Significance
Peroxisomes are eukaryotic organelles crucial for development. Peroxisomal matrix proteins are imported by the peroxisome import machinery composed of peroxins (PEX proteins), but how the function of these PEX proteins is regulated is largely unknown. We discovered in Arabidopsis that the ubiquitin–proteasome system regulates peroxisome protein import via an E3 ubiquitin ligase, SP1 (suppressor of ppi1 locus1), which targets PEX13 and possibly several other components of the peroxisome matrix protein import machinery for degradation. Our data demonstrate that the same E3 ubiquitin ligase can be shared by metabolically linked peroxisomes and chloroplasts to promote the destabilization of distinct components of the two import machineries, suggesting that the ubiquitin–proteasome system may represent an important regulatory mechanism coordinating the biogenesis of functionally associated organelles.
Keywords: peroxisome biogenesis, E3 ubiquitin ligase, protein import, peroxin, SP1
Abstract
Peroxisomes are ubiquitous eukaryotic organelles that play pivotal roles in a suite of metabolic processes and often act coordinately with other organelles, such as chloroplasts and mitochondria. Peroxisomes import proteins to the peroxisome matrix by peroxins (PEX proteins), but how the function of the PEX proteins is regulated is poorly understood. In this study, we identified the Arabidopsis RING (really interesting new gene) type E3 ubiquitin ligase SP1 [suppressor of plastid protein import locus 1 (ppi1) 1] as a peroxisome membrane protein with a regulatory role in peroxisome protein import. SP1 interacts physically with the two components of the peroxisome protein docking complex PEX13–PEX14 and the (RING)-finger peroxin PEX2. Loss of SP1 function suppresses defects of the pex14-2 and pex13-1 mutants, and SP1 is involved in the degradation of PEX13 and possibly PEX14 and all three RING peroxins. An in vivo ubiquitination assay showed that SP1 has the ability to promote PEX13 ubiquitination. Our study has revealed that, in addition to its previously reported function in chloroplast biogenesis, SP1 plays a role in peroxisome biogenesis. The same E3 ubiquitin ligase promotes the destabilization of components of two distinct protein-import machineries, indicating that degradation of organelle biogenesis factors by the ubiquitin–proteasome system may constitute an important regulatory mechanism in coordinating the biogenesis of metabolically linked organelles in eukaryotes.
Peroxisomes are single-membrane organelles that are present in virtually all eukaryotic cells and host critical metabolic reactions including fatty acid β-oxidation and H2O2 degradation (1). In plants, peroxisomes are essential to many metabolic processes such as lipid mobilization, the glyoxylate cycle, photorespiration, detoxification, biosynthesis, and metabolism of plant hormones (2, 3). Some of these metabolic processes are coordinated by peroxisomes and other organelles, e.g., mitochondria and chloroplasts for photorespiration, lipid bodies and mitochondria for lipid mobilization, and chloroplasts for jasmonic acid biosynthesis (3–6). The enzymatic composition of plant peroxisomes is dynamic, depending on developmental and environmental cues. For example, in young seedlings of oilseed plants such as Arabidopsis, the major enzymatic content of peroxisomes shifts from the glyoxylate cycle enzymes to photorespiratory enzymes within a few days after germination. This process is induced by light, is achieved through proteome remodeling, and occurs simultaneously with chloroplast development (3, 7–10). The turnover of the peroxisome proteome is correlated with the spatial distribution of peroxisomes, because they are found next to lipid bodies during seed germination but become physically associated with chloroplasts when plants reach photoautotrophic growth in the light (11–13). One can speculate that the development of peroxisomes and chloroplasts may be coregulated to accommodate the functional association of these two organelles in photoautotrophic growth.
The peroxisome proteome is encoded entirely in the nucleus. Peroxisome matrix proteins are imported from the cytosol by the evolutionarily conserved peroxins (PEX proteins) (3, 14). In Arabidopsis, PEX5 and PEX7 are cytosolic receptors for peroxisome proteins that contain C-terminal PTS1 (peroxisome targeting signal type 1) and N-terminal PTS2 sequences, respectively (15, 16). In Arabidopsis, PEX5 is also required for PTS2 protein import (16). Two membrane proteins, PEX13 and PEX14, form the docking site for PEX5 and PEX7 (17, 18). After receptor docking, cargo proteins translocate into the matrix before receptors are recycled to the cytosol (19–21). These processes require the RING (really interesting new gene)-finger peroxins PEX2, PEX10, and PEX12 (22–25), the ATPases PEX1 and PEX6 and their membrane tether APEM9 (aberrant peroxisome morphology 9) and the ubiquitin-conjugating enzyme PEX4 and its membrane anchor PEX22 (26–29). Studies from yeast reveal that PEX12-mediated PEX5 monoubiquitination precedes PEX5 recycling (30). Although there is no direct evidence so far for PEX5 ubiquitination in plants, the RING domain of Arabidopsis PEX2, PEX10, and PEX12 was shown to possess E3 activity in vitro; the in vivo targets for their activities remain unclear (31). Disruption of the function of plant PEX proteins causes embryonic lethality or compromises peroxisome function such as β-oxidation (3, 32, 33). Moreover, maintaining the balance between cargo translocation into the peroxisome/receptor docking and receptor recycling back to the cytosol appears to be important for the functional integrity of peroxisomes in plants. For example, as is consistent with both PEX13 and PEX14 being involved in receptor docking at the peroxisome membrane, the peroxisomal defects of pex14-2 are enhanced by pex13-1, a weak allele with mildly reduced PEX13 mRNA levels (17, 18, 34). However, the same pex13-1 allele partially suppressed the peroxisomal phenotypes of the late-acting peroxin mutant pex4-1, which is deficient in the translocation of PEX5 out of the peroxisome. This observation led to the conclusion that the inefficiency in both cargo translocation into the peroxisome (pex13-1) and PEX5′s recycling back to the cytosol (pex4-1) restored the balance between the import and export of PEX5 in the pex13-1 pex4-1 double mutant (34).
How the function of the peroxins is regulated remains poorly understood. We have been investigating the role of the ubiquitin–proteasome system (UPS) in the biogenesis of peroxisomes and mitochondria (25, 31, 35). The UPS is a key regulatory mechanism that controls various cellular pathways in eukaryotic cells, in which polyubiquitinated proteins are degraded by the 26S proteasome (36, 37). Proteins involved in the UPS pathway were estimated to make up ∼5–6% of the Arabidopsis proteome; the majority (∼1,400) of these proteins are, or are predicted to be, E3 ubiquitin ligases (38). The triad cascade of protein ubiquitination consists of the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (36). E3s associate with both E2s and substrate proteins to promote substrate-specific ubiquitination; therefore the tremendous diversity of E3 ligases in plant genomes is consistent with the key roles of this class of proteins in defining substrate specificity. The UPS is involved in organelle biogenesis and/or morphogenesis in plants. Arabidopsis SP1 [suppressor of ppi1 (plastid protein import locus 1) 1] is a RING-type ubiquitin ligase that binds to and promotes the degradation of a few components of the TOC (translocon at the outer envelope of chloroplasts) complex (39, 40). We also identified an Arabidopsis mitochondrial outer membrane-associated ubiquitin-specific protease, UBP27, which is involved in mitochondrial morphogenesis, possibly through division (35).
This study is part of our continuing effort to identify proteins involved in UPS-mediated regulation of organelle biogenesis. We show that SP1, a RING-type E3 ubiquitin ligase known to regulate chloroplast protein import, is also associated with the peroxisome membrane and targets PEX13 and possibly several other components of the peroxisome protein import machinery for degradation. Thus, the same E3 ligase targets distinct components of the peroxisome and chloroplast protein import apparatuses, indicating that UPS-mediated protein degradation may constitute an important regulatory mechanism by which eukaryotic cells coordinate the biogenesis of metabolically linked organelles.
Results
The Arabidopsis E3 Ubiquitin Ligase SP1 Is also a Peroxisome Membrane Protein.
While searching the Arabidopsis genome for putative peroxisome membrane-localized E3 ubiquitin ligases, we identified a small family of proteins comprising SP1, SPL1 (SP1-like 1), and SPL2 as candidates because of the presence of transmembrane domains (TMDs) in addition to the RING domain on these proteins (Fig. 1A and Fig. S1A). In a recent study, all three members were found to be associated with chloroplasts, and SP1 was shown to regulate chloroplast protein import (40).
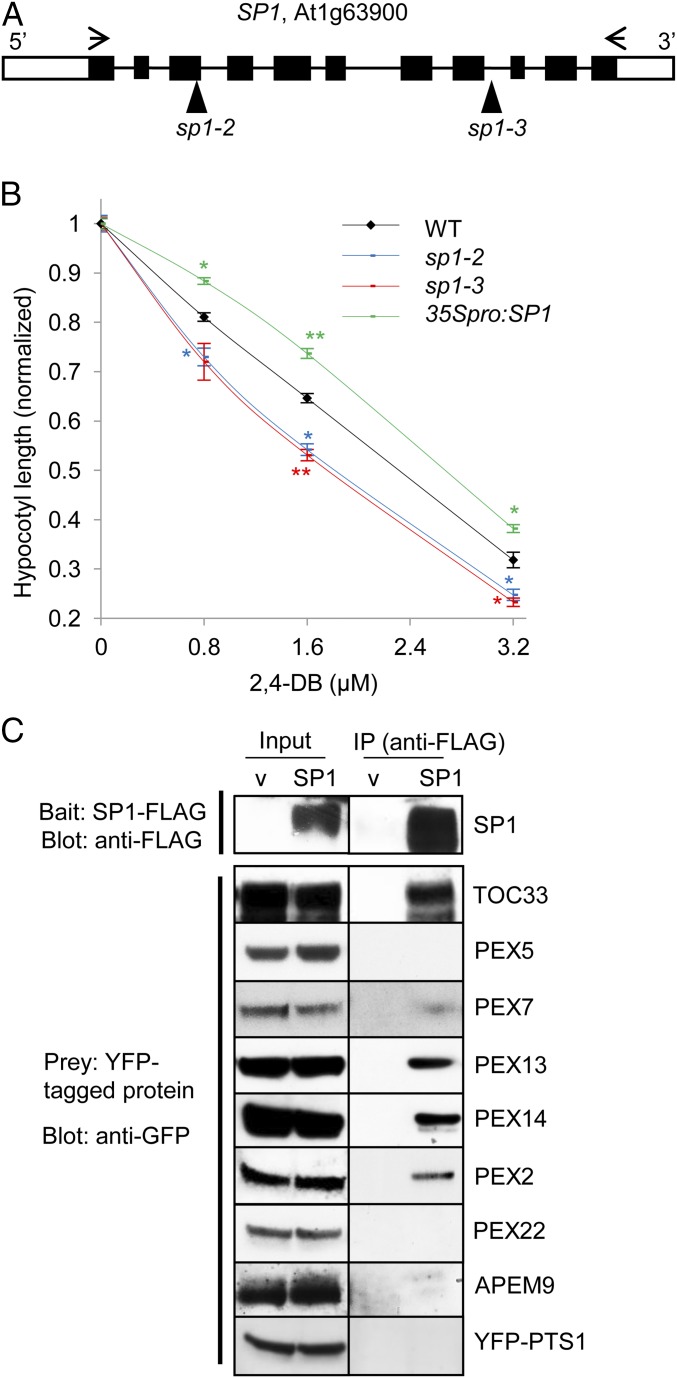
Fig. 1.
Localization of the SP1 protein. (A) Domain structure of the SP1 protein. TM, transmembrane domain. RING, RING domain. C-term, C terminus. Amino acid numbers are indicated. T-DNA insertion sites of in the two sp1 mutants are also indicated. (B) Peroxisome localization of SP1-YFP. Confocal images were taken in leaf epidermal cells of 2-wk-old Arabidopsis T2 plants coexpressing SP1-YFP and CFP-PTS1. (Scale bar, 10 μm.) (C) Assessment of the purity of peroxisomes isolated from the leaf tissue of transgenic Arabidopsis plants coexpressing SP1-YFP and CFP-PTS1. Organelle-specific antibodies used were against Arabidopsis VDAC (Voltage-dependent anion channel) (mitochondrial), FtsZ1 (Filamenting temperature-sensitive mutant Z) (chloroplast), and PEX11d (peroxisomal) proteins. CE, crude extract. (D) Peroxisomal membrane association of SP1-YFP. Purified peroxisomes were treated with TE, NaCl, or Na2CO3 (pH 11.0) and were separated into soluble (S) and pellet (P) fractions by centrifugation. SP1-YFP and CFP-PTS1 were detected by α-GFP. CFP-PTS1 and PEX11d are controls for matrix and integral membrane proteins, respectively. The matrix protein CFP-PTS1 is often detected in the pellet as well as in the soluble fraction, as has been observed in previous studies.
Fig. S1.
Localization of SPL1 and SPL2 proteins. (A) Domain structure of SPL1 and SPL2. TM, transmembrane domain; RING, RING domain. Amino acid numbers are indicated. (B) SP1-YFP is localized to peroxisomes and chloroplasts. Confocal images were taken in leaf mesophyll cells of 2-wk-old Arabidopsis T2 plants coexpressing SP1-YFP and CFP-PTS1. (Scale bar, 10 μm.) (C) SPL1 and SPL2 localize to chloroplasts. Confocal images were taken in leaf mesophyll cells from 2-wk-old T2 lines expressing SPL1-YFP or SPL2-YFP. Chloroplasts are marked by chlorophyll autofluorescence. (Scale bar, 5 μm.) (D) Subcellular localization of SPL1-YFP and SPL2-YFP with reference to peroxisomes. Confocal images were taken in leaf epidermal cells from 2-wk-old T2 lines expressing SPL1-YFP and CFP-PTS1 or SPL2-YFP and CFP-PTS1. (Scale bar, 10 μm.) (E and F) SP1-YFP is localized to peroxisomes and chloroplasts. SP1pro:SP1-YFP was transiently expressed in Arabidopsis using the FAST method. Confocal images were taken in leaf epidermal (E) and mesophyll (F) cells of Arabidopsis seedlings stably expressing CFP-PTS1. (Scale bars, 10 μm.)
To test the possibility that these E3s are also associated with peroxisomes, translational fusions of the coding region of each protein and YFP were generated. Arabidopsis transgenic lines expressing the 35S constitutive promoter-driven fusion proteins were generated in a wild-type Col-0 background that already contained the peroxisomal marker CFP-PTS1 (SKL) (25, 41). Confocal microscopic analysis of T2 plants revealed that, in agreement with the previous report (40), SP1, SPL1, and SPL2 are associated with chloroplasts (Fig. S1 B and C). Interestingly, all three seemed to be concentrated on small (potentially developing) chloroplasts (Fig. S1 B and C); this localization may be related to their proven (SP1) or potential (SPL1 and SPL2) roles in chloroplast protein import. Among the three fusion proteins, only SP1 showed clear peroxisomal localization in different cell types, such as epidermal cells (Fig. 1B) and mesophyll cells (Fig. S1B). In contrast, SPL1 and SPL2 showed very weak and possibly partial (SPL1) or no (SPL2) peroxisomal localization (Fig. S1D). To confirm SP1’s localization to peroxisomes, we expressed SP1-YFP by SP1’s native promoter (SP1pro:SP1-YFP) in leaves of Arabidopsis seedlings by the Fast Agro-mediated Seedling Transformation (FAST) method (42) and observed the same pattern of localization to peroxisomes and chloroplasts (Fig. S1 E and F). Based on these observations, we decided to focus on SP1 to investigate its possible role in peroxisomes.
To assess SP1’s peroxisome localization further and to determine the membrane association of SP1 on peroxisomes, we isolated peroxisomes from the leaf tissue of Arabidopsis plants stably expressing SP1-YFP and CFP-PTS1. The purity of the isolated peroxisomes was assessed by immunoblot analysis using organelle-specific protein antibodies (Fig. 1C). To separate integral membrane proteins from total proteins, the isolated organelles were treated with Tris–EDTA (TE), a high-concentration salt solution (1 M NaCl), and alkaline carbonate (Na2CO3, pH 11.0), respectively. SP1-YFP, along with the peroxisomal membrane protein PEX11d, was detected only in the pellet after each treatment (Fig. 1D), suggesting that SP1 is an integral membrane protein of the peroxisome.
SP1 Represses Peroxisome Function.
To analyze the function of SP1 in peroxisomes, we obtained from the Arabidopsis Biological Resource Center two transferred DNA (T-DNA) insertion mutants that previously had been designated sp1-2 and sp1-3 (Fig. 2A) (40). Homozygous mutants were obtained after PCR genotyping of genomic DNA, and SP1 transcript levels were analyzed using RT-PCR, which showed the absence of full-length SP1 transcripts in both mutant alleles (Fig. S2 A and B). As previously reported (40), the growth and development of these mutants were similar to that in wild-type plants under normal growth conditions (Fig. S2C).
Fig. 2.
SP1 affects peroxisome function and interacts with several peroxins. (A) SP1 gene structure. The open box represents the UTR; the black boxes represent exons; the solid line represents the intron. T-DNA insertion sites in the two sp1 mutants are indicated. Arrows indicate the primers used to amplify full-length cDNA. (B) 2,4-DB response assays. Seedlings were grown on Murashige and Skoog (MS) medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average hypocotyl length of 7-d-old dark-grown seedlings is shown. All data on 2,4-DB are normalized to the data obtained from plants grown on 0.5% sucrose medium with no 2,4-DB. Data are shown as mean ± SEM; n = 3 (>30 seedlings were used for each biological replicate). **P < 0.01; *P < 0.05 from wild type. (C) Co-IP of PEX proteins with SP1. YFP-tagged candidate substrate proteins were coexpressed in tobacco leaves with either SP1-FLAG or empty vector (v). The top panel is a representative blot showing the level of SP1-FLAG in co-IP experiments performed for different YFP-tagged proteins.
Fig. S2.
Phenotypes of sp1 mutants. (A) SP1 gene structure. The open box represents the UTR; the black boxes represent exons; the solid line represents the intron. T-DNA insertion sites in the mutants are indicated. Arrows indicate the primers used in RT-PCR. (B) RT-PCR analysis of RNA from 4-wk-old sp1 T-DNA insertion mutant lines. Two individual plants were analyzed for each mutant. P, wild-type; N, water only (negative control). Ubiquitin 10 (UBQ10) was used as a loading control. White lines separate noncontiguous gel lanes. The negative control for UBQ10 in sp1-2 was run on a separate gel. (C) Four-week-old sp1 mutants. (D) 2,4-DB response assays of 7-d-old light-grown seedlings. Seedlings were grown on MS medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average root length normalized to the data obtained from plants grown on 0.5% sucrose medium without 2,4-DB is shown. Error bars indicate SEM; n = 3 (>30 seedlings for each biological replicate). **P < 0.01; *P < 0.05 from wild type. (E) RT-PCR analysis of RNA from a 4-wk-old 35Spro: SP1 line. P, wild-type; N, negative control (water only). Ubiquitin 10 was used as a loading control. White lines separate discontinuous gel lanes. (F) Epifluorescence images showing peroxisome morphology in mesophyll cells of 2-wk-old plants expressing the peroxisome marker YFP-PTS1. (Scale bar, 10 μm.) drp3A-2 (SALK_147485), which is defective in fission, is included as an example of abnormal peroxisome morphology.
To assess peroxisome functions in the mutants, we used the 2,4-DB (2,4-dichlorophenoxybutryic acid) response assay, which is often used to identify mutants in β-oxidation enzymes or peroxisome biogenesis. 2,4-DB undergoes β-oxidation in peroxisomes to form the active auxin-like derivative 2,4-D, resulting in the inhibition of primary root and hypocotyl elongation when applied to seedlings (32). Both sp1 mutant alleles were more sensitive than wild type to 2,4-DB (Fig. 2B and Fig. S2D). We also generated transgenic lines stably expressing SP1 under the 35S promoter (Fig. S2E), which were more resistant to 2,4-DB’s inhibitory effect on hypocotyl and root elongation (Fig. 2B and Fig. S2D). Hence, peroxisomal β-oxidation activities appeared to be elevated in sp1 mutants and deficient when SP1 was overexpressed, suggesting that SP1 plays a repressive role in peroxisomal function, given that peroxisomes are the sole sites of β-oxidation in plants.
Physical and Genetic Interaction Between SP1 and Members of the Peroxisome Protein Import Machinery.
To determine whether SP1 affects peroxisome morphology, we introduced the peroxisome marker YFP-PTS1 into sp1 mutants through transformation. Confocal microscopic analysis of the mutants expressing YFP-PTS1 did not reveal obvious abnormalities in peroxisome morphology (Fig. S2F), suggesting that SP1 does not play a major role in peroxisome morphogenesis. Because SP1 was shown to regulate chloroplast biogenesis by targeting components of the TOC protein import complex for degradation (40), we tested the idea that SP1 may target the peroxisomal protein import machinery. First, we performed coimmunoprecipitation (co-IP) assays using FLAG-tagged SP1 and several YFP-tagged PEX proteins. Similar to the chloroplast TOC33 positive control, PEX13, PEX14, and PEX2 clearly coprecipitated with SP1-FLAG (Fig. 2C). Because of protein instability, we were unable to test the other two RING peroxins, PEX10 and PEX12, in this assay. Based on the co-IP results, we considered PEX13, PEX14, and possibly the RING peroxins as potential substrates for SP1.
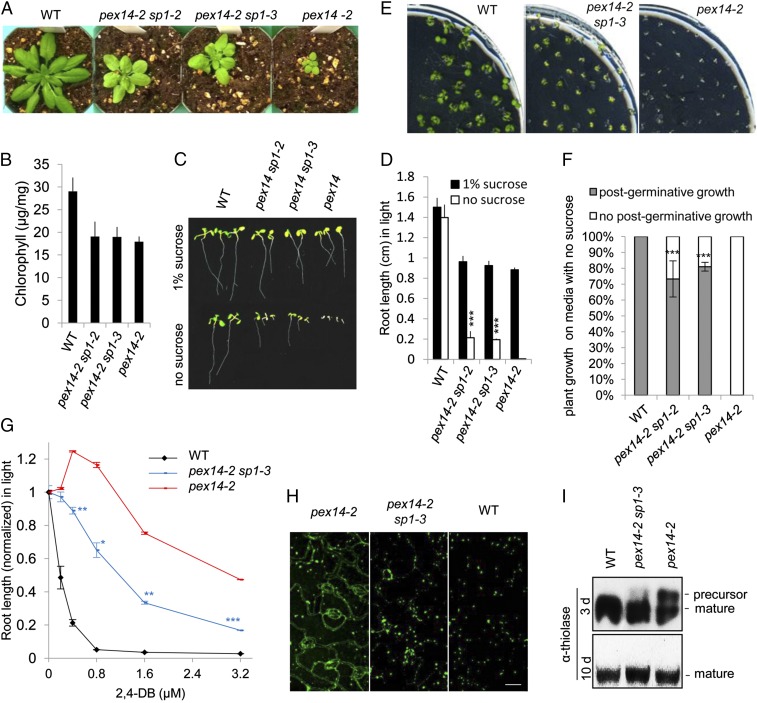
Next we examined genetic interactions between SP1 and the PEX proteins. We crossed sp1 with pex14-2, a null allele that carries a T-DNA insertion 41 bp downstream of PEX14’s start codon (17). The sp1 pex14-2 double mutants exhibited marked rescue of the small stature of pex14-2 (Fig. 3A), although the chlorophyll content was not significantly different from that of pex14-2 (Fig. 3B). Because peroxisomes are the sole site of fatty acid β-oxidation in plants, mutants defective in β-oxidation, such as pex14-2, are often dependent upon exogenous fixed carbon sources, such as sucrose, for seedling establishment (43). On medium without sucrose, pex14-2 sp1 seedlings overall had longer roots and hypocotyls than pex14-2 seedlings (Fig. 3 C and D and Fig. S3 A and B). Although all pex14-2 seedlings died at 10 d after germination because of their dependence on sucrose, 70–80% of the double mutant seedlings continued to grow (Fig. 3 E and F). Compared with pex14-2 seedlings, the pex14-2 sp1 seedlings were less resistant to 2,4-DB’s inhibitory effect on root and hypocotyl elongation (Fig. 3G and Fig. S3C). Together these data demonstrated that the mutant phenotype of pex14-2 was significantly rescued by the sp1 mutation.
Fig. 3.
The loss of SP1 function suppresses pex14-2 mutant phenotypes. (A) Four-week-old plants that had been grown for 1 wk in sucrose-containing Linsmaier and Skoog (LS) medium and for 3 wk in soil. (B) Leaf chlorophyll contents of the plants shown in A. Data are shown as mean ± SD; n = 3. (C) Ten-day-old seedlings grown on MS medium without or supplemented with 1% sucrose. (D) Root length measurement of 10-d-old seedlings grown on MS medium without or supplemented with 1% sucrose. Data are shown as mean ± SD; n = 3 (>30 seedlings were used for each biological replicate). ***P < 0.001 from pex14-2. (E) Ten-day-old seedlings grown on LS medium with no sucrose. (F) Quantitative analysis of postgerminative growth of the seedlings shown in E. Error bars indicate the SD; n = 3 (>100 seedlings for each biological replicate). ***P < 0.001 from pex14-2. (G) Root length measurement of 7-d-old light-grown seedlings grown on MS medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average root length normalized to the data obtained from plants grown on 0.5% sucrose medium without 2,4-DB is shown. Data are shown as mean ± SEM; n = 3 (>30 seedlings were measured for each biological replicate). ***P < 0.001; **P < 0.01; *P < 0.05 from pex14-2. (H) Confocal microscopic images of leaf epidermal cells from 2-wk-old light-grown plants stably expressing 35Spro:YFP-PTS1. Images were acquired with identical microscopic settings. (Scale bar, 20 μm.) (I) Immunoblots of total protein extracts from seedlings 3 and 10 days after germination using the anti-thiolase antibody.
Fig. S3.
Phenotypes of sp1 pex14-2 and sp1 pex13-1 double mutants. (A and B) Sucrose-dependence assays of 10-d-old dark-grown seedlings grown on MS medium without or supplemented with 1% sucrose. Average hypocotyl length is shown. Error bars indicate SD; n = 3 (>30 seedlings for each biological replicate). **P < 0.01 from pex14-2. (C) 2,4-DB response assays of 7-d-old dark-grown seedlings grown on MS medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average hypocotyl length is normalized to the data obtained from seedlings grown on 0.5% sucrose medium without 2,4-DB. Error bars indicate SEM; n = 3 (>30 seedlings for each biological replicate). **P < 0.01; *P < 0.05 from pex14-2. (D) Sucrose-dependence assays of 1-wk-old dark-grown seedlings grown on 0.25× MS medium with or without 1% sucrose. The average hypocotyl length is shown. Error bars indicate the SD; n = 3 (>30 seedlings for each biological replicate). **P < 0.01 from pex13-1. (E) 2,4-DB response assays on 7-d-old dark-grown seedlings grown on MS medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average hypocotyl length is normalized to the data obtained from seedlings grown on 0.5% sucrose medium without 2,4-DB. Error bars indicate SEM; n = 3 (>30 seedlings for each biological replicate). **P < 0.01 from pex13-1.
The strong suppression of the pex14 phenotype by sp1 prompted us to check the effect of sp1 mutation on peroxisomal matrix protein import in the pex14-2 background. We crossed pex14-2 plants to transgenic lines stably expressing 35Spro:YFP-PTS1 (44, 45). In contrast to wild-type leaf cells, in which YFP-labeled peroxisomes appeared as punctate structures, YFP-PTS1 was partially cytosolic in pex14-2 plants (Fig. 3H), similar to observations in a previous report (17), indicating deficient import of PTS1-containing proteins in the mutant (17). In the pex14-2 sp1 double mutant, the cytosolic localization of YFP-PTS1 was largely diminished (Fig. 3H), suggesting that PTS1 protein import in pex14-2 plants was significantly restored as a result of the sp1 mutation. To assess the impact of sp1 on peroxisome protein import in pex14-2 plants further, we also performed immunoblot analysis of a PTS2-containing protein, 3-ketoacyl-CoA thiolase (thiolase), whose precursors are retained for a longer time in mutants defective in PTS2 protein import than in wild-type plants (17). After PTS2-containing proteins are delivered into the matrix, the N-terminal PTS2 peptides are removed by proteases such as DEG15 (46, 47). By evaluating PTS2 protein processing, we could indirectly assess the import of PTS2-containing proteins or the import of PTS1-containing proteases such as DEG15 that are required for PTS2 processing. In 3-d-old seedlings there was a strong accumulation of the thiolase precursor in pex14-2 plants, but this accumulation was greatly reduced in the pex14-2 sp1 double mutant (Fig. 3I). In summary, these data provided evidence that the amelioration of the physiological defects of pex14-2 plants by the sp1 mutation correlated with improved matrix protein import, further supporting the idea that SP1 plays a negative role in peroxisome protein import.
We also crossed sp1 with pex13-1, a weak allele with mildly reduced PEX13 mRNA levels (34). The weak sucrose-dependent and 2,4-DB–resistant phenotypes in pex13-1 plants were both suppressed in the sp1 pex13-1 double mutant (Fig. S3 D and E). Hence, the peroxisomal β-oxidation deficiencies in pex14-2 and pex13-1 plants were at least partially recovered by the lack of SP1 function, as is consistent with the hypothesis that SP1 negatively regulates the function of these two components of the peroxisome protein import machinery. Because the PEX14 protein is absent in pex14-2 plants, the observed rescue of the mutant phenotype in sp1 pex14-2 plants is unlikely to be caused by the stabilization or enhancement of the function of PEX14. Instead, the stabilization/enhancement of other proteins, such as PEX13, may compensate for the loss of PEX14.
SP1’s Negative Role in Peroxisome Protein Import Is Restricted to Certain Steps of the Process.
PEX13 and PEX14 mediate receptor docking and cargo translocation, which are early steps of peroxisome protein import (3, 17, 18, 34). To determine whether SP1 also impacts peroxins that act in other steps of matrix protein import, we generated additional double mutants, namely sp1 pex5-10 and sp1 lon2-2. PEX5 and LON2 (LON protease-like 2) are involved in different phases of peroxisome protein import: PEX5 is the receptor for matrix proteins, and LON2 is a peroxisomal protease involved in sustained protein import in older seedlings (48, 49). The pex5-10 mutant allele produces a PEX5 protein that has a deletion in the middle region, has a germination defect, and is dependent on exogenous sucrose for early seedling establishment (48). The lon2-2 mutant has matrix protein import defects in older seedlings and is partially dependent on exogenous sucrose for seedling development (49). Neither pex5-10 sp1 nor lon2-2 sp1 mutants exhibited significant differences from pex5-10 and lon2-2 mutants in growth (Fig. S4 A and B) or sucrose dependence (Fig. S4 C–F). Thus, the suppression of peroxisome-related physiological defects by sp1 is restricted to specific peroxisomal biogenesis mutants.
Fig. S4.
Phenotypes of sp1 pex5-10, sp1 lon2-2 and sp1 pex4-1 double mutants. (A and B) Four-week-old plants grown on LS medium with sucrose for 1 wk followed by 3 wk of growth on soil. (C and D) Sucrose-dependence assays of seedlings grown on MS medium supplemented with 0%, 0.2%, or 1% sucrose. The average root length of 7-d-old light-grown seedlings (C) and the average hypocotyl length of 7-d-old dark-grown seedlings (D) are shown. Error bars indicate the SD; n = 3 (>30 seedlings were used for each biological replicate). The seed coats of wild-type, pex5-10, and pex5-10 sp1-3 seeds were nicked before seeds were plated in this assay. (E and F) Sucrose-dependence assays of seedlings grown on MS medium with or without 1% sucrose. The average root length of 7-d-old light-grown seedlings (E) and the average hypocotyl length of 7-d-old dark-grown seedlings (F) are shown. Error bars indicate the SD; n = 3 (>30 seedlings were used for each biological replicate). (G and H) Plants grown on LS medium with sucrose for 1 wk followed by 3 wk (G) or 4 wk (H) of growth in soil. (I) 2,4-DB response assays of 7-d-old light-grown seedlings grown on MS medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. The average root length is normalized to the data obtained from plants grown on 0.5% sucrose medium without 2,4-DB. Error bars indicate the SEM; n = 3 (>30 seedlings for each biological replicate). *P < 0.05 from pex4-1.
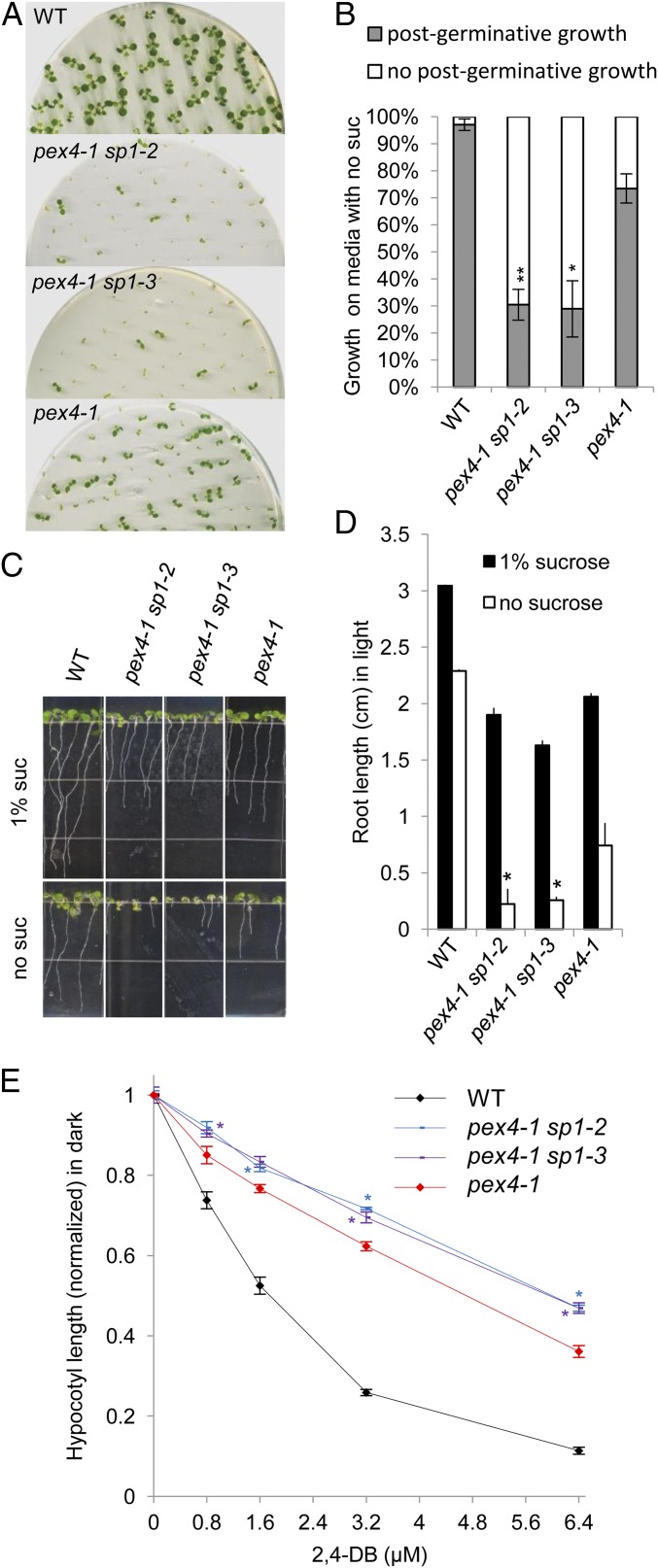
The sp1 mutant was also crossed with pex4-1, a mutant of the E2 ubiquitin-conjugating enzyme involved in the recycling of PEX5 back to the cytosol, a late step in protein import (29). Lack of SP1 function did not significantly alter the growth of the pex4-1 mutant (Fig. S4 G and H). However, on medium without sucrose, more pex4-1 sp1 seedlings than pex4-1 seedlings exhibited inhibited postgerminative growth (Fig. 4 A and B), and, in the seedlings that were established, the double mutants had shorter roots than pex4-1 seedlings (Fig. 4 C and D) and were more resistant than pex4-1 seedlings to 2,4-DB’s inhibition of hypocotyl and root elongation (Fig. 4E and Fig. S4I). Thus, sp1 appeared to enhance the physiological defects in pex4-1; this enhancement is the opposite of its effect on pex14 and pex13. Because the removal of PEX5 from the peroxisome is inefficient in pex4-1 plants, we reasoned that the lack of SP1 function in the pex4-1 sp1 double mutant may lead to the stabilization of peroxins such as PEX13 and PEX14, causing more translocation of PEX5 into the peroxisome and thus increasing the imbalance between the translocation of PEX5 into and out of the peroxisome. Based on these findings, we concluded that SP1’s repressive role in peroxisome protein import is restricted mainly to the earlier steps.
Fig. 4.
Loss of SP1 function enhances pex4 mutant phenotypes. (A) Ten-day-old seedlings grown on LS medium without sucrose. (B) Quantitative analysis of postgerminative growth of the seedlings in A. Error bars indicate SD; n = 3 (>100 seedlings for each biological replicate). **P < 0.01; *P < 0.05 from pex4-1. (C) Ten-day-old seedlings grown on MS medium without or supplemented with 1% sucrose. (D) Average root length of 10-d-old light-grown seedlings. Data are shown as mean ± SD; n = 3 (>30 seedlings for each biological replicate). *P < 0.05 from pex4-1. (E) 2,4-DB response assays. Seedlings were grown on MS medium supplemented with 0.5% sucrose and various concentrations of 2,4-DB. Shown is the average hypocotyl length of 7-d-old dark-grown seedlings normalized to that of seedlings grown on 0.5% sucrose medium with no 2,4-DB. Data are shown as mean ± SEM; n = 3 (>30 seedlings for each biological replicate). *P < 0.05 from pex4-1.
SP1’s Function in Regulating Peroxisome Biogenesis Depends on Its RING Domain.
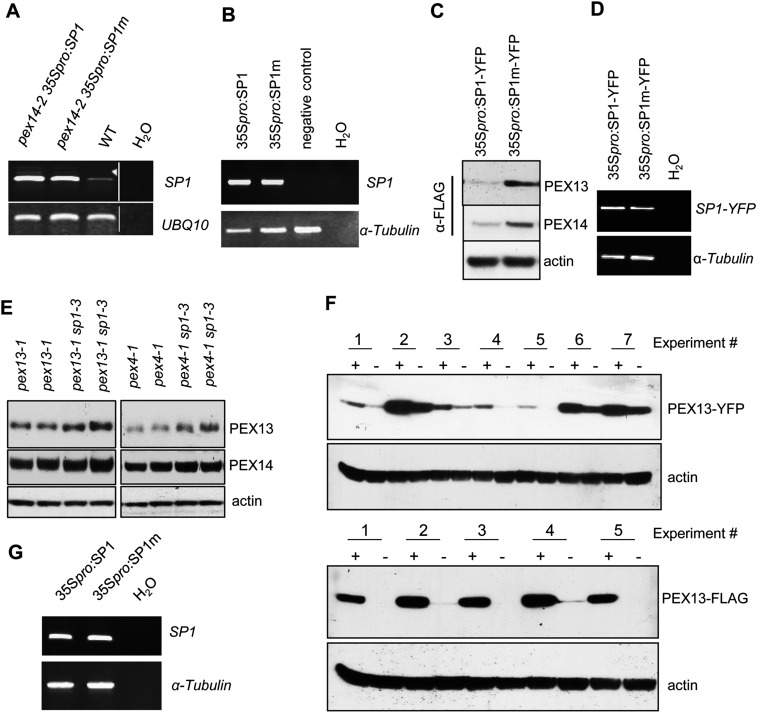
SP1 was shown to possess E3 ubiquitin ligase activities both in vivo and in vitro (40). The RING domain, which confers ubiquitin ligase activity for this E3, contains well-conserved Cys and His residues (Fig. 5A) (39, 40). To determine the importance of the RING domain for SP1’s function, we made five amino acid substitutions (Cys → Ser and His → Tyr) in the RING domain to disrupt SP1’s E3 activity (Fig. 5A). The mutant SP1m and wild-type SP1 proteins, both driven by the 35S constitutive promoter, were stably overexpressed in the pex14-2 mutant background. Transgenic lines that showed high expression of SP1m or SP1 were selected for further analysis (Fig. S5A). Overexpression of SP1m, but not of SP1, significantly rescued the growth defect (Fig. 5B), sucrose dependence (Fig. 5 C and D), and 2,4-DB resistance (Fig. 5E) in pex14-2 plants, similar to observations in the pex14-2 sp1 double mutant (Fig. 3). We also noticed that pex14-2 35Spro:SP1 plants were generally slightly smaller (Fig. 5 C–E) and were even more sucrose dependent (Fig. 5 C and D) than pex14-2 plants, as is consistent with our earlier conclusion that SP1 plays a repressive role in peroxisome function. Thus, overexpression of SP1m can cause a dominant negative effect on SP1, and the RING domain is indeed required for SP1’s function in regulating peroxisome biogenesis.
Fig. 5.
SP1’s function depends on its RING domain. (A) Sequence of SP1’s RING domain. The conserved active Cys and His residues are in red. The underlined residues were replaced in SP1m by the residues shown below in green. Amino acid numbers are indicated. (B) Four-week-old plants that had grown for 1 wk on LS medium and 3 wk on soil. (C and D) Sucrose-dependence assays. Seedlings were grown on MS medium supplemented with different concentrations of sucrose. Average root lengths of 10-d-old light-grown seedlings (C) and average hypocotyl length of 10-d-old dark-grown seedlings (D) are shown. Data are shown as mean ± SD; n = 3 (>30 seedlings for each biological replicate). ***P < 0.001; **P < 0.01; *P < 0.05 from pex14-2. (E) 2,4-DB response assays. Seedlings were grown on MS medium supplemented with 0.5% sucrose with or without 1.6 μM 2,4-DB. The average root length of 7-d-old light-grown seedlings is shown. Data are shown as mean ± SD; n = 3 (>30 seedlings for each biological replicate).
Fig. S5.
Expression of SP1 and SP1m proteins in tobacco. (A) RT-PCR analysis of RNA from 4-wk-old Arabidopsis leaves. Ubiquitin 10 was used as a loading control. White lines separate samples from separate gels. (B) RT-PCR analysis of RNA from tobacco leaf tissue expressing 35Spro:SP1 or 35Spro:SP1m. The negative control is RNA from tissue expressing the empty vector. The α-tubulin gene was used as a loading control. The image shown is representative of RT-PCR performed in the experiments for different PEX proteins (see Fig. 6A). (C) Immunoblot analysis of protein extracts from tobacco leaves co-overexpressing SP1-YFP or SP1m-YFP and PEX13-FLAG or PEX14-FLAG. FLAG-tagged proteins and actin were detected by anti-FLAG and anti-actin antibodies, respectively. (D) RT-PCR analysis of RNA from tobacco leaf tissue expressing either 35Spro:SP1-YFP or 35Spro:SP1m-YFP. The α-tubulin gene was used as a loading control. This image is representative of RT-PCR performed in the experiments for different PEX proteins (C). (E) Immunoblot analysis of total protein from 8-d-old Arabidopsis seedlings. Endogenous PEX13 and PEX14 protein levels were detected by PEX13 and PEX14 antibodies, respectively; α-actin was used as the loading control. (F) Immunoblot analysis of protein extracts from tobacco leaves co-overexpressing SP1 or SP1m and PEX13 tagged with YFP or FLAG. YFP-tagged protein and FLAG-tagged protein were detected by anti-GFP and anti-FLAG antibodies, respectively. +, SP1m overexpression; −, SP1 overexpression. In each experiment, the SP1- and SP1m-containing samples were from two sections of the same leaf. α-Actin was used as the loading control. (G) RT-PCR analysis of RNA from tobacco leaf tissue expressing 35Spro:SP1 or 35Spro:SP1m. The α-tubulin gene was used as a loading control. This image is representative of RT-PCR performed in the experiments for different PEX proteins (see Fig. 6E).
SP1 Targets PEX13 and Possibly Several Other PEX Proteins for Degradation.
The findings in co-IP assays that SP1 interacts with PEX13, PEX14, and PEX2 and that sp1 suppresses the mutant phenotypes of pex14 and pex13 led us to propose that these PEX proteins are the potential substrates for SP1. To test this hypothesis, we first used the tobacco (Nicotiana tabacum) transient protein expression system to examine whether SP1 has the ability to affect the stability of the interacting PEX proteins. To this end, various YFP-tagged PEX proteins and wild-type SP1 or the dominant negative SP1m protein were coexpressed (Fig. 6A and Fig. S5B). When coexpressed with wild-type SP1, not only PEX13, PEX14, and PEX2 but also PEX10 and PEX12 were destabilized (Fig. 6 A and B). In a similar experiment, YFP-tagged SP1 also promoted the destabilization of PEX13 and PEX14 (Fig. S5 C and D). These results were consistent with our co-IP data on PEX13, PEX14, and PEX2 (PEX10 and PEX12 could not be tested in co-IP because of protein instability), suggesting that PEX13, PEX14, and the RING peroxins are possible targets of SP1.
Fig. 6.
SP1 promotes the destabilization of several peroxins, and this function depends on the RING domain. (A) Immunoblot analysis of protein extracts from tobacco leaves co-overexpressing SP1 or SP1m and a YFP-tagged candidate substrate protein. YFP-tagged proteins and actin were detected by anti-GFP and anti-actin antibodies, respectively. The actin blot is a representative loading control (for the PEX13 blot). (B) Quantification of the levels of the YFP-tagged proteins shown in A. YFP and actin protein bands were quantified using ImageJ. The amount of each candidate protein coexpressed with SP1 was normalized to the data obtained from the sample coexpressed with SP1m. Data are shown as mean ± SD; n = 3. ***P < 0.001; **P < 0.01; *P < 0.05 from actin. (C) Immunoblot analysis of total leaf protein from 3-wk-old Arabidopsis plants. Endogenous PEX13 and PEX14 proteins were detected by α-PEX13 and α-PEX14 antibodies, respectively, and α-actin was used as the loading control. (D) Immunoblot analysis of total protein from 9-d-old Arabidopsis seedlings. Lanes 3 and 4 contain samples from two independent lines overexpressing SP1. (E) Co-IP of YFP-tagged PEX13 or PEX14 with FLAG-tagged ubiquitin. Each YFP-tagged PEX protein, FLAG-UBQ10, and SP1 (or SP1m) were co-overexpressed in tobacco leaves.
We reasoned that if SP1 promotes the destabilization of its targets, we would see an increased level of the target proteins in sp1 mutant background and a decrease in the target protein level in SP1 overexpressors. To test this prediction, we performed immunoblot analysis of proteins from various genetic backgrounds using the available PEX13 and PEX14 antibodies. As predicted, the endogenous PEX13 protein level was negatively correlated with the level of SP1 expression (Fig. 6 C and D and Fig. S5E). However, the endogenous PEX14 protein level was not very sensitive to changes in SP1 dosage; we observed only mild decreases in the level of PEX14 protein in lines overexpressing SP1 (Fig. 6D). In addition, we found that PEX13-FLAG was generally more susceptible to SP1 overexpression than PEX13-YFP in tobacco leaves (Fig. S5F), possibly because the FLAG tag is much smaller than the YFP tag, making PEX13-FLAG more efficient or more accurate in peroxisome targeting than PEX13-YFP. In summary, SP1 can promote the degradation of PEX13 in tobacco and Arabidopsis, and it also can promote the degradation of PEX14 and the RING peroxins, at least in the tobacco transient expression system. Although we did not observe more accumulation of endogenous PEX14 in the SP1 loss-of-function background, PEX14 still could be a substrate of SP1. It is possible that in the SP1 loss-of-function mutant background, the PEX14 level may be subjected to feedback regulatory mechanisms, such as increased activity of proteins functionally redundant with SP1, increased degradation of overly accumulated PEX14, and/or decreased PEX14 transcription. Also, our immunoblot method may not be sensitive enough to detect subtle changes in the PEX14 protein level in the plant. However, in the tobacco transient protein expression system, the capacity of such feedback mechanisms may be exceeded by the overexpression of SP1m (or SP1) and substrate proteins, thus demonstrating a much stronger SP1-mediated destabilization of the substrates.
To test whether SP1 promotes the association of ubiquitin with its potential substrate proteins, we performed in vivo ubiquitination assays. Because PEX2, PEX10, and PEX12 are ubiquitin E3 ligases, which often auto-ubiquitinate themselves, we tested only PEX13 and PEX14. FLAG-tagged ubiquitin, YFP-tagged PEX13 (or PEX14), and SP1 (or SP1m) were coexpressed in tobacco leaves, and the proteins pulled down by the α-FLAG antibody were analyzed by immunoblot analysis. In total protein input, PEX13 and PEX14 were much less stable when they were coexpressed with wild-type SP1 than when they were coexpressed with SP1m (Fig. 6E, Input and Fig. S5G). However, PEX13 and PEX14 apparently were more concentrated in the pull down when they were coexpressed with SP1 rather than with SP1m (Fig. 6E, IP). Hence, SP1 appears to be able to increase the association of ubiquitin with PEX13 and PEX14. More importantly, modified forms of PEX13, which likely represent ubiquitinated PEX13 because FLAG-UBQ10 was the bait for the co-IP, were strongly intensified in the presence of overexpressed SP1 (Fig. 6E). Therefore, our data show that SP1 is able to promote the ubiquitination of PEX13, at least when both proteins are overexpressed in tobacco. Given that SP1 interacts with PEX13 in co-IP, sp1 suppresses the mutant phenotype of pex13 and pex14, and SP1 promotes the destabilization of PEX13 in both the tobacco transient protein expression system and Arabidopsis pex14-2 plants, we conclude that PEX13 is most likely a direct target of SP1 for ubiquitination and destabilization in Arabidopsis.
Discussion
The molecular mechanisms that regulate the function of components of the peroxisome import machinery are largely unknown. In this study, we identified the Arabidopsis RING-type E3 ubiquitin ligase SP1 as a peroxisome membrane protein that regulates peroxisome biogenesis. A previous study on SP1 by Ling et al. (40) used transient expression of 35Spro:SP1-YFP in protoplast to show SP1’s association with chloroplasts. In our study, we used both 35Spro:SP1-YFP and SP1pro:SP1-YFP and the peroxisomal marker protein to demonstrate the association of SP1 with peroxisomes and chloroplast in both tobacco and Arabidopsis. Results from our co-IP, genetic, cell biological, pull-down, and in vivo ubiquitination analyses led us to a working model in which SP1 plays a negative regulatory role in peroxisome import and function by promoting the destabilization of components of the peroxisome import machinery. Specifically, SP1 interacts with PEX13 and prompts PEX13’s destabilization through the UPS and likely targets PEX14 and the RING peroxins PEX2, PEX10, and PEX12 as well (Fig. 7). Lack of functional SP1 may stabilize its target PEX proteins, such as PEX13, thus partially compensating for the loss of PEX14 in pex14-2 plants and the reduction of PEX13 in pex13-1 plants. Lack of SP1 function is expected to increase PEX5 translocation into peroxisomes and enhance pex4-1 defects that stem from the retention of PEX5 in the peroxisome membrane.
Fig. 7.
A working model for SP1’s role in peroxisome protein import in Arabidopsis. PEX5 and PEX7 are cytosolic receptors for peroxisome matrix proteins. The cargo-receptor complexes dock to the docking site formed by two membrane proteins, PEX13 and PEX14. The exact function of the E3 ubiquitin ligases PEX2, PEX10, and PEX12 in Arabidopsis is still unclear. The export of PEX5 to the cytosol is dependent on the ubiquitin-conjugating enzyme PEX4, which needs PEX22 as its membrane anchor, and the AAA ATPases PEX6 and PEX1, which are tethered to the membrane by APEM9. The balance between cargo translocation and receptor recycling is also important for the function of the machinery. SP1 interacts with PEX13, PEX14, and PEX2 in co-IP. SP1 promotes the ubiquitination and destabilization of PEX13 and promotes the destabilization of PEX14, PEX2, PEX10, and PEX12, at least in tobacco. Therefore, SP1 regulates peroxisome biogenesis by associating with and destabilizing PEX13 and possibly several other peroxins in peroxisome matrix protein import. Double-ended arrows indicate a physical interaction in co-IP. Dashed arrows indicate hypothesized SP1-mediated ubiquitination. PEX13 is shown in bold because it is the most likely target of SP1.
The sp1 single mutant was never identified from forward genetic screens because it does not display obvious growth- or organelle-specific phenotypes. This absence of obvious phenotypes in the single mutant is consistent with SP1 being a negative regulator instead of a major component of peroxisome and chloroplast protein import. SP1 was identified in a suppressor screen of ppi1 (or toc33) by its role in chloroplast protein import (40) and from our in silico study followed by in vivo targeting and functional analyses of its role in peroxisome protein import. The role of SP1 in protein import in both chloroplasts and peroxisomes could be revealed clearly only in the background of an import mutant, such as toc33 and pex14. Consistent with this result and with our findings of the partial suppression of pex14 in the pex14 sp1 double mutant, Ling et al. (40) showed direct evidence of the increased chloroplast protein import caused by sp1 mutation only in toc33 mutants but not in the wild-type background.
Unlike PEX13, no modified PEX14 bands representing ubiquitinated PEX14 were detected in our in vivo ubiquitination assay (Fig. 6E). This result is similar to that reported in a similar in vivo ubiquitination experiment using TOC75, in which the authors speculated that the lack of detectable ubiquitinated TOC75 suggests that TOC75 is less ubiquitinated than TOC33 and TOC159 or is more quickly degraded through UPS when ubiquitinated (40). This scenario may apply to PEX14 as well. However, if PEX14 is indeed much less ubiquitinated by SP1, SP1’s promotion of PEX14 degradation in the tobacco protein transient expression system may be caused indirectly by the degradation of PEX13. Because PEX13 and PEX14 form the receptor docking complex, they may stabilize each other, a notion that was supported by the reduction of the level of PEX13 protein in pex14-2 mutants (17). Therefore, even if PEX14 is not or is less ubiquitinated by SP1, PEX13 ubiquitination mediated by SP1 may be sufficient to facilitate the turnover of both PEX13 and PEX14, because the destabilization of one protein can cause the destabilization of the other component of the same protein complex. Last, the coprecipitation of unmodified TOC159, TOC75, and TOC33 with FLAG-ubiquitin in in vivo ubiquitination assays (40) is similar to the coprecipitation of unmodified PEX13 and PEX14 with FLAG-ubiquitin in our study. We thus speculate that ubiquitin and substrate protein may interact physically even before the ubiquitination reaction occurs or that unmodified PEX14 and PEX13 coprecipitate with modified PEX13 because these two proteins are in the same complex. Nevertheless, whether SP1 can mediate the ubiquitination of PEX14 and the RING peroxins remains to be clarified.
In plants, peroxisomes and chloroplasts function closely in a few metabolic pathways, including photorespiration (3, 50). As a result, the level of enzymes housed in these two organelles may need to be coordinately adjusted during developmental and environmental changes. For example, after germination, or when dark-grown seedlings are exposed to light, chloroplasts develop and actively import photosynthetic proteins, and seed peroxisomes turn into photorespiratory peroxisomes to metabolize phosphoglycolate, a product of the oxygenase activity of the photosynthetic enzyme Rubisco (7, 12). Our study demonstrated that SP1 plays a dual role in protein import for both chloroplasts and peroxisomes. The sp1 toc33 double-mutant plants are greener than the toc33 single-mutant plants (40), but the chlorophyll content in pex14-2 sp1 plants is not significantly different from that of pex14-2 plants (Fig. 3B), possibly because the pale leaf color in pex14-2 plants is an indirect consequence of disrupted PEX14 function and thus is not highly responsive to a partial rescue of the peroxisome defects in these plants. E3 often has multiple substrates (37, 51). SP1 was found to interact with and promote the degradation of multiple TOC proteins required for chloroplast protein import (40). Likewise, SP1 may target multiple peroxins at the peroxisome membrane.
The previous study of SP1’s role in chloroplasts showed that sp1 mutants are less efficient in meeting the developmental requirement for chloroplast proteome change, such as when etiolated seedlings are exposed to light or when leaf senescence is induced (40). Another study reported that SP1 (DAL1) and SPL1 (DAL2) are involved in the regulation of reactive oxygen species (ROS) accumulation and programmed cell death (52). A more recent study showed that SP1 promotes stress tolerance by depleting TOC components, leading to diminished ROS produced in photosynthesis (53). We speculate that SP1’s function in coping with developmental changes and stress conditions also may be attributed to its regulatory role in peroxisome biogenesis. ROS are key signaling molecules in plant stress response (54), and peroxisomes possess numerous ROS-producing and -decomposing enzymes (3). Hence, developmental or environmental cues that induce changes in the chloroplast proteome may also trigger proteome adjustment of the peroxisome. This notion is particularly interesting, given that these two organelles also share a major component of the division machinery, the dynamin-like GTPase DRP5B (55). Thus, both the organelle division and the protein import processes of peroxisomes and chloroplasts share key factors that ultimately may contribute to their linked metabolism.
All three members of the Arabidopsis SP1 protein family share limited sequence similarities with the human MAPL (mitochondrial-anchored protein ligase) protein, a regulator of mitochondrial fission that targets the dynamin-related protein DRP1 (56). Neither SP1 nor MAPL has obviously similar sequences in yeast or nematode, suggesting that this family of E3 enzymes may be present only in higher eukaryotes. The plant SP1-like E3s seem to be more divergent and can be divided into two subclades, with SP1 and SPL1 in one subclade and SPL2 in the other (Fig. S6 and Dataset S1). Human MAPL is localized to both mitochondria and peroxisomes, but its function in peroxisomes is unclear (57). It is possible that this family of E3 enzymes targets to and regulates the biogenesis of multiple organelles across diverse species. Whether the animal MAPL-like proteins also govern organelle import and the roles of the Arabidopsis SPL1 and SPL2 proteins in organelle biogenesis or morphogenesis remain to be elucidated.
Fig. S6.
Phylogenetic analysis of SP1-related and human MAPL-related proteins. Protein sequences were subjected to phylogenetic analysis using Phylogeny.fr (www.phylogeny.fr/). Plant species include Arabidopsis thaliana, Zea mays, Oryza sativa, and N. tabacum. Nonplant species include Homo sapiens, Mus musculus, Xenopus laevis, and Gallus gallus. (Scale bar, 0.5 amino acid substitutions per site.) Branch support values (%) are shown in red.
Materials and Methods
Arabidopsis plants were grown at 22 °C with 70% humidity and a 14-h day of white light at 70–80 μmol⋅m−2⋅s−1. N. tabacum plants were grown at 24 °C with 70% humidity and a 14-h day of white light treatment at 50 μmol⋅m−2⋅s−1. The Arabidopsis Col-0 ecotype was the wild-type reference. The sources of Arabidopsis transgenic lines and detailed methods are described in SI Materials and Methods. Primers used in this study are listed in Table S1; vectors used in this study are listed in Table S2.
Table S1.
Primers used in this study
| Primer | Sequence | Purpose |
| SP1-Primer 1 | ggggacaagtttgtacaaaaaagcaggcttcATGATTCCTTGGGGTGGAGTTAC | Cloning full-length SP1 and RT-PCR |
| SP1-Primer 4 | ggggaccactttgtacaagaaagctgggtcGTGACGATATGTCTTAACCGCCAG | Cloning full-length SP1 and RT-PCR |
| SP1-Primer 2 | ATTACATGGACACGGCTTGTC | RT-PCR |
| SP1-Primer 3 | TTGTTGGTGAGGCTGTCAAG | RT-PCR |
| FLAG-MAPL1-R | CTTGTCGTCATCGTCTTTGTAGTCCGCCGCGTGACGATATGTCTTAACCGCCAG | Cloning full-length SP1 with FLAG |
| AttB2-stop-FLAG-R | ggggaccactttgtacaagaaagctgggtcTCACTTGTCGTCATCGTCTTTGTAG | To add a FLAG tag at the C terminus |
| SP1-RingMutant-F | GTTTGTCCCGAgtggtTatatgAgcAgcAgcACCGCATGCTC | To generate SP1m |
| SP1-RingMutant-R | GAGCATGCGGTgcTgcTgcTcatatAaccacTCGGGACAAAC | To generate SP1m |
| SPL1-Fw-AttB1 | ggggacaagtttgtacaaaaaagcaggcttcATGATACATTTGGCTGGATTTAC | Cloning full-length SPL1 |
| SPL1-Re-AttB2 | ggggaccactttgtacaagaaagctgggtcATGGCGGTAAATTTTCAAAAC | Cloning full-length SPL1 |
| SPL2-Fw-AttB1 | ggggacaagtttgtacaaaaaagcaggcttcATGTCCTCGCCGGAGCGTGCTC | Cloning full-length SPL2 |
| SPL2-Re-AttB2 | ggggaccactttgtacaagaaagctgggtcAGAGTAATATACACGCATAGATC | Cloning full-length SPL2 |
| UBQ10-1 | TCAATTCTCTCTACCGTGATCAAGATG | RT-PCR analysis of UBQ10 |
| UBQ10-2 | GGTGTCAGAACTCTCCACCTCAAGAG | RT-PCR analysis of UBQ10 |
| α-Tubulin-F | AGTTGGAGGAGGTGATGATG | RT-PCR analysis of α-Tubulin |
| α-Tubulin-R | TATGTGGGTCGCTCAATGTC | RT-PCR analysis of α-Tubulin |
| SALK_063571-LP | AGGATCATTCAATTCCAACCC | Genotyping of sp1-2 |
| SALK_063571-RP | ATTACATGGACACGGCTTGTC | Genotyping of sp1-2 |
| SALK_002099-LP | TATTCGCTGAATCGAGCAAAC | Genotyping of sp1-3 |
| SALK_002099-RP | GCTGCCATGTATAACAGGCTG | Genotyping of sp1-3 |
| RP-1 | ACGGCCAGTGCCAAGCTTGCATGCCTGCAGGGTTGTCCACAAAACAACTAAGTTGAAACC | Cloning SP1pro:SP1-YFP |
| RP-2 | CAGACTCTGAAGTCACTCTTACTTGTTTGC | Cloning SP1pro:SP1-YFP |
| RP-3 | TAAGAGTGACTTCAGAGTCTGATGATTCCTTGGGGTGGAGTTACTTG | Cloning SP1pro:SP1-YFP |
| RP-4 | GTGACGATATGTCTTAACCGCCAGATC | Cloning SP1pro:SP1-YFP |
| RP-5 | GCGGTTAAGACATATCGTCACGGTTCTGGTTCT GGTTCTATGGTGAGCAAGGGCGAGG | Cloning SP1pro:SP1-YFP |
| RP-6 | CAAATGTTTGAACGATCGGGGAAATTCGAGCTCTTACTTGTACAGCTCGTCCATGCC | Cloning SP1pro:SP1-YFP |
For primers SP1-RingMutant-F and SP1-RingMutant-R, lowercase letters indicate region of sequences containing designed mutations. For other primers, lowercases indicate AttB sequences for gateway reaction.
Table S2.
Vectors used in this study
| Vector | Purpose | Genes |
| pDonor 207 (Invitrogen) | Donor vector | All donor plasmids |
| pEarleyGate 100 | Gene overexpression | SP1; SP1m; SP1-FLAG |
| pEarleyGate 101 | Fusing YFP to the N-terminal end of the gene | TOC33; PEX4; PEX5; PEX7; PEX22; APEM9 |
| pEarleyGate 104 | Fusing YFP to the C-terminal end of the gene | SP1; SP1m; SPL1; SPL2; PEX13; PEX14; PEX2; PEX10; PEX12 |
| pEarleyGate 202 | Fusing FLAG to the N-terminal end of the gene | FLAG-UBQ10 |
| pCAMBIA1300-20 | Cloning SP1pro:SP1-YFP | SP1pro:SP1-YFP |
SI Materials and Methods
Plant Materials.
Arabidopsis plants were grown at 22 °C with 70% humidity and white light for 14 h/d at 70–80 μmol⋅m−2⋅s−1. N. tabacum plants were grown at 24 °C with 70% humidity and white light for 14 h/d at 50 μmol⋅m−2⋅s−1. The Arabidopsis Col-0 ecotype was the wild-type reference. The T-DNA insertion mutants sp1-2 (SALK_063571), sp1-3 (SALK_002099), pex14-2 (SALK_007441), pex13-1 (SALK_006744), and lon2-2 (SALK_043857) were provided by the Arabidopsis Biological Resource Center (58). Homozygous mutants were identified by PCR of genomic DNA followed by RT-PCR analysis as described previously (55). Peroxisome marker lines CFP-PTS1 and YFP-PTS1 were generated in our previous study (25).
Physiological Assays.
To analyze postgerminative growth, seeds were grown on LS medium solidified with 0.8% (wt/vol) agar after 4 d of stratification at 4 °C. The plates were kept in a growth chamber with continuous light. After 2 wk, the number of plants that continued or did not continue to grow was quantified. For each experiment, >100 seeds per genotype were quantified, and three biological replicates were included.
For sucrose-dependence assays, seeds were grown on MS medium without or supplemented with 1% (wt/vol) sucrose and were stratified at 4 °C for 4 d. For 2,4-DB response assays, seeds were grown on plates containing 0.5% (wt/vol) sucrose and various concentrations of 2,4-DB (Sigma-Aldrich) as indicated in the figures. The plates then were stored at 4 °C for 4 d. Hypocotyl and root lengths were measured using ImageJ (https://imagej.nih.gov/ij/. For dark-grown seedlings, the first 24 h of growth were in the light. For each experiment, >30 seeds per genotype were quantified, and three biological replicates were included.
Plant Transformation.
All organelle markers and other constructs used in this study were transformed into Arabidopsis plants via the simplified floral dipping method (www.plantpath.wisc.edu/fac/afb/protocol.html), using the Agrobacterium tumefaciens strain GV3101 (pMP90). To screen for transformants with kanamycin, T1 seeds were plated on 1/2 LS medium containing 50 µg/mL kanamycin. To obtain Basta-resistant transformants, T1 seeds were sewn on soil and, after 7 d of growth, were sprayed at least two times with 0.1% (vol/vol) Basta (Finale; Farnam Companies) that contained 0.025% (vol/vol) Silwet L-77.
Gene Cloning, Plasmid Construction, and RT-PCR.
With the exception of the SP1pro:SP1-YFP construct, the coding sequences of genes were amplified with Gateway compatible primers by Phusion polymerase (New England Biolabs). The cDNA made from total RNA extracts of Col-0 plants was used as the template. The PCR products were cloned into pDonor207 vectors and destination vectors via the standard Gateway cloning system (Invitrogen, www.thermofisher.com/). Various pEarley vectors were used as destination vectors (59).
SP1pro:SP1-YFP was generated using Gibson assembly reactions (60). A 2-kb genomic fragment (SP1’s native promoter) upstream of SP1’s start codon was amplified with primer set RP-1/RP-2, using genomic DNA extracted from wild-type Col-0 as template (Promega). The SP1 fragment that covers the genomic sequence of the coding region without the stop codon was amplified from the genomic DNA sequence with primer set RP-3/RP-4, and the EYFP fragment was amplified from pEarley 101 with the primer set RP-5/RP-6. Through Gibson assembly reactions, all fragments were cloned into pCAMBIA1300-20 (61) that had been digested with PstI/SacI. The final construct was verified by sequencing.
To generate mutant forms of SP1, overlapping PCR was performed (https://gfp.dpb.carnegiescience.edu/protocol/index5.html) with primers containing the mutation sites.
To analyze gene-expression level in plants, RT-PCR was performed as previously described (62). Generally, 0.5 µg of total RNA was used to make cDNA using high-capacity RNA-to-cDNA master mix (Applied Biosystems), followed by PCR amplification using GoTaq Green Master Mix (Promega) and gene-specific primers.
Information about the primers and vectors used in this study is given in Tables S1 and S2, respectively.
Protein Preparation and Immunoblot Analysis.
To extract crude plant protein from Arabidopsis and tobacco plants, 30 mg of fresh tissue was ground by plastic pestles using liquid nitrogen and then was homogenized with 300 µL extraction buffer [60 mM Tris⋅HCl (pH 8.8), 2% SDS, 2.5% glycerol, 0.13 mM EDTA (pH 8.0), and protease inhibitor mixture complete (Roche)]. The tissue lysates were vortexed for 30 s, heated at 75 °C for 10 min, and centrifuged at 13,000 × g twice for 5 min at room temperature. The supernatants were transferred to new tubes, mixed with lithium dodecyl sulfate (LDS) sample buffer (15 µL supernatant:5 µL LDS) before being loaded onto SDS/PAGE gel. To prepare peroxisome proteins for SDS/PAGE analysis, 15 µL of purified peroxisomes was mixed with 5 µL of LDS sample buffer and then heated at 75 °C for 10 min before being loaded onto SDS/PAGE. After separation on the gel, proteins were transferred to a PVDF membrane, which then was blocked with 5% milk in 1× TBST [Tris-buffered saline plus Tween 20 (50 mM Tris-base, 150 mM NaCl, 0.05% Tween 20, pH 8.0)] for 1 h at room temperature, followed by incubation for 1 h at room temperature with primary antibody prepared in the blocking buffer as follows: 1:20,000 for α-GFP (Abcam), 1:1,000 for α-PEX11D (44), 1:5,000 for α-Actin (63), 1:5,000 for a-VDAC (63), 1:3,000 for a-FtsZ1 (64) (provided by Kathy Osteryoung, Michigan State University, East Lansing, MI), 1:100 for α-PEX13 (18), and 1:1,000 for α-FLAG (Sigma). Then the membrane was washed three times (5 min each time) with 1× TBST, incubated with the secondary antibody (i.e., 1:20,000 goat anti-rabbit IgG or 1:20,000 goat anti-mouse IgG; Millipore) at room temperature, washed four times (10 min each time) with 1× TBST, and then visualized using the SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology).
Peroxisome Isolation and Determination of Protein Membrane Association.
Rosette leaves from 4-wk-old transgenic Arabidopsis plants expressing SP1-YFP and the peroxisomal marker CFP-PTS1 were used for peroxisome purification using a previously published protocol (65). Peroxisome membrane association was tested using a previously described method (35, 44).
Fluorescence Microscopy.
A Zeiss Axio Imager Upright Microscope and an Olympus Fluoview FV1000 confocal laser-scanning microscope were used for fluorescence microscopy. For confocal microscopy, CFP, YFP, and chlorophyll autofluorescence were excited with 458-, 515-, and 515-nm lasers, respectively, and were detected at 465–490 nm, 505–555 nm, and 655–755 nm, respectively.
Co-IP.
Co-IP was performed as previously described (62). In general, tobacco leaves (∼1 g fresh weight) transiently expressing the fusion proteins were homogenized in 4 mL radioimmunoprecipitation assay (RIPA) buffer (Thermo) with complete protease inhibitor mixture (Roche) and were lysed on a rotator for 1 h at 4 °C. After the lysed tissue was centrifuged at 13,000 × g for 10 min, the supernatant (∼3 mL) was incubated with 30 µL agarose-conjugated anti-FLAG (Sigma) on a rotator at 4 °C for 1 h to pull down the fusion proteins. The agarose beads were spun down at 3,000 × g for 15 s and were washed three times with RIPA buffer. Interacting proteins were eluted by adding 40 µL 2× NuPAGE LDS sample buffer (Invitrogen) and heating at 75 °C for 10 min. Then 15 µL of the total protein extract and 15 µL of the immunoprecipitated sample were subjected to immunoblot analysis as described above. Note: Because SP1 can destabilize PEX13, PEX14, and PEX2, as shown in Fig. 6A, the concentration of Agro cells infiltrated into tobacco leaves for PEX13, PEX14, and PEX2 was adjusted to produce similar amounts of prey proteins in the SP1-FLAG sample and the empty vector sample.
Tobacco Infiltration.
To perform transient expression in tobacco plants, leaves of 6- to 8-wk-old N. tabacum plants were infiltrated with resuspended A. tumefaciens cells that contained specific plasmids. Agrobacterium cells were grown for 24 h at 28 °C and with shaking at 200 rpm in an incubator shaker. Then the cells were spun down and resuspended in water to an A600 of 0.05. Leaf areas subjected to infiltrations were cut off and used for imaging or protein extraction. Note: For the tobacco infiltration experiments in Fig. 6 A and E, the same amount of Agro cells for the SP1-containing and SP1m-containing samples were infiltrated into different sections between veins on the same leaf to reduce differences in gene expression caused by tissue variance.
Phylogenetic Analysis.
The phylogenetic tree was created using the online tools in Phylogeny.fr (66, 67). Protein sequences were aligned using the MUSCLE 3.7 program (68) with the maximum number of iterations at 16, followed by manual adjustment. The alignment curation was done with the Gblocks 0.91b program (69) (see Dataset S1). The phylogeny analysis was done using the PhyML v3.0 program (70). The statistical test for branch support was done using the Approximate Likelihood-Ratio Test (aLRT) with the setting of SH-Like (71). In aLRT, the model of amino acids substitution was WAG, the number of taxa was 13, the log-likelihood was −1,256.66891, the discrete gamma model was yes, the number of categories was four, the gamma shape parameter was 2.396, and the proportion of invariant was 0.028. Tree rendering was done using the TreeDyn program (72). Midpoint rooting was used; the scale bar represents 1.0 amino acid substitutions per site.
Statistical Analysis.
Quantitative analysis results are presented as means ± SD (or SEM) from repeated experiments as indicated in the figure legends. A pairwise Student’s t test was used to analyze statistical significance.
Accession Numbers.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis SP1 (At1g63900), SPL1 (At1g59560), and SPL2 (At1g54150).
Supplementary Material
Acknowledgments
We thank Cheng Chen for technical assistance; the Arabidopsis Biological Resource Center for providing the T-DNA insertion mutants sp1-2, sp1-3, pex14-2, pex13-1, and lon2-2; Dr. Bonnie Bartel for pex4-1 seeds and the PEX13 and PEX14 antibodies; Dr. Steven Smith for the thiolase antibody; Dr. Kathy Osteryoung for the FtsZ1 antibody; and Dr. Danny Schnell for comments on the manuscript. This work was supported by National Science Foundation Grant MCB 1330441 (to J.H.) for personnel and supplies; and Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy Grant DE-FG02-91ER20021 (to J.H.) for infrastructure.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613530113/-/DCSupplemental.
References
- 1.Schrader M, Fahimi HD. The peroxisome: Still a mysterious organelle. Histochem Cell Biol. 2008;129(4):421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michels PAM, et al. Peroxisomes, glyoxysomes and glycosomes (review) Mol Membr Biol. 2005;22(1-2):133–145. doi: 10.1080/09687860400024186. [DOI] [PubMed] [Google Scholar]
- 3.Hu J, et al. Plant peroxisomes: Biogenesis and function. Plant Cell. 2012;24(6):2279–2303. doi: 10.1105/tpc.112.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timm S, et al. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell. 2008;20(10):2848–2859. doi: 10.1105/tpc.108.062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham IA. Seed storage oil mobilization. Annu Rev Plant Biol. 2008;59(1):115–142. doi: 10.1146/annurev.arplant.59.032607.092938. [DOI] [PubMed] [Google Scholar]
- 6.Acosta IF, Farmer EE. Jasmonates. Arabidopsis Book. 2010;8:e0129. doi: 10.1199/tab.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Titus DE, Becker WM. Investigation of the glyoxysome-peroxisome transition in germinating cucumber cotyledons using double-label immunoelectron microscopy. J Cell Biol. 1985;101(4):1288–1299. doi: 10.1083/jcb.101.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura M, Yamaguchi J, Mori H, Akazawa T, Yokota S. Immunocytochemical analysis shows that glyoxysomes are directly transformed to leaf peroxisomes during greening of pumpkin cotyledons. Plant Physiol. 1986;81(1):313–316. doi: 10.1104/pp.81.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sautter C. Microbody transition in greening watermelon cotyledons Double immunocytochemical labeling of isocitrate lyase and hydroxypyruvate reductase. Planta. 1986;167(4):491–503. doi: 10.1007/BF00391225. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura M, Hayashi M, Kato A, Yamaguchi K, Mano S. Functional transformation of microbodies in higher plant cells. Cell Struct Funct. 1996;21(5):387–393. doi: 10.1247/csf.21.387. [DOI] [PubMed] [Google Scholar]
- 11.Trelease RN, Becker WM, Gruber PJ, Newcomb EH. Microbodies (glyoxysomes and peroxisomes) in cucumber cotyledons: Correlative biochemical and ultrastructural study in light- and dark-grown seedlings. Plant Physiol. 1971;48(4):461–475. doi: 10.1104/pp.48.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hondred D, Wadle DM, Titus DE, Becker WM. Light-stimulated accumulation of the peroxisomal enzymes hydroxypyruvate reductase and serine:glyoxylate aminotransferase and their translatable mRNAs in cotyledons of cucumber seedlings. Plant Mol Biol. 1987;9(3):259–275. doi: 10.1007/BF00166462. [DOI] [PubMed] [Google Scholar]
- 13.Oikawa K, et al. Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat Plants. 2015;1(4):15035. doi: 10.1038/nplants.2015.35. [DOI] [PubMed] [Google Scholar]
- 14.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14(12):803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanyon-Hogg T, Warriner SL, Baker A. Getting a camel through the eye of a needle: The import of folded proteins by peroxisomes. Biol Cell. 2010;102(4):245–263. doi: 10.1042/BC20090159. [DOI] [PubMed] [Google Scholar]
- 16.Nito K, Hayashi M, Nishimura M. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43(4):355–366. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]
- 17.Monroe-Augustus M, et al. Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol Biol. 2011;77(1-2):1–15. doi: 10.1007/s11103-011-9782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodward AW, et al. A viable Arabidopsis pex13 missense allele confers severe peroxisomal defects and decreases PEX5 association with peroxisomes. Plant Mol Biol. 2014;86(1-2):201–214. doi: 10.1007/s11103-014-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105(2):187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 20.Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: Evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135(6 II):1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167(4):599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, et al. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science. 2002;297(5580):405–409. doi: 10.1126/science.1073633. [DOI] [PubMed] [Google Scholar]
- 23.Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2003;100(16):9626–9631. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sparkes IA, et al. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 2003;133(4):1809–1819. doi: 10.1104/pp.103.031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan J, et al. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 2005;139(1):231–239. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geisbrecht BV, Collins CS, Reuber BE, Gould SJ. Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci USA. 1998;95(15):8630–8635. doi: 10.1073/pnas.95.15.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiel JAKW, et al. Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast. 1999;15(11):1059–1078. doi: 10.1002/(SICI)1097-0061(199908)15:11<1059::AID-YEA434>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 28.Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA. 2004;101(6):1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17(12):3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platta HW, et al. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29(20):5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur N, Zhao Q, Xie Q, Hu J. Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins(F) J Integr Plant Biol. 2013;55(1):108–120. doi: 10.1111/jipb.12014. [DOI] [PubMed] [Google Scholar]
- 32.Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156(3):1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48(6):763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- 34.Ratzel SE, Lingard MJ, Woodward AW, Bartel B. Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic. 2011;12(1):121–134. doi: 10.1111/j.1600-0854.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan R, Kaur N, Hu J. The Arabidopsis mitochondrial membrane-bound ubiquitin protease UBP27 contributes to mitochondrial morphogenesis. Plant J. 2014;78(6):1047–1059. doi: 10.1111/tpj.12532. [DOI] [PubMed] [Google Scholar]
- 36.Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012;196(1):13–28. doi: 10.1111/j.1469-8137.2012.04266.x. [DOI] [PubMed] [Google Scholar]
- 37.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 38.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 39.Stone SL, et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137(1):13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling Q, Huang W, Baldwin A, Jarvis P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science. 2012;338(6107):655–659. doi: 10.1126/science.1225053. [DOI] [PubMed] [Google Scholar]
- 41.Aung K, Hu J. The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell. 2011;23(12):4446–4461. doi: 10.1105/tpc.111.090142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J-F, Park E, von Arnim AG, Nebenführ A. The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods. 2009;5:6. doi: 10.1186/1746-4811-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18(3):665–675. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orth T, et al. The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell. 2007;19(1):333–350. doi: 10.1105/tpc.106.045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desai M, Hu J. Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 2008;146(3):1117–1127. doi: 10.1104/pp.107.113555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helm M, et al. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA. 2007;104(27):11501–11506. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuhmann H, Huesgen PF, Gietl C, Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 2008;148(4):1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan BR, Zolman BK. pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 2010;154(4):1602–1615. doi: 10.1104/pp.110.162479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lingard MJ, Bartel B. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 2009;151(3):1354–1365. doi: 10.1104/pp.109.142505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peterhansel C, et al. 2010. Photorespiration. The Arabidopsis Book (The American Society of Plant Biologists, Rockville, MD), p e0130.
- 51.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 52.Basnayake BMVS, et al. Arabidopsis DAL1 and DAL2, two RING finger proteins homologous to Drosophila DIAP1, are involved in regulation of programmed cell death. Plant Cell Rep. 2011;30(1):37–48. doi: 10.1007/s00299-010-0941-6. [DOI] [PubMed] [Google Scholar]
- 53.Ling Q, Jarvis P. Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr Biol. 2015;25(19):2527–2534. doi: 10.1016/j.cub.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gadjev I, Stone JM, Gechev TS. Programmed cell death in plants: New insights into redox regulation and the role of hydrogen peroxide. Int Rev Cell Mol Biol. 2008;270:87–144. doi: 10.1016/S1937-6448(08)01403-2. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Hu J. The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell. 2010;22(2):431–442. doi: 10.1105/tpc.109.071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10(7):748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neuspiel M, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18(2):102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 58.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 59.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 60.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 61.Zhang M, Schmitz AJ, Kadirjan-Kalbach DK, Terbush AD, Osteryoung KW. Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in arabidopsis through interaction with FtsZ2. Plant Cell. 2013;25(5):1787–1802. doi: 10.1105/tpc.113.111047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan R, Jones AD, Hu J. Cardiolipin-mediated mitochondrial dynamics and stress response in Arabidopsis. Plant Cell. 2014;26(1):391–409. doi: 10.1105/tpc.113.121095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reumann S, et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 2009;150(1):125–143. doi: 10.1104/pp.109.137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 2009;149(2):1196–1204. doi: 10.1104/pp.108.131508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reumann S, Singhal R. Isolation of leaf peroxisomes from Arabidopsis for organelle proteome analyses. Methods Mol Biol. 2014;1072:541–552. doi: 10.1007/978-1-62703-631-3_36. [DOI] [PubMed] [Google Scholar]
- 66.Dereeper A, Audic S, Claverie J-M, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 70.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 71.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 72.Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.