Abstract
Idiopathic pulmonary fibrosis (IPF) is a lethal interstitial lung disease characterized by airway remodeling, inflammation, alveolar destruction, and fibrosis. We utilized single-cell RNA sequencing (scRNA-seq) to identify epithelial cell types and associated biological processes involved in the pathogenesis of IPF. Transcriptomic analysis of normal human lung epithelial cells defined gene expression patterns associated with highly differentiated alveolar type 2 (AT2) cells, indicated by enrichment of RNAs critical for surfactant homeostasis. In contrast, scRNA-seq of IPF cells identified 3 distinct subsets of epithelial cell types with characteristics of conducting airway basal and goblet cells and an additional atypical transitional cell that contributes to pathological processes in IPF. Individual IPF cells frequently coexpressed alveolar type 1 (AT1), AT2, and conducting airway selective markers, demonstrating “indeterminate” states of differentiation not seen in normal lung development. Pathway analysis predicted aberrant activation of canonical signaling via TGF-β, HIPPO/YAP, P53, WNT, and AKT/PI3K. Immunofluorescence confocal microscopy identified the disruption of alveolar structure and loss of the normal proximal-peripheral differentiation of pulmonary epithelial cells. scRNA-seq analyses identified loss of normal epithelial cell identities and unique contributions of epithelial cells to the pathogenesis of IPF. The present study provides a rich data source to further explore lung health and disease.
Keywords: Inflammation
Single cell RNA-sequencing of epithelial cells from lung tissue from patients with idiopathic pulmonary fibrosis identified abnormal states of differentiation and gene expression.
Introduction
Idiopathic pulmonary fibrosis (IPF) is a common lethal disorder representing a form of interstitial lung disease (ILD) resulting from alveolar tissue remodeling and fibrosis leading to respiratory failure (1–3). While pulmonary inflammation and loss of lung architecture in IPF involve interactions among multiple cell types, recent studies provide increasing support for the concept that injury to the respiratory epithelium plays an important role in IPF pathogenesis (4, 5). Loss of normal alveolar architecture in IPF is accompanied by fibrotic remodeling, loss of AT1 and AT2 cells, and the presence of atypical epithelial cells expressing differentiated cell markers characteristic of proximal airways and submucosal glands (e.g., basal cell and goblet cell markers) in the normal lung (6, 7). Basal cells in conducting airways, and AT2 cells in the alveoli, serve as progenitor cells, with critical roles in regeneration of the respiratory epithelium following both acute and chronic injury. In experimental models, severe injury to the respiratory epithelium is associated with pathological features of IPF, with alveolar remodeling and the presence of atypical basal-like cells in alveolar regions (8, 9). Mutations in genes affecting AT2 cell function or survival, e.g., TEL, TERC, ABCA3, SFTPB, SFTPC, SFTPA, RTELI, and PARN are associated with ILD, further implicating alveolar cell injury and abnormal repair processes in these disorders (for review, see refs. 10–21). Tissue remodeling seen in peripheral airways supports the concept that the pathogenesis of IPF is influenced by complex interactions among multiple cell types, including epithelial, stromal, and inflammatory cells, leading to fibrosis and loss of alveolar architecture. The contributions and responses of individual cell types to the pathogenesis of IPF are unknown.
Organ formation and homeostasis are dependent on a precise temporal and spatial progression of progenitor cells from undifferentiated to differentiated states as individual cell identities are established. During morphogenesis of the respiratory tract, endodermal progenitors differentiate into distinct epithelial cell types that are regionally specified along the proximal-peripheral/cephalocaudal axis of the lung (22). At maturity, conducting airways are lined by well-defined basal, ciliated, goblet, neuroendocrine, and other secretory cells, while the peripheral alveoli are lined exclusively by AT2 and AT1 cells. At homeostasis, each cell maintains unique cell morphologies, gene expression patterns, and functions. Early in lung morphogenesis, epithelial cell type specification is firmly established, and patterns of gene expression and cell types are not overlapping in conducting versus alveolar regions of the lung. While histopathological analyses of lung tissue from patients with IPF demonstrate abnormalities in the morphology of epithelial cells lining remodeled regions of the peripheral lung parenchyma (6, 7), it is presently unclear what mechanisms lead to tissue remodeling and altered epithelial cell fates. Interpretation of proteomic and transcriptomic data obtained from lung tissue in IPF is complicated by the complexity and heterogeneity of tissue changes, obscuring identification of the roles of individual cell types in disease pathogenesis (23). To overcome these limitations, we utilized single-cell RNA sequencing (scRNA-seq) and high-resolution confocal microscopy to identify unique differentiation states and gene expression patterns of epithelial cells isolated from the peripheral regions of the normal and IPF lung.
Results
Features of usual interstitial pneumonia in IPF.
Patchy interstitial fibrosis, loss of alveolar structure, and honeycombing, hallmarks of usual interstitial pneumonia (UIP), were present in all IPF explant tissues evaluated after transplant (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/jci.insight.90558DS1). Uniformly thin alveolar septae lined by AT2 and AT1 cells were characteristic of normal lungs. IPF tissues consisted of heterogeneous lesions with dense connective tissue, fibroblastic foci, and cystic lesions, many containing mucus. “Honeycomb” cysts were lined by diverse epithelial cell types, including cuboidal “hyperplastic” AT2 cells, goblet cells, and ciliated cells, the latter two cell types normally primarily restricted to tracheal, bronchial, and bronchiolar epithelium lining cartilaginous airways. Heterogeneous lesions containing disorganized epithelial cells and inflammatory infiltrates were present in all IPF samples.
Gene expression patterns in pulmonary epithelial cells obtained by cell sorting.
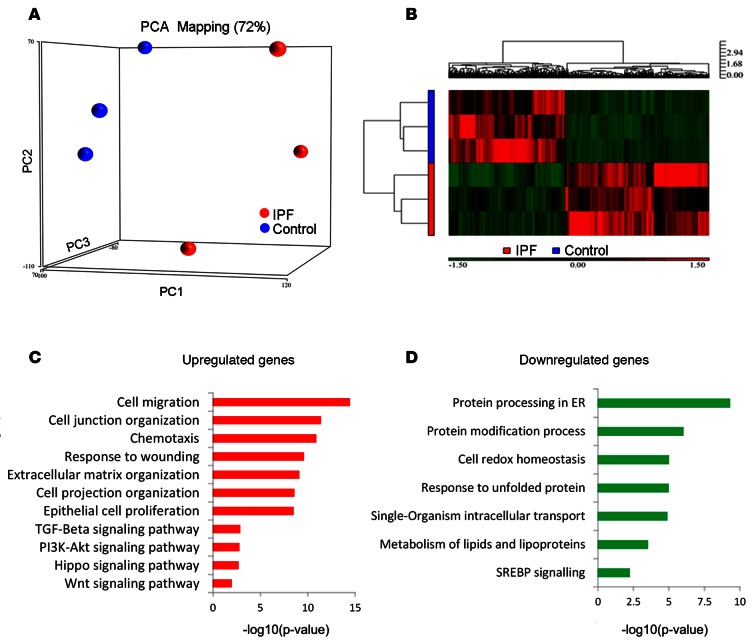
Lung cells were isolated from peripheral control and IPF lung tissue after protease digestion and viable cells sorted on the basis of their 7AAD–, CD45–, CD31–, CD326+ (EPCAM), HTII-280+ phenotype (herein referred to as HTII-280+ epithelial cells); HTII-280 is a selective surface marker of normal AT2 cells (24). Consistent differences were observed between distal normal donor lung and distal IPF explant lung tissue in the relative abundance of epithelial cell types recognized by anti-CD326 and HTII-280 monoclonal antibodies. Control lung tissue consistently yielded >90% HTII-280 surface reactive epithelial cells, indicative of an abundant AT2 cell fraction; and relatively few NGFR+ or double negative epithelial cells, indicating few airway basal or luminal cell types, respectively. In contrast, distal lung tissue from IPF patients demonstrated a remarkable decline in HTII-280+ cells, decreasing to approximately 5% of total epithelial cells, with a corresponding increase in the abundance of NGFR+ and double negative epithelial cells. Differences in the relative abundance of HTII-280+ cells between control and IPF tissues were associated with disease-dependent changes in their molecular phenotype. RNA sequencing demonstrated clear separation of gene expression between populations of HTII-280+ epithelial cells isolated from IPF and control tissues (Figure 1A). A heatmap illustrates differentially expressed genes based on unsupervised hierarchical clustering (Figure 1B). Transcriptome profiles from normal lung HTII-280+ cells were consistent with expression profiles of AT2 cells that mediate surfactant protein and lipid homeostasis, with this extensive RNA data providing a useful resource for further investigation of human AT2 cell biology. Transcriptome profiles of HTII-280+ epithelial cells from IPF lungs were surprisingly enriched in transcripts normally associated with conducting airway epithelial cells, but also expressed AT2 cell–associated transcripts. Genes related to “cell migration, cell junction/extracellular matrix organization, response to wounding, and epithelial cell proliferation” were induced in IPF (Figure 1, C and D). Ingenuity pathway analysis (IPA) predicted activation of TGF-β–, PI3K/AKT-, HIPPO/YAP-, p53-, and WNT-mediated pathways, whereas those associated with lipid synthesis and metabolism, endosomal protein processing, and unfolded protein responses were suppressed in IPF epithelial cells (Figure 1, C and D). TGFB1, TP53, IGF1, NFKB, and ETS family transcription factors (ETV5, SPDEF, EHF) were predicted to be driving forces in IPF epithelial cells, influencing their transcriptional responses (Supplemental Figure 2). The most upregulated or suppressed RNAs in the CD326+HTII-280+ sorted IPF cells are shown in Supplemental Tables 1 and 2. AT2 cell “signature” genes, including surfactant-associated genes, were highly expressed in sorted cells from both control and IPF tissue samples; however, their expression levels were moderately decreased (P > 0.05) in IPF cells compared with controls. A number of AT1-associated transcripts, on the other hand, were present at relatively high levels in HTII-280+ cells of IPF samples. The finding that the HTII-280+ cells from both control and IPF samples expressed AT2 cell gene signatures suggests that some IPF cells either maintain or acquire some AT2-like identity despite or possibly as a result of extensive tissue remodeling, respectively. AT1 cell–associated transcripts were either absent or were present at low levels in epithelial cells sorted from normal lung. Since the transcriptome of epithelial cells within remodeled lung tissue of IPF patients is likely to be impacted by the altered inflammatory and stromal milieu, we identified transcripts encoding cytokines, chemokines, growth factors, and associated genes that were selectively expressed in IPF epithelial cells. Supplemental Figure 3 demonstrates expression of mediators selectively expressed by IPF cells that are known to influence cell migration, chemotaxis, and epithelial cell growth, supporting their potential roles in the inflammatory and fibrotic processes in IPF.
Figure 1. Heatmap, principal component analysis, and predicted function in sorted normal and IPF epithelial cells.
EPCAM+ (CD326+) and HTII-280+ epithelial cells from control and IPF donors were isolated from peripheral lung tissue by FACS and subjected to RNA sequencing (RNA-seq). (A) Principal component analysis (PCA) RNA-seq data from IPF and control donors (n = 3 per group) shows the primary separation of samples by disease status. (B) Heatmap represents 2D hierarchical clustering of genes and samples and shows differentially expressed genes in IPF versus control samples. (C) Functional enrichment of predicted biological processes and genes induced in IPF is shown. (D) Functional enrichment of predicted biological processes and genes suppressed in IPF is shown. x axis represents the –log10 transformed enrichment P value
Differential gene expression in normal and IPF HTII-280+ cells.
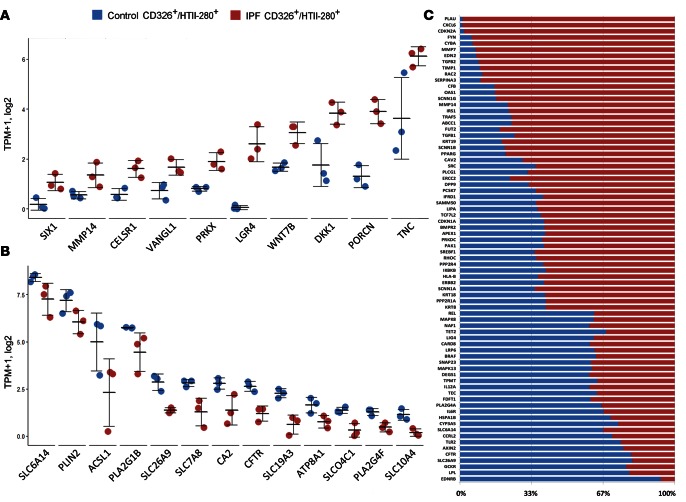
Expression of genes involved in early lung morphogenesis (SOX9, CELSR1, SIX1, LGR4) and Wnt signaling PRKX, WNT7B, DKK1, PORCN) were largely induced in IPF epithelial cells, being negligibly expressed in normal (CD326/HTII-280) AT2 cells (Figure 2A). Expression of genes involved in “ion transport” (SLC26A9, SLCO4C1, CA2, SLC6A14, CFTR) were reduced in IPF HTII-280+ cells in comparison with the controls (Figure 2B). Genes associated with “fibrosis, pulmonary fibrosis, and idiopathic pulmonary fibrosis” were compiled from the disease-centered database HuGE Navigator (25) and OMIM (http://www.omim.org/). The overlap among known fibrosis-related genes and genes differentially expressed in CD326/HTII-280 cells from IPF tissues are identified. Relative expression levels of known fibrosis markers in control and IPF CD326/HTII-280 sorted cells were calculated and are shown in Figure 2C.
Figure 2. Representative genes and their relative expression in Control-CD326/HTII-280 versus IPF CD326/HTII-280 cell populations.
Genes involved in (A) “branching morphogenesis” and Wnt signaling and (B) “anion transport” were induced and suppressed in IPF epithelial cells, respectively. RNA sequencing data from IPF and control donors (n = 3 per group); data are presented in dot plot with mean ± SEM. (C) Genes associated with fibrosis, pulmonary fibrosis, and idiopathic pulmonary fibrosis were compiled from the disease-centered database HuGE Navigator (ref. 25) and OMIM (http://www.omim.org/). The overlap between the known fibrosis genes and genes induced or suppressed in IPF HTII-280 was identified. Relative expression of known fibrosis-associated transcripts in control and IPF CD326/HTII-280–sorted cells was calculated as shown in the bar graph. The portion of fibrosis related genes in control is noted in blue, and IPF in red.
scRNA-seq analysis identifies distinct epithelial cell types in IPF.
We utilized scRNA-seq to test whether the differences in gene expression between sorted IPF and control epithelial cells were related to admixtures of cells from conducting airways and peripheral lung or to fundamental changes in the differentiation state of individual epithelial cells. Viable epithelial cells were FACS enriched following lung tissue dissociation by virtue of their 7AAD–CD31–CD45–CD326+ phenotype and further separated into single cells using the Fluidigm C1 microfluidics system (26). scRNA-seq was obtained from 3 control and 6 IPF patient samples (Figure 3A). The average DNA fragment read per cell was greater than 4 million, average alignment 0.7, average sequencing quality score 34, and average coverage depth 125. In total, 540 cells from control (n = 215) and IPF patients (n = 325) passed quality control and were used for further analysis. We applied our newly developed analytic pipeline Sincera (27) to the dataset and identified 4 distinct cell clusters that we defined as AT2 (C1), “indeterminate” (C2), basal (C3), and goblet/club (C4) cells. The landscape of all cells in 3D and 2D space is shown by PCA and hierarchical clustering (Figure 3, A–C). Expression patterns of a subset of known lung epithelial cell markers are shown in Figure 3B. Epithelial-specific transcripts EPCAM and CDH1 were expressed in virtually all IPF and normal cells, demonstrating successful isolation of single epithelial cells and removal of stromal, vascular, and immune cells. Transcripts associated with AT2 cells, including SFTPC, SLC34A2, and ABCA3 were highly expressed in all AT2 cells from control lungs, consistent with RNA expression profiles in single AT2 cells from the mouse lung (26, 28), as well as those from HTII-280+ cells obtained from normal lung (Figure 1).
Figure 3. Single-cell RNA sequencing analysis from human IPF and normal lung epithelial cells.
CD326+ epithelial cells were isolated from peripheral lung tissues by FACS as described in Methods, followed by single-cell isolation using Fluidigm C1 system and RNA sequencing. (A) Hierarchical clustering and principal component analysis (PCA) of 540 single cells from control (n = 3) and IPF patients (n = 6) reveals 4 major cell types (C1–C4), termed as normal AT2 (C1, green), indeterminate (C2, yellow), basal (C3, red), and club/goblet (C4, blue) cells. Single cells are colored by cluster on a 3D space. (B) Heatmap represents the expression of distinct RNAs that identify each of the 4 cell types. (C) Hierarchical clustering of all IPF and control cells using differentially expressed genes involved in epithelial proliferation (GO:0050673), “response to cytokines” (GO:0034097), and “response to growth factors” (GO:0034097) is shown. Minimum expression values were set to 0.01 TPM. Genes (n = 9,154) with specificity >0.7 and with TPM >1 in at least 10 cells in at least 6 samples were selected for hierarchical clustering using Z score–transformed expression.
All 3 IPF predominant cell types expressed increased levels of genes involved in epithelial proliferation (GO:0050673), genes “response to cytokines” (GO:0034097), and “response to growth factors (GO:0034097) (Figure 3C). Heterogeneity and variation among different patients were readily detectable, as shown by distribution of the 4 major epithelial cell types from IPF and from control lungs (Supplemental Figure 4). The relative abundance of the 4 major cell types varied somewhat among IPF patients, e.g., cells from sample 003 selectively expressed RNAs typical of goblet cells and lacked a clear basal cell signature.
Transcripts for signature genes defining normal AT2 cells and IPF-related basal, goblet, and indeterminate cells are shown in Figure 3C. Cluster C1 consisted of 95% of the control cells, sharing gene expression patterns consistent with AT2 cells. AT2 cells were well defined by expression of known AT2 cell-selective transcripts, including those encoding surfactant proteins and related proteins (SFTPB, SFTPC, NAPSA, LPCAT1, SLC34A2, and ABCA3) known to play critical and cell-selective roles in surfactant homeostasis in the alveolus. Potential regulators for cells within the C1 category include FOXA2, NKX2-1, CEBPA, and SREBF1. Functional classes selectively enriched in AT2 cells were absent in IPF, including genes mediating cytoprotection/detoxification and response to oxidative stress (e.g., SRXN1, CAT, FBVLN5, PRDX6, SOD2, SOD3, and GPX4) (Figure 3, B and C, and Table 1). We hypothesize that abnormal responses to oxidative stress may play a role in IPF pathogenesis. Interestingly, only 9 of 325 IPF epithelial cells clustered with normal epithelial cells in the C1 category, a finding that was consistent with dramatic loss of AT2 cells in IPF tissue.
Table 1. Enriched functional annotations for cell clusters.
Three distinct cell types were identified in the IPF samples by signature gene expression patterns that were consistent with conducting airway epithelial cells, e.g., basal (C3), goblet/club (C4), and indeterminate (C2) cells. Cells belonging to the C3 category harbored transcripts for TP63, KRT5, KRT14, BMP7, LAMB3, LAMC2, and ITGB, known markers of human basal cells (Figure 3B). Signature genes of the C3 cell cluster were involved in “alpha6-beta4 integrin signaling,” “wound healing,” “cell migration,” and “laminin interaction,” consistent with an airway origin for these cells (Table 1). Transcripts for SOX2, TP63, and TGFB1 were predicted as upstream regulators of cells belonging to the C3 cluster. IPF goblet-like cells (C4) selectively expressed SPDEF, a transcription factor regulating goblet cell differentiation (29, 30), MUC5AC, MUC5B, PIGR, AQP3, and SCGB1A1, transcripts that are characteristically expressed by airway secretory cells. Genes associated with “goblet cell morphology” (e.g., SPDEF, TCF7L2, and ELF3) and “O-linked glycosylation of mucins” (e.g., MUC16, MUC20, MUC4, MUC5AC, MUC5B, GALNT6, and GALNT5) were enriched in C4 IPF goblet cells (Table 1 and Figure 3, B and C).
Cells belonging to the C2 cluster express CD326 and CDH1 signature genes, but were not typical of any known epithelial cell type present in the normal lung. C2 cluster cells generally expressed AT2 cell–associated markers at higher levels than IPF basal or goblet/club cells and shared “lipid transport” and “innate immunity” functions with normal AT2 cells; however, C2 cells also expressed markers normally restricted to the proximal airway epithelium. The uniquely enriched functions predicted to be active in the C2 cluster included “activation of myofibroblasts,” “flux of anion,” and “T cell proliferation” (Table 1). The predicted driving forces (key regulators) for the C2 cluster include CTGF, GF11, and FL11) (see below). Although all of the cells expressed CDH1 and EPCAM, the C2 cells did not express clear signature genes associated with any known lung epithelial subtypes, and we termed these cells “indeterminate” IPF cells.
scRNA-seq analysis reveals “bronchiolization” and novel epithelial cell types in IPF.
Through the single-cell analysis, we noted that RNAs associated with AT2 cells, including SFTPC, SLC34A2, and ABCA3, were highly expressed in all normal AT2 cells present within the C1 cluster (26, 28). Conducting airway epithelial cell–selective marker transcripts, including SOX2, PAX9, TP63, KRT5, KRT14, MUC5B, and MUC5AC, were generally absent from single cells from normal lung but were present in many IPF cells (Figure 4A). While AT1-associated transcripts (e.g., AQP5, AQP3, AGER, CLIC5) were not enriched in control samples, remarkably, these transcripts were present in relative higher levels in IPF cells. The distinct, squamous morphology and fragility of AT1 cells likely exclude them from the isolation process. Although ciliated cells are readily detected in IPF samples by light and confocal microscopy, there was clear absence of multiciliated cell signature genes, indicating their loss in the cell isolation process. The presence of AT2 and AT1 transcripts and their coexpression with conducting airway and other alveolar markers seen in the single-cell RNA profiles support the hypothesis that the epithelial cells of the remodeled distal lung in IPF acquire atypical mixed differentiation states. While conducting airway-associated genes (SOX2, MUC5B, and PAX9) were rarely or not expressed in single cells from control tissue, these transcripts were frequently expressed in a subset of single IPF cells, even in cells expressing normally AT2-restricted RNAs (Figure 4B). Likewise, single cells expressing goblet cell–associated transcripts — e.g., MUC5AC, SPDEF, LTE, DUSP4, and KRT6A, genes normally selectively expressed in conducting airway epithelial cells — were identified in IPF, but not in control AT2 cells, consistent with the “bronchiolization” typical of IPF. Transcriptomes seen in individual cells demonstrate (a) the loss of normal regional proximal-peripheral cellular identity and (b) coexpression of normally cell type–restricted transcripts in some IPF epithelial cells, indicating loss of normal epithelial cell type identity.
Figure 4. Single-cell RNA analysis identifies altered epithelial gene expression and epithelial cell types in IPF.
(A) Single cells from human IPF (n = 6) and donor (n = 3) distal lung (CD326+) were prepared using the Fluidigm C1 system. RNA was prepared and analyzed from a total of 325 single cells from IPF and 215 cells from donor lungs. Shown are lung epithelial cell markers: EPCAM and CDH1; alveolar type 1 cell markers: AGER and HOPX; alveolar type 2 cell markers: SFTPC, SLC34A2, and ABCA3; proximal lung epithelial cell markers: SOX2, PAX9, TP63, KRT5, KRT14, MUC5B, and SCGB1A1. Expression values were measured in TPM and square root (sqrt) normalized. Cells are shown in solid colors if the expressions of the markers were greater than 1 (TPM). (B) MUC5B, PAX9, and SOX2 were selectively expressed in subsets of IPF cells (MUC5B: n = 24, PAX9: n = 65; SOX2: n = 24) but not present in C1 control cells. Representative genes clustering with MUC5B, PAX9, and SOX2 in IPF cells are shown in the heatmaps. Equal numbers of control cells were randomly selected. IPF cells expressed a diversity of conducting airway epithelial markers not present in control cells, the latter expressing RNAs characteristic of AT2 cells. (C) Only 9 of 325 IPF cells clustered with control cells, the heatmap indicating “AT2”-like expression patterns; however, these 9 normal IPF cells also coexpressed some of the of IPF-associated disease markers. Expression data (TPM) were log10 transformed.
scRNA-seq analysis identifies potential biomarkers for IPF based on the distinct IPF epithelial cell subtypes.
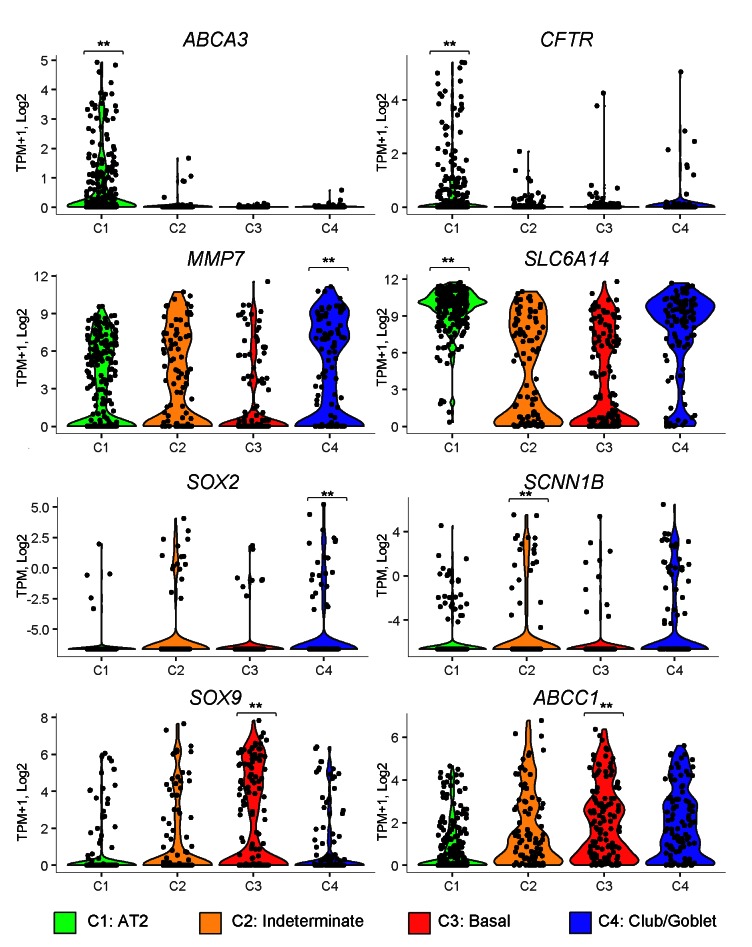
Through the single-cell analysis, we identified signature genes associated with each epithelial cell subtype. Upon validation, these genes may serve as new and cell-selective biomarkers to monitor and predict IPF disease processes that would be useful for diagnosis, prognosis, or therapeutic monitoring. For example, MMP-7, a biomarker indicating IPF prognosis and disease activity in IPF (31, 32), was most highly expressed in the IPF goblet cells (Figure 5). While SOX2 and SOX9 are normally expressed in a mutually exclusive pattern in endoderm of proximal and distal tubules of the developing lung, SOX2 transcripts were enriched in goblet (C4 cluster) and SOX9 transcripts in basal cells (C3 cluster) in IPF. Remarkably, transcripts for SOX2 and SOX9 were frequently coexpressed in indeterminate (C2) cells in IPF, perhaps indicating disruption of proximal-distal patterning (Figure 5). Expression of genes regulating epithelial fluid and electrolyte transport was disrupted in IPF. Chloride transporters, including CLCN2/4/5, SLC26A4, SLC6A14, and CFTR, were significantly decreased in IPF, while the expression of amiloride-sensitive sodium transporter subunits SCNN1G and SCNN1B and bicarbonate transporters CA14, SLC26A8, SLC4A1, and CA5A was induced. Dramatic alternations in expression of associated genes and loss of CFTR are likely to influence mucociliary clearance in a manner similar to that in cystic fibrosis (CF). Expression of ABCA3, a gene essential for AT2 cell lipid transport, was decreased in IPF C2 indeterminate cells and absent in C3 and C4 IPF cells.
Figure 5. Expression of predicted IPF marker genes in 4 epithelial cell types from IPF and control single-cell samples.
Violin plots show the expression of the gene markers in all 540 cells from the 4 cell types. Cell types are color coded. Green: AT2 (n = 219); orange: indeterminate (n = 91); red: basal (n = 131); blue: club/goblet (n = 101). One-tailed Welch’s t test was used to identify cell type–specific gene markers. **P < 0.05.
Prediction of active cell signaling pathways in IPF epithelial cells.
To identify signaling pathways involved in the pathological changes in IPF, we performed enrichment analysis of the KEGG pathways (http://www.genome.jp/kegg/) using differentially expressed genes in the AT2 cell cluster (C1) and each of the IPF cell clusters (C2, C3, and C4). KEGG pathways enriched or suppressed in IPF epithelium were determined by the following criteria: (a) at least 5 genes in the pathway are expressed (transcripts per kilobase million [TPM] ≥1); (b) at least 30% of expressed genes were differentially expressed; and (c) the ratio between the number of C1 differentially expressed genes and the number of IPF differentially expressed genes in the pathway was ≥1.5 or ≤0.67. Pathways were ranked based on the ratios. The average expression of pathway genes in each individual cell was calculated for each significantly altered KEGG pathway. A heatmap (Figure 6A) represents single-cell gene expression profiles of the top 25 ranked KEGG pathways significantly altered in IPF epithelial cells. HIPPO/YAP, TGF-β, and PI3K/AKT signaling pathways were induced in single cells in IPF, findings consistent with RNA data from FACS-sorted cells (Figure 1). Likewise, processes related to normal AT2 cell functions including lipid synthesis and metabolism were suppressed in IPF. Figure 6B shows the relative expression of representative TGF-β signaling pathway genes (BMP1, BMPR1B, INHBA, INHBB, TGFBR1, TGFB1, TGFB2, and SMAD3) in control (n = 215) and IPF (n = 316) cells, and in the 9 IPF cells that clustered together with control AT2 cells. As shown, even the 9 relatively “normal” IPF AT2-like cells expressed significantly higher levels of TGFBR1 and VIM that may represent less advanced stages of IPF pathology (Figure 6B and Supplemental Table 3). Although all IPF cells maintained epithelial cell identities, vimentin, normally selectively expressed in mesenchymal cells, was increased in basal and indeterminate cells in IPF, with highest expression in the relatively “normal” IPF AT2-like cells, perhaps indicating epithelial-mesenchymal transition (EMT) in the early transitional stage of IPF (Supplemental Table 3).
Figure 6. Expression of altered KEGG pathways in human IPF and control single cells.
(A) The heatmap shows the top 25 pathways and differentially expressed genes identified using a 1-tailed Welch’s t test of gene expression between the control AT2 cells (C1) and IPF cell clusters (C2, C3, and C4) using the following criteria: P < 0.01, expressed (TPM ≥1) in at least 80% of cell type with induced gene expression. KEGG pathways enriched or suppressed in IPF epithelium were determined by the following criteria: (a) at least 5 genes in the pathway were expressed (TPM ≥1), (b) at least 30% of expressed genes were differentially expressed, and (c) the ratio between the number of C1 differentially expressed genes and the number of IPF differentially expressed genes in the pathway was ≥1.5 or ≤0.67. 4). Pathways were ranked based on the ratios. The expression of a pathway in a cell was measured by the average expression (TPM + 1, log2 transformed) of differentially expressed genes associated in the pathway. Pathways were clustered using hierarchical clustering analysis with Spearman’s correlation–based distance measure and complete linkage. Cancer- or disease-related pathways were excluded. (B) Representative TGF-β signaling pathway genes (BMP1, BMPR1B, INHBA, INHBB, TGFBR1, TGFB1, TGFB2, and SMAD3) in control (n = 215), IPF (n = 316), and relatively normal IPF cells that clustered with control AT2 cells (n = 9). Data are presented as dot plot with mean ± SEM. P values were determined by Student’s t test. **P < 0.05.
Single-cell transcriptome analysis predicted the activation of EMT from basal to indeterminate cells (Supplemental Figure 5). EMT-related signaling molecules (TGFBR1, Wnt/β-catenin, EGFR, PI3K/AKT), transcription factors (ZEB1, SNAl2, SMAD2/3), and an mRNA splicing factor involved in EMT (ESRP1) were selectively induced in basal cells. While E-cadherin (CDH1) was suppressed in basal cells, mesenchymal markers acquired in EMT including MMPs, vimentin, N-cadherin (CDH12), and fibronectin (FN1) were selectively induced in either basal or indeterminate cells, indicating that basal cells may be the progenitor of the indeterminate cell type. A number of genes regulating planar polarity or EMTs, e.g., PRICKLE1, VANGL1, SNAI2, and VIM, also were induced in IPF cells, consistent with cell shape alterations, and pan-cytokeratin, E-cadherin, and vimentin staining in IPF epithelial cells (Figure 4, Supplemental Figure 5, Supplemental Figure 6A, and Supplemental Table 3). In spite of the enrichment of transcripts related to epithelial cell proliferation in KEGG cells observed in the pathway analysis of IPF cells (Figure 1 and 3), phospho–histone 3 staining was rarely detected in epithelial cells in either normal or IPF lung tissue (Supplemental Figure 6B).
Expression of conducting airway and alveolar cell markers in IPF epithelial cells.
Since single-cell transcriptome analysis identified remarkable changes in epithelial cell gene expression patterns, we utilized immunofluorescence confocal microscopy to distinguish epithelial cells in normal and IPF lungs. In normal lung, cuboidal AT2 epithelial cells were identified by coexpression of both HTII-280 (24) and ABCA3. HOPX intensely stained the cytoplasm and nuclei of squamous AT1 cells and was detected at lower levels in normal AT2 cells (Figure 7). Neither ABCA3 nor HTII-280, AT2 cell markers present in normal lungs, was detected in conducting airways. In contrast to normal alveolar and bronchial tissue, lesions in the periphery of IPF lungs contained epithelial cells of atypical shapes and staining characteristics. Abnormally shaped cells intensely costaining for HOPX, ABCA3, and/or HTII-280 were present in all IPF samples, and normal squamous AT1 cells were rarely detected (Figure 7). Ciliated, basal, and goblet cell markers were frequently observed in IPF lesions, found in close proximity to ABCA3-stained cuboidal cells, indicating a loss of regional specification and epithelial cell gene expression (Figure 7A). The epithelial lining of IPF cysts frequently contained p63-positive basal cells located in close proximity to cells expressing ABCA3 or HOPX. Clusters of KRT14+ “basal” cells were also present in these IPF lesions (Figure 7B). Thus, differentiation-specific cell markers that are normally spatially restricted in conducting versus alveolar regions were frequently found in close contiguity within the IPF lesions. Similarly, individual IPF cells coexpressing SFTPB and MUC5B, markers normally restricted to distinct alveolar and goblet epithelial cell types, were readily identified in IPF tissues, findings consistent with the “abnormal” cell differentiation characteristics seen in the single-cell RNA analyses (Supplemental Figure 7). Although α-SMA was not detected in IPF epithelial cells, vimentin, which normally stains only mesenchymal cells, was detected in subsets of pan-cytokeratin– and E-cadherin–stained epithelial cells, perhaps indicating a partial epithelial to mesenchymal transition, findings consistent with Vim and pathway analysis of the single-cell IPF RNA profiles (Supplemental Figure 6A).
Figure 7. Immunofluorescence confocal microscopy identifies atypical epithelial cell differentiation in IPF.
(A) Peripheral samples of normal and IPF lung tissue were stained for epithelial cell markers used to identify AT2 (HTII-280 and ABCA3), AT1 (HOPX), ciliated (TUBA4A), goblet (MUC5B) cells. Yellow staining indicates coexpression of the proteins. HTII-280 and ABCA3, normally restricted to peripheral/alveolar epithelial cells in normal lung, were expressed in IPF lesions; cystic lesions were variably lined by hyperplastic AT2 cells that stained for ABCA3 in close proximity to MUC5B (goblet) or TUBA4A (ciliated) stained cells. Abnormally shaped epithelial cells variably staining for HOPX, ABCA3, and HTII-280 were characteristic of IPF tissues that generally lacked normal squamous AT1 cells. (B) Epithelial cells expressing conducting airway and alveolar epithelial cell markers were found in close proximity in the IPF lesions (e.g., TP63, KRT14 and MUC5B) and (ABCA3 and HOPX) respectively are shown. Figures are representative of n = 3–5 control and 9 IPF samples, except for KRT14 (n = 3). Images were obtained at ×10 magnification (scale bars: 200 μm). Insets in yellow boxes are at ×60 magnification
Discussion
Formation and repair of the alveolar gas exchange region of the lung requires the precise orchestration of interactions among diverse epithelial, stromal, and immune cells. In spite of extensive pathological, cellular, and genetic studies, the molecular and cellular processes underlying tissue injury and remodeling in IPF remain enigmatic. Molecular analyses in IPF have primarily utilized whole tissue and in vitro studies for genetic and epigenetic analyses. In the present study, we identify profound changes in epithelial cell gene expression at the single-cell level of resolution in IPF. Our findings identified the activation of a number of canonical signaling and transcriptional pathways associated with cell injury and repair in IPF and identify IPF-related epithelial cells of indeterminate differentiation. The respiratory epithelium is capable of robust regeneration from rapid amplifying progenitor cells and various other progenitor cell types, including basal cells in conducting airways and AT2 cells in the alveoli (22). Recent studies support the activation and/or migration of p63/Sox2-expressing basal cells in the alveoli after severe, chronic epithelial cell injury caused by bleomycin and influenza viruses (8, 9). Our present single-cell data support the concept that an abnormal differentiation program has been initiated in the tissue microenvironment of IPF in which the proximal-peripheral patterns of cell differentiation are disrupted, with many respiratory epithelial cells acquiring aberrant, multilineage-like states and some individual cells sharing the characteristics of both conducting airway and alveolar epithelial cells. As such, these cells represent cell types not previously identified during normal lung morphogenesis or at maturity (26, 28).
Present bioinformatics and immunofluorescence evidence strongly supports the concept that the multilineage-like state of IPF epithelial cells is related to the tissue microenvironment in IPF. Since the capture of more than one cell is possible in the Fluidigm C1 apparatus, we carefully considered this possibility in our analyses. Based on our previous single-cell analysis of lung transcript (28), we noted capture of doublets of 7%–9% using present 10- to 17-μm settings on the Fluidigm C1 chip. Doublets were readily identified by shared expression of marker genes crossing two distinct cell types. In the present study, many indeterminate cells (n = 89 cells), coexpressed multilineage markers, including AT2, AT1, and conducting airway epithelial and mesenchymal cell (Fn1, Vim, Col1a1) markers, combinations of which are not expressed by normal AT2 cells. Likewise, gene signatures of AT2 cells (C1) were highly consistent and did not include non-AT2 epithelial or mesenchymal cell markers. This concept is supported by immunofluorescence studies demonstrating the loss of normal proximal-peripheral patterning of epithelial cell types and the presence of individual cells costaining for usually cell type–restricted markers. Whether these cells originate from alveolar or conducting airway progenitors or from subpopulations of distinct progenitors is presently unclear. IPF cells are not typical of normal undifferentiated lung progenitors related to a block in normal alveolar or airway epithelial cell development, but rather represent a failure to suppress multilineage differentiation programs via normal epigenetic mechanisms.
During branching morphogenesis, the proximal-peripheral axis of the lung is demarcated by the mutually exclusive expression of SOX2 (in conducting) and SOX9 in acinar (peripheral) buds of the embryonic lung (22, 26). Consistent with present single-cell RNA studies, we found a paucity of SOX2 and other conducting airway markers in the normal alveolar epithelium (Figure 4) (26, 28, 33–36). In sharp contrast, SOX2 RNA was coexpressed with many AT2 cell–related gene markers (e.g., SFTPA2, CTSH, LPCAT, SCD), including goblet and basal cell–associated RNAs (e.g., P63, KRT5, 6AB, ITGB4, PAX9, MUC5AC, and MUC5B) in IPF. Remarkably, cells coexpressing both SOX2 and SOX9 RNAs were seen in some single cells (mostly in the indeterminate [C2] cells) (Figure 4 and 5). Present immunofluorescence studies demonstrated the loss of normal spatial restriction of alveolar versus conducting airway epithelial cell types. Coexpression of normally relatively restricted AT1 (HOPX) with AT2 (ABCA3 and HTII-280) marker genes and frequent coexpression of SP-B with MUC5B are consistent with loss of region-specific cellular identities in IPF (Figure 7). Prediction of EZH2 as a candidate regulator of altered fate in IPF epithelial cells (Supplemental Figure 2) may reflect changes in epigenetic states associated with widespread and dramatic changes in gene expression, a concept previously supported by methylation studies in IPF (37–39).
Activation of canonical regulatory pathways in IPF epithelial cells.
The cellular origins of the abnormal signaling and transcriptional networks are confounded by the complex tissue remodeling seen in IPF, which includes recruitment of inflammatory and stromal cells, obscuring the role of individual cell types in its pathogenesis. Neutrophilic, lymphocytic, and monocytic infiltrates are prominent in IPF. In the present study, expression of a number of cytokines, chemokines, and growth factors was increased in IPF epithelial cells, supporting a unique role in the secretion of molecules influencing the activity of myofibroblasts and recruitment of inflammatory cells (Supplemental Figure 3). As such, IPF epithelial cells may not be dissimilar from cancer cells that contribute to a tumorigenic microenvironment. Further similarities between cancer and IPF are indicated by the enhancement of RNAs related to cell migration or “metastatic” activities (Figure 1 and Supplemental Figure 3). Alternatively, signals from inflammatory cells and fibroblasts may activate epithelial cell gene expression in IPF. CD326/HTII-280 (sorted cells and single-cell isolates) IPF cells expressed genes associated with activation of canonical TGF-β, HIPPO/YAP, PI3K/AKT, p53, and WNT signaling cascades consistent with extensive crosstalk among these canonical pathways that likely function in an integrated network (Figure 6 and Supplemental Figures 2 and 3). This abnormal cell signaling signature was most strongly expressed in the basal IPF cells, perhaps indicating their importance in the pathogenesis of IPF. The diverse, interacting signaling pathways activated in IPF support the concept that strategies for treatment of IPF may require pharmaceutical targeting of multiple molecular pathways. Taken together, the profound changes in gene expression seen in epithelial cells in IPF suggest global epigenetic changes in chromatin that may influence diverse cellular functions, including cell differentiation, proliferation, survival, and the production of effector molecules in the pathogenesis of IPF. The finding that EZH2 was predicted to be a strong transcriptional driver regulating genes involved in cell growth, migration, and chemotaxis in IPF, as shown in Supplemental Figure 2, supports the concept that changes in chromatin organization may influence the abnormal gene expression patterns involved in the pathogenesis of IPF.
Dysregulation of CF-regulated genes in IPF.
Analysis of RNAs differentially regulated in IPF epithelial cells identified a remarkable loss of CFTR, SLC26A9, SLC6A14, and SLC9A3 (Figure 2B and Figure 5) and mutations in genes causing (CFTR) or modifying the severity of CF lung diseases (40, 41). In sharp contrast to the loss of CFTR and associated solute carriers, expression of amiloride-sensitive sodium transporter subunits SCNN1G and SCNN1B, whose hyperactivity in CF contributes to airway dehydration (42), was increased in IPF cells. Taken together, these data strongly suggest that the dysregulation of epithelial ion transport properties are shared in CF- and IPF-related pathologies that include pulmonary inflammation and goblet cell metaplasia.
Activation of TGF-β–regulated pathways in IPF epithelia.
RNA profiles from both FACS-sorted and single-cell isolates demonstrate the strong expression of TGF-β–related gene networks in IPF, findings consistent with the role of TGF-β and its pathogenesis (see ref. 43 for review). Present findings support a role for the respiratory epithelium in the activation of pathways that are predicted to interact with fibroblasts and myofibroblasts, which are implicated in fibrotic remodeling in IPF. Predicted regulators and biological processes in normal AT2 cells were highly distinct from those in IPF basal, goblet, and indeterminate cells, and these were most pronounced in the IPF basal cells, which were enriched in genes mediating “cell migration,” “wound healing,” “EMT,” and “TGF-β signaling” (Figure 8). Increased expression of TGF-βR1, SMAD1/2, SNA1/2, EGFR, and PI3K pathways, support the concept that the abnormal activation and differentiation of basal progenitor cells play an important role in IPF pathogenesis. The diverse proximal airway-like cells in IPF selectively expressed cytokines, chemokines, and genes associated with the distinct biological processes, indicating that each cell type likely contributes uniquely to the complex pathological microenvironment and cell-cell communications by which epithelial, inflammatory, and stromal cells interact in IPF (Supplemental Figure 3).
Figure 8. Bioprocesses and gene networks influenced by IPF.
Single-cell RNA-sequencing analysis of human IPF and control epithelial cells identified 4 distinct lung epithelial cell subtypes. Cell-specific gene signatures and associated pathways, bioprocesses, and predicted driving forces (key regulators) of each cell type are shown. The analysis predicts crosstalk among individual cell types at the level of upstream regulators, bioprocesses, and genes, as illustrated in this summary chart.
Activation of the HIPPO/YAP pathway in IPF.
Present RNA expression data indicate an unrecognized role for the HIPPO/YAP pathway in epithelial cells in IPF. Activation of YAP by genetic deletion of the genes encoding serine/threonine-specific kinases (STKs) Mst1 and Mst2 or increased expression of YAP caused pulmonary lesions with features shared with IPF (44). Increased CTGF and AJUBA (both targets of YAP activation) and alterations in epithelial cell shape, planar polarity, and differentiation were seen in conducting airways of the mice in which YAP was activated (44). Epithelial shape changes and increased expression of SNAI1, VIM, and planar polarity genes indicate a partial EMT-like phenotype in IPF epithelial cells, likely related in part to increased TGF-β and HIPPO/YAP signaling (Figure 6 and Supplemental Figures 5 and 6). A recent study implicated a YAP/miR-130/301–mediated pathway in stretch responses and fibrotic remodeling, although the cell types involved were not identified (45).
The present study provides what we believe to be the first in-depth transcriptomes from normal human AT2 cells and those from IPF epithelial cells at the single-cell level. We identify previously undetected subpopulations of epithelial cells, revealing relationships between individual cells, the activation of cell-specific regulatory processes by cell-specific transcriptional regulators, and potential signaling interactions among diverse lung epithelial cell types (Figure 8). As summarized in Figure 8, each of the 4 distinct epithelial cell types (C1–C4) expresses distinct combinations of regulators and genes mediating their unique contributions to the complex cellular microenvironment involved in the pathogenesis of IPF.
Conclusions.
Rapid advances in RNA/DNA sequencing, cell isolation, imaging, and systems biology are enabling ever more detailed insight into the genes and processes controlling cell behaviors and functions. Single-cell transcriptomic analyses are readily applied to small tissue samples and are capable of providing new insights into the biological processes active within and among specific cells that are obscured in analysis of whole tissues. Single-cell transcriptomic analyses revealed a diversity of transcriptional “states” of individual IPF cells, blurring present concepts regarding precise epithelial cell identities, and the biological processes involved in lung health and disease.
Methods
Study population.
Deidentified lung tissue samples were obtained from Cedars Sinai Medical Center, Duke University Medical Center, or the University of North Carolina at Chapel Hill. IPF tissues (n = 3) were collected from lung explants of IPF patients undergoing lung transplantation, and lung lobes from adult donors that were resected on the basis of size incompatibility or that were deemed unsuitable for transplant were utilized as control tissues (n = 3) for cell sorting by FACS. Distinct control (n =3) and IPF (n =6) lung samples were used for single-cell isolation and analysis. Diagnostic criteria for IPF established by the American Thoracic Society/European Respiratory Society were used. Additional IPF and control samples were provided by Andreas Gunther (Pulmonary and Critical Care Medicine, Justus-Liebig-University, Giessen, Germany).
Lung histology and immunostaining.
Paraffin sections (5 μm) were stained with H&E or deparaffinized in xylene and rehydrated in a series of graded alcohols. For immunohistochemical staining, endogenous peroxidase activity was blocked in a solution of methanol/3% hydrogen peroxide. Antigen retrieval was performed in 0.1 M citrate buffer (pH 6.0) by microwaving. Slides were blocked for 2 hours at room temperature using 4% normal donkey serum in 0.1 M PBS containing 0.1% Triton X-100 (T). Tissue sections were incubated with primary antibodies diluted in blocking buffer for an extended incubation period, approximately 48 hours. Primary antibodies are listed in Supplemental Table 4. Appropriate secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 555 or 568, or Alexa Fluor 647, were used at a dilution of 1:200 in blocking buffer. Nuclei were counterstained with DAPI (1 μg/ml) (Invitrogen). Sections were mounted using ProLong Gold mounting medium and coverslipped.
Confocal microscopy.
Tissue sections stained for immunofluorescence were imaged on an inverted Nikon A1R confocal microscope (×10, ×20, and ×60 WI) NA 1.27 objective using an 1.2 AU pinhole. Maximum intensity projections of multilabeled Z stack images obtained sequentially using channel series across the 5-μm-thick sections were generated using Nikon NIS-Elements software. Brightfield images were captured using an Zeiss Axio ImagerA2 microscope utilizing Axiovision software.
RNA extraction and sequencing.
Peripheral human lung tissue with airways no larger than 2 mm in diameter and lacking visceral pleura was mechanically minced and enzymatically dissociated to generate a single-cell suspension. Briefly, finely minced tissues were washed in Ham’s F12 (Corning) at 4°C for 5 minutes with rocking, followed by centrifugation for 5 minutes at 600 g and 4°C. The minced cleaned tissue was then incubated in DMEM/F12 (Life Technologies) containing 2 mg/ml Dispase (Corning), incubated at 4°C with rocking for 1 hour, and incubated for another 30 minutes at 37°C. Further elastase/trypsin digestion was then accomplished by the addition of an equal volume of Ham’s F12 medium containing 5 U/ml elastase (Worthington Biochemical Corp.) and 0.125% trypsin/EDTA (Corning). The reaction was arrested by addition of HBSS containing 0.01% HEPES, 0.5 M EDTA, and 0.02% FBS. Triturated tissue solution was subjected to a final protease digestion of 1 volume DNase I for 15 minutes at 37°C. Cells were plated overnight on collagen-coated plates in bronchial epithelial growth medium (Lonza) and harvested by mild trypsin digestion.
Epithelial cell FACS.
Dissociated single-cell preparations from peripheral lung of IPF patients (n = 3) and controls (n = 3) from cohort 2 were enriched for AT2 epithelial cells by FACS for CD326 (CD326) double positive, CD45 (hematopoietic) negative, CD31 (endothelial) negative cells, and HTII-280 after dissociation by proteases (Supplemental Figure 8). Cells were plated overnight and sorted. After sorting, cells were preserved in either RNAprotect Cell (QIAGEN) or RNAlater RNA stabilization (QIAGEN) reagent. Total RNA was isolated using an RNeasy Micro Kit (QIAGEN) by incorporating an on-column DNAsel digestion step. Amplified cDNA was generated using the Ovation RNA-Seq System V2 kit (NuGEN) to amplify both polyA+ and non-polyadenylated RNA transcripts. Sequencing libraries were prepared using the Nextera XT DNA Sample Preparation Kit (Illumina). Next-generation sequencing of equimolar pools of cDNA libraries was performed using a single read 50 rapid flow cell on a HiSeq 2500 sequencing platform (Illumina), generating greater than 30 million raw reads per sample. Distinct “basal” cell populations were prepared by FACS, selecting CD326 (EPCAM) NGFR double positive cells from peripheral lung tissue from patients with IPF (n = 3). Due to low abundance of basal cells in normal lung tissue, this procedure failed to extract sufficient RNA from the normal donor lung.
Single-cell isolation.
Cells were isolated from 2 normal controls and 6 IPF tissue samples prepared as described for FACS, except that cells were sorted initially with CD326 prior to single-cell isolation. For single-cell analysis cell capture, lysis, reverse transcription, and cDNA amplification were performed on the C1 integrated fluidic circuit (IFC) for mRNA-seq on a Fluidigm C1 Single-Cell Auto Prep System following the manufacturer’s protocol. Medium-sized C1 mRNA-Seq chips (10–17 μm) were used to capture each cell cycle fraction. The C1 Auto Prep System captures the dissociated single cells across 96 wells and performs cell lysis, cDNA synthesis with reverse transcription, and PCR reaction using the SMARTer Ultra Low Input RNA Kit v3 (ClonTech). Cells captured across the 96 wells are manually inspected as a quality control measure to remove empty well, doublets, or debris-containing wells. cDNA from several representative cells are checked by High Sensitivity DNA chips using Fragment Analyzer (Advanced Analytical), and all cDNA libraries are quantitated via Qubit (Thermo Fisher Scientific). Libraries for each of the 96 captured cells are prepared using the Illumina Nextera XT DNA sample preparation kit with 96 dual barcoded indices. Off-chip controls were prepared in the same manner, except with a standard 96-well thermal cycler as opposed to on the C1. Single-cell libraries are multiplexed and sequenced across 4 lanes of a NextSeq 500 platform (Illumina) using 75-bp single-end sequencing. On average, about 4–5 million reads were generated from each single-cell library.
RNA-seq analysis on sorted cell populations.
The raw sequenced reads were aligned from FASTQ files to the human genome build GRCh37/hg19 and the UCSC reference transcriptome (http://ccb.jhu.edu/software/tophat/index.shtml) using “Cufflinks” (46). Samples were further processed via trimmed mean normalization (47). Differentially expressed genes in control CD326/HTII-280 and IPF-CD326/HTII-280 sorted lung epithelial cell types were identified using Smyth’s moderated t test and Benjamini-Hochberg procedure for adjusted P value (FDR). Genes with FPKM (fragments per kilobase of exon per million fragments mapped) of <1 in all samples were removed from further analysis. A gene was considered to be differentially expressed when the P value was ≤0.05 (with FDR correction) and expression fold change ≥1.5 was observed in at least one condition. Differentially expressed genes were subject to hierarchical clustering using Euclidean distance and average linkage to measure the cluster similarity/dissimilarity. Gene set enrichment and pathway analysis were performed using ToppGene software (https://toppgene.cchmc.org/) and Ingenuity Pathway Analysis (http://www.ingenuity.com).
RNA-seq analysis of single-cell isolates.
For single-cell RNA-seq analysis, we developed an analytic pipeline to identify major cell types and unique gene expression signatures for individual cells (27). Briefly, the analytic pipeline consisted of 4 components that included: gene prefiltering and normalization, reiterative cell type mapping (hierarchical clustering followed by PCA), cell-specific signature gene identification, and driving force prediction for each cell type. Detailed methods and code for this analytic pipeline are found in ref. 27 (https://research.cchmc.org/pbge/sincera.html). Signature genes of each cell type were subject to functional enrichment analysis, pathway analysis, and potential upstream regulators prediction using the ToppGene Suite and Ingenuity Pathway Analysis.
For scRNA-seq pathway analysis, differentially expressed genes were identified using 1-tailed Welch’s t test of gene expression between the AT2 cell cluster (C1) and IPF cell clusters (C2, C3, and C4) using the following criteria: P < 0.01 for at least one condition, expression value TPM ≥1 in at least 80% of cell type with induced gene expression. KEGG pathways enriched or suppressed in IPF epithelium were determined by the following criteria: (a) at least 5 genes in the pathway had expression value of TPM ≥1, (b) at least 30% of expressed genes were differentially expressed, and (c) the ratio between the number of C1 differentially expressed genes and the number of IPF differentially expressed genes in the pathway was ≥1.5 or ≤0.67. (d) Pathways were ranked based on the ratios. The expression of a pathway in a cell was measured by the average expression (TPM + 1, log2 transformed) of differentially expressed genes of the pathway in the cell. Pathways were clustered using hierarchical clustering analysis with Spearman’s correlation–based distance measure and complete linkage. Cancer or disease-related pathways were excluded.
Statistics.
All data are expressed as mean ± SEM. For sorted cell RNA-seq analysis, statistical significance was determined using the moderated t statistics by Smyth et al. (48), followed by the Benjamini-Hochberg procedure for adjusted P value (FDR); a P value of less than 0.05 was considered statistically significant. For scRNA-seq analysis, 1-tailed Welch’s t test with a P value less than 0.01 was used for cell type–specific signature gene selection.
Study approval.
Deidentified lung tissue samples were obtained from Cedars Sinai Medical Center, Duke University Medical Center, or the University of North Carolina at Chapel Hill and approved by the Human Research Committee at Cedars Sinai Medical Center. Additional IPF and control samples provided by Andreas Gunther were reviewed and approved by the Ethics Committee of Justus-Liebig-University of Giessen, with sample and patient characteristics reported previously (6).
Data availability.
The full data set is available in the NCBI’s Gene Expression Omnibus (GEO GSE86618 and GSE94555). The analytic and interpreted results from this study will be incorporated into LungGENS website and database hosted by our group (https://research-test.cchmc.org/pbge/lunggens/mainportal.html) (28).
Author contributions
YX designed and completed RNA-seq analyses and contributed to writing, editing, and interpretation of expression data. TM designed and performed single-cell isolation procedures, FACS sorting, and RNA preparations. AS designed and completed confocal microscopy and fluorescence and RNA studies, and contributed to writing and editing. YD contributed to RNA-seq analysis, interpretation, and integration of data with knowledge bases. MG developed the scRNA-seq analytic pipeline, and performed single-cell analysis and interpretation of data. JT contributed to RNA sequencing and analysis. VAF developed RNA libraries and DNA sequencing. KAWB reviewed lung pathology and contributed to writing and editing. AKTP reviewed data and contributed to writing and editing. JJG contributed to data analysis and writing. BRS co-designed cell experiments, supervised cell isolation and RNA preparation, and edited the manuscript. JAW co-designed experiments, reviewed all data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We appreciate support from Ann Maher and Joseph Kitzmiller in manuscript preparation, Shawn Grant and Gail Macke for tissue sectioning, the CCHMC Gene Expression Core for technical assistance with RNA-seq, and Andreas Günther and coworkers of Giessen University, who provided IPF tissues. We are grateful to Jordan Brown and Lindsay Spurka of the Cedars-Sinai Genomics Core for technical assistance with scRNA-seq, Adrianne Kurkciyan for assistance with human lung tissue procuring and processing, and Dr. Scott Randell, UNC-Chapel Hill, for providing normal human lung tissue for isolation of HTII-280 cells. Supported by: HL122642, HL110967, HL108793, and CIRM LA1-06915.
Version 1. 12/08/2016
Electronic publication
Version 2. 03/16/2017
The accession number for the RNA-seq data was incorrectly noted. The correct sentence has been updated in this version as: The full data set is available in the NCBI's Gene Expression Omnibus (GEO GSE86618 and GSE94555). The authors regret the error.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2016;1(20):e90558. doi:10.1172/jci.insight.90558.
Contributor Information
Yan Xu, Email: yan.xu@cchmc.org.
Takako Mizuno, Email: takako.mizuno@cshs.org.
Anusha Sridharan, Email: anusha.sridharan@cchmc.org.
Yina Du, Email: yina.du@cchmc.org.
Minzhe Guo, Email: Minzhe.Guo@cchmc.org.
Jie Tang, Email: jie.tang@cshs.org.
Barry R. Stripp, Email: barry.stripp@cshs.org.
References
- 1.Blackwell TS, et al. Future directions in idiopathic pulmonary fibrosis research. An NHLBI workshop report. Am J Respir Crit Care Med. 2014;189(2):214–222. doi: 10.1164/rccm.201306-1141WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodwin AT, Jenkins G. Molecular endotyping of pulmonary fibrosis. Chest. 2016;149(1):228–237. doi: 10.1378/chest.15-1511. [DOI] [PubMed] [Google Scholar]
- 3.Steele MP, Schwartz DA. Molecular mechanisms in progressive idiopathic pulmonary fibrosis. Annu Rev Med. 2013;64:265–276. doi: 10.1146/annurev-med-042711-142004. [DOI] [PubMed] [Google Scholar]
- 4.Barkauskas CE, Noble PW. Cellular mechanisms of tissue fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am J Physiol, Cell Physiol. 2014;306(11):C987–C996. doi: 10.1152/ajpcell.00321.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camelo A, Dunmore R, Sleeman MA, Clarke DL. The epithelium in idiopathic pulmonary fibrosis: breaking the barrier. Front Pharmacol. 2014;4:173. doi: 10.3389/fphar.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plantier L, et al. Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax. 2011;66(8):651–657. doi: 10.1136/thx.2010.151555. [DOI] [PubMed] [Google Scholar]
- 7.Seibold MA, et al. The idiopathic pulmonary fibrosis honeycomb cyst contains a mucocilary pseudostratified epithelium. PLoS One. 2013;8(3):e58658. doi: 10.1371/journal.pone.0058658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517(7536):621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuo W, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517(7536):616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alder JK, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. 2008;105(35):13051–13056. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronkhite JT, et al. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178(7):729–737. doi: 10.1164/rccm.200804-550OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.d’Adda di Fagagna F, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426(6963):194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 13.Fingerlin TE, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45(6):613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kropski JA, Blackwell TS, Loyd JE. The genetic basis of idiopathic pulmonary fibrosis. Eur Respir J. 2015;45(6):1717–1727. doi: 10.1183/09031936.00163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84(1):52–59. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulugeta S, Maguire JA, Newitt JL, Russo SJ, Kotorashvili A, Beers MF. Misfolded BRICHOS SP-C mutant proteins induce apoptosis via caspase-4- and cytochrome c-related mechanisms. Am J Physiol Lung Cell Mol Physiol. 2007;293(3):L720–L729. doi: 10.1152/ajplung.00025.2007. [DOI] [PubMed] [Google Scholar]
- 17.Mulugeta S, Nguyen V, Russo SJ, Muniswamy M, Beers MF. A surfactant protein C precursor protein BRICHOS domain mutation causes endoplasmic reticulum stress, proteasome dysfunction, and caspase 3 activation. Am J Respir Cell Mol Biol. 2005;32(6):521–530. doi: 10.1165/rcmb.2005-0009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nogee LM, Dunbar AE, Wert S, Askin F, Hamvas A, Whitsett JA. Mutations in the surfactant protein C gene associated with interstitial lung disease. Chest. 2002;121(3 Suppl):20S–21S. doi: 10.1378/chest.121.3_suppl.20s. [DOI] [PubMed] [Google Scholar]
- 19.Nogee LM, Dunbar AE, Wert SE, Askin F, Hamvas A, Whitsett JA. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N Engl J Med. 2001;344(8):573–579. doi: 10.1056/NEJM200102223440805. [DOI] [PubMed] [Google Scholar]
- 20.Stuart BD, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet. 2015;47(5):512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitsett JA, Wert SE, Weaver TE. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–393. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan BL, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusko RL, et al. Integrated Genomics reveals convergent transcriptomic networks underlying chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;194(8):948–960. doi: 10.1164/rccm.201510-2026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez RF, Allen L, Gonzales L, Ballard PL, Dobbs LG. HTII-280, a biomarker specific to the apical plasma membrane of human lung alveolar type II cells. J Histochem Cytochem. 2010;58(10):891–901. doi: 10.1369/jhc.2010.956433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40(2):124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 26.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo M, Wang H, Potter SS, Whitsett JA, Xu Y. SINCERA: a pipeline for single-cell RNA-seq profiling analysis. PLoS Comput Biol. 2015;11(11):e1004575. doi: 10.1371/journal.pcbi.1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Y, Guo M, Whitsett JA, Xu Y. ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax. 2015;70(11):1092–1094. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G, et al. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest. 2009;119(10):2914–2924. doi: 10.1172/JCI39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajavelu P, Chen G, Xu Y, Kitzmiller JA, Korfhagen TR, Whitsett JA. Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J Clin Invest. 2015;125(5):2021–2031. doi: 10.1172/JCI79422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosas IO, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008;5(4):e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Kaminski N. Biomarkers in idiopathic pulmonary fibrosis. Curr Opin Pulm Med. 2012;18(5):441–446. doi: 10.1097/MCP.0b013e328356d03c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Artinian N, Cloninger C, Holmes B, Benavides-Serrato A, Bashir T, Gera J. Phosphorylation of the Hippo pathway component AMOTL2 by the mTORC2 kinase promotes YAP signaling, resulting in enhanced glioblastoma growth and invasiveness. J Biol Chem. 2015;290(32):19387–19401. doi: 10.1074/jbc.M115.656587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackett NR, et al. RNA-Seq quantification of the human small airway epithelium transcriptome. BMC Genomics. 2012;13:82. doi: 10.1186/1471-2164-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiling K, et al. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med. 2013;187(9):933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, et al. Transcriptional programs controlling perinatal lung maturation. PLoS One. 2012;7(8):e37046. doi: 10.1371/journal.pone.0037046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabinovich EI, et al. Global methylation patterns in idiopathic pulmonary fibrosis. PLoS One. 2012;7(4):e33770. doi: 10.1371/journal.pone.0033770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabinovich EI, Selman M, Kaminski N. Epigenomics of idiopathic pulmonary fibrosis: evaluating the first steps. Am J Respir Crit Care Med. 2012;186(6):473–475. doi: 10.1164/rccm.201208-1350ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders YY, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186(6):525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corvol H, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright FA, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livraghi-Butrico A, et al. Loss of Cftr function exacerbates the phenotype of Na(+) hyperabsorption in murine airways. Am J Physiol Lung Cell Mol Physiol. 2013;304(7):L469–L480. doi: 10.1152/ajplung.00150.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolters PJ, Collard HR, Jones KD. Pathogenesis of idiopathic pulmonary fibrosis. Annu Rev Pathol. 2014;9:157–179. doi: 10.1146/annurev-pathol-012513-104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange AW, Sridharan A, Xu Y, Stripp BR, Perl AK, Whitsett JA. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J Mol Cell Biol. 2015;7(1):35–47. doi: 10.1093/jmcb/mju046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertero T, et al. A YAP/TAZ-miR-130/301 molecular circuit exerts systems-level control of fibrosis in a network of human diseases and physiologic conditions. Sci Rep. 2015;5:18277. doi: 10.1038/srep18277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia-Escudero LA, Gordaliza A. Robustness properties of k means and trimmed k means. J Am Stat Assoc. 1999;94(447):956–969. [Google Scholar]
- 48.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full data set is available in the NCBI’s Gene Expression Omnibus (GEO GSE86618 and GSE94555). The analytic and interpreted results from this study will be incorporated into LungGENS website and database hosted by our group (https://research-test.cchmc.org/pbge/lunggens/mainportal.html) (28).