Significance
Bacteria use small molecules to mediate their relationships with nearby microbes, and these molecules represent both a promising source of therapeutic agents and a model system for the evolution and dissemination of molecular diversity. This study deals with one such molecule, selvamicin, which is produced by ant-associated bacteria. These bacteria protect the ants’ nests against fungal pathogens. Selvamicin is an atypical member of a clinically important class of antifungal agents, and it appears to have both better therapeutic properties and a different mechanism of action. Further, the genes for producing it are found on the bacteria’s chromosome in one ant nest but on a plasmid in another, illustrating the likely path by which it has spread.
Keywords: antifungal, horizontal gene transfer, biosynthesis, symbiosis, natural products
Abstract
The bacteria harbored by fungus-growing ants produce a variety of small molecules that help maintain a complex multilateral symbiosis. In a survey of antifungal compounds from these bacteria, we discovered selvamicin, an unusual antifungal polyene macrolide, in bacterial isolates from two neighboring ant nests. Selvamicin resembles the clinically important antifungals nystatin A1 and amphotericin B, but it has several distinctive structural features: a noncationic 6-deoxymannose sugar at the canonical glycosylation site and a second sugar, an unusual 4-O-methyldigitoxose, at the opposite end of selvamicin’s shortened polyene macrolide. It also lacks some of the pharmacokinetic liabilities of the clinical agents and appears to have a different target. Whole genome sequencing revealed the putative type I polyketide gene cluster responsible for selvamicin’s biosynthesis including a subcluster of genes consistent with selvamicin’s 4-O-methyldigitoxose sugar. Although the selvamicin biosynthetic cluster is virtually identical in both bacterial producers, in one it is on the chromosome, in the other it is on a plasmid. These alternative genomic contexts illustrate the biosynthetic gene cluster mobility that underlies the diversity and distribution of chemical defenses by the specialized bacteria in this multilateral symbiosis.
Fungus-growing ants range from central Argentina to New York State, and in tropical regions they are the dominant herbivores. The complex web of interactions involving these ants, their fungal crops, specialized pathogens, and symbiotic bacteria has become both a model system for chemical ecology and a productive source of naturally occurring small molecules (1–4).
Fungus-growing ants collect plant material to feed their fungal crop, which metabolizes the plant matter to provide nutrients for the ants. Pathogenic ascomycetous fungi, especially members of the genus Escovopsis, threaten the fungal crop. In response, the ants maintain antibiotic-producing Actinobacteria (genus Pseudonocardia) to provide chemical defenses (5). These bacteria produce antifungal and/or antibacterial agents, including representatives of both nonribosomal peptide synthetase and polyketide synthase (PKS) biosynthetic pathways (6–9).
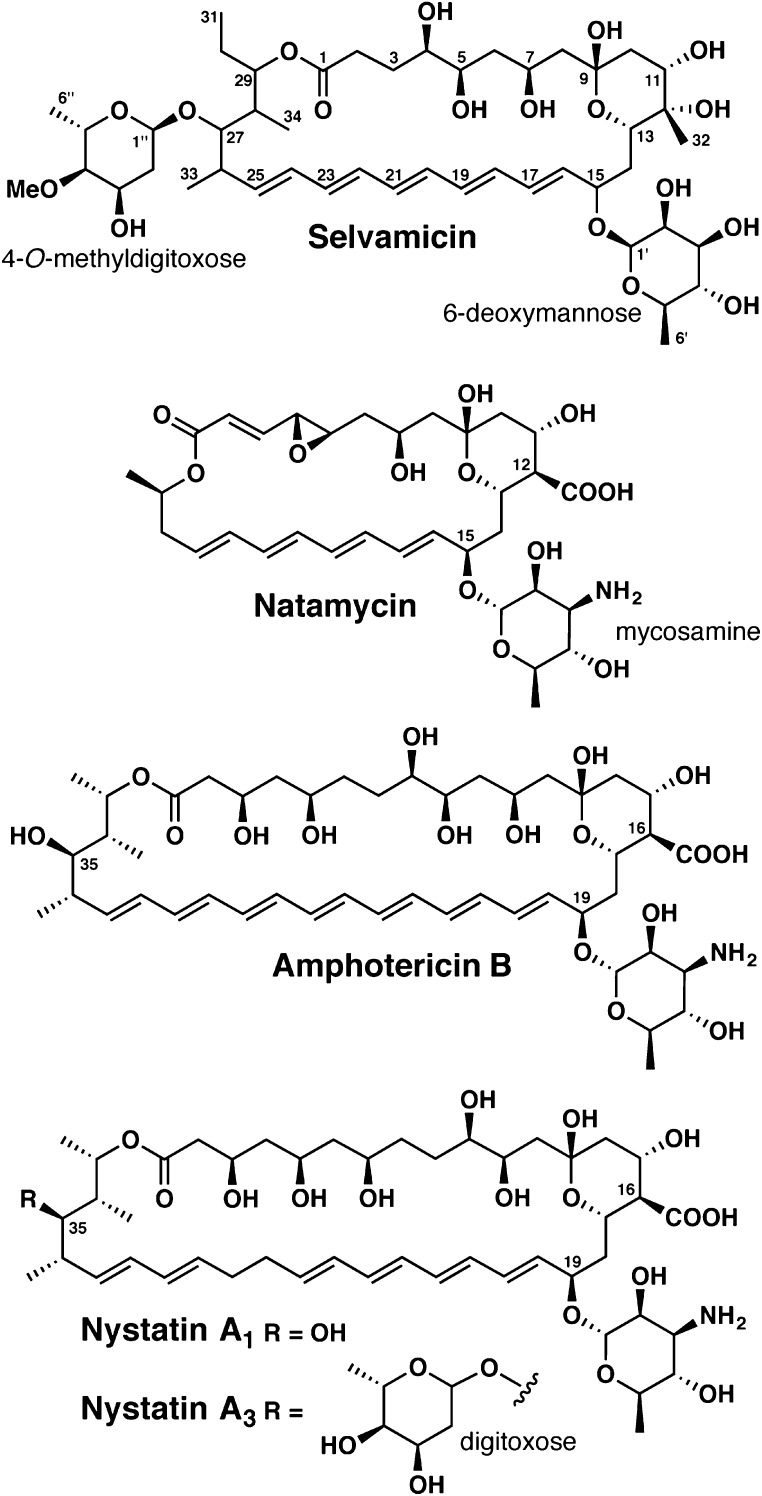
As part of a systematic study of Pseudonocardia isolates derived from the basal fungus-growing ant genus Apterostigma, we discovered an unusual antifungal polyene, which we have named selvamicin (Fig. 1). Selvamicin was found in two bacterial isolates from nearby ant nests. Selvamicin shares features with the clinically important antifungal agents amphotericin B and nystatin A1, both of which are on the World Health Organization’s List of Essential Medicines (10), and with the food preservative and topical antifungal natamycin (Fig. 1). Amphotericin B and nystatin A1 have serious liabilities including high toxicity and negligible oral bioavailability, and their continued use reflects the lack of better alternatives (11, 12).
Fig. 1.
Structures of selvamicin and antifungal polyene natural products currently in clinical use.
Several features distinguish selvamicin from nystatin A1 and amphotericin B: a second sugar, a truncated macrocyclic core, and missing carboxylate and ammonium groups. The biosynthetic gene cluster for selvamicin, which includes a subcluster of biosynthetic genes for the distinctive second sugar, was identified bioinformatically. Whole genome sequencing revealed that in one isolate the cluster resides on the chromosome while in the other it resides on a plasmid. These divergent genomic contexts highlight the gene cluster mobility that introduces and disseminates molecular diversity in this system.
Results and Discussion
Discovery and Structure Elucidation.
We examined two Pseudonocardia isolates from ants in the genus Apterostigma collected at La Selva Biological Station, Costa Rica, HH130629-09 and HH130630-07 (hereafter LS1 and LS2, respectively). We evaluated antifungal activity of organic-soluble extracts of both cultured strains against the common human fungal pathogen Candida albicans. The LS1 extract was active and we used activity-guided fractionation through a C18 cartridge followed by reverse-phase HPLC to trace this activity to a molecule with a previously unreported molecular formula of C47H76O18 [high-resolution electrospray ionization mass spectrometry (HR-ESI-MS) [M+Na]+ calcd 951.4924, expt 951.4928]. We examined the LS2 extract by high-resolution liquid chromatography–mass spectrometry (LC-MS) and observed the same compound, although at approximately fivefold lower abundance. The active compound’s UV-vis spectrum is characteristic of a polyene, with three prominent peaks (319, 334, 352 nm) consistent with a chromophore of five conjugated double bonds (SI Appendix, Fig. S1). Subsequent NMR analysis using a variety of 2D methods (COSY, TOCSY, HMBC, H2BC, and ROESY) revealed this compound to be an unreported polyene macrolide, which we have named selvamicin after the site of our original collection.
Selvamicin production can be greatly up-regulated by adding high concentrations of sodium butyrate (150 mM) to the culture medium (SI Appendix, Fig. S7), consistent with reports that butyrate can regulate secondary metabolism (13). We observed 13C labeling of selvamicin when [1-13C] sodium butyrate was used, indicating that butyrate can also act as a metabolic precursor, and similar results were obtained with [1-13C] propionate (SI Appendix, Fig. S8).
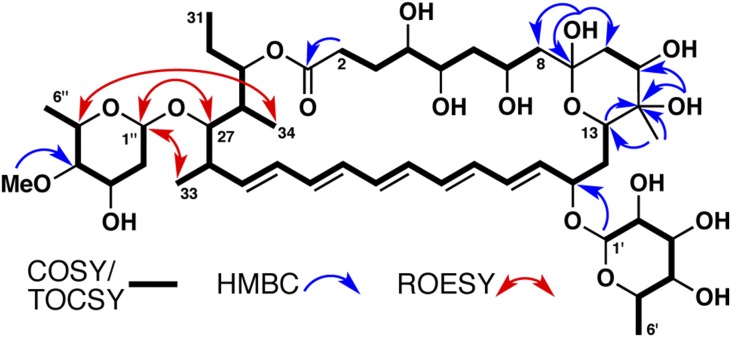
COSY and TOCSY correlations allowed us to construct two major fragments of the selvamicin macrolide: one from C2–C8 and another from C13 across the pentaene to the molecule’s terminus at C31 (overlap of the polyene resonances prevented definitive assignments of C19–C24, Fig. 2). HMBC couplings link the C2–C8 fragment to quaternary carbons at either end: an ester carbonyl at C1 (172.7 ppm) and a hemiketal at C9 (97.3 ppm). The hemiketal forms a six-membered ring established by a series of HMBC couplings from the hemiketal OH at C9, a tertiary alcohol and methyl substituent at C12, and the other bridgehead carbon at C13. H2BC correlations support the placement of substituents along the macrolide core of selvamicin (SI Appendix, Fig. S2). A series of ROESY correlations establishes an extended geometry for the C2–C8 aliphatic chain and a chair conformation for the hemiketal ring (SI Appendix, Fig. S2). These correlations, corroborated by available scalar coupling constants, allowed the assignment of relative stereochemistry from C4 to C13.
Fig. 2.
Key NMR correlations establishing the planar structure of selvamicin.
Our NMR analysis also revealed two sugars in the structure of selvamicin. COSY and HMBC couplings revealed their planar structures as 6-deoxy and 2,6-dideoxy hexoses, as shown in Fig. 2. To better resolve the crowded sugar CH signals and reveal additional peak fine structure, we reacted selvamicin with acetic anhydride to modify its free hydroxyl groups. In the acetylation product, the hemiketal at C9 was instead observed as a ketone, and with the exception of the tertiary alcohol at C12, all OH groups were acetylated (SI Appendix, Fig. S3). Scalar couplings and ROESY correlations allowed the acetylated sugars in this product to be assigned as (Ac)3-β-6-deoxymannose and Ac-α-4-O-methyldigitoxose (SI Appendix, Fig. S4). The absolute configurations of the sugars were not determined.
A clear HMBC coupling from the anomeric proton of the β-6-deoxymannose places this sugar at C15 of selvamicin (Fig. 2). Whereas no HMBC couplings were observed for the anomeric proton of 4-O-methyldigitoxose, a series of ROESY correlations (1′′-H/27-H, 1′′-H/33-H, 5′′-H/34′′-H) locates this sugar on the opposite side of the macrolide at C27. The 1H and 13C chemical shifts of the CH at position 27 support an oxygen substituent linking this sugar. From C25–C31, we observed broadened 1H and 13C resonances, which obscured the couplings needed to establish relative stereochemistry in this region. This peak broadening could reflect conformational flexibility near the 4-O-methyldigitoxose attachment.
Selvamicin’s structure diverges from the antifungal polyenes amphotericin B, nystatin A1, and natamycin in several key respects. Its 30-membered polyene macrolide core is intermediate between that of the smaller antifungal natamycin and those of amphotericin B and nystatin A1. Selvamicin’s unusual glycosylation is also noteworthy. The 6-deoxymannose replaces the mycosamine sugar common to most antifungal polyenes, and a second glycosylation, observed here at C27, is also unusual.
Although uncommon, several diglycosylated antifungal polyenes have been reported in the literature, some derived from Pseudonocardia. A diglycosylated nystatin analog named NPP was isolated from Pseudonocardia autotrophica, although the additional sugar, an N-acetylglucosamine, is appended to the 4′ position of the mycosamine (14). A yet-unidentified nystatin analog from the ant-associated Pseudonocardia strain P1 also appears to have an additional sugar appendage at this same 4′ position (15).
A second glycosylation located instead on the opposite end of the macrolide, as in selvamicin, has been observed among the minor fermentation products of the nystatin A1 producer Streptomyces noursei (nystatin A3, Fig. 1, and NYST1070), and the candidin producer Streptomyces viridoflavus (candidoin), with the second sugar located at C35, the position corresponding to selvamicin’s 4-O-methyldigitoxose attachment (16–18). Whereas structurally distinct from 4-O-methyldigitoxose, these are also 2,6-dideoxy sugars (digitoxose, mycarose, and 2,6-dideoxy-l-erythro-hexopyranos-3-ulose, respectively). Notably, in contrast to fermentations of S. noursei and S. viridoflavus, we observe the diglycosylated polyene selvamicin as the major polyene species, and neither monoglycosylated analog is detectable by LC-MS in extracts of LS1 or LS2.
The presence of 4-deoxymannose in place of mycosamine represents the only example to our knowledge of a noncationic sugar at that position in a glycosylated polyene natural product. Correspondingly, the usual paired carboxylate substituent (C16 in nystatin and amphotericin B and C12 in natamycin) is absent in selvamicin. Instead, C12 bears a methyl group and a tertiary alcohol.
Antifungal Activity and Solubility.
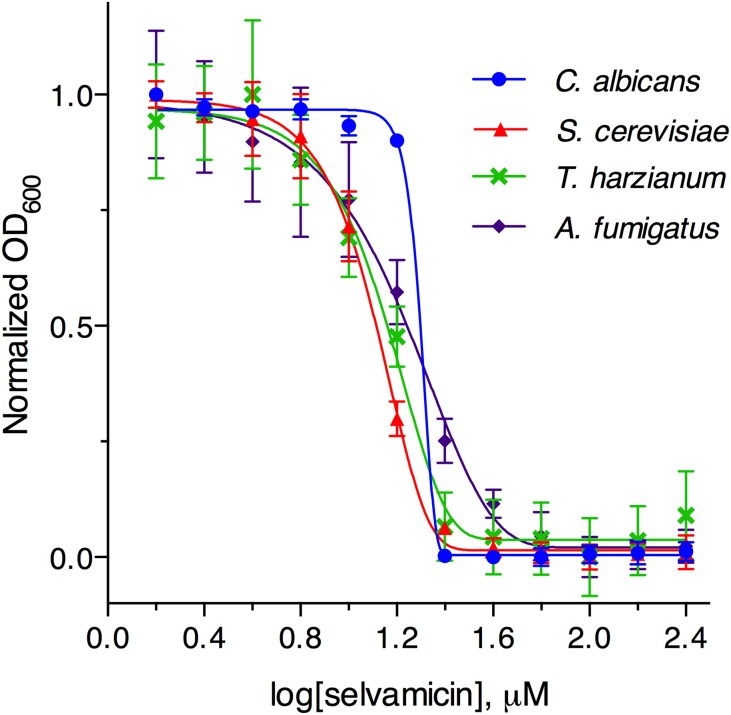
Liquid broth-based activity testing confirmed selvamicin’s antifungal activity against C. albicans (minimum inhibitory concentration, MIC, = 23 μM), with similar activity observed across a panel of fungi (Saccharomyces cerevisiae, Aspergillus fumigatus, and Trichoderma harzianum, Fig. 3 and SI Appendix, Table S3). No activity was detected against either Gram-negative (Escherichia coli) or Gram-positive (Bacillus subtilis, Micrococcus luteus) bacteria. We note that selvamicin has more modest antifungal activity than clinically used antifungal polyenes such as nystatin A1 (MIC = 1.0 μM against C. albicans). However, its improved aqueous solubility (2.3 mM compared with 0.3 mM for nystatin A1) addresses a major limitation of clinically available antifungal polyenes. Selvamicin’s improved solubility, despite its lack of charged carboxylate and ammonium groups, is probably contributed by its second sugar moiety. Indeed, glycosylation has been reported to improve solubility dramatically in analogs of nystatin; NPP, a diglycosylated analog bearing N-acetylglucosamine, has more than 300-fold greater aqueous solubility than nystatin A1 (14).
Fig. 3.
Growth inhibition of C. albicans, S. cerevisiae, T. harzianum, and A. fumigatus by selvamicin.
The activity of known antifungal polyenes derives from interactions with ergosterol, the primary sterol of fungal plasma membranes. Such interactions can compromise membrane integrity and inhibit the function of membrane proteins (19, 20). Recent studies suggest that ergosterol sequestration into extracellular aggregates may be the dominant mechanism of action (21, 22), although several polyenes, including nystatin and amphotericin B, have also long been known to permeabilize membranes by the formation of ergosterol-dependent transmembrane channels (23). The presumed geometry of these channels situates the charged end of the molecule at the lipid–water interface, with the polyene and polyol interacting with ergosterol within the plasma membrane. The dramatically different electrostatic nature of selvamicin would likely preclude channel formation, with a hydrophilic yet uncharged sugar at each end of the molecule. We probed for an interaction with ergosterol using an established isothermal calorimetry assay for binding to liposome-embedded ergosterol (21, 24). These experiments showed no evidence for binding, in stark contrast to control experiments using nystatin A1, suggesting that this interaction is much attenuated if present at all (SI Appendix, Fig. S9). Further investigation of selvamicin’s mechanism of action is currently underway.
Biosynthetic Gene Cluster.
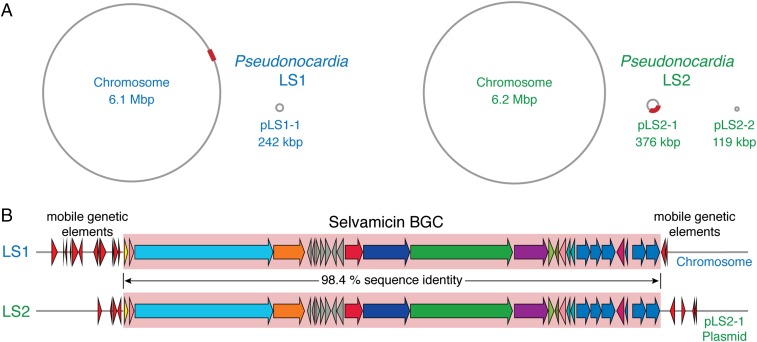
To understand the genetic origins of selvamicin biosynthesis, we turned to the genomes of Pseudonocardia isolates LS1 (7) and LS2, which were sequenced using PacBio technology (25, 26). We readily identified a large type I PKS gene cluster in both genomes that matches the biosynthetic requirements for selvamicin (Fig. 4). The 109-kbp selvamicin biosynthetic gene clusters (BGC) from each isolate share perfect synteny and 98.4% nucleotide identity over their length. In contrast, the whole genomes differ more substantially. The average nucleotide identity (27) calculated across conserved replicons on both chromosomes is only 83% and a comparison of housekeeping gene sequences places LS1 and LS2 into distinct clades previously established for ant-associated Pseudonocardia (28, 29). Overall, the two BGCs are much more similar to one another than are their bacterial hosts.
Fig. 4.
(A) Genomes of Pseudonocardia isolates LS1 and LS2. The selvamicin BGC in each is marked with a red box. (B) Selvamicin BGCs from LS1 and LS2. Mobile genetic element genes flanking the selvamicin clusters are shown as red arrows.
Surprisingly, the selvamicin BGC is situated in completely different genomic contexts in the two selvamicin producers; in LS1 it resides on the 6.1 Mbp circular chromosome, whereas in LS2 it is on a 376-kbp plasmid, pLS2-1 (Fig. 4A). The presence of an identical BGC in two divergent Pseudonocardia isolates, and in different genomic contexts, points to horizontal transfer. In keeping with BGC transfer, numerous mobile genetic elements including transposases and integrases flank it in both genomes (Fig. 4B). Mobile genetic elements are prominent features of both genomes. On the pLS2-1 plasmid containing the selvamicin BGC, an impressive 24% of all RAST-annotated genes are mobile genetic elements.
Selvamicin provides the most striking example yet for the emerging theme that plasmids drive the genetic, chemical, and functional diversity found in Pseudonocardia symbionts. Plasmid-derived BGCs for an antibacterial rebeccamycin analog and for the gerumycin depsipeptides feature in other ant-associated Pseudonocardia (7, 8). A rearranged variant of the gerumycin BGC also appears on the LS1 chromosome, suggesting that plasmid-mediated exchange also links the two gerumycin BGCs (7), but at greater evolutionary distance than for the virtually identical selvamicin BGCs.
Biosynthesis.
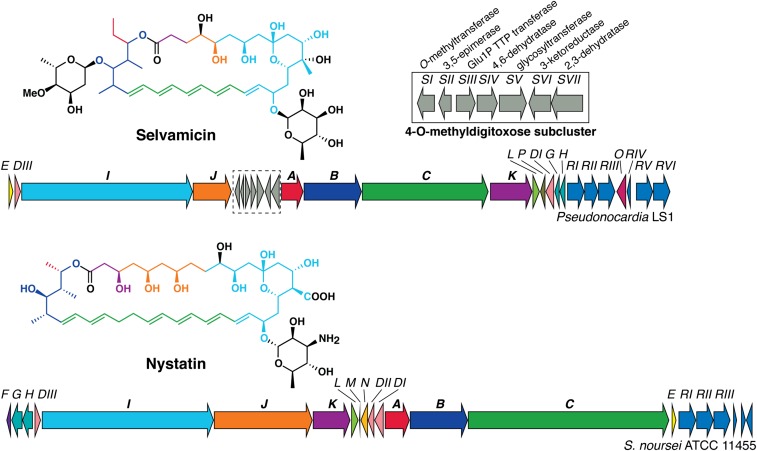
The bioinformatically identified selvamicin cluster resembles known type I PKS-derived polyene BGCs (30–35), and a side-by-side comparison with the well-characterized nystatin BGC (36, 37) readily reveals the origins of selvamicin’s unusual structural features (Fig. 5). Both natural products derive from type I iterative PKSs with polyketide elongation modules spread across five genes (sel/nysB, -C, -I, -J, and -K). Relative to the corresponding genes for nystatin, selC and selJ each lack two PKS modules, corresponding to the observed four-carbon truncations of selvamicin’s polyene and polyol moieties opposite one another on the macrolide. The polyketide backbone of selvamicin can be traced through 14 PKS modules with ketoreductase (KR), dehydratase (DH), and enoylreductase (ER) domains dictating the oxidation state of each malonyl or methylmalonyl unit (SI Appendix, Figs. S10 and S11). As often observed in type I PKS modules, there are several presumably inactive vestigial domains with mutations and/or truncations at their active sites: a DH and ER in module 13 and a KR in module 11.
Fig. 5.
Nystatin and selvamicin BGCs. Polyketide synthase genes are labeled with bold font. Coloring of the polyketide-derived portions of the nystatin A1 and selvamicin structures corresponds to the genes encoding their biosynthesis.
SelA, the putative PKS loading module for selvamicin’s propionate starter unit, shares several unusual features with known polyene loading modules (36). Unlike most type I PKS loading modules, SelA is a separate protein distinct from the first elongation module and a serine is found in place of the canonical KS active site cysteine. Like NysA, the nystatin loading module critical for initiation of that molecule’s biosynthesis,17 SelA contains a presumably inactive DH domain with no obvious function. Atypically for type I PKS loading modules, the SelA AT domain lacks the critical active site histidine and has a large truncation of ∼65 amino acids in the middle of the domain (SI Appendix, Fig. S10), suggesting that an alternative means of loading the initial acyl starter unit may be operative.
Tailoring of the polyketide core of selvamicin requires hydroxylations at C4 and C12. SelL, a cytochrome p450 with homology to the p450 NysL that installs nystatin’s C10 hydroxyl, is the most probable oxidase for C4 (38). We also identified SelP, a 2-oxoglutarate–dependent oxygenase with homology to phytanoyl-CoA dioxygenases. No homologous enzyme has been observed in other polyene clusters and this oxidase could be responsible for selvamicin’s unusual C12 hydroxylation.
The canonical paired carboxylate and ammonium in antifungal polyenes are both lacking in selvamicin. Notably, both the cytochrome P450 NysN and ferredoxin NysM believed to install nystatin’s carboxylate at C16 (37) are absent in the selvamicin cluster, consistent with selvamicin’s unoxidized methyl substituent at C12. The aminotransferase responsible for ammonium installation on the mycosamine sugar, NysDII, is also absent from the selvamicin cluster. The remaining sugar-related enzymes in the nystatin BGC, the mannose 4,6-dehydratase NysDIII and the glycosyltransferase NysDI, both have homologs in the selvamicin cluster and are consistent with the 6-deoxymannose found at C15.
The most significant divergence from nystatin’s BGC is a subcluster of seven sugar biosynthesis genes, selSI through selSVII, found in the middle of the selvamicin BGC. These include a glycosyltransferase gene, selSV, and six genes consistent with 4-O-methyldigitoxose biosynthesis as a TDP-sugar from glucose-1-phosphate (SI Appendix, Fig. S12) (39). The putative 4-O-methyldigitoxose biosynthesis proteins are homologous to a similar suite of proteins responsible for digitoxose biosynthesis in the BGC for jadomycin B in Streptomyces venezuelae ISP5230 (40). However, the selvamicin sugar subcluster contains an additional O-methyltransferase gene (selSI) and lacks an NDP-sugar 4-ketoreductase, which would normally be required for digitoxose formation. Recently, 4-ketoreductase activity has been reported for a bifunctional S-adenosylmethionine–dependent methyltransferase involved in the biosynthesis of mithramycin’s sugars (41–43). Similar bifunctional activity could be operative for the SelSI methyltransferase or alternatively this activity could require a separate 4-ketoreductase outside the selvamicin BGC in both the LS1 and LS2 genomes.
This sugar subcluster’s insertion within a cluster of familiar polyene biosynthetic genes fits well with the paradigm of modular subclusters recombining over the course of natural product evolution to generate new products (44). Presumably, a similar suite of genes synthesizes and attaches the digitoxose sugar to nystatin A3, although no such subcluster occurs in the nystatin BGC from Streptomyces noursei. Whole genome sequencing of this Streptomyces strain may eventually reveal the location of these genes. We note that nystatin A3 is a minor product of the nystatin BGC whereas selvamicin is the principal product of the selvamicin cluster. The 4-O-methyldigitoxose subcluster’s incorporation into the selvamicin BGC likely reflects selection for diglycosylation in the principal product. If this subcluster is truly modular, it presents an opportunity for appending 4-O-methyldigitoxose to other polyene scaffolds to create diglycoslated nystatin analogs, which are currently available only as minor products from S. noursei fermentation but have comparable anti-Candida potency to nystatin A1 (18). A solubility boost from an additional sugar would address a major pharmacological limitation of antifungals such as nystatin A1 and amphotericin B.
Conclusion
The systematic study of bacterially produced small molecules from well-defined ecological niches continues to uncover potentially useful molecules and to provide examples of the evolution of molecular diversity. Selvamicin, a previously unknown antifungal agent with activity against the human pathogen C. albicans, represents a significant structural departure from the known antifungal polyenes. Preliminary studies also indicate that selvamicin’s atypical structure might be reflected in an unusual mechanism of action. In other ant-associated Pseudonocardia we have reported plasmid-encoded chemical defenses, and circumstantial evidence for the integration of plasmid-encoded biosynthetic pathways into chromosomes (7, 8). However, the variable genomic contexts for selvamicin’s biosynthetic gene cluster—on a plasmid or on a chromosome—provide the most convincing evidence to date for horizontal gene transfer, illustrating an environment in which frequently exchanged plasmids are the platforms for the formation and transmission of a dynamic suite of chemical defenses.
Materials and Methods
Selvamicin Production and Purification.
Spores of Pseudonocardia LS1 were diluted into sterile double-distilled water and spread onto plates of ISP2 agar (BD Difco ISP2; 60 mL agar per 150 × 15 mm Petri dish) supplemented with sodium butyrate (Aldrich, 150 mM final concentration, added after autoclaving), which were incubated at 30 °C for 14 d. Agar was then cut into squares and soaked in ethyl acetate overnight to extract organic components from the solid media. This extract was decanted and the agar was soaked in an additional volume of ethyl acetate for 3 h. The combined ethyl acetate extracts were concentrated in vacuo and adsorbed onto celite for dry packing onto a 10-g C18 SepPak column (Waters) that had been conditioned with acetonitrile and preequilibrated with 30% acetonitrile in water. Fractions were eluted with a step gradient of 30%, 50%, 70%, and 100% acetonitrile in water and concentrated to dryness. Consecutive fractions from elution at 50% acetonitrile were most active in inhibition of C. albicans. Semipure material from these fractions was purified by reversed-phase HPLC (Agilent 1200 series preparative HPLC equipped with a diode array detector; Phenomenex Luna 10-μm phenyl-hexyl preparative column, 250 × 21.20 mm, 10 mL/min) with a gradient of 40–63% acetonitrile in water over 20 min. Selvamicin eluted at 12.5 min. The overall yield of pure selvamicin (isolated as an amorphous pale-yellow solid) was 50 mg/L of agar. Selvamicin: [α]D26 +128° (MeOH); UV (MeOH) λmax (log ε) 305 (4.4), 319 (4.7), 334 (4.9), 352 (4.9) nm; NMR spectral data, see SI Appendix, Table S1; HR-ESI-MS m/z 951.4928 [M+Na]+ (calcd for C47H76NaO18: 951.4924).
Determination of Minimum Inhibitory Concentration.
Fresh DMSO solutions of selvamicin and nystatin were prepared as serial dilutions and dispensed into clear flat-bottom 96-well plates in 4 replicates. A starting inoculum of the appropriate test strain in media was added to each well to yield a final concentration of 1% DMSO by volume. The plates were incubated at 30 °C with shaking at 200 rpm. Growth was assayed by OD600 readings taken on an M5 plate reader (Molecular Devices). For E. coli, B. subtilis, and M. luteus, the starting inoculum consisted of an overnight culture in LB diluted into LB media at 10 μL/mL and final OD readings were taken at 22 h. For C. albicans and S. cerevisiae, the starting inoculum consisted of an overnight culture in yeast extract peptone dextrose (YPD) broth diluted to an OD600 of 0.05 in YPD broth and final OD readings were taken at 14 h. For T. harzianum and A. fumigatus, the starting inoculum consisted of a stock of concentrated conidia diluted into potato dextrose broth at 2 μL/mL and final OD readings were taken at 22 h. Using Prism (GraphPad), the OD data were normalized and fit to a Gompertz function, from which MIC values were extracted as described by Lambert and Pearson (45).
Genome Sequencing and Data Deposition.
DNA isolation and genome sequencing was performed as described previously (7). The complete genome for Pseudonocardia LS2 (HH130630-07) has been deposited in the GenBank database (accession nos. CP013854, CP013855, and CP013856) and raw sequence data have been deposited in the Sequence Read Archive (accession no. SRP075179). The Pseudonocardia LS1 (HH130629-09) genome (7) can be accessed using GenBank accession nos. CP011868 and CP011869.
Additional experimental details are available in the SI Appendix, including preparation of Ac9-selvamicin, solubility determination, isothermal calorimetry sterol binding assay, induction with propionate and butyrate, and sequence comparisons and analysis.
Supplementary Material
Acknowledgments
We thank Emily Mevers for measurement of optical rotation. We are grateful to the Organization for Tropical Studies and to Ronald Vargas Castro for logistical support and facilities at the La Selva Biological Station, Costa Rica. We thank the Duke GCB Genome Sequencing Shared Resource, which provided PacBio sequencing service, and the Harvard Medical School Information Technology Department for access to the Orchestra High Performance Computing Cluster. Collecting permits were granted by the “Comisión Institucional de Biodiversidad” (Institutional Biodiversity Committee, University of Costa Rica; Resolution 020; Material Transfer Agreement MTA VI-4307-2013) and authorized by La Selva Biological Station. E.B.V. was supported by an NIH Postdoctoral Fellowship F32 GM117661. C.S.S. was supported by an Alberta Innovates Health Solutions Fellowship and a Banting Postdoctoral Fellowship; A.C.R. was supported by Harvard Medical School−Merck and CIHR Fellowships. The work was also supported by NIH Grant R01 GM086258 (to J.C.), NIH Grant U19 AI09673 (to E.B.V., J.C., and C.R.C.), and the University of Costa Rica (to A.A.P.-T.).
Footnotes
Conflict of interest statement: E.B.V., A.C.R., C.S.S., A.A.P.-T., C.R.C., and J.C. have filed a patent based on the work presented in this paper.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CP013854, CP013855, and CP013856); and National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession no. SRP075179).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613285113/-/DCSupplemental.
References
- 1.Hölldobler B, Wilson EO. The Leafcutter Ants: Civilization by Instinct. W. W. Norton & Company; New York: 2010. [Google Scholar]
- 2.Currie CR. A community of ants, fungi, and bacteria: A multilateral approach to studying symbiosis. Annu Rev Microbiol. 2001;55:357–380. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 3.Ramadhar TR, Beemelmanns C, Currie CR, Clardy J. Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery. J Antibiot (Tokyo) 2014;67(1):53–58. doi: 10.1038/ja.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Curr Biol. 2009;19(11):R437–R441. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311(5757):81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 6.Oh D-C, Poulsen M, Currie CR, Clardy J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5(6):391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sit CS, et al. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci USA. 2015;112(43):13150–13154. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Arnam EB, Ruzzini AC, Sit CS, Currie CR, Clardy J. A rebeccamycin analog provides plasmid-encoded niche defense. J Am Chem Soc. 2015;137(45):14272–14274. doi: 10.1021/jacs.5b09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr G, Derbyshire ER, Caldera E, Currie CR, Clardy J. Antibiotic and antimalarial quinones from fungus-growing ant-associated Pseudonocardia sp. J Nat Prod. 2012;75(10):1806–1809. doi: 10.1021/np300380t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization . WHO Model List of Essential Medicines: 19th List, April 2015. WHO; Geneva: 2015. [Google Scholar]
- 11.Ashley ESD, Lewis R, Lewis JS, Martin C, Andes D. Pharmacology of systemic antifungal agents. Clin Infect Dis. 2006;43(Supplement 1):S28–S39. [Google Scholar]
- 12.Ngo HX, Garneau-Tsodikova S, Green KD. A complex game of hide and seek: The search for new antifungals. MedChemComm. 2016;7(7):1285–1306. doi: 10.1039/C6MD00222F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore JM, Bradshaw E, Seipke RF, Hutchings MI, McArthur M. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Methods Enzymol. 2012;517:367–385. doi: 10.1016/B978-0-12-404634-4.00018-8. [DOI] [PubMed] [Google Scholar]
- 14.Lee M-J, et al. Structural analysis and biosynthetic engineering of a solubility-improved and less-hemolytic nystatin-like polyene in Pseudonocardia autotrophica. Appl Microbiol Biotechnol. 2012;95(1):157–168. doi: 10.1007/s00253-012-3955-x. [DOI] [PubMed] [Google Scholar]
- 15.Barke J, et al. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010;8(1):109. doi: 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zieliński J, Golik J, Pawlak J, Borowski E, Falkowski L. The structure of nystatin A3, a component of nystatin complex. J Antibiot (Tokyo) 1988;41(9):1289–1291. doi: 10.7164/antibiotics.41.1289. [DOI] [PubMed] [Google Scholar]
- 17.Synak R, Zieliński J, Golik J, Borowski E. The structure of candidoin a component of the candidin antibiotic complex. J Antibiot (Tokyo) 1983;36(10):1415–1417. doi: 10.7164/antibiotics.36.1415. [DOI] [PubMed] [Google Scholar]
- 18.Bruheim P, et al. Chemical diversity of polyene macrolides produced by Streptomyces noursei ATCC 11455 and recombinant strain ERD44 with genetically altered polyketide synthase NysC. Antimicrob Agents Chemother. 2004;48(11):4120–4129. doi: 10.1128/AAC.48.11.4120-4129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamiński DM. Recent progress in the study of the interactions of amphotericin B with cholesterol and ergosterol in lipid environments. Eur Biophys J. 2014;43(10-11):453–467. doi: 10.1007/s00249-014-0983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.te Welscher YM, van Leeuwen MR, de Kruijff B, Dijksterhuis J, Breukink E. Polyene antibiotic that inhibits membrane transport proteins. Proc Natl Acad Sci USA. 2012;109(28):11156–11159. doi: 10.1073/pnas.1203375109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray KC, et al. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci USA. 2012;109(7):2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson TM, et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol. 2014;10(5):400–406. doi: 10.1038/nchembio.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szpilman AM, Manthorpe JM, Carreira EM. Synthesis and biological studies of 35-deoxy amphotericin B methyl ester. Angew Chem Int Ed Engl. 2008;47(23):4339–4342. doi: 10.1002/anie.200800590. [DOI] [PubMed] [Google Scholar]
- 24.te Welscher YM, et al. Natamycin blocks fungal growth by binding specifically to ergosterol without permeabilizing the membrane. J Biol Chem. 2008;283(10):6393–6401. doi: 10.1074/jbc.M707821200. [DOI] [PubMed] [Google Scholar]
- 25.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 26.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 27.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 28.Cafaro MJ, et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc Biol Sci. 2011;278(1713):1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldera EJ, Currie CR. The population structure of antibiotic-producing bacterial symbionts of Apterostigma dentigerum ants: Impacts of coevolution and multipartite symbiosis. Am Nat. 2012;180(5):604–617. doi: 10.1086/667886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M. Amphotericin biosynthesis in Streptomyces nodosus: Deductions from analysis of polyketide synthase and late genes. Chem Biol. 2001;8(7):713–723. doi: 10.1016/s1074-5521(01)00046-1. [DOI] [PubMed] [Google Scholar]
- 31.Chen S, et al. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem Biol. 2003;10(11):1065–1076. doi: 10.1016/j.chembiol.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Aparicio JF, Fouces R, Mendes MV, Olivera N. A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis. Chem Biol. 2000;7(11):895–905. doi: 10.1016/s1074-5521(00)00038-7. [DOI] [PubMed] [Google Scholar]
- 33.Seco EM, Pérez-Zúñiga FJ, Rolón MS, Malpartida F. Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem Biol. 2004;11(3):357–366. doi: 10.1016/j.chembiol.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Kim B-G, et al. Identification of functionally clustered nystatin-like biosynthetic genes in a rare actinomycetes, Pseudonocardia autotrophica. J Ind Microbiol Biotechnol. 2009;36(11):1425–1434. doi: 10.1007/s10295-009-0629-5. [DOI] [PubMed] [Google Scholar]
- 35.Caffrey P, De Poire E, Sheehan J, Sweeney P. Polyene macrolide biosynthesis in streptomycetes and related bacteria: Recent advances from genome sequencing and experimental studies. Appl Microbiol Biotechnol. 2016;100(9):3893–3908. doi: 10.1007/s00253-016-7474-z. [DOI] [PubMed] [Google Scholar]
- 36.Brautaset T, et al. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: Analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol. 2000;7(6):395–403. doi: 10.1016/s1074-5521(00)00120-4. [DOI] [PubMed] [Google Scholar]
- 37.Fjaervik E, Zotchev SB. Biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei. Appl Microbiol Biotechnol. 2005;67(4):436–443. doi: 10.1007/s00253-004-1802-4. [DOI] [PubMed] [Google Scholar]
- 38.Volokhan O, Sletta H, Ellingsen TE, Zotchev SB. Characterization of the P450 monooxygenase NysL, responsible for C-10 hydroxylation during biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei. Appl Environ Microbiol. 2006;72(4):2514–2519. doi: 10.1128/AEM.72.4.2514-2519.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thibodeaux CJ, Melançon CE, 3rd, Liu H-W. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed Engl. 2008;47(51):9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, White RL, Vining LC. Biosynthesis of the dideoxysugar component of jadomycin B: Genes in the jad cluster of Streptomyces venezuelae ISP5230 for L-digitoxose assembly and transfer to the angucycline aglycone. Microbiology. 2002;148(Pt 4):1091–1103. doi: 10.1099/00221287-148-4-1091. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Kharel MK, Pahari P, Rohr J. Investigating Mithramycin deoxysugar biosynthesis: Enzymatic total synthesis of TDP-D-olivose. ChemBioChem. 2011;12(17):2568–2571. doi: 10.1002/cbic.201100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, et al. Cooperation of two bifunctional enzymes in the biosynthesis and attachment of deoxysugars of the antitumor antibiotic mithramycin. Angew Chem Int Ed Engl. 2012;51(42):10638–10642. doi: 10.1002/anie.201205414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J-M, Hou C, Wang G, Tsodikov OV, Rohr J. Structural insight into MtmC, a bifunctional ketoreductase-methyltransferase involved in the assembly of the mithramycin trisaccharide chain. Biochemistry. 2015;54(15):2481–2489. doi: 10.1021/bi501462g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. A systematic computational analysis of biosynthetic gene cluster evolution: Lessons for engineering biosynthesis. PLOS Comput Biol. 2014;10(12):e1004016. doi: 10.1371/journal.pcbi.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert RJ, Pearson J. Susceptibility testing: Accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J Appl Microbiol. 2000;88(5):784–790. doi: 10.1046/j.1365-2672.2000.01017.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.