Significance
Symbiosis between animals and bacteria enables the exploitation of new ecological niches and promotes diversification. Many insects have obligate heritable bacteria that provision nutrients enabling new lifestyles, such as sap-feeding. A model system for obligate symbiosis is the pea aphid and its heritable bacterial symbiont Buchnera aphidicola. Using this system, we address the extent of variation in host regulation of symbiont populations and find that pea aphids have extensive genetic variation in the ability to regulate Buchnera numbers. In addition, higher abundance of Buchnera is associated with lower reproductive rates in aphid hosts. Thus, we provide experimental evidence demonstrating that genetic variation in hosts affects regulation of symbiosis, suggesting fitness costs and potential negative consequences of obligate symbiosis.
Keywords: coevolution, maternal transmission, cytonuclear interactions, Acyrthosiphon pisum, Buchnera aphidicola
Abstract
Symbiotic relationships promote biological diversification by unlocking new ecological niches. Over evolutionary time, hosts and symbionts often enter intimate and permanent relationships, which must be maintained and regulated for both lineages to persist. Many insect species harbor obligate, heritable symbiotic bacteria that provision essential nutrients and enable hosts to exploit niches that would otherwise be unavailable. Hosts must regulate symbiont population sizes, but optimal regulation may be affected by the need to respond to the ongoing evolution of symbionts, which experience high levels of genetic drift and potential selection for selfish traits. We address the extent of intraspecific variation in the regulation of a mutually obligate symbiosis, between the pea aphid (Acyrthosiphon pisum) and its maternally transmitted symbiont, Buchnera aphidicola. Using experimental crosses to identify effects of host genotypes, we measured symbiont titer, as the ratio of genomic copy numbers of symbiont and host, as well as developmental time and fecundity of hosts. We find a large (>10-fold) range in symbiont titer among genetically distinct aphid lines harboring the same Buchnera haplotype. Aphid clones also vary in fitness, measured as developmental time and fecundity, and genetically based variation in titer is correlated with host fitness, with higher titers corresponding to lower reproductive rates of hosts. Our work shows that obligate symbiosis is not static but instead is subject to short-term evolutionary dynamics, potentially reflecting coevolutionary interactions between host and symbiont.
Symbioses between bacteria and animals enable the exploitation of novel environments by providing the genetic and phenotypic advantages needed to use previously unavailable niches (1–3). Some symbioses involve bacteria that are universal within individuals of a host species. Prominent examples are found in sap-feeding insects in the order Hemiptera, which depend on bacterial symbionts to synthesize essential amino acids and other nutrients that are absent or rare in their specialized diets of plant sap (1, 4, 5). In many of these insects, the vertical transmission of bacterial symbionts from mother to offspring over millions of years has resulted in very intimate associations such that neither organism is capable of growth and reproduction without the other (6–9). Although heritable symbionts play an integral role in the success of many major animal groups, obligate symbiosis also presents a major challenge to hosts. In addition to the metabolic costs of maintaining symbionts, hosts must organize and regulate symbionts within their bodies, and exchange molecules with them (10, 11).
For maternally transmitted symbioses, selection on both hosts and symbionts promotes cooperation and efficiency of the symbiosis so as to increase host fecundity, which is the basis for symbiont transmission. However, maternally transmitted symbionts are clonal and subject to high levels of genetic drift, leading to degradation of their genomes and potentially imposing fitness costs on hosts (9, 12–14). In addition, heritable symbiosis can be impacted by within-host selection that can lead to symbiont selfishness (15, 16). For example, a mutation in a symbiont that speeds replication could spread because of the increase in proportional representation in subsequent progeny of its host, even if the overall fecundity of the host is lowered. Thus, obligate symbiosis entails a balancing act between conflicting evolutionary forces and could instigate a coevolutionary arms race between host and symbiont (15). Potentially, genetic interactions involving symbiosis accelerate reproductive isolation and thus diversification (17). Indeed, interactions between organellar genomes (mitochondria and plastids) and host genotypes appear to often affect host fitness (e.g., refs. 17–20).
A model for heritable, mutually obligate symbiosis is the association of pea aphids (Acyrthosiphon pisum) and their maternally transmitted symbiont Buchnera aphidicola (Gammaproteobacteria). Buchnera is found in almost all aphids and provisions hosts with essential amino acids that are lacking or rare in the diet of plant phloem sap (5, 21, 22). Buchnera cells are located in the cytosol of specialized host cells called bacteriocytes, where each is enclosed in a host-derived membrane (4, 22–24). Host and symbiont functions must be coordinated to maintain an integrated system of nutrient biosynthesis and to ensure transmission to progeny, and some evidence supports host-control of the symbiosis (10, 21, 25). One might expect tight control of Buchnera numbers, given the essentiality of this symbiosis, but several reports suggest that regulation of the symbiosis is highly variable. Total number and pooled volume of bacteriocytes vary among field collected A. pisum clones differing in Buchnera haplotype (26), and Buchnera genome numbers vary among sibling A. pisum genotypes (27). Whether or how this variation affects host fitness has not been determined.
In this study, we examined how obligate symbiosis varies across genotypes within a single host species and how this variation is linked to host fitness. Specifically, we performed sexual crosses to generate distinct A. pisum clones of known relatedness and possessing known Buchnera haplotypes. We determined whether symbiont titer varies across these genotypes, and whether symbiont titer is related to host fitness. We find that Buchnera titer is significantly impacted by host genotype. We also find that symbiont titer is negatively correlated with overall host reproductive rate, suggesting a fitness cost associated with greater symbiont abundance. Thus, our findings show that the regulation of obligate symbiosis is dynamic within a host species rather than a static optimum. This variation in regulation of symbiosis is consistent with expected consequences of a coevolutionary arms race between host and symbiont.
Results
Establishment of Aphid Clones.
To estimate the genetic basis of symbiont titer and its relationship to host fitness, we induced sexuals from four A. pisum clones, performed crosses, and generated sexually produced progeny belonging to full-sibling and half-sibling cohorts (Materials and Methods). Each progeny was an asexual female that gave rise to an asexual aphid line (clone), maintained long term in the laboratory under constant long-day conditions. To focus on the obligate symbiosis with Buchnera, any other (secondary) symbionts were eliminated before our experiments. Following sexual induction through exposure to short day length (27), clones LSR1 and 5AY, containing Buchnera haplotype B, produced both males and sexual females, whereas clones AustinC and TucsonC, containing Buchnera haplotype A, produced only sexual females. Thus, sexual females from all four clones were mated with either LSR1 or 5AY males to produce an F1 generation that was then maintained as individual asexual clones under long-day conditions (Table 1). Each parental and progeny clone was grown as three replicate sublines for measurements of symbiont titer and host fitness.
Table 1.
Experimental sexual crosses conducted and average Buchnera titer and host fitness measurements of progeny clones produced from each cross
| Parentals | Progeny | Symbiont | Average | ||||||
| Mother | Father | Total | Buchnera haplotype | Buchnera titer | Days to adult | Adult weight (mg) | Days to first reproduction | Total offspring (over 6 d) | Reproductive rate |
| AustinC | 5AY | 16 | A | 47.4 ± 5.1 | 7.45 ± 0.13 | 0.239 ± 0.01 | 9.45 ± 0.17 | 51.97 ± 1.72 | 5.02 ± 1.21 |
| AustinC | LSR1 | 13 | A | 59.1 ± 6.1 | 7.24 ± 0.12 | 0.264 ± 0.01 | 9.26 ± 0.14 | 54.00 ± 1.73 | 5.29 ± 0.19 |
| TucsonC | 5AY | 1 | A | 16.2 ± 6.5 | 7.00 ± 0 | 0.260 ± 0.08 | 8.83 ± 0.43 | 56.17 ± 8.17 | 7.72 ± 0.86 |
| TucsonC | LSR1 | 3 | A | 18.9 ± 8.5 | 7.17 ± 0.25 | 0.276 ± 0.02 | 8.92 ± 0.33 | 60.45 ± 3.95 | 6.10 ± 0.39 |
| 5AY | 5AY | 0 | B | — | — | — | — | — | – |
| 5AY | LSR1 | 4 | B | 36.2 ± 6.7 | 6.79 ± 0.22 | 0.297 ± 0.02 | 8.46 ± 0.28 | 57.96 ± 1.54 | 6.14 ± 0.32 |
| LSR1 | LSR1 | 0 | B | — | — | — | — | — | — |
| LSR1 | 5AY | 2 | B | 34.4 ± 12.4 | 6.8 ± 0.25 | 0.305 ± 0.05 | 8.7 ± 0.31 | 60.67 ± 5.37 | 6.28 ± 0.56 |
Buchnera Titer.
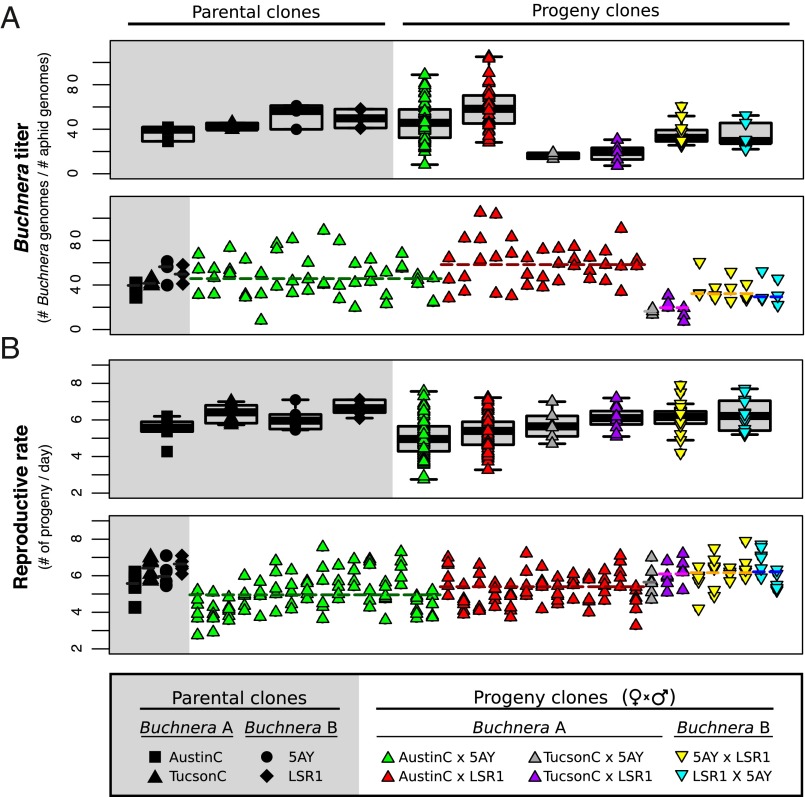
To determine Buchnera titers for each clone, we used quantitative PCR (qPCR) to measure the ratio of the number of symbiont genome copies to the number of host genome copies, measured in fourth-instar juveniles, just before maturity. Buchnera titers varied among individuals representing each clone; this likely reflects various factors, including differences in quality of the food plants for each subline, differences in our exact timing of sampling, and environmental noise. Our sampling of fourth-instar juveniles corresponds to a phase of rapid growth of symbiont populations (28) and potentially adds to the sampling variation, because a few hours difference in sampling time may affect titer. Nonetheless, titers differed significantly among all clones (one-way ANOVA, F41, 84 = 2.13, P < 0.01) (Fig. 1A). Average titer varied by over 10-fold across clones, ranging from 7.2 in a TucsonC × LSR1 clone to 105.1 in an AustinC × LSR1 clone.
Fig. 1.
Measurements of Buchnera titer and of reproductive rate of A. pisum parental clones and F1 progeny clones generated from experimental crosses. (A) Titer for parental clones and for progeny clones grouped by sibships (Upper) and for each subline (Lower). Dashed line represents median titer for each sibship. (B) Reproductive rate of parental clones and progeny grouped by sibships (Upper) and for each subline (Lower). Dashed line represents the median for each sibship.
To determine whether Buchnera titer differs because of aphid genotype, we compared titer among the 32 progeny clones with Buchnera haplotype A (from maternal AustinC and TucsonC) and among the 6 progeny clones with Buchnera haplotype B (from maternal LSR1 and 5AY). Clones with Buchnera haplotype A differed in Buchnera titer (one-way ANOVA, F31, 64 = 2.02, P < 0.01). Clones with Buchnera haplotype B did not differ significantly (one-way ANOVA, F5, 12 = 0.31, P > 0.05), potentially because of the small sample size. Therefore, aphid genotype may have an effect on symbiont titer.
We further tested progeny lines with Buchnera haplotype A, for effects of parentage, either maternal or paternal, on symbiont titer. We found significant effects of maternal clone and of paternal clone (two-way ANOVA, maternal: F1, 64 = 33.75, P < 0.0001, paternal: F1, 64 = 4.98, P < 0.03). The effect of parentage supports some heritability of titer. We tested for differences among full-sibling progeny clones, using the two largest sets of full-sibling clones (from AustinC × 5AY and AustinC × LSR1 crosses) and found no significant differences (one-way ANOVA, F15, 32 = 0.92, P > 0.05 and F12, 26 = 0.62, P > 0.05, respectively).
Clone Fitness.
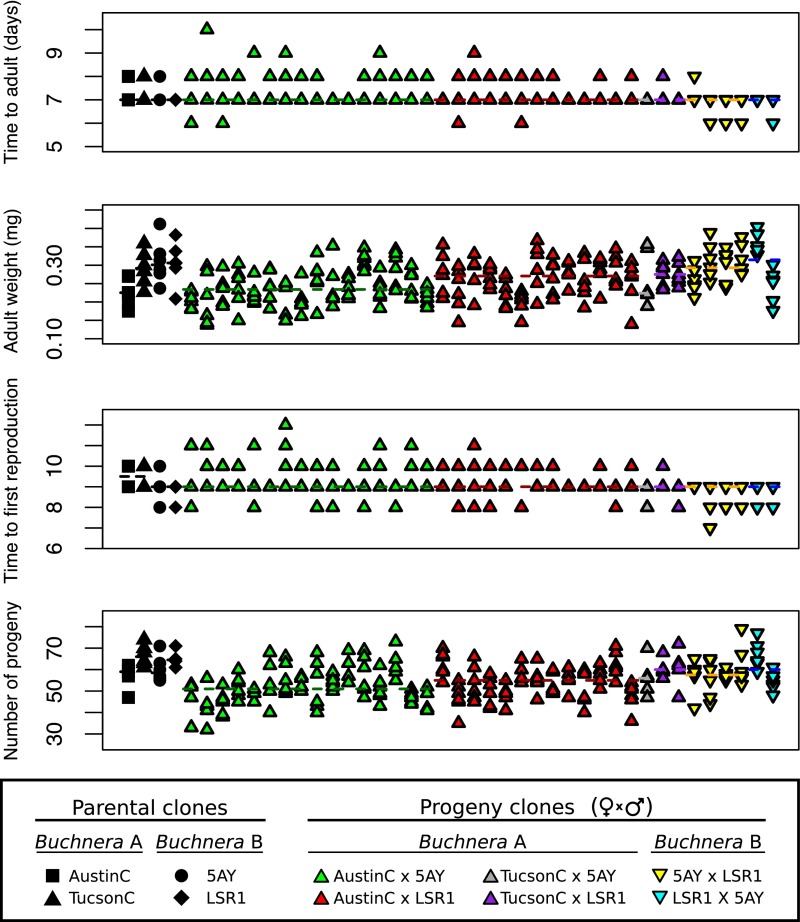
To estimate differences in host fitness, we measured developmental time, adult mass, and fecundity of all A. pisum progeny clones (Fig. S1 and Table S1), and calculated reproductive rates (Fig. 1B and Table 1). Comparing across all progeny clones, there were significant differences in all fitness measures: adult weight (Kruskal–Wallis, χ2 = 94.81, df = 37, P < 0.0001), development time (Kruskal–Wallis, χ2 = 70.41, df = 37, P < 0.001), time to first reproduction (Kruskal–Wallis, χ2 = 83.01, df = 37, P < 0.0001), fecundity measured as number of offspring produced in 6 d (Kruskal–Wallis, χ2 = 107.33, df = 37, P < 0.0001), and reproductive rate (as calculated from developmental time and fecundity; Kruskal–Wallis, χ2 = 121.39, df = 37, P < 0.0001). All fitness parameters also significantly differed among progeny from different maternal clones and among progeny from different paternal clones (Table S1). Full-sibling clones from AustinC × 5AY and AustinC × LSR1 crosses varied significantly for multiple fitness parameters, including reproductive rate (Kruskal–Wallis, χ2 = 45.91, df = 15, P < 0.0001 and χ2 = 30.03, df = 12, P < 0.01, for AustinC × 5AY and AustinC × LSR1 crosses, respectively).
Fig. S1.
Fitness measurements of each subline for parental and progeny Acyrthosiphon pisum clones. Dashed line represents median for each sibship.
Table S1.
Summary statistics for effects of parent on aphid fitness parameters of progeny clones
| Parentals | Factor | Test statistic* | P value |
| Matriline | Time to adult (d) | χ2 = 26.283, df = 3 | <0.0001 |
| Adult weight (mg) | χ2 = 21.045, df = 3 | <0.001 | |
| Time to first reproduction (d) | χ2 = 39.515, df = 3 | <0.0001 | |
| Number of progeny (n) | χ2 = 22.424, df = 3 | <0.0001 | |
| Reproduction rate (n progeny/n days to first reproduction +1) | χ2 = 40.907, df = 3 | <0.0001 | |
| Patriline | Time to adult (d) | χ2 = 6.506, df = 1 | <0.02 |
| Adult weight (mg) | χ2 = 14.259, df = 1 | <0.001 | |
| Time to first reproduction (d) | χ2 = 5.596, df = 1 | <0.02 | |
| Number of progeny (n) | χ2 = 6.145, df = 1 | <0.02 | |
| Reproduction rate (n progeny/ n days to first reproduction +1) | χ2 = 8.02, df = 1 | <0.01 |
Significance of matriline (LSR1, 5AY, AustinC, TucsonC) and patriline (LSR1 and 5AY) were tested using Kruskal–Wallis.
Relationship of Symbiont Titer to Host Fitness.
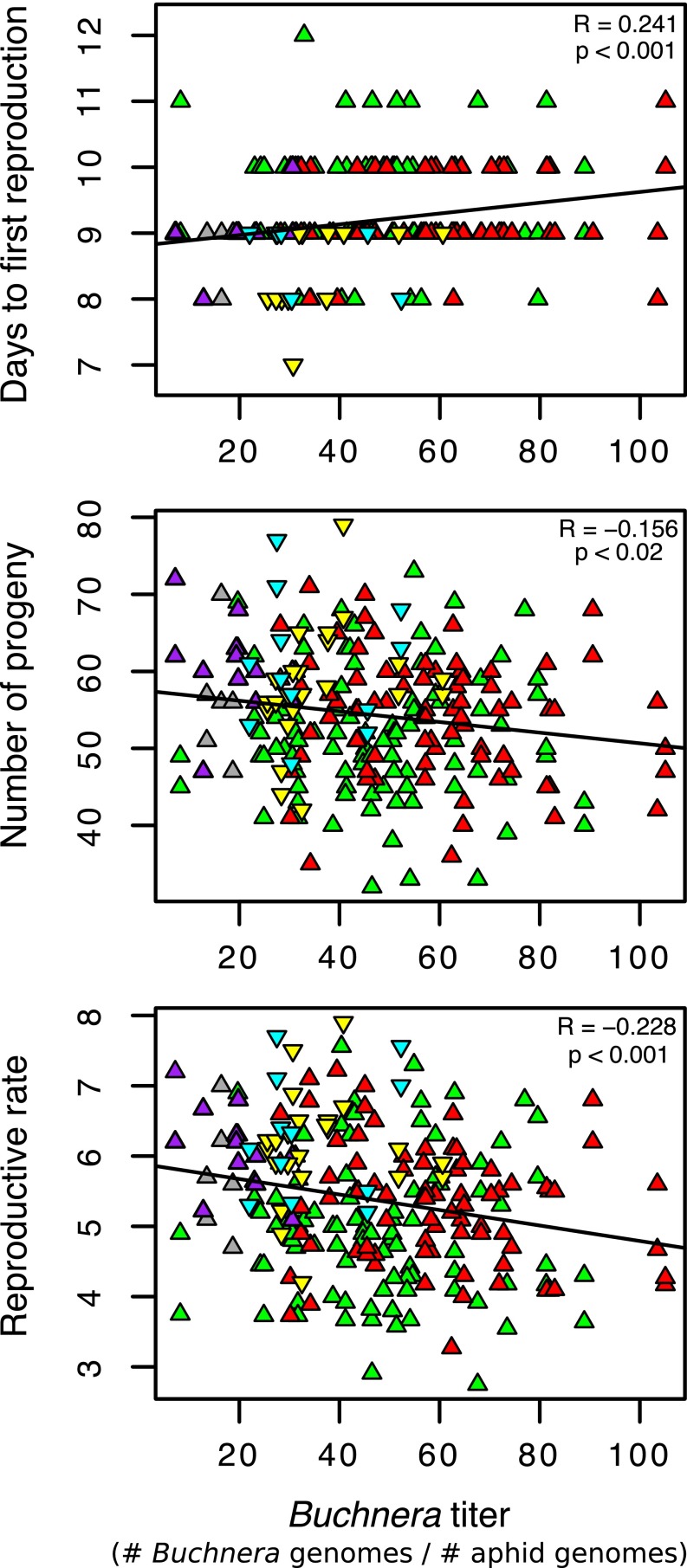
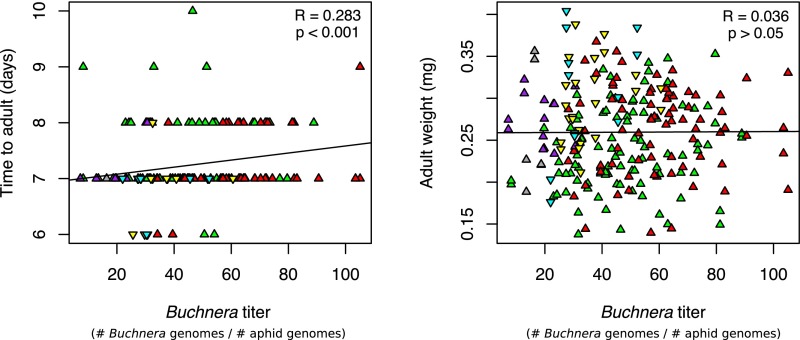
To determine how symbiont titer may relate to host fitness, we tested for correlations between titer and development time, adult mass, and fecundity of all A. pisum progeny lines (Fig. 2 and Fig. S2). Comparing across progeny clones, symbiont titer was significantly correlated with development time (Spearman’s R = 0.238, P < 0.001), time to first reproduction (Spearman’s R = 0.241, P < 0.001), and number of progeny produced (Spearman’s R = −0.156, P < 0.02), but not to adult mass (Spearman’s R = 0.036, P > 0.05). We combined developmental time and fecundity to obtain a single measure of reproductive rate and observed a highly significant relationship between titer and reproductive rate (Spearman’s R = −0.228, P < 0.001) (Fig. 2). Thus, a higher Buchnera titer is associated with longer time to reproductive maturity as well as fewer offspring.
Fig. 2.
Correlation of Buchnera titer and host fitness measurements, including days to reproduction, number of progeny, and reproductive rate for sublines of A. pisum clones. Refer to Fig. 1 for key.
Fig. S2.
Correlation of Buchnera titer and host fitness parameters, including time to adult and adult weight for sublines of A. pisum clones.
Discussion
As for any obligate, heritable symbiosis, aphids are strongly selected to maintain their Buchnera and to ensure transmission to progeny. Past studies have made it clear that severe reduction or loss of Buchnera is highly detrimental to aphid growth and reproduction (29, 30). Similarly, selection generally favors Buchnera mutations that increase host fecundity, because Buchnera relies on host reproduction for its own persistence. Such selection for mutualistic traits appears to underlie many observed features of the Buchnera genome, including retention of amino acid biosynthetic pathways (24), and amplification of some amino acid biosynthetic genes on plasmids (31). Aphids also exhibit symbiotic adaptations including bacteriocyte-specific expression of genes involved in amino acid metabolism and altered function of specific amino acid transporters within bacteriocytes (21, 25, 32). Nonetheless, there is a zone of potential coevolutionary conflict. A Buchnera mutant with elevated replication rate could spread by being transmitted disproportionately to progeny, even if it depresses host fecundity. This potential for the spread of “selfish” symbiont mutations is enhanced if the inoculum size is large (16, 33). This conflict between levels of selection could lead to a coevolutionary arms race, because selection will act on hosts to counter the impact of symbiont selfishness.
If these dynamics are affecting an obligate symbiosis, one expectation is intraspecific variation affecting the regulation of the symbiosis. Because aphids have a clonal life-cycle phase, genotypes can be reared in independent clonal sublines, providing an unusual opportunity to measure genotypic variation. We found that symbiont titer, measured as genome copy number, is surprisingly variable among host genotypes, and that average titer can vary several-fold among clones harboring the same Buchnera haplotype (Fig. 1).
A second expectation, if coevolutionary dynamics are affecting an obligate symbiosis, is that symbiont regulation affects host fitness, and specifically that overreplication of symbionts can lower host fitness. In our experiments, higher Buchnera titer is associated with lower performance of aphids for all three of our performance measures (fecundity, developmental time, and time to first reproduction) (Fig. 2 and Fig. S2). The negative correlation between symbiont titer and overall reproductive rate (Fig. 2) raises the possibility of a fitness cost of greater symbiont abundance, at least under our laboratory conditions. Thus, our findings are consistent with the possibility that the intimate obligate relationship between heritable symbionts and hosts entails a coevolutionary arms race.
However, our results do not demonstrate a causal relationship between high symbiont titer and aphid fitness, and several alternative explanations for the correlation are possible. Specifically, higher Buchnera titers might increase aphid fitness under conditions of low nutrient availability or heat exposure, which can kill Buchnera cells (29, 34). Our laboratory conditions may remove the pressure to have large numbers of Buchnera, as we provide highly nutritious host plants (fava bean seedlings) and a constant favorable temperature. Potentially, the correlation could reflect differences in compartmentalization of Buchnera between maternal and embryonic bacteriocytes, which might vary with aphid growth rate, thus impacting the ratio of Buchnera to aphid genomes. Weighing against this explanation are the results of Simonet et al. (28), who found that only a small proportion of the Buchnera cells within a fourth-instar A. pisum (the stage we measured) are within the embryos. Nonetheless, explanations such as these, in which the aphid genotype has independent effects on Buchnera titer and on fecundity, cannot be ruled out as possible explanations for the correlation.
We were unable to test directly for an effect of Buchnera genotype on titer, or on host fitness, because any effect of the Buchnera haplotype is confounded with aphid genetic contributions. We note that the three progeny clones with the TucsonC Buchnera haplotype A show the lowest titers and some of the highest reproductive rates (Fig. 1 and Table 1). (Reciprocal crosses to distinguish effects of host and symbiont genomes were not possible because TucsonC parental clones failed to produce males.)
Our findings reveal that symbiont titer is variable and depends at least in part on host genotype. The mechanisms by which aphids control their symbiont populations are not known. In some other symbioses, modified elements of the innate immune system play a role, as for the symbionts of Sitophilus grain weevils, in which antimicrobial peptides, typically associated with defense against pathogens, function to confine mutualistic symbionts within bacteriocytes (35). In A. pisum, short peptides resembling antimicrobial peptides are the most highly overexpressed genes in bacteriocytes, and show specific patterns of expression in these cells or in the nearby sheath cells, suggesting a role in controlling Buchnera (36). Other genes with potential roles in controlling Buchnera are several genes that were laterally transferred from bacterial genomes into the genome of an ancestral aphid, and that are highly overexpressed in bacteriocytes (32, 37). Some of these laterally transferred genes encode lysozymes that could act to lyse symbiont cells as part of a control mechanism. Another bacterial origin aphid gene up-regulated in bacteriocytes encodes the protein RlpA, which is imported into Buchnera cells, where it may act as an effector for symbiont regulation (38). Together, these aphid genes may form part of a system for recognizing, controlling, and transmitting Buchnera. On the Buchnera side, surface molecules that may interact with host systems include outer-membrane porins and the outer components of the flagellar apparatus. Buchnera lacks a flagellum but retains a large set of genes that underlie the flagellar secretion system, which is abundantly expressed on the symbiont cell surface (39). Furthermore, some of the flagellar proteins show unusual evolutionary acceleration in Buchnera (40, 41), potentially reflecting coevolution with host control mechanisms.
Buchnera cells are highly polyploid, and can vary in ploidy across aphid life stages (42, 43). A recent study (28) highlighted the cellular dynamics of Buchnera across the A. pisum life cycle using Buchnera cell counts rather than Buchnera genomic copy number, as in our experiments. It is likely that cell numbers and genomic copy numbers are strongly correlated; and it is not clear which is a better indicator of functional capacity of symbionts within a host individual. In any case, our measurement of symbiont titer reveals substantial variation in regulation of the symbiosis.
With the establishment of a maternally inherited obligate symbiosis, partner lineages enter into an irreversible coevolutionary relationship where symbionts provide significant benefits to hosts, which coadapt to maintain the interaction (12, 15). This relationship is distinct from those of hosts and their pathogens, which can also exhibit within-host evolution (44); for obligate, heritable symbiotic partners, hosts must maintain a viable symbiosis even if it involves a fitness cost. This study shows that intraspecific variation in hosts affects the regulation of obligate symbiosis and sheds light on its potential long-term evolutionary consequences.
Materials and Methods
Aphid Clones and Sexual Crosses.
All A. pisum clones used in this study were reared on seedlings of the broad bean Vicia faba in laboratory growth chambers under long-day (16-h of light: 8-h of dark) conditions at 20 °C. These clones represent A. pisum females sampled from alfalfa or broad bean in the United States between 1998 and 2014 and maintained as laboratory clones since collection. For this study, a single female from each clone was used to establish clonal lineages and maintained under constant conditions. Of the four North American A. pisum clones used in this study, LSR1 and 5AY share nearly identical Buchnera (16 single-nucleotide differences and no frameshifts in coding genes in a 640-kb genome) and are divergent from Buchnera found in AustinC and TucsonC clones, which have similar Buchnera (12 single-nucleotide differences, no frameshifts) We refer to Buchnera from AustinC and TucsonC as Buchnera haplotype A, and to Buchnera from 5AY and LSR1 as Buchnera haplotype B. These groups differ by 1,428 single-nucleotide differences, including many that affect amino acid residues, as well as 455 insertions/deletions, of which some disrupt protein-coding sequences. Clone AustinC was cured of secondary symbionts using antibiotic treatments (45). TucsonC was cured of secondary symbionts in a previous study (34). Both LSR1 and 5AY lack secondary symbionts. Thus, our sampling of clones eliminates potential effects that the presence of secondary symbionts may have on obligate symbiont titer.
To examine genetic variation affecting symbiont titer and aphid host fitness, we crossed four A. pisum clones to generate full-sibling and half-sibling progeny clones. We used standard methods to induce sexual female and male forms by exposing clones to a gradual reduction in daylight over 6 wk to reach short-day conditions (10-h of light:14-h of dark); conditions under which most A. pisum clones are known to produce both male and sexual females (oviparae) (46). Crosses were set up on caged plants, and the resulting eggs were “overwintered,” and subsequently monitored for hatching fundatrices to establish new clonal lines caged on individual host plants (23, 27, 46). For all experiments, progeny clones and parental clones were divided into three separate sublines and allowed to reproduce for at least three generations before data collection, to control for maternal and environmental effects.
Real-Time qPCR.
Real-time qPCR was used to estimate symbiont titer, the number of Buchnera genome copies compared with the number of aphid genome copies using single-copy genes from both Buchnera and the aphid nuclear genome. A single individual was measured for each subline within a clone. For each clone, nymphs were allowed to develop for 6 d to reach their fourth instar and then collected for titer measurements. Each individual was collected in a tube, frozen in liquid nitrogen, crushed with a pestle, and the resulting homogenate was treated using the Qiagen DNEasy kit to isolate DNA for qPCR.
Aphid genome copy number was estimated by measuring copy number of the single-copy gene encoding elongation factor-1 alpha (ef1α) using the primers ApEF1a 107F and ApEF1a 246R, yielding a product of 139 bp (27). Buchnera genome copy number was estimated by measuring copy number of the single-copy symbiont gene encoding a heat-shock protein (groES) using primers designed in this study: groES 18F (5′–CATGATCGTGTGCTTGTTAAG–3′) and groES 98R (5′– CTGTTCCTCGAGTCGATTTCC–3′), yielding a product of 80 bp. We prepared standards ranging from102 to 108 copies for each gene to generate a standard curve, which was used to estimate absolute copy number. All qPCR samples were prepared using BioRad iTaq SYBR Green master mix and performed on an Eppendorf Realplex Mastercycler. All reactions used the same “touchdown” cycling conditions, which included: 95 °C for 2 min, [95 °C for 10 s, 65(−1) °C for 15 s, 72 °C for 20 s] × 5 cycles, and (95 °C for 10 s, 60 °C for 15 s, 72 °C for 20 s) × 35 cycles. For each sample, the number of copies of ef1α and groES were determined using the Realplex Mastercycler software based on comparisons with serial dilutions of the qPCR standards. Reactions were performed in triplicate for each sample, and the average was used to represent that subline.
Measurement of Aphid Fitness.
Average fitness was measured for sublines of new progeny clones and for sublines of the original parental clones. Adults of each clone were placed on plants overnight at 20 °C and removed, resulting in even-aged nymphs on each plant. Aphids were monitored daily, and dates of molting to adulthood and weights of new adults were recorded. Newly eclosed adults were placed individually on new plants, and the date of first reproduction and total number of nymphs produced during the following 6 d was recorded. Fecundity for each subline was estimated as the total number of progeny produced in the first 6 d following first reproduction. This measure accounts for a majority of the offspring an adult A. pisum will produce in a generation, as daily production declines a few days after first reproduction (47, 48). In field populations, aphids undergo high mortality, further decreasing the contribution of fecundity of older females. Reproductive rate was calculated as the total number of offspring produced in the first 6 d divided by number of days from birth to first reproduction plus 1 d. All fitness parameters were measured twice for each subline.
Statistical Analyses.
All statistical analyses were performed using R 3.2.3, with a statistical significance threshold of P < 0.05. Buchnera titer was estimated by the relative number of Buchnera gene-copies divided by the number of the single-copy host gene. Significance of Buchnera titer variation among progeny clones and parental clones was tested using ANOVA. Aphid fitness measurements were analyzed using Kruskal–Wallis tests. Correlations between Buchnera titer and host fitness parameters were tested using Spearman’s rank correlation coefficient.
Acknowledgments
We thank members of the N.A.M. and H. Ochman laboratory groups at University of Texas at Austin for discussion and feedback; K. Hammond, C. Cole, and D. Tran for assistance with aphid rearing and plantings; and C. Cole for help with laboratory work and fitness measurements. This project was supported by Agriculture and Food Research Initiative Competitive Grant 2016-67012-24676 (to R.A.C.) from the US Department of Agriculture National Institute of Food and Agriculture; National Science Foundation Award IOS1347116 (to N.A.M.); and the University of Texas at Austin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610749113/-/DCSupplemental.
References
- 1.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126(3):453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: Insights from genomics. Trends Ecol Evol. 2000;15(8):321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P, et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 6.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106(36):15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323(5912):379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 8.Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect host. Proc Biol Sci. 1993;253(1337):167–171. [Google Scholar]
- 9.Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3(11):850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 10.Koga R, Meng XY, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA. 2012;109(20):E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 12.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 13.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Ham RCHJ, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100(2):581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett GM, Moran NA. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112(33):10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller’s Ratchet with two levels of selection. Am Nat. 2000;156(4):425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 17.Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF. Genetic incompatibilities are widespread within species. Nature. 2013;504(7478):135–137. doi: 10.1038/nature12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloan DB, Triant DA, Wu M, Taylor DR. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol Biol Evol. 2014;31(3):673–682. doi: 10.1093/molbev/mst259. [DOI] [PubMed] [Google Scholar]
- 19.Willett CS, Burton RS. Evolution of interacting proteins in the mitochondrial electron transport system in a marine copepod. Mol Biol Evol. 2004;21(3):443–453. doi: 10.1093/molbev/msh031. [DOI] [PubMed] [Google Scholar]
- 20.Mossman JA, Biancani LM, Zhu CT, Rand DM. Mitonuclear epistasis for development time and its modification by diet in Drosophila. Genetics. 2016;203(1):463–484. doi: 10.1534/genetics.116.187286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108(7):2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shigenobu S, Wilson AC. Genomic revelations of a mutualism: The pea aphid and its obligate bacterial symbiont. Cell Mol Life Sci. 2011;68(8):1297–1309. doi: 10.1007/s00018-011-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran NA, Degnan PH. Functional genomics of Buchnera and the ecology of aphid hosts. Mol Ecol. 2006;15(5):1251–1261. doi: 10.1111/j.1365-294X.2005.02744.x. [DOI] [PubMed] [Google Scholar]
- 24.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407(6800):81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 25.Price DR, et al. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. 2014;111(1):320–325. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglas AE, Wilkinson TL. Host cell allometry and regulation of the symbiosis between pea aphids, Acyrthosiphon pisum, and bacteria, Buchnera. J Insect Physiol. 1998;44(7-8):629–635. doi: 10.1016/s0022-1910(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 27.Vogel KJ, Moran NA. Effect of host genotype on symbiont titer in the aphid—Buchnera symbiosis. Insects. 2011;2(3):423–434. doi: 10.3390/insects2030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonet P, et al. Direct flow cytometry measurements reveal a fine-tuning of symbiotic cell dynamics according to the host developmental needs in aphid symbiosis. Sci Rep. 2016;6:19967. doi: 10.1038/srep19967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunbar HE, Wilson AC, Ferguson NR, Moran NA. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007;5(5):e96. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson TL. The elimination of intracellular microorganisms from insects: An analysis of antibiotic-treatment in the pea aphid (Acyrthosiphon pisum) Comp Biochem Physiol. 1998;119:871–881. [Google Scholar]
- 31.Lai CY, Baumann L, Baumann P. Amplification of trpEG: Adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA. 1994;91(9):3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102(15):5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersson ME, Berg OG. Muller’s ratchet in symbiont populations. Genetica. 2007;130(2):199–211. doi: 10.1007/s10709-006-9007-7. [DOI] [PubMed] [Google Scholar]
- 34.Burke G, Fiehn O, Moran N. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. 2010;4(2):242–252. doi: 10.1038/ismej.2009.114. [DOI] [PubMed] [Google Scholar]
- 35.Login FH, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334(6054):362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 36.Shigenobu S, Stern DL. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc Biol Sci. 2013;280(1750):20121952. doi: 10.1098/rspb.2012.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikoh N, et al. Bacterial genes in the aphid genome: Absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6(2):e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima SY. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr Biol. 2014;24(14):R640–R641. doi: 10.1016/j.cub.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 39.Maezawa K, et al. Hundreds of flagellar basal bodies cover the cell surface of the endosymbiotic bacterium Buchnera aphidicola sp. strain APS. J Bacteriol. 2006;188(18):6539–6543. doi: 10.1128/JB.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toft C, Fares MA. The evolution of the flagellar assembly pathway in endosymbiotic bacterial genomes. Mol Biol Evol. 2008;25(9):2069–2076. doi: 10.1093/molbev/msn153. [DOI] [PubMed] [Google Scholar]
- 41.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296(5577):2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 42.Komaki K, Ishikawa H. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol. 1999;48(6):717–722. doi: 10.1007/pl00006516. [DOI] [PubMed] [Google Scholar]
- 43.Komaki K, Ishikawa H. Genomic copy number of intracellular bacterial symbionts of aphids varies in response to developmental stage and morph of their host. Insect Biochem Mol Biol. 2000;30(3):253–258. doi: 10.1016/s0965-1748(99)00125-3. [DOI] [PubMed] [Google Scholar]
- 44.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 2016;14(3):150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koga R, Tsuchida T, Sakurai M, Fukatsu T. Selective elimination of aphid endosymbionts: Effects of antibiotic dose and host genotype, and fitness consequences. FEMS Microbiol Ecol. 2007;60(2):229–239. doi: 10.1111/j.1574-6941.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- 46.Via S. Inducing the sexual forms and hatching the eggs of pea aphids. Entomol Exp Appl. 1992;65:119–127. [Google Scholar]
- 47.Barribeau SM, Sok D, Gerardo NM. Aphid reproductive investment in response to mortality risks. BMC Evol Biol. 2010;10:251. doi: 10.1186/1471-2148-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez AJ, Ritter SG, Doremus MR, Russell JA, Oliver KM. Aphid-encoded variability in susceptibility to a parasitoid. BMC Evol Biol. 2014;14:127. doi: 10.1186/1471-2148-14-127. [DOI] [PMC free article] [PubMed] [Google Scholar]