al., 2015).
P. gingivalis and oral epithelial cells engage in an intricate molecular dialog, one consequence of which is entry of bacterial cells into the cytoplasm of the host cell (Lamont and Hajishengallis, 2015, Lamont et al., 1995). Primary cultures of epithelial cells containing P. gingivalis do not undergo apoptotic cell death and indeed P. gingivalis can suppress several proapoptotic pathways. In response to P. gingivalis infection Jak1/Akt/Stat3 signaling is activated with resultant increase in Bcl2 and inhibition of intrinsic mitochondrial apoptotic pathways (Yilmaz et al., 2004, Mao et al., 2007). By an independent mechanism P. gingivalis upregulates the level of miR-203 which suppresses expression of SOCS3, consequently impeding apoptosis (Moffatt and Lamont, 2011). In tandem with suppression of apoptosis, P. gingivalis promotes acceleration of primary epithelial cells through the S-phase of the cell cycle by impacting cyclin/CDK activities and reducing the amount of p53 (Kuboniwa et al., 2008). The process is dependent on the major fimbriae of P. gingivalis as a mutant deficient in FimA, the structural fimbrial subunit protein, does not induce increased cell proliferation.
While inhibition of apoptosis and enhanced replication of cells can contribute directly to tumor development, it is unknown if P. gingivalis is capable of initiating the malignant transformation or oncogenic progression of epithelial cells. The epithelial-mesenchymal transition (EMT) is a process by which epithelial cells change shape and acquire a motile phenotype (Lamouille et al., 2014). The EMT is required for normal development and wound healing; however, it is also associated with the generation of self-renewing tumor-initiating cells, and in a malignant tumor it gives rise to a population of migratory and invasive cancer cells (Lamouille et al., 2014). This switch in cell differentiation and behavior is controlled by a group of transcription factors including the zinc-finger E-box-binding homeobox 1 and 2 proteins (ZEB1/2), SNAIL and TWIST (Vandewalle et al., 2009, Scanlon et al., 2013). The ZEB1 (δEF1, Zfhx1a, Zfhep) and ZEB2 (SIP1) transcription factors are critical EMT activators that bind to 5′-CACCTG sequences and repress transcription of epithelial specific genes such as E-cadherin (cdh1) (Vandewalle et al., 2009). ZEB can also positively regulate genes associated with the mesenchymal phenotype such as those encoding vimentin and matrix-metalloproteinases (Vandewalle et al., 2009, Lamouille et al., 2014). ZEB1/2 are in the TGFβ signaling pathway, binding SMADs and having essential effects on embryonic development (Gheldof et al., 2012). ZEB1 has been implicated in activating EMT and metastasis in several type of cancers (Sanchez-Tillo et al., 2012, Jia et al., 2012). Moreover, ZEB1/2 are linked to the miR-200 family in a reciprocal negative feedback loop whereby each regulates the expression of the other (Brabletz and Brabletz, 2010).
H. pylori has been shown to upregulate expression of ZEB1 which can initiate an EMT and cancer stem-cell properties in infected gastric epithelial cells (Baud et al., 2013, Bessede et al., 2014). In this study we show that P. gingivalis can increase ZEB1 levels in gingival epithelial cells in a fimbriae dependent manner. Upregulation of ZEB1 was dependent on increased promoter activity. Elevated expression of ZEB1 was associated with a partial mesenchymal phenotype in P. gingivalis-infected gingival epithelial cells, including increased migration. We also detected P. gingivalis antigens in oral carcinoma in situ and poorly differentiated cancer, and mice orally infected with P. gingivalis had an increase in ZEB1 mRNA expression in gingival tissues. The results suggest a novel mechanism by which oral bacteria such as P. gingivalis can contribute to a mesenchymal phenotype, and potentially drive the progression of cancer.
Results
P. gingivalis upregulates ZEB1 in gingival epithelial eells
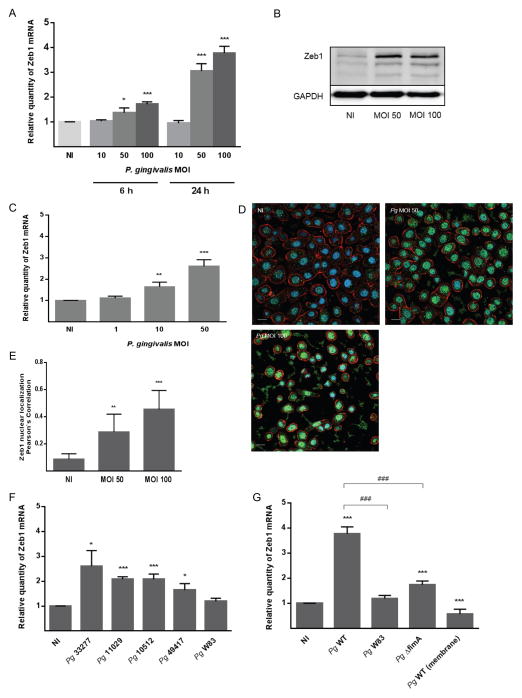
We investigated the impact of P. gingivalis on ZEB1 expression in TIGK cells using qRT-PCR and immunoblotting. As shown in Figure 1A, P. gingivalis increased ZEB1 mRNA levels in a time and dose dependent manner, with maximal induction occurring after 24 h infection with a MOI of 100. An increase in the amount of ZEB1 protein was also observed at 24 h following P. gingivalis infection at both MOI 50 and 100 (Fig 1B). As P. gingivalis infections of oral tissue are chronic conditions, we further examined ZEB1 activity 72 h after P. gingivalis infection. MOIs of 1, 10 and 50 were used as at MOI 100 the proteases of P. gingivalis can cause detachment of cells from the substratum. While an MOI 1 did not affect ZEB1 expression, mRNA levels were increased by P. gingivalis at MOI 10 and 50 (Figure 1C). The ability of P. gingivalis at MOI 10 to increase ZEB1 expression after 72 h, but not earlier, indicates that infection of epithelial cells with low numbers of the organism has the potential to elevate ZEB1 over extended times, possibly as a result of intracellular P. gingivalis replication and cell to cell spread (Lamont et al., 1995, Yilmaz et al., 2006). To corroborate the nuclear location of the ZEB1 transcription factor following P. gingivalis infections, TIGKs were examined by CLSM with quantitative image analysis (Figures 1D and E). After P. gingivalis infection there was increased expression of ZEB1 protein in the nucleus where it is functionally active.
Figure 1. P. gingivalis up-regulates ZEB1 expression in TIGK cells in a FimA-dependent manner.
A. TIGKs were infected with P. gingivalis 33277 at the MOI and time indicated. ZEB1 mRNA levels were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD; n = 3; * P < 0.05; *** P < 0.001.
B. Immunoblot of lysates of TIGK cells infected with P. gingivalis 33277 for 24 h at the MOI indicated. Control cells were uninfected (NI). Duplicate blots were probed with antibodies to ZEB1 or GAPDH (loading control).
C. ZEB1 mRNA levels in TIGKs after 72 h infection with P. gingivalis 33277 at MOI indicated. qRT-PCR data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; ** P < 0.01; *** P < 0.001.
D. Fluorescent confocal microscopy of TIGK cells infected with P. gingivalis 33277 (Pg) at MOI 50 or MOI 100 for 24 h. Control cells were noninfected (NI). Cells were fixed and probed with ZEB1 antibodies (green). Actin (red) was stained with Texas Red-phalloidin, and nuclei (blue) stained with DAPI. Cells were imaged at magnification ×63, and shown are representative merged images of projections of z-stacks obtained with Volocity software. Bar = 10 μm.
E. Nuclear localization of ZEB1 calculated by Pearson’s correlation coefficient from images in C (n=100 cells) using Volocity software. Results are mean ± SD; ** P < 0.01; *** P < 0.001.
F. qRT-PCR of ZEB1 mRNA expression in TIGK cells infected with P. gingivalis (Pg) strains at MOI 100 for 24 h. qRT-PCR data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; * P < 0.05; *** P < 0.001.
G. qRT-PCR of ZEB1 mRNA expression in TIGK cells infected with P. gingivalis 33277 (Pg WT), ΔfimA mutant, or W83, or Pg WT in the presence of membrane insert. qRT-PCR data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; *** P < 0.001 compared to NI; ### P < 0.001 compared to Pg WT.
ZEB1 responses to P. gingivalis are strain and fimbriae dependent
P. gingivalis is a host adapted organism with a nonclonal population structure, and isolates from different individuals often vary considerably (Tribble et al., 2013). Hence, we next examined the ability of different strains of P. gingivalis to enhance ZEB1 mRNA levels. As shown in Figure 1F, an additional ATCC strain (49417) and two low passage clinical isolates (11029 and 10512) induced ZEB1 expression to a similar degree as the type strain 33277. In contrast, the commonly used laboratory strain W83 did not significantly increase ZEB1 expression. One of the major differences among P. gingivalis strains is in the expression of fimbriae (Nadkarni et al., 2014). The two ATCC strains, along with the two low passage isolates, all expressed FimA, the structural subunit protein of the major fimbriae (Supporting Information Figure S1). Strain W83 does not expresses FimA (Nishikawa and Duncan, 2010), which prompted us to speculate that FimA may be an effector protein for ZEB1 induction. This concept was corroborated by the failure of an isogenic fimA mutant of 33277 to increase ZEB1 mRNA levels (Figure 1G). We also found that induction of ZEB1 expression required direct contact between P. gingivalis and epithelial cells (Figure 1G), consistent with a role for the FimA adhesin. Fimbriated P. gingivalis activate JNK signaling (Watanabe et al., 2001) which has been reported to increase transcription of ZEB1 (Zhang et al., 2012). However, siRNA knockdown of JNK in TIGK cells did not impede the ability of P. gingivalis to upregulate ZEB1 (Supporting Information Figure S2). In addition, pharmacological inhibition of Akt with LY294002 also failed to reduce P. gingivalis-mediated ZEB1 induction (Supporting Information Figure S3). Hence the signaling pathways activated by P. gingivalis fimbriae that converge on Zeb1 remain to be determined, and this topic is under active investigation in our laboratory.
The FimA fimbriae are required for maximal invasion of P. gingivalis into gingival epithelial cells (Lamont and Jenkinson, 1998). However, invasion per se was not required for ZEB1 induction as a mutant of P. gingivalis that is invasion-deficient, due to disruption of the gene encoding the serine phosphatase SerB (Takeuchi et al., 2013), retained the ability to upregulate ZEB1 (Supporting Information Figure S4). Nonetheless, the spatial definition of P. gingivalis initiation of ZEB1 activation requires further study. Moreover, when P. gingivalis was separated from the epithelial cells by a semi-permeable membrane, ZEB1 levels were lower than the control uninfected condition, indicating that there may be components secreted by P. gingivalis that can antagonize ZEB1 regulation in the absence of FimA mediated contact. Thus, multiple effectors of P. gingivalis may be capable of impacting ZEB1 expression, with the effect of whole cells representing the collective output of several distinct interactions and signaling pathways.
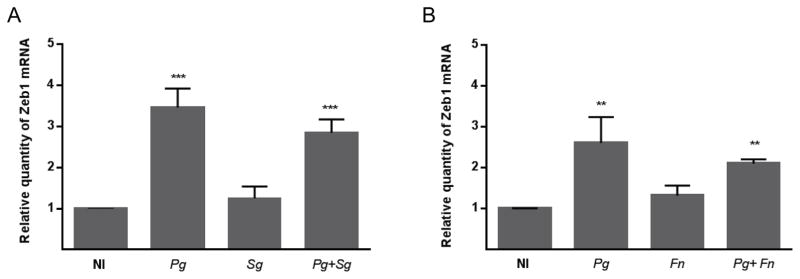
P. gingivalis communities regulate ZEB1 expression
On mucosal surfaces bacteria rarely exist as monospecies accumulations but rather as complex multispecies communities. P. gingivalis engages in synergistic community formation with S. gordonii and F. nucleatum, common inhabitants of the oral microbiota, and in vivo these organisms can be found in close association (Benitez-Paez et al., 2014, Valm et al., 2011, Wright et al., 2014, Hendrickson et al., 2014). Individually, neither S. gordonii nor F. nucleatum were capable of regulating ZEB1 expression, indicating that of these three widespread oral species, P. gingivalis has the most potential to effect an EMT through ZEB1 (Figure 2). Importantly, P. gingivalis remained effective at elevating ZEB1 mRNA in the context of a community with either S. gordonii or F. nucleatum, consistent with recent reports demonstrating that a community of P. gingivalis and F. nucleatum can promote tumor progression in animal models (Gallimidi et al., 2015). Thus, the tumorigenic properties of P. gingivalis can prevail in the presence of co-colonizing organisms, an important principle for in vivo relevance.
Figure 2. P. gingivalis communities regulate ZEB1 expression.
A. qRT-PCR of ZEB1 mRNA expression in TIGK cells infected with P. gingivalis 33277 (Pg), S. gordonii (Sg) or a combination of Pg and Sg at MOI 100 for 24 h.
B. qRT-PCR of ZEB1 mRNA expression in TIGK cells infected with P. gingivalis 33277 (Pg), F. nucleatum (Fn) or a combination of Pg and Fn at MOI 100 for 24 h.
Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD; n = 3; ** P < 0.01; *** P < 0.001.
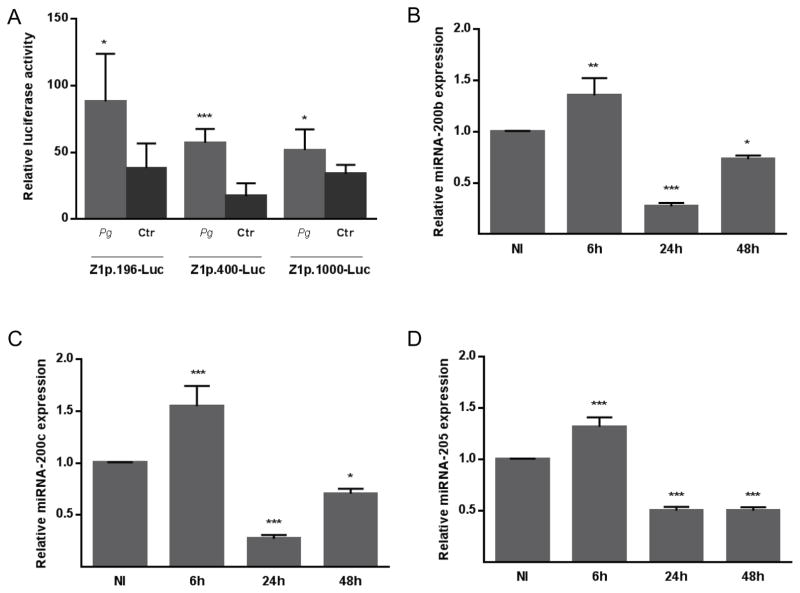
P. gingivalis upregulates ZEB1 promoter activity and downregulates miR200
An increase in the amount of steady state mRNA levels can result from an increase in transcription or a decrease in degradation. To begin to distinguish between these possibilities, we first examined transcriptional activity from the ZEB1 promoter using a series of ZEB1 upstream regulatory regions promoting transcription of the luc gene. These human ZEB1 promoter constructs contain sequences important for regulation of ZEB1 expression in several cell types (Manavella et al., 2007, Liu et al., 2007). P. gingivalis stimulated the activity of all of these promoter constructs (Figure 3A), indicating that the increase of ZEB1 mRNA induced by P. gingivalis can occur through an elevated transcription rate. These data also localize the response element(s) within the first 400 bp of the promoter. An additional mechanism by which ZEB1 is controlled posttranscriptionally is through the action of the miR-200 family of microRNAs (Brabletz and Brabletz, 2010). miR-200 family members target conserved recognition sites on the 3′ UTR of ZEB1 mRNA (Brabletz and Brabletz, 2010), and thus a decrease in miR-200 leads to higher levels of ZEB1 mRNA. However, we did not observe a reduction in the amount of miR-200b, miR-200c or miR-205 in cells at 6 h after P. gingivalis infection when levels of ZEB1 mRNA begin to rise (Figure 3B–D). Indeed there was a slight increase in miR-200 family expression, indicating that the increase in ZEB1 levels at 6 h is not the consequence of decreased miR-200 expression. The action of ZEB1, in turn, represses the transcription of the miR-200 family, and consistent with this at 24 and 48 h after P. gingivalis infection, miR-200b, miR-200c and miR-205 levels were reduced. A control microRNA, miR-21, which is not involved in ZEB1 feedback regulation, did not show a significant decrease in expression (Supporting Information Figure S5). Hence, the pattern of miRNA expression is consistent with the results from the promoter-reporter constructs in pointing toward increased mRNA synthesis as the initial cause of the elevated levels of steady state mRNA for ZEB1.
Figure 3. P. gingivalis regulates ZEB1 promoter activity and increases in ZEB1 levels are not dependent on the miRNA 200 family.
A. TIGK cells were transiently transfected with ZEB1 promoter-luciferase plasmids: Z1p 1000-Luc (−912 bp to +43 bp), Z1p.400-Luc (−367 bp to +43 bp) or Z1p.196-Luc (−212 bp to +43 bp); or with a constitutively-expressing Renilla luciferase reporter. Transfected cells were infected with P. gingivalis 33277 (Pg) at MOI 100 for 24 h. Control cells were noninfected (Ctr). Luciferase activity was normalized to the level of Renilla luciferase. Results are mean ± SD, n = 3; * P < 0.01; *** P < 0.001.
B–D. Expression of mature miRNA-200b (B), miRNA-200c (C), or miRNA-205 (D) in TIGK cells infected with P. gingivalis 33277 MOI 100 for the times indicated. miRNA levels were measured by qRT-PCR, normalized to RNU48 miRNA, and expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; * P < 0.05; ** P < 0.01; *** P < 0.001.
Changes in epithelial and mesenchymal marker expression upon P. gingivalis infection
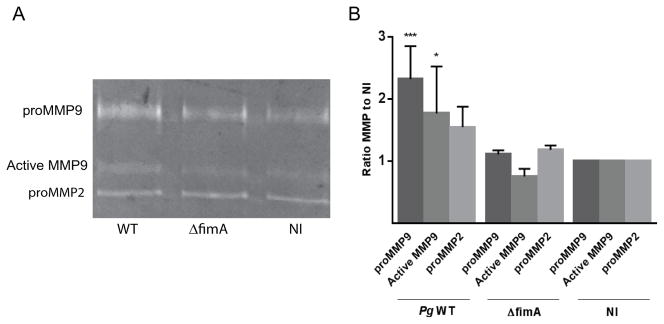
The expression pattern of ZEB1 targets in TIGKs infected with P. gingivalis was assessed by qRT-PCR (Table 1). Mesenchymal markers N-cadherin, vimentin and matrix metalloproteinase (MMP)-9 were upregulated at 24 h after infection by P. gingivalis at MOI 50 and 100, while fibronectin levels were increased by P. gingivalis at MOI 100. An increase in vimentin protein expression was confirmed by immunoblotting (Supporting Information Figure S6), and elevated MMP-9 amounts following infection with fimbriated P. gingivalis was corroborated by zymography (Figure 4). While both pro and active forms of MMP-9 were increased by P. gingivalis wild type, there was no difference in the ratio of active MMP-9 to total (cleaved and pro-MMP9) between parental and fimbrial deficient mutant strains. Under these infection conditions, therefore, fimbriated P. gingivalis elevate the amount of MMP-9 produced by TIGK cells but do not modulate MMP-9 activation. In contrast expression of MMP-2, which may be more predominantly regulated by Twist (Yang et al., 2013), was not impacted by P. gingivalis infection. Of the epithelial markers tested, collagen 1 (COL1A1) and cytokeratin 19 (KRT19) were suppressed by P. gingivalis. Collectively, these results support the concept that infection by P. gingivalis can contribute to the process of transition toward a mesenchymal phenotype. Although the mesenchymal marker integrin α5 (ITGA5) and the epithelial marker cytokeratin 7 (KRT7) were unaffected by P. gingivalis, variability in expression of important cell proteins is not unexpected as control of expression by ZEB1 is cell and context dependent (Lamouille et al., 2014). One of the major targets of ZEB1 is E-cadherin, and ZEB1 mediated repression of E-cadherin, with associated disruption of E-cadherin dependent junctions, is an important marker of EMT. We did not observe differential regulation of E-cadherin following P. gingivalis infection (not shown). However, the gingival epithelium is highly porous with only sparse interconnections (Bosshardt and Lang, 2005) and expression of E-cadherin is very low (Heymann et al., 2001). Thus a reduction of E-cadherin may not be as important for the EMT of gingival epithelial cells as in other cell types.
Table 1.
Changes in expression of mesenchymal and epithelial markers in TIGK cells infected with P. gingivalis 33277.
| Fold change induced by P. gingivalis 33277 | |||
|---|---|---|---|
| MOI 50 | MOI 100 | ||
| Mesenchymal markers | Vimentin | 1.72 ± 0.14 ** | 3.4 ± 0.41 ** |
| ITGA5 | 1.06 ± 0.12 | 1.29 ± 0.23 | |
| MMP-9 | 8.943 ± 0.38 *** | 12.77 ± 0.75 *** | |
| Fibronectin | 1.16 ± 0.05 | 3.13 ± 0.16 *** | |
| N-cadherin | 2.55 ± 0.34** | 2.4 ± 0.17** | |
| Epithelial markers | KRT7 | 0.97 ± 0.09 | 1.29 ± 0.09 |
| KRT19 | 1.2 ± 0.07 | 0.63 ± 0.003 *** | |
| COL-1A1 | 0.63 ± 0.06 ** | 0.67 ± 0.035 *** | |
Data represent qRT-PCR results of the individual genes normalized to that of GAPDH mRNA and expressed relative to noninfected cells. Results are means ± SD, n = 6;
P < 0.01;
P < 0.001
Figure 4. P. gingivalis induces MMP9 expression in a FimA-dependent manner.
TIGKs were infected with P. gingivalis 33277 (WT) or ΔfimA mutant at MOI 10 for 24 h, or left uninfected. A. Culture supernatants were analyzed for MMP9 and MMP2 by gelatin zymography. B. Quantitative analysis of 4 independent zymograms using ImageJ. * P < 0.05; *** P < 0.001.
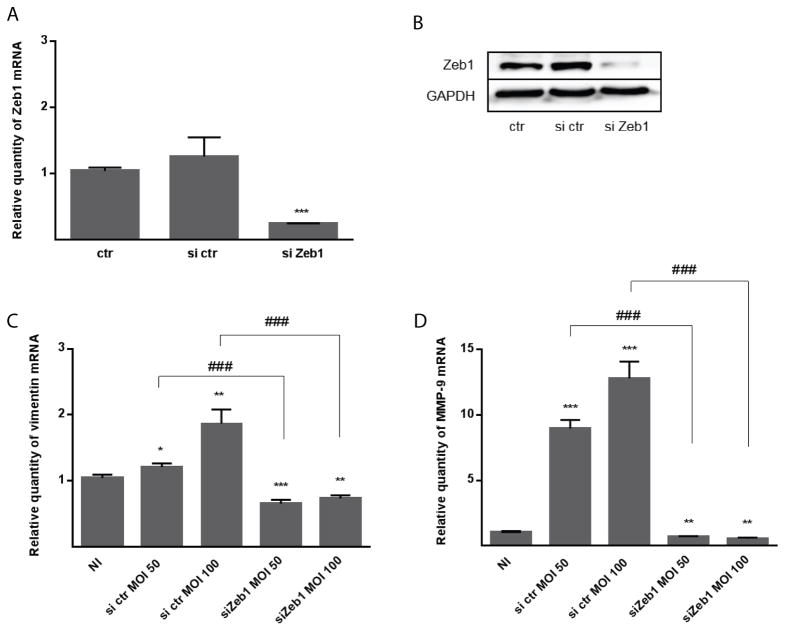
To corroborate the role ZEB1 in the differential regulation of mesenchymal markers, siRNA mediated knockdown was performed. Reduction of ZEB1 mRNA and protein following siRNA transfection was confirmed by qRT-PCR and immunoblotting, respectively (Figure 5A,B). TIGKs with diminished ZEB1 expression were then infected with P. gingivalis MOI 50 or 100 over 24 h. As shown in Figure 5C and D, ZEB1 deficiency prevented P. gingivalis induced modulation of expression of vimentin and MMP-9.
Figure 5. ZEB1 knockdown suppresses TIGK responses to P. gingivalis.
A. TIGK cells were transiently transfected with siRNA to ZEB1 (si Zeb1) or scrambled siRNA (si ctr). Control (ctr) cells were nontransfected. ZEB1 mRNA levels in transfected cells were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to ctr. Results are means ± SD, n = 3; *** P < 0.001.
B. Immunoblot of lysates of TIGK cells transfected (as in A) and probed with antibodies to ZEB1 or GAPDH (loading control).
C–D. Transfected TIGK cells (as in A) were infected with P. gingivalis 33277 for 24 h at the MOI indicated. The Effect of ZEB1 knockdown on expression of vimentin (B) and MMP9 (C) was measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to the noninfected (NI) control. Results are means ± SD, n = 3; * P < 0.05; ** P < 0.01; *** P < 0.001 compared to NI. ### P < 0.001 compared to si ctr.
P. gingivalis promotes migration of epithelial cells
Cells that acquire an EMT phenotype display an invasive behavior in vitro, and thus we tested the ability of P. gingivalis to increase the migration of TIGK cells into matrigel. Figure 6 shows that P. gingivalis infection resulted in a greater than 2-fold increase in TIGK cell invasion into the gel compared to control cells. Knockdown of ZEB1 prevented P. gingivalis-induced TIGK cell migration, verifying the importance of ZEB1 in this aspect of the P. gingivalis-dependent partial mesenchymal phenotype.
Figure 6. P. gingivalis increases TIGK migration in a ZEB1-dependent manner.
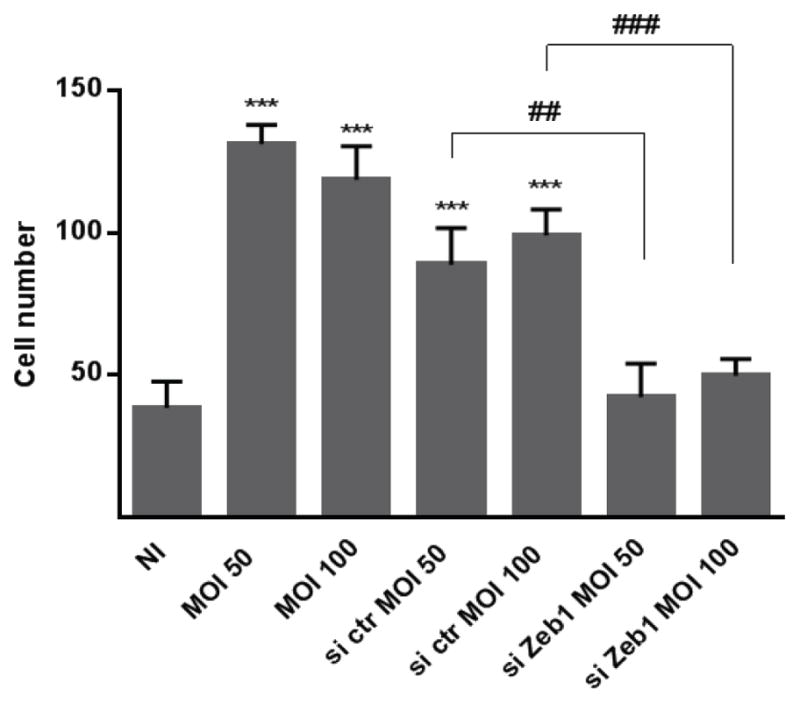
Quantitative analysis of TIGK migration through matrigel-coated transwells. TIGK cells were transiently transfected with siRNA to ZEB1 (si Zeb1) or scrambled siRNA (si ctr). Control cells were nontransfected. TIGKs were infected with P. gingivalis 33277 for 24 h at the MOI indicated. Data are presented as the average number of cells invading through inserts coated with matrigel. Results are means ± SD, n = 3; *** P < 0.001 compared to NI. ### P < 0.001 compared to si ctr.
P. gingivalis elevates ZEB1 levels in vivo
To determine whether P. gingivalis’ ability to increase ZEB1 levels also occurs in vivo, mice were orally infected with P. gingivalis, and gingival tissue recovered 1, 3 and 8 days following the final inoculation. Levels of P. gingivalis on the gingival tissues were determined by qPCR (Supporting Information Figure S7) and remained constant over the 8-day period. As shown in Figure 7, colonization with P. gingivalis induced an increase in gingival tissue expression of ZEB1 mRNA over 8 days compared to sham infected animals. Thus, P. gingivalis has the potential to stimulate ZEB1 and contribute to an EMT in an animal model.
Figure 7. P. gingivalis induces ZEB1 expression in vivo.
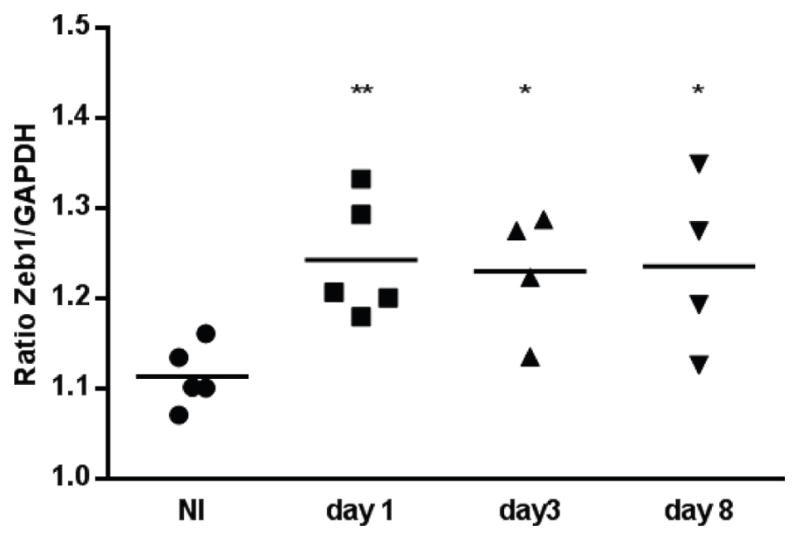
A. qRT-PCR of ZEB1 mRNA expression relative to GAPGH control in gingival tissues from mice infected with P. gingivalis or sham infected (NI). Tissue samples were collected at days 1, 3 and 8 after infection. Each point represents the determination from a single animal. * P < 0.05; ** P < 0.01.
Presence of P. gingivalis in human OSCC
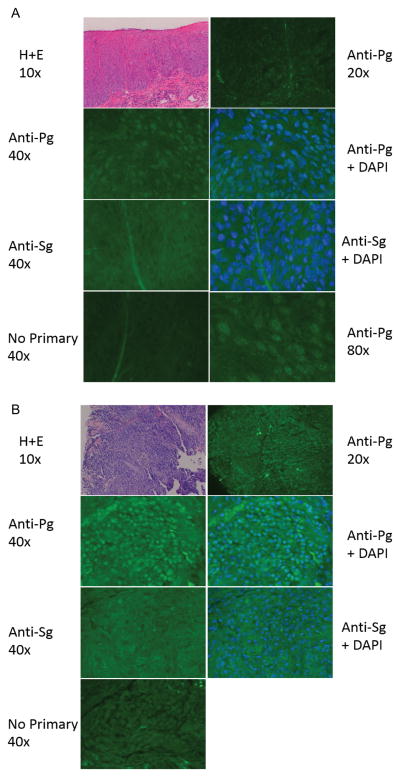
P. gingivalis could exacerbate carcinogenesis at several stages through its ability to increase ZEB1 expression, but only if the bacteria are physically associated with the developing cancer. We investigated whether P. gingivalis bacteria are present within oral squamous cell carcinoma biopsy samples. Immunofluorescence microscopy with a specific polyclonal P. gingivalis antiserum labeled discrete speckles in the cells of a poorly differentiated OSCC sample and a carcinoma in situ, whereas antibodies to S. gordonii showed little or no labeling (Figure 8A–B). Similar results were seen with two carcinoma in situ cases and two poorly differentiated carcinomas. Confocal microscopy more clearly detected the particles which were fluorescently labeled by the P. gingivalis antibodies. Serial optical sections were taken at 0.4 microns, and individual particles were found to persist in 5 to 7 adjacent optical slices (Supporting Information Figure S8). This estimates the fluorescent particles to be 2.0 to 2.8 microns in size, consistent with intact P. gingivalis. As described in primary gingival epithelial cells, P. gingivalis was observed in both the cytoplasm and in the nuclei (Belton et al., 1999).
Figure 8. P. gingivalis antigens are present in OSCC.
Tissue biopsy of (A) poorly differentiated carcinoma, and (B) a tongue carcinoma in situ. Biospsy sections were stained with H&E, P. gingivalis 33277 antibodies (Anti-Pg 1:1000) or S. gordonii antibodies (Anti-Sg 1:1000) in the presence or absence of DAPI. Controls had no primary antibody. Red blood cells in the connective tissue are autofluorescent. Samples were imaged with a Zeiss Axiocam MRc5 fluorescence microscope at the magnification indicated.
Discussion
Typically, the gingival epithelium provides a major physical barrier to oral pathogens. Disruption of the gingival barrier by inducing an EMT may enhance the ability of P. gingivalis to invade the tissue and enhance access to nutrients derived from inflammatory tissue breakdown (Hajishengallis, 2014). Hence, up-regulation of ZEB1 by P. gingivalis can be seen as providing an evolutionary advantage to the organism. Beyond this, the ability of P. gingivalis to stimulate ZEB1 expression could have several distinct clinically relevant effects. ZEB1 influences multiple stages of carcinogenesis, including the initial transformation, progression, EMT leading to metastasis, and resistance to therapy (Sanchez-Tillo et al., 2012, Zhang et al., 2014). Therefore, the presence of P. gingivalis, interacting with other environmental effectors, may enhance the initiation of oral cancer within the pre-cancerous field, or increase carcinogenic progression.
The ability to manipulate ZEB1 location and function constitutes an important attribute of bacteria with a potential role in carcinogenesis. P. gingivalis is a keystone member of dysbiotic oral communities, which in combination with its ability to spread systemically, and enhance cell survival and proliferation, supports epidemiological evidence of an association with cancers such as OSCC (Whitmore and Lamont, 2014, Lamont and Hajishengallis, 2015). Moreover, in established invasive OSCC lines, P. gingivalis activates the ERK1/2-Ets1, p38/HSP27, and PAR2/NF-κB pathways to promote cellular invasion (Inaba et al., 2014). The results of the present study indicate the P. gingivalis may induce nontransformed gingival epithelial cells to undergo a partial EMT through the upregulation of ZEB1. We show that P. gingivalis increases the transcriptional activity of the ZEB1 gene and increases ZEB1 protein levels in the nucleus. Infection of epithelial cells with P. gingivalis upregulated expression of genes associated with the mesenchymal phenotype and knockdown of ZEB1 attenuated this effect. P. gingivalis also induced a migratory phenotype in epithelial cells which was ZEB1-dependent. Oral infection of mice with P. gingivalis stimulated ZEB1 expression in the gingival tissues and biopsy tissue from human OSCC carcinoma in situ and poorly differentiated cancer showed the presence of P. gingivalis.
On the hard and soft tissues of the oral cavity P. gingivalis is an inhabitant of multispecies communities. Organisms such as S. gordonii and F. nucleatum provide mutual physiological support, and P. gingivalis in the context of a community is phenotypically distinct from single species accumulations (Wright et al., 2013). In addition, infection of epithelial cells with the early colonizing streptococci can reprogram specific signaling pathways such that they do not respond to the later colonizing P. gingivalis (Handfield et al., 2005). We found here that while neither S. gordonii nor F. nucleatum modulated ZEB1 mRNA levels, combinations of P. gingivalis with either species retained the capacity to upregulate ZEB1. As porphyromonads, fusobacteria and streptococci are all found in higher numbers on the surfaces of OSCC compared to contiguous healthy mucosa (Nagy et al., 1998), it is likely therefore that these microbial communities contribute to the EMT.
P. gingivalis strains exhibit extensive genetic variation as a result of genomic rearrangements (Naito et al., 2008), and horizontal gene transfer is considered an adaptive strategy for long term survival in the oral environment (Nadkarni et al., 2014, Tribble et al., 2007). Most strains of P. gingivalis expresses fimbriae comprised of FimA major fimbrial subunit proteins, although in some strains, such as the commonly used lab strain W83, FimA production is very low due to a mutation in the FimS histidine kinase component of the FimS/FimR TCS that controls transcription of the fimA operon (Nishikawa and Duncan, 2010). Results with a variety of strains and isogenic mutants of P. gingivalis indicate that FimA is the effector of P. gingivalis responsible for upregulation of ZEB1. FimA is a major antigen on the P. gingivalis surface and can also incite the production of proinflammatory cytokines (Lamont and Jenkinson, 1998, Bostanci and Belibasakis, 2012). FimA is capable of manipulating a number of signal transduction pathways and transcription factors in different cell types (Zhou and Amar, 2007, Hajishengallis et al., 2012), and the processes that lead to increased ZEB1 promoter activity require further study. The potential importance of FimA expressing P. gingivalis lineages in the events that can lead to tumor development is corroborated by the role of this protein in the acceleration of the epithelial cell cycle (Kuboniwa et al., 2008). FimA fimbriae, which do not share significant homology to other fimbrial proteins (Enersen et al., 2013), may thus constitute an attractive target for novel biomarkers or therapeutics.
ZEB1 can be regulated at the transcriptional level and posttranscriptionally regulated by the miR-200 family through a double negative feedback loop (Brabletz and Brabletz, 2010). In epithelial cells miR-200s inhibit ZEB expression and maintain the epithelial phenotype. By contrast, in mesenchymal cells elevated ZEB activity suppresses expression of the miR-200s. Our results show that the increased levels of ZEB1 were associated with a reduction in the amounts of the miR200 family, potentially facilitating a stable transition to the partial mesenchymal phenotype. The initial upregulation of ZEB1 in epithelial cells, however, was not associated with a decrease in the amounts of the miR200 family, but with an increase in ZEB1 promoter activity. NF-κB activation also promotes ZEB1 transcription (Vandewalle et al., 2009); however, in epithelial cells P. gingivalis suppresses the activation of NF-κB by dephosphorylation of the p65 subunit at the S536 residue (Takeuchi et al., 2013). It is unlikely, therefore, that NF-κB is involved in P. gingivalis-induced ZEB1 upregulation.
P. gingivalis induced expression of the mesenchymal markers and decreased expression of the epithelial markers. siRNA knockdown of ZEB1 abrogated the ability of P. gingivalis to regulate epithelial and mesenchymal gene expression, establishing ZEB1 as a major transcriptional effector of the P. gingivalis-induced partial EMT. Epithelial markers down regulated by P. gingivalis included cytokeratin 19 which is characteristically expressed in cells of the junctional epithelium within the gingival tissues, and has also been reported to be suppressed in OSCC (Khanom et al., 2012). The mesenchymal relevant genes induced by P. gingivalis included N-cadherin, vimentin, fibronectin and MMP-9. N-cadherin is a calcium dependent cell-cell adhesion glycoprotein, which is upregulated in EMT and some studies have found associated with OSCC (Zhao et al., 2012). Vimentin is a cytoskeletal intermediate filament protein involved in maintaining cell shape and stabilizing cytoskeletal interactions. Expression of vimentin is associated with OSCC tumorigenesis (Lee et al., 2015), and vimentin has been proposed as a predictor of the malignant potential of high risk oral lesions (Sawant et al., 2014). Fibronectin is a component of the cell matrix involved in cell migration processes including metastasis, and expression of alternatively spliced segments of fibronectin is related to OSCC tumorigenesis (Kamarajan et al., 2010). MMP-9 is secreted as inactive proproteins which are activated by proteolytic cleavage. As a gelatinase, MMP-9 can degrade collagen IV in the basement membrane and extracellular matrix facilitating tumor growth, invasion, metastasis, and angiogenesis (Westermarck and Kahari, 1999). MMP-9 plays a crucial role in the development of several human malignancies, including OSCC (Kruger et al., 2005, Bedal et al., 2014). Moreover, epithelial cells infected with P. gingivalis showed ZEB1-dependent increased migration into matrigel, a phenotype consistent with increased MMP-9 activity and with an overall partial mesenchymal phenotype.
To begin to translate our results from reductionist in vitro models to the in vivo situation, we orally infected mice with P. gingivalis and examined ZEB1 expression in gingival tissues. Although P. gingivalis is not a normal member of the mouse oral microbiota, it does colonize transiently and causes alveolar bone loss (Hajishengallis et al., 2015). Our results show for the first time that P. gingivalis colonization of the gingival tissues in vivo leads to upregulation of ZEB1. Further in vivo evidence for a role of P. gingivalis in oral tumor development was provided by IF analysis of OSCC biopsy samples. Antigenic based detection of P. gingivalis within biopsy samples from OSCC poorly differentiated cancer and carcinoma in situ corroborates a similar study in which P. gingivalis antigens were detected in ten gingival squamous cell carcinomas of differing degrees of differentiation (Katz et al., 2011). The use of optical sectioning in the current study established that the size of particles detected by P. gingivalis antibodies was in the 2–3 μm range, consistent with whole organisms, rather than shed antigens or outer membrane vesicles. Intimate association of P. gingivalis with OSCC lesions shows that the organism has the opportunity as well as the capability, to contribute to EMT in vivo.
Oral cancers are among the most prevalent (Jemal et al., 2008), and despite considerable advances in diagnosis and therapeutic options, the 5-year survival rate has remained stable at approximately 50% among all tumor stages during the past decades (Wikner et al., 2014). The early phase of OSCC is often asymptomatic, therefore the identification of both novel biomarkers and contributing etiological agents is important for improving survival rates. The results of the current study suggest that infection with FimA-positive P. gingivalis can induce a ZEB1 dependent partial EMT. The detection of P. gingivalis, or of FimA, in early erythroplakia or leukoplakia lesions, therefore, may have utility for the early detection of lesion likely to progress to malignant status.
Materials and Methods
Bacterial strains, eukaryotic cells, and growth and infection conditions
Porphyromonas gingivalis strain ATCC 33277 and its isogenic mutant ΔfimA, strains ATCC 49417, W83, and low passage clinical isolates 11029, 10512 (laboratory strains), were cultured in trypticase soy broth (TSB) supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml) and menadione (1 μg/ml). Tetracycline (1 μg/ml) was incorporated into the medium for the growth of ΔfimA. Fusobacterium nucleatum ATCC 25586 was cultured in brain heart infusion (BHI) broth supplemented with hemin (5 μg/ml) and menadione (1 μg/ml). Streptococcus gordonii DL1 was grown in BHI supplemented with yeast extract (5 μg/ml). All bacterial strains were cultured anaerobically at 37°C. Human telomerase immortalized keratinocytes (TIGKs) derived from gingival epithelium were maintained at 37°C and 5% CO2 in Dermalife-K serum free culture medium (Lifeline Cell Technology, Carlsbad, CA) as described (Moffatt-Jauregui et al., 2013). TIGKs between passages 10 and 20 at 70% confluence were stimulated with bacteria as described for individual experiments. For transwell (Corning, Corning NY) assays, TIGKs were cultured in the lower compartment and P. gingivalis added to the upper chamber.
Immunoblotting
TIGK cells were solubilized in cold cell lysis reagent (Cell Signaling, Danvers, MA) containing Protease Inhibitor and PhosSTOP Phosphatase Inhibitor (Roche, Indianapolis, IN). Proteins (40 ng) were separated by 10% SDS-polyacrylamide gel electrophoresis, blotted onto a PVDF membrane and blocked in 5% BSA in TBS with 0.1% Tween20. Blots were probed at 4°C overnight with primary antibodies followed by 1 h with HRP-conjugated secondary antibody at room temperature. Antigen-antibody binding were detected using ECL Substrate (Thermoscientific, Hudson NH). Primary antibodies targeted ZEB1 (Novus, Littleton, CO) or vimentin (Cell Signaling). Duplicate blots were probed with GAPDH antibodies (Cell Signaling) as a loading control
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
Total RNA from TIGK cells and from homogenized gingival tissue was isolated and purified with PerfectPure RNA kit (5Prime, Gaithersburg, MD). miRNA was isolated and purified from TIGKs with PureLink miRNA isolation kit (Invitrogen, Carlsbad, CA). RNA concentrations were determined by spectrophotometry (NanoDrop Technology, Wilmington, DE). cDNA from total RNA and miRNA was synthesized (2 μg RNA/reaction volume) using a High Capacity cDNA Reverse Transcription kit and a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Grand Island, NY), respectively. Real time RT-PCRs used TaqMan Fast universal PCR mastermix and gene expression assays for Zeb1, vimentin, MMP-9, ITGA5, fibronectin, KRT7, COL-1A1 and GAPDH. Negative RT reactions were included in each assay. TaqMan microRNA assays were used to quantify the mature miRNA-200b, mi-RNA-200c, miRNA-205, miRNA-21 and RNU48. Real time qPCR was performed on an Applied Biosystems StepOne Plus cycler with StepOne software V2.2.2 and autocalculated threshold cycle selected. The cycle threshold (Ct) values were determined, mRNA and miRNA expression levels were normalized to GAPDH and RNU48, respectively, and expressed relative to controls following the 2−ΔΔCT method.
Luciferase reporter assay
TIGK cells were transfected with ZEB1 promoter constructs: Z1p 1000-Luc (−912 bp to +43 bp of the ZEB1 gene), Z1p.400-Luc (−367 bp to +43 bp) and Z1p.196-Luc (−212 bp to +43 bp); at 2 μg/105 cells using FuGENE6 Transfection Reagent (Promega, Madison, WI). Following 48 h in transfection media, cells were returned to TIGK medium for further 24 h, prior to the stimulation with P. gingivalis. Cells were lysed and the reporter activity was determined with the Dual-Glo Luciferase Assay System (Promega). Firefly luciferase activity was normalized on the basis of Renilla luciferase activity in the same samples.
Zymography
The activities of MMP2 and MMP9 in culture supernatant collected from control uninfected and P. gingivalis-infected TIGK cells, were determined using gelatin zymography as described (Inaba et al., 2014). Samples were mixed with SDS sample buffer without reducing reagents, then separated on 10% SDS-polyacrylamide gels containing 0.1% gelatin. The gels were incubated at 37°C with 2.5% Triton X-100 for 1 h, and then in 20 mM Tris-HCl (pH 7.5) containing 200 mM NaCl and 5 mM Ca Cl2 for 48 h. After staining with 5% Coomassie Brilliant Blue R-250, gelatinolytic activities were visualized as clear bands against a blue background and quantified using ImageJ.
RNA interference
TIGKs were transfected with predesigned 30 nM siRNA (siGENOME SMARTpool siRNA) targeting ZEB1 or control siRNA (GE Healthcare Dharmacon, Lafayette, CO) for 24 h using LipoJet (SignaGen, Gaithersburg, MD) transfection reagent. At 48 h after transfection, the medium was replaced and cells were incubated for a further 24 h prior to infection.
Immunofluorescence and confocal laser scanning microscopy
TIGK cells were grown on glass coverslips, washed twice in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 10 min. Permeabilization was with 0.2% TritonX-100 for 10 min at room temperature prior to blocking in 10% goat serum for 20 min. ZEB1 was detected by reacting with primary antibodies at 1:100 overnight at 4°C, followed by Alexa Fluor 488-conjugated anti-rabbit secondary antibodies (Life Technologies) at 1:200 for 3 h in the dark. Following a 20 min blocking in 0.1% goat serum, actin was labeled with Texas Red-phalloidin (Life Technologies) for 2 h at room temperature in the dark. Coverslips were mounted with on glass slides using ProLong Gold with DAPI (4′6-diamidino-2-phenylindole) mounting medium (Invitrogen) prior to imaging with a Leica SP8 confocal inverted fluorescence microscope. Images were analyzed using Volocity 6.3 Software (PerkinElmer, Waltham, MA).
Matrigel invasion assay
Cell motility was measured by assessment of the migration rate of TIGKs using a BD BioCoat Matrigel Invasion Chamber (BD Biosciences, San Jose, CA). Cells (2 × 105) were plated on transwell filters coated with matrigel. The lower compartment of the invasion chambers contained cell culture medium. After 18 h incubation at 37°C, cells remaining on the upper surface of the filter were removed, and the cells that migrated through the filter were fixed with 1% methanol and stained with toluidine blue. Cells were enumerated from three random 20× fields for each filter using a Nikon Eclipse TS100 microscope.
Animal infection
BALB/c mice were orally infected with 107 cfu P. gingivalis 33277 five times at 2-day intervals as described previously (Daep et al., 2011) and approved by the University of Louisville Institutional Animal Care and Use Committee. The levels of P. gingivalis colonization were determined by qPCR with the P. gingivalis 16SrRNA gene as described (Daep et al., 2011). On days 1, 3 and 8 after the last infection, mice were euthanized and the upper and lower jaw with gingival tissue were recovered. After RNA extraction, the ratio of ZEB1 mRNA to GAPDH mRNA for each mouse was determined by qRT-PCR using the 2− ΔCT method.
Human oral tissue immunofluorescent and immunohistochemical staining
Paraffin embedded human tongue biopsy samples were sectioned at 4 μm, dewaxed and rehydrated. The slides were blocked with 10% goat serum for 1 h, and reacted with P. gingivalis 33277 or S. gordonii antibodies 1:1000 for 2 h at room temperature. Secondary Alexa Fluor 488 conjugated anti-rabbit antibody (1:500) was for 1 h, following which slides were treated with DAPI (1:2000). Slides were mounted with VectaShield (Vector Labs, Burlingame, CA) and imaged with a Zeiss Axiocam MRc5 fluorescence microscope. Procedures were approved by the University of Louisville Institutional Review Board.
Statistical analyses
Statistical analyses were conducted using the GraphPad Prism software. Data were evaluated by one-way ANOVA with Tukey’s multiple comparison test. Experimental data presented are representative of at least three biological replicates.
Supplementary Material
Figure S1. Immunoblot of whole cell lysates of P. gingivalis strains probed with polyclonal antibodies to the FimA protein of strain 33277.
Figure S2. JNK knockdown does not affect regulation of Zeb1 by P. gingivalis. A. TIGK cells were transiently transfected with siRNA to JNK1/2 (si Jnk, 100 nM, Sigma) or scrambled siRNA (si ctr). Control (ctr) cells were nontransfected. JNK mRNA levels in transfected cells were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to ctr. Results are means ± SD, n = 3; *** P < 0.001. B. Transfected TIGKs cells were infected with P. gingivalis 33277 for 24 h at MOI 100. ZEB1 mRNA was measured by qRT-PCR, the data were normalized to GAPDH mRNA and are expressed relative to the noninfected (NI) control. Results are means ± SD, n = 3; *** P < 0.001 compared to NI; NS: not significant.
Figure S3. Pharmacological inhibition of Akt does not affect regulation of Zeb1 by P. gingivalis. TIGK cells were preincubated with 10 μM LY294002 or vehicle (DMSO) only for 1 h and infected with P. gingivalis 33277 MOI 50 or100 for 6 h. ZEB1 mRNA levels were measured by qRT-PCR, normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; * P < 0.05; *** P < 0.001; NS: not significant.
Figure S4. A non-invasive mutant of P. gingivalis can induce ZEB1 expression. qRT-PCR of ZEB1 mRNA expression in TIGK cells infected with P. gingivalis 33277 (Pg WT) or a ΔserB mutant. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; *** P < 0.001 compared to NI; NS: not significant.
Figure S5. Expression of miRNA-21 is not down-regulated by P. gingivalis. TIGK cells were infected with P. gingivalis 33277 (Pg) at MOI 100 for the time indicated. miRNA levels were measured by qRT-PCR, normalized to RNU48 miRNA, and expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; *** P < 0.001 compared to NI.
Figure S6. P. gingivalis increases expression of vimentin. Immunoblot of lysates of TIGK cells infected with P. gingivalis 33277 for 24 h at the MOI indicated. Control cells were uninfected (NI). Duplicate blots were probed with antibodies to vimentin or GAPDH (loading control).
Figure S7. Colonization of mice. Mice were orally infected with 107 cfu P. gingivalis five times at 2-days intervals. Bacterial samples were collected along the gingiva of the upper molars. Samples were lysed, DNA extracted and qPCR performed with primers specific for P. gingivalis 16S DNA. For enumeration, genomic DNA was isolated from laboratory cultures of P. gingivalis 33277 (numbers determined by viable counting) and a series of dilutions prepared. The number of gene copies in the oral samples was determined by comparison with the standard curve. In the sham infected animals, 2 of 5 mice were colonized with low levels of organisms sufficient similar to P. gingivalis to give a positive result. P. gingivalis levels from day 1, 3 and 8 were statistically greater than sham infected (P < 0.0001) but were not statistically different from each other.
Figure S8. Fluorescent confocal microscopy of a carcinoma in situ biopsy sample probed with P. gingivalis antibodies (green) and stained with DAPI (blue). Cells were imaged at magnification ×63. Red arrows point to a discrete fluorescent spot, yellow arrows indicate the same position where that spot is absent. Numbers are the slice number in an optical stack of 40 slices at 0.4 μm. Fluorescent spots are present in typically 5 to 7 adjacent optical slices (0.4 μm slices), indicating that the fluorescent particles are about 2.0 to 2.8 μm in size, consistent with the size of P. gingivalis.
Acknowledgments
Grant Sponsors: We thank the NIH/NIDCR for support through DE011111, DE017921, DE016690 and DE023193.
Footnotes
Conflict of Interest
The authors have no conflict of interest to declare.
References
- Baud J, Varon C, Chabas S, Chambonnier L, Darfeuille F, Staedel C. Helicobacter pylori initiates a mesenchymal transition through ZEB1 in gastric epithelial cells. PLoS One. 2013;8:e60315. doi: 10.1371/journal.pone.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedal KB, Grassel S, Oefner PJ, Reinders J, Reichert TE, Bauer R. Collagen XVI induces expression of MMP9 via modulation of AP-1 transcription factors and facilitates invasion of oral squamous cell carcinoma. PLoS One. 2014;9:e86777. doi: 10.1371/journal.pone.0086777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton CM, Izutsu KT, Goodwin PC, Park Y, Lamont RJ. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. 1999;1:215–223. doi: 10.1046/j.1462-5822.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- Benitez-Paez A, Belda-Ferre P, Simon-Soro A, Mira A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genomics. 2014;15:311. doi: 10.1186/1471-2164-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessede E, Staedel C, Acuna Amador LA, Nguyen PH, Chambonnier L, Hatakeyama M, et al. Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene. 2014;33:4123–4131. doi: 10.1038/onc.2013.380. [DOI] [PubMed] [Google Scholar]
- Bosshardt DD, Lang NP. The junctional epithelium: from health to disease. J Dent Res. 2005;84:9–20. doi: 10.1177/154405910508400102. [DOI] [PubMed] [Google Scholar]
- Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. J Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.20265. doi:10.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimidi AB, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6:22613–22623. doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof A, Hulpiau P, van Roy F, De Craene B, Berx G. Evolutionary functional analysis and molecular regulation of the ZEB transcription factors. Cell Mol Life Sci. 2012;69:2527–2541. doi: 10.1007/s00018-012-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014;29:248–257. doi: 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Krauss JL, Liang S, McIntosh ML, Lambris JD. Pathogenic microbes and community service through manipulation of innate immunity. Adv Exp Med Biol. 2012;946:69–85. doi: 10.1007/978-1-4614-0106-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ, Graves DT. The enduring importance of animal models in understanding periodontal disease. Virulence. 2015;6:229–235. doi: 10.4161/21505594.2014.990806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield M, Mans JJ, Zheng G, Lopez MC, Mao S, Progulske-Fox A, et al. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- Hendrickson EL, Wang T, Beck DA, Dickinson BC, Wright CJ, Lamont RJ, Hackett M. Proteomics of Fusobacterium nucleatum within a model developing oral microbial community. Microbiologyopen. 2014;3:729–751. doi: 10.1002/mbo3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann R, Wroblewski J, Terling C, Midtvedt T, Obrink B. The characteristic cellular organization and CEACAM1 expression in the junctional epithelium of rats and mice are genetically programmed and not influenced by the bacterial microflora. J Periodontol. 2001;72:454–460. doi: 10.1902/jop.2001.72.4.454. [DOI] [PubMed] [Google Scholar]
- Inaba H, Sugita H, Kuboniwa M, Iwai S, Hamada M, Noda T, et al. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014;16:131–145. doi: 10.1111/cmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Jia B, Liu H, Kong Q, Li B. Overexpression of ZEB1 associated with metastasis and invasion in patients with gastric carcinoma. Mol Cell Biochem. 2012;366:223–229. doi: 10.1007/s11010-012-1299-6. [DOI] [PubMed] [Google Scholar]
- Kamarajan P, Garcia-Pardo A, D’Silva NJ, Kapila YL. The CS1 segment of fibronectin is involved in human OSCC pathogenesis by mediating OSCC cell spreading, migration, and invasion. BMC Cancer. 2010;10:330. doi: 10.1186/1471-2407-10-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3:209–215. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanom R, Sakamoto K, Pal SK, Shimada Y, Morita K, Omura K, et al. Expression of basal cell keratin 15 and keratin 19 in oral squamous neoplasms represents diverse pathophysiologies. Histol Histopathol. 2012;27:949–959. doi: 10.14670/HH-27.949. [DOI] [PubMed] [Google Scholar]
- Kim SS, Ruiz VE, Carroll JD, Moss SF. Helicobacter pylori in the pathogenesis of gastric cancer and gastric lymphoma. Cancer Lett. 2011;305:228–238. doi: 10.1016/j.canlet.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger A, Arlt MJ, Gerg M, Kopitz C, Bernardo MM, Chang M, et al. Antimetastatic activity of a novel mechanism-based gelatinase inhibitor. Cancer Res. 2005;65:3523–3526. doi: 10.1158/0008-5472.CAN-04-3570. [DOI] [PubMed] [Google Scholar]
- Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz O. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Chan A, Belton CM, Izutsu KT, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Chang JS, Syu SH, Wong TS, Chan JY, Tang YC, et al. IL-1beta promotes malignant transformation and tumor aggressiveness in oral cancer. J Cell Physiol. 2015;230:875–884. doi: 10.1002/jcp.24816. [DOI] [PubMed] [Google Scholar]
- Liu Y, Costantino ME, Montoya-Durango D, Higashi Y, Darling DS, Dean DC. The zinc finger transcription factor ZFHX1A is linked to cell proliferation by Rb-E2F1. Biochem J. 2007;408:79–85. doi: 10.1042/BJ20070344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Roqueiro G, Darling DS, Cabanillas AM. The ZFHX1A gene is differentially autoregulated by its isoforms. Biochem Biophys Res Commun. 2007;360:621–626. doi: 10.1016/j.bbrc.2007.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34:2193–2197. doi: 10.1093/carcin/bgt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt-Jauregui CE, Robinson B, de Moya AV, Brockman RD, Roman AV, Cash MN, et al. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J Periodontal Res. 2013;48:713–721. doi: 10.1111/jre.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt CE, Lamont RJ. Porphyromonas gingivalis induction of microRNA-203 expression controls suppressor of cytokine signaling 3 in gingival epithelial cells. Infect Immun. 2011;79:2632–2637. doi: 10.1128/IAI.00082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni MA, Chhour KL, Chapple CC, Nguyen KA, Hunter N. The profile of Porphyromonas gingivalis kgp biotype and fimA genotype mosaic in subgingival plaque samples. FEMS Microbiol Lett. 2014;361:190–194. doi: 10.1111/1574-6968.12631. [DOI] [PubMed] [Google Scholar]
- Nagy KN, Sonkodi I, Szoke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34:304–308. [PubMed] [Google Scholar]
- Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 2008;15:215–225. doi: 10.1093/dnares/dsn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Duncan MJ. Histidine kinase-mediated production and autoassembly of Porphyromonas gingivalis fimbriae. J Bacteriol. 2010;192:1975–1987. doi: 10.1128/JB.01474-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahingur SE, Yeudall WA. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol. 2015;6:214. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Tillo E, Liu Y, de Barrios O, Siles L, Fanlo L, Cuatrecasas M, et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant SS, Vaidya M, Chaukar DA, Alam H, Dmello C, Gangadaran P, et al. Clinical significance of aberrant vimentin expression in oral premalignant lesions and carcinomas. Oral Dis. 2014;20:453–465. doi: 10.1111/odi.12151. [DOI] [PubMed] [Google Scholar]
- Scanlon CS, Van Tubergen EA, Inglehart RC, D’Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114–121. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H, Hirano T, Whitmore SE, Morisaki I, Amano A, Lamont RJ. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-kappaB RelA/p65. PLoS Pathog. 2013;9:e1003326. doi: 10.1371/journal.ppat.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble GD, Kerr JE, Wang BY. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8:607–620. doi: 10.2217/fmb.13.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble GD, Lamont GJ, Progulske-Fox A, Lamont RJ. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J Bacteriol. 2007;189:6382–6388. doi: 10.1128/JB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Yilmaz O, Nakhjiri SF, Belton CM, Lamont RJ. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect Immun. 2001;69:6731–6737. doi: 10.1128/IAI.69.11.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermarck J, Kahari VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781–792. [PubMed] [Google Scholar]
- Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10:e1003933. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikner J, Grobe A, Pantel K, Riethdorf S. Squamous cell carcinoma of the oral cavity and circulating tumour cells. World J Clin Oncol. 2014;5:114–124. doi: 10.5306/wjco.v5.i2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Xue P, Hirano T, Liu C, Whitmore SE, Hackett M, Lamont RJ. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen. 2014;3:383–394. doi: 10.1002/mbo3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Yuan J, Li K. EMT transcription factors: implication in osteosarcoma. Med Oncol. 2013;30:697. doi: 10.1007/s12032-013-0697-2. [DOI] [PubMed] [Google Scholar]
- Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72:3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74:703–710. doi: 10.1128/IAI.74.1.703-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Cai Y, Soofi A, Dressler GR. Activation of Wnt11 by transforming growth factor-beta drives mesenchymal gene expression through non-canonical Wnt protein signaling in renal epithelial cells. J Biol Chem. 2012;287:21290–21302. doi: 10.1074/jbc.M112.357202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wei Y, Wang L, Debeb BG, Yuan Y, Zhang J, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–875. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Tang XF, Yang K, Liu JY, Ma XR. Over-expression of integrin-linked kinase correlates with aberrant expression of Snail, E-cadherin and N-cadherin in oral squamous cell carcinoma: implications in tumor progression and metastasis. Clin Exp Metastasis. 2012;29:957–969. doi: 10.1007/s10585-012-9485-1. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Amar S. Identification of signaling pathways in macrophage exposed to Porphyromonas gingivalis or to its purified cell wall components. J Immunol. 2007;179:7777–7790. doi: 10.4049/jimmunol.179.11.7777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immunoblot of whole cell lysates of P. gingivalis strains probed with polyclonal antibodies to the FimA protein of strain 33277.
Figure S2. JNK knockdown does not affect regulation of Zeb1 by P. gingivalis. A. TIGK cells were transiently transfected with siRNA to JNK1/2 (si Jnk, 100 nM, Sigma) or scrambled siRNA (si ctr). Control (ctr) cells were nontransfected. JNK mRNA levels in transfected cells were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to ctr. Results are means ± SD, n = 3; *** P < 0.001. B. Transfected TIGKs cells were infected with P. gingivalis 33277 for 24 h at MOI 100. ZEB1 mRNA was measured by qRT-PCR, the data were normalized to GAPDH mRNA and are expressed relative to the noninfected (NI) control. Results are means ± SD, n = 3; *** P < 0.001 compared to NI; NS: not significant.
Figure S3. Pharmacological inhibition of Akt does not affect regulation of Zeb1 by P. gingivalis. TIGK cells were preincubated with 10 μM LY294002 or vehicle (DMSO) only for 1 h and infected with P. gingivalis 33277 MOI 50 or100 for 6 h. ZEB1 mRNA levels were measured by qRT-PCR, normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; * P < 0.05; *** P < 0.001; NS: not significant.
Figure S4. A non-invasive mutant of P. gingivalis can induce ZEB1 expression. qRT-PCR of ZEB1 mRNA expression in TIGK cells infected with P. gingivalis 33277 (Pg WT) or a ΔserB mutant. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; *** P < 0.001 compared to NI; NS: not significant.
Figure S5. Expression of miRNA-21 is not down-regulated by P. gingivalis. TIGK cells were infected with P. gingivalis 33277 (Pg) at MOI 100 for the time indicated. miRNA levels were measured by qRT-PCR, normalized to RNU48 miRNA, and expressed relative to noninfected (NI) controls. Results are means ± SD, n = 3; *** P < 0.001 compared to NI.
Figure S6. P. gingivalis increases expression of vimentin. Immunoblot of lysates of TIGK cells infected with P. gingivalis 33277 for 24 h at the MOI indicated. Control cells were uninfected (NI). Duplicate blots were probed with antibodies to vimentin or GAPDH (loading control).
Figure S7. Colonization of mice. Mice were orally infected with 107 cfu P. gingivalis five times at 2-days intervals. Bacterial samples were collected along the gingiva of the upper molars. Samples were lysed, DNA extracted and qPCR performed with primers specific for P. gingivalis 16S DNA. For enumeration, genomic DNA was isolated from laboratory cultures of P. gingivalis 33277 (numbers determined by viable counting) and a series of dilutions prepared. The number of gene copies in the oral samples was determined by comparison with the standard curve. In the sham infected animals, 2 of 5 mice were colonized with low levels of organisms sufficient similar to P. gingivalis to give a positive result. P. gingivalis levels from day 1, 3 and 8 were statistically greater than sham infected (P < 0.0001) but were not statistically different from each other.
Figure S8. Fluorescent confocal microscopy of a carcinoma in situ biopsy sample probed with P. gingivalis antibodies (green) and stained with DAPI (blue). Cells were imaged at magnification ×63. Red arrows point to a discrete fluorescent spot, yellow arrows indicate the same position where that spot is absent. Numbers are the slice number in an optical stack of 40 slices at 0.4 μm. Fluorescent spots are present in typically 5 to 7 adjacent optical slices (0.4 μm slices), indicating that the fluorescent particles are about 2.0 to 2.8 μm in size, consistent with the size of P. gingivalis.