Significance
Some viruses, including lymphocytic choriomeningitis virus clone 13, shut down the ability of CD4 T lymphocytes to produce IL-2, a cytokine required for the survival and function of T lymphocytes. This shutdown contributes to exhaustion of CD4 and CD8 T lymphocytes and chronic viral infection of the host. The underlying mechanism responsible for the loss of cytokine production by CD4 T cells remains poorly understood. We demonstrate that the expression of a protein tyrosine phosphatase, PTPN22, contributes to chronic viral infection. PTPN22 increases the production of IFN-β following infection, resulting in increased expression of the cAMP response element modulator (CREM) in CD4 T lymphocytes. CREM prevents production of IL-2, thereby contributing to T-cell exhaustion and chronic viral infection.
Keywords: chronic infection, T-cell exhaustion, PTPN22, LCMV, CREM
Abstract
The protein encoded by the autoimmune-associated protein tyrosine phosphatase nonreceptor type 22 gene, PTPN22, has wide-ranging effects in immune cells including suppression of T-cell receptor signaling and promoting efficient production of type I interferons (IFN-I) by myeloid cells. Here we show that mice deficient in PTPN22 resist chronic viral infection with lymphocytic choriomeningitis virus clone 13 (LCMV cl13). The numbers and function of viral-specific CD4 T lymphocytes is greatly enhanced, whereas expression of the IFNβ-induced IL-2 repressor, cAMP-responsive element modulator (CREM) is reduced. Reduction of CREM expression in wild-type CD4 T lymphocytes prevents the loss of IL-2 production by CD4 T lymphocytes during infection with LCMV cl13. These findings implicate the IFNβ/CREM/IL-2 axis in regulating T-lymphocyte function during chronic viral infection.
Chronic viral infections including HIV, hepatitis B, and hepatitis C affect more than 500 million people worldwide. Viral persistence is associated with loss of effector functions by CD4 and CD8 T cells, generally referred to as “exhaustion” (1, 2). Exhausted T-cell responses have been reported in tumor settings as well as in parasitic and bacterial infection (3–7). The clone 13 strain of lymphocytic choriomeningitis virus (LCMV cl13) is an excellent tool for studying chronic infection because the exhausted T-cell phenotype is molecularly similar to that observed in serious human chronic infections and tumor immunity (1).
CD8 T cells are required for elimination of LCMV (8). Reduced cytokine production and proliferation and the up-regulated expression of inhibitory cell surface markers, such as PD-1 (programmed death 1), LAG-3 (lymphocyte activation gene 3), and TIM-3 (T-cell immunoglobulin and mucin domain-3), are characteristic of the exhausted state of CD8 cells that contributes to viral persistence following infection with LCMV cl13 (1, 9, 10). Strategies to invigorate exhausted CD8 T cells have proved successful in some cases, suggesting that immune modulation is a viable and efficacious therapy for these diseases (11, 12). CD4 T cells, in particular, are a critical aspect of antiviral immunity, and their impairment occurs during a number of chronic viral infections (13–16). Depletion of CD4 T cells before LCMV cl13 infection leads to increased viral persistence, whereas adoptive transfer of antigen-specific CD4 T cells promotes viral clearance (16–18). Loss of IL-2 production by CD4 cells is important in establishing exhaustion, because treatment with low-dose IL-2 leads to increased CD8 number, cytokine production, and reduced expression of the negative costimulatory molecule PD-1 on CD8 T cells during LCMV cl13 infection (19). These results suggest that functional CD4 cells have a critical role in CD8 function in this model.
Recent reports suggest that induction of T-cell exhaustion following infection with LCMV cl13 is related to excessive type I IFN (IFN-I) production immediately postinfection (20–23). Specifically, blocking IFNβ signaling before infection with LCMV cl13 resulted in enhanced viral clearance that was associated with increased CD4 T-cell function early postinfection (23). There is, as yet, no mechanism that explains the decrease in CD4 T-cell function associated with IFN-I signaling.
PTPN22 (protein tyrosine phosphatase nonreceptor type 22) encodes lymphoid tyrosine phosphatase (LYP) in humans and PEST (proline, glutamic acid, serine, threonine)-enriched protein phosphatase (PEP) in mice (collectively referred to as PTPN22). PTPN22 is expressed in all hematopoietic cells with highest expression in activated T cells. Genome-wide association studies demonstrated the association of a minor allele of PTPN22 (R620W) with a number of autoimmune diseases including type I diabetes, rheumatoid arthritis, and systemic lupus erythematosus, among others (24–26). PTPN22-deficient mice have been instrumental in uncovering the role of this gene in T-cell function (27–32). Aged mice develop splenomegaly and enlarged lymph nodes that exhibit the accumulation of effector/memory T cells, enlarged spontaneous germinal centers, and increased (non-autoreactive) serum IgG. Following T-cell receptor (TCR) activation there are increases in phosphorylation of ZAP-70 (70-kDa zeta-associated protein) and LCK (lymphocyte-specific protein tyrosine kinase), molecules that are direct substrates of PTPN22 in T cells, in the PTPN22−/− mice compared with WT mice (33).
A recent study has demonstrated a novel role for PTPN22 in myeloid cell production of IFN-I through interaction with TRAF3 (TNF receptor associated factor-3) in a non–phosphatase-dependent manner (34). The absence of PTPN22 or the expression of the R620W mutant human form of PTPN22 results in reduced IFN-I production by myeloid cells. We recently have confirmed these findings in plasmacytoid dendritic cells (pDCs) stimulated with TLR7 (Toll-like receptor 7) ligands (35). Based on the reported functions of PTPN22 in T cells and innate immune cells, we hypothesized that deficiency in PTPN22 would result in improved clearance of chronic viral infection because of enhanced T-cell responses and reduced IFN-I production. In this report we have used the LCMV cl13 chronic infection model to test this hypothesis.
Results
PTPN22−/− Mice Show Improved Clearance of LCMV cl13 Infection.
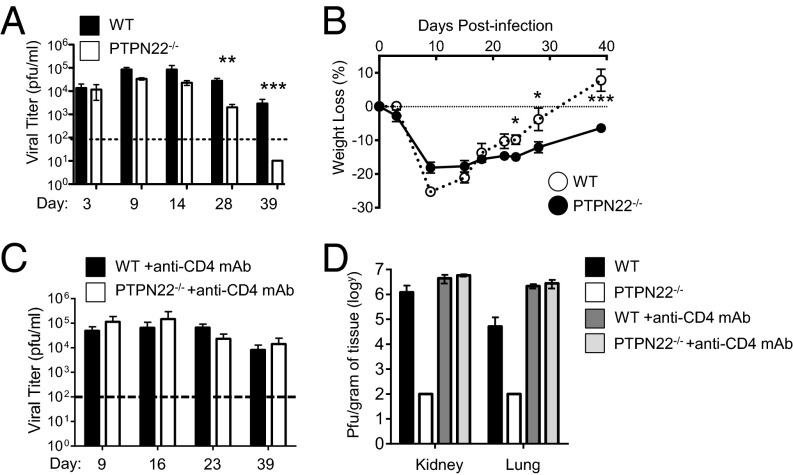
To investigate whether the loss of PTPN22 promotes antiviral activity in chronic infection, we infected PTPN22−/− and WT mice with 106 pfu of LCMV cl13 i.v. and determined viral titers in serum at the indicated time points (Fig. 1A). Titers reached similar levels and peaked between day 9 and 14 in both groups; however a significant reduction (∼1 log) was observed by day 28 in PTPN22−/− mice as compared with WT mice. By day 39 all PTPN22−/− mice had cleared the infection, whereas significant levels of virus persisted in all WT mice (Fig. 1A). In addition, PTPN22−/− mice, but not WT mice, regained the weight loss associated with LCMV cl13 infection (Fig. 1B). Viral clearance was also observed in the organs of PTPN22−/− mice by day 14, whereas WT mice had detectable virus in the spleen, lung, liver, and kidney at this time point (Fig. S1 A and B).
Fig. 1.
PTPN22 deficiency leads to improved clearance of LCMV cl13. PTPN22−/− (n = 10) and WT (n = 10) mice were infected i.v. with 106 pfu of LCMV cl13, and serum was collected at the specified time points. (A) Plaque assays were carried out to determine viral titer in the serum. (B) Weight loss over time postinfection. Each experiment was done at least twice, and data were combined. (C and D) PTPN22−/− and WT mice were injected with CD4-depleting antibody before infection with LCMV cl13, and viral titer was measured in the serum (C) and in kidney and lung (D) on day 40. The data in the graphs are shown as the mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S1.
PTPN22−/− mice clear LCMV cl13 infection (related to Fig. 1). (A and B) PTPN22−/− and WT mice were infected with 106 pfu of LCMV cl13, and organ viral load was measured at day 8 (A) and day 14 (B). (C) PTPN22−/− and WT mice were infected i.v. with 105 pfu of LCMV cl13, and serum was collected at the specified time points. Plaque assays were carried out to determine the viral titer in the serum. ND, not detected. The data in graphs are shown as the mean ± SEM.
At a lower dose of LCMV cl13 (105 pfu) clearance was more rapid, and virus was undetectable in the serum as early as 14 d postinfection (Fig. S1C). Overall these results indicate that deficiency in PTPN22 enhances viral clearance. Because our goal was to assess immune function in WT and PTPN22−/− mice during comparable levels of infection, we chose to focus on the 106 pfu dose in future studies.
Normally, functional CD4 cells are required for clearance of LCMV. However, because PTPN22 deficiency has been reported to enhance CD8 function (27, 31), it was of interest to determine whether PTPN22−/− CD8 cells alone were capable of viral clearance. To determine whether CD4 T cells were necessary for the enhanced clearance of LCMV cl13 by PTPN22−/− mice, we injected a group of mice with CD4-depleting antibody (Fig. 1 C and D). In the PTPN22−/− group treated with CD4-depleting antibody, LCMV cl13 persistence was similar to that in WT mice, suggesting that CD4 T cells are necessary for the viral clearance mechanism in PTPN22−/− mice.
PTPN22 Is Necessary for Efficient IFN-I Production During LCMV cl13 Infection.
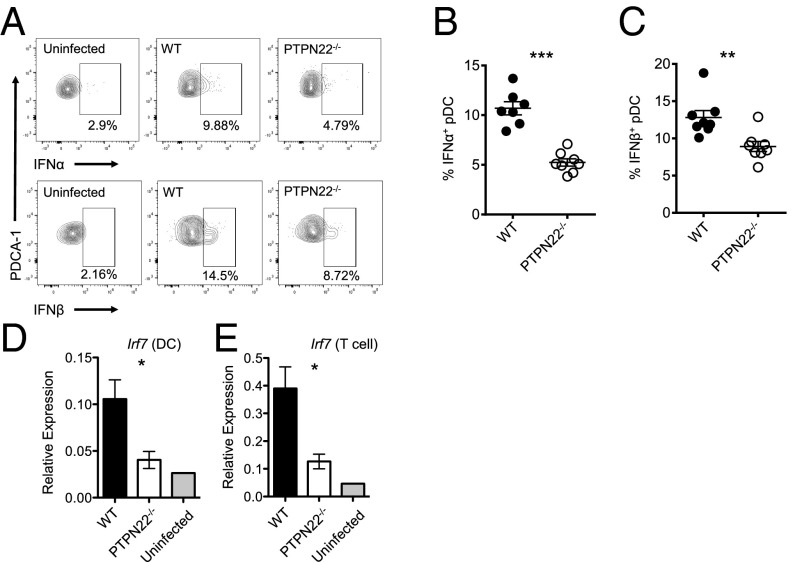
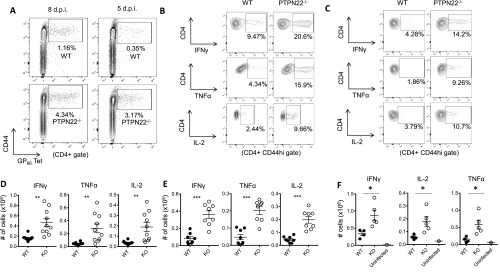
IFN-I is produced rapidly and in large quantities immediately following LCMV cl13 infection and recently has been found to contribute to T-cell exhaustion and viral persistence (21, 22). Because PTPN22 affects the level of IFN-I production by myeloid cells, we investigated this pathway following LCMV cl13 infection. Twenty-four hours after infection with LCMV cl13 the frequency of IFNα-producing pDCs was reduced significantly (∼50%) in spleen cells from PTPN22−/− mice as compared with WT mice (Fig. 2 A and B). In addition a significant reduction in the percentage of IFNβ+ pDCs was observed in PTPN22−/− mice compared with WT mice (Fig. 2 A and C). Consistent with previous reports (21), we also observed that pDCs, rather than conventional dendritic cells (cDCs), macrophages, or B cells, were the cell type producing the most IFN-I at this time point following infection with LCMV cl13.
Fig. 2.
PTPN22 deficiency results in reduced IFN-I. PTPN22−/− and WT mice were infected with LCMV cl13 (106 pfu i.v.). Twenty-four hours later spleens were removed, cultured in vitro with Brefeldin A for 3 h, and stained for IFNα and IFNβ. (A) Representative flow cytometric plots of pDC (CD19− CD11clo PDCA-1+) expression of IFNα and IFNβ. (B) Combined data showing pDC expression of IFNα. (C) Combined data showing pDC expression of IFNβ. B and C show combined data from two independent experiments combined, each symbol represents one mouse. (D and E) Eight days postinfection spleens were removed from WT and PTPN22-KO mice, and DCs (D) and T cells (E) were isolated by MACS. mRNA was purified, and quantitative RT-PCR was performed for irf7. D and E are two independent experiments. Each bar has four data points; each point is representative of two pooled mouse spleens. The data in graphs are shown as the mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
To assess the consequences of reduced IFN-I production in PTPN22−/− mice, the levels of transcription of the IFN-inducible gene irf7 (interferon regulatory factor f) were measured by quantitative RT-PCR in DCs and T cells 8 d postinfection of WT and PTPN22−/− mice. There was significantly less expression of irf7 in DCs and T cells of PTPN22−/− mice (Fig. 2 D and E).
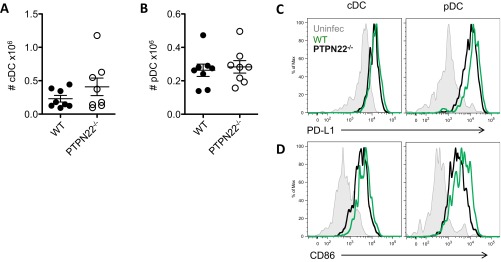
PTPN22 deficiency does not alter the numbers of pDCs or cDCs 24 h postinfection (Fig. S2 A and B). PTPN22-deficient pDCs, but not cDCs, have slightly lower expression of PD-L1 (programmed death ligand 1) (Fig. S2C), which is generally up-regulated on DCs during chronic infection, and both PTPN22-KO pDCs and cDCs have slightly lower CD86 expression compared with the WT cells (Fig. S2D).
Fig. S2.
The effect of PTPN22 on DC numbers and costimulatory phenotype (related to Fig. 2). PTPN22−/− and WT mice were infected with 106 pfu of LCMV cl13. Twenty-four hours later spleens were removed and stained for DC markers. (A) The number of cDCs (CD19−/PDCA-1−/CD11c+/MHC class II+) in the spleen. (B) The number of pDCs (CD19−/PDCA-1+/CD11clo) in the spleen. (C) Representative histograms of PD-L1 expression on cDCs and pDCs. (D) Representative histograms of CD86 expression on cDCs and pDCs. Data in A and B are pooled from at least two independent experiments. Each symbol represents one mouse. The data in graphs are shown as the mean ± SEM.
Overall, these data indicate that PTPN22 is required for optimal IFN-I production in response to LCMV cl13, resulting in a reduced IFN-I signature in DCs and T cells in PTPN22−/− mice. No significant differences in the expression of costimulatory molecules were observed in DCs in WT and PTPN22−/− mice.
PTPN22 Deficiency Increases IL-10 Production During LCMV Infection.
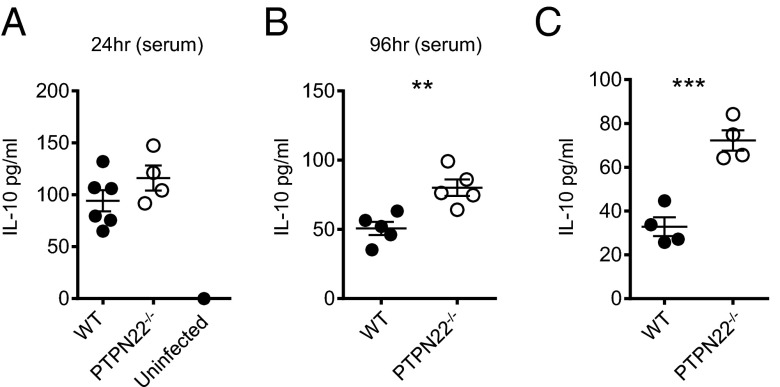
Reports have indicated that IL-10 contributes to the induction and maintenance of T-cell exhaustion (36, 37). In addition, blockade of IFN-I signaling results in reduced IL-10 production following LCMV cl13 infection (21, 22). We analyzed IL-10 levels in the serum at 24 h (Fig. 3A) and 96 h (Fig. 3B) postinfection by ELISA. At 24 h postinfection IL-10 is elevated in both WT and PTPN22−/− mice compared with uninfected controls, and by 96 h postinfection the concentration of IL-10 in the serum is significantly higher in PTPN22−/− mice than in WT mice. To measure IL-10 production by antiviral CD4 T cells, we cultured day 8 infected splenocytes with GP61–80 peptide for 48 h and measured IL-10 in the supernatant (Fig. 3C). PTPN22−/− mice produced significantly more IL-10 than WT mice, suggesting that the mechanism of enhanced viral clearance in PTPN22-deficient mice does not involve a reduction of IL-10.
Fig. 3.
PTPN22 deficiency results in increased IL-10 production. (A and B) PTPN22−/− and WT mice were infected i.v. with LCMV cl13 (106 pfu), and serum levels of IL-10 were measured by ELISA at 24 h (A) and 96 h (B) postinfection. (C) At day 8 postinfection splenocytes were cultured with GP66 peptide in vitro for 48 h, and the IL-10 concentration in the culture supernatant was measured by ELISA. Experiments were performed twice, and data were pooled. Each symbol represents one mouse. The data in graphs are shown as the mean ± SEM; **P < 0.01; ***P < 0.001.
PTPN22 Deficiency Enhances Numbers and Functions of CD4 T Cells Early Postinfection.
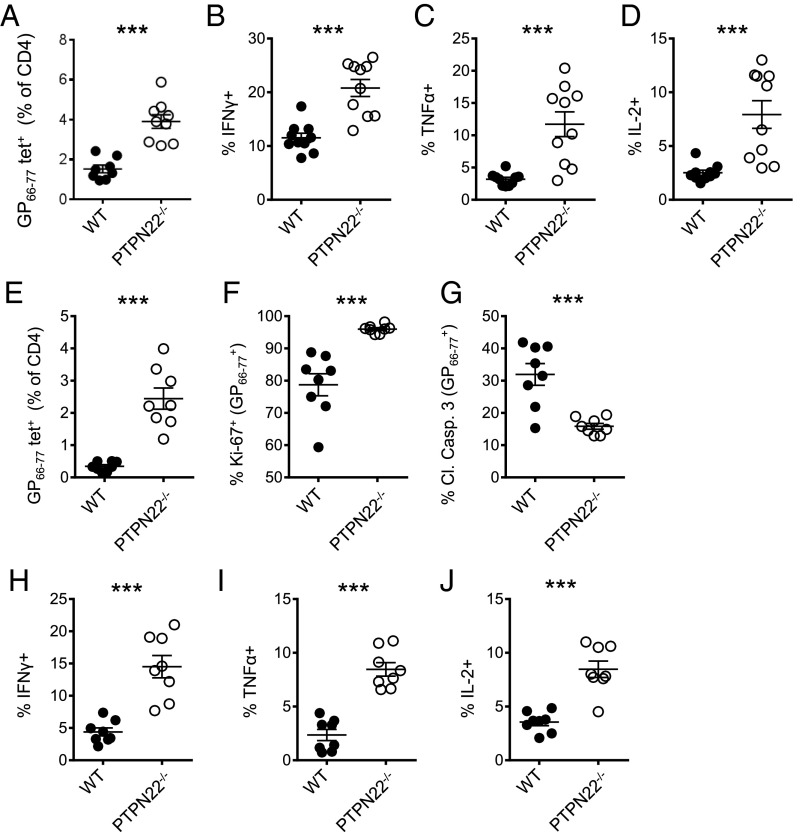
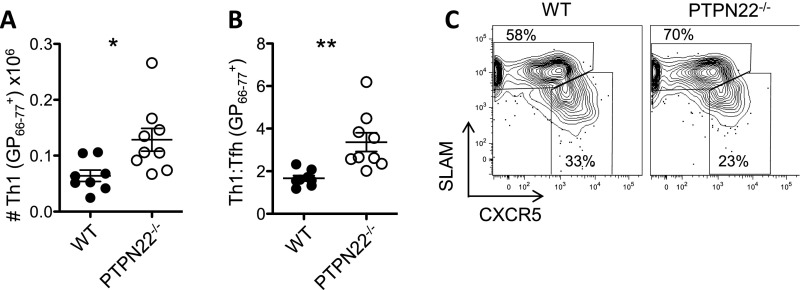
CD4 cells are required to retain the function of CD8 cells during chronic infection with LCMV cl13 (16, 38). Th1 cytokines such as TNFα and IL-2 are particularly important in this regard (19). Specifically, depletion of CD4 T cells in PTPN22−/− mice leads to viral persistence, indicating that CD4 T cells are critical to viral clearance (Fig. 1C). Therefore we decided to investigate CD4 cells in more detail. Eight days after infection with LCMV cl13, the numbers of LCMV GP66–77 tetramer-positive CD4 T cells were significantly increased in PTPN22−/− mice compared with WT mice (Fig. 4A and Fig. S3A). IFNγ, TNFα, and IL-2 production in response to GP61–80 peptide was strongly increased in both frequency and absolute number in CD4/CD44hi T cells from PTPN22-deficient mice compared with cells from WT mice (Fig. 4 B–D and Fig. S3 B and D). This trend was also observed at the low dose of LCMV cl13 (Fig. S3F). In addition, GP66–77 tetramer binding PTPN22−/− CD4 T cells showed more of a bias toward a Th1 phenotype (SLAMhi CXCR5−) than a follicular helper T cell (Tfh) phenotype (SLAMlo CXCR5+) than did WT cells (Fig. S4).
Fig. 4.
PTPN22−/− enhances numbers and function of viral-specific CD4 T cells. PTPN22−/− and WT mice were infected i.v. with LCMV cl13 (106 pfu), and spleens were collected and restimulated with GP61–80 peptide. (A) The number of GP66–77 tetramer-positive CD4 T cells in the spleen on day 8. (B–D) Intracellular staining for IFNγ (B), TNFα (C), and IL-2 (D) was performed and is shown for CD4/CD44hi T cells on day 8. (E) Viral-specific CD4 T cells were stained with the GP66–77 tetramer and counted by flow cytometry on day 5. (F and G) Tetramer-positive CD4 T cells were stained with Ki-67 (F) and cleaved caspase 3 (G) for flow cytometry analysis on day 5. (H–J) Spleens were restimulated with GP61–80 peptide on day 5 postinfection, and intracellular staining of CD4/CD44hi cells for IFNγ (H), TNFα (I), and IL-2 (J) was performed. Data in A–D are combined from three independent experiments; data in E–J are combined from two independent experiments. Each symbol represents one mouse. The data in graphs are shown as the mean ± SEM; ***P < 0.001.
Fig. S3.
Representative FACS plots of CD4 tetramer-positive cells and cytokine production (related to Fig. 4). PTPN22−/− and WT mice were infected with 106 pfu of LCMV cl13 8. (A) Five days later spleens were removed, and GP66–77 tetramer-positive CD4 T cells were stained. (B and C) Spleens were collected and on day 8 (B) and on day 5 (C) and were restimulated with GP66–80 peptide. Representative FACS plots (gated on CD4/CD44hi cells) of intracellular cytokine staining for IFNγ, IL-2, and TNFα are shown. (D) The absolute number of CD4/CD44hi cytokine-positive cells following peptide stimulation on day 8. (E) The absolute number of CD4/CD44hi cytokine-positive cells following peptide stimulation on day 5. (F) Intracellular cytokine staining following GP66–80 peptide stimulation on day 8 postinfection with low-dose LCMV cl13. Gated on CD4/CD44hi cells. Each symbol represents one mouse. The data in graphs are shown as the mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. S4.
PTPN22-deficient CD4 T cells are biased toward a Th1 phenotype (related to Fig. 4). PTPN22−/− and WT mice were infected with 106 pfu of LCMV cl13. Eight days later spleens were removed, and GP66–77 tetramer-positive CD4 T cells were stained for Th1 (SLAMhi CXCR5−) and Tfh (SLAMlo CXCR5+) markers. (A) The number of Th1 phenotype cells in WT and PTPN22−/− mice. (B) The ratio of Th1:Tfh phenotype GP66–77 tetramer-positive CD4 T cells. Each symbol in A and B represents one mouse. (C) Representative dot plot staining to define the Th1 and Tfh populations. Data were pooled from two independent experiments. The data in graphs are shown as the mean ± SEM. *P < 0.05; **P < 0.01.
To understand the basis for the greatly increased frequency of GP66–77 tetramer -positive CD4 T cells in PTPN22−/− mice (Fig. 4A), we analyzed the expression of proliferation and apoptotic markers in GP66–77 tetramer-positive T cells 5 d after infection with LCMV cl13. The frequency of GP66–77 tetramer-positive cells in PTPN22−/− mice was increased significantly, and these cells divided more frequently (based on the expression of Ki-67) than did the cells from WT mice (Fig. 4 E and F and Fig. S3A). In addition, cleaved caspase 3, a marker of apoptosis, was significantly lower among GP66–77 tetramer-positive CD4 cells in PTPN22−/− mice than in WT mice (Fig. 4G). Overall these data suggest that the greater frequency of GP66–77 tetramer-positive CD4 T cells in PTPN22−/− mice can be attributed to both increased proliferation and reduced cell death.
In addition to the increased frequency of GP66–77 tetramer-positive cells at day 5 postinfection, the increased cytokine production observed at day 8 by PTPN22−/−/CD4/CD44hi T cells is clearly evident at 5 d postinfection. At this time point CD4/CD44hi T cells from PTPN22−/− mice had a significantly higher percentage and absolute numbers of IFNγ-, TNFα-, and IL-2–positive cells in response to GP61–80 peptide (Fig. 4 H–J and Fig. S3 C and E). Overall, these data indicate that PTPN22 deficiency has an early effect on the expansion and effector function of CD4 T cells.
PTPN22 Deficiency Affects CD4 T-Cell Number and Function in a T-Cell–Extrinsic Manner.
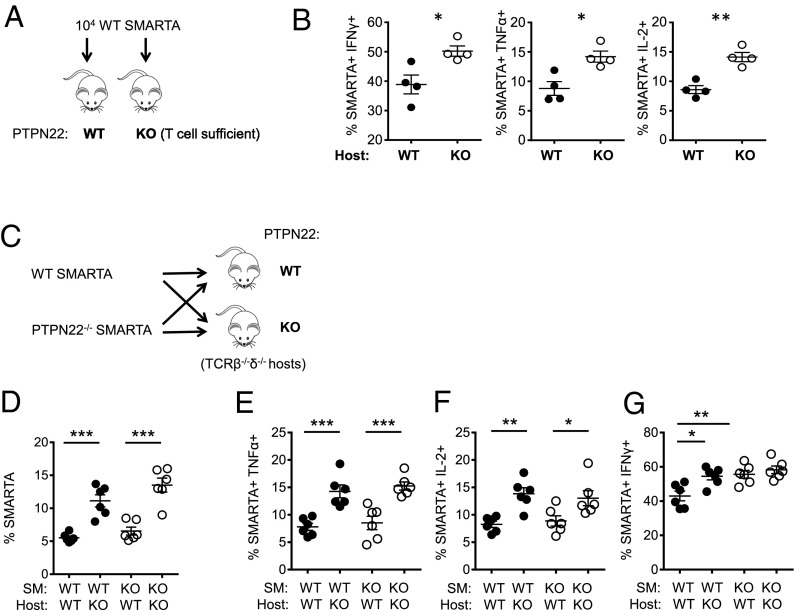
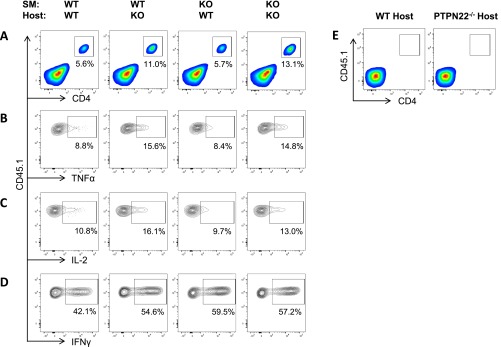
PTPN22 is expressed in all hematopoietic cells, and LCMV cl13 infection of PTPN22-deficient mice reveals a number of phenotypic differences in both antigen-presenting cells and T cells (Figs. 2 and 4). To determine whether the increased number and function of viral-specific CD4 T cells observed in PTPN22−/− mice is intrinsic or extrinsic to the T cell, we purified CD4 T cells (CD45.1+) from SMARTA TCR transgenic mice [expressing a TCR specific for the immuno-dominant CD4 LCMV epitope (GP66–80) (39)] and transferred these cells into either WT or PTPN22−/− (CD45.2+) hosts (Fig. 5A). Eight days postinfection spleens were harvested and were restimulated with GP61–80 peptide, and SMARTA production of IFNγ, TNFα, and IL-2 was measured by flow cytometry (Fig. 5B). Significant increases in SMARTA cytokine production were observed in PTPN22−/− hosts compared with WT hosts. These results suggest that host expression of PTPN22 affects the outcome of infection. A similar experiment using SMARTA PTPN22−/− cells could not be performed, because the cells were rejected by WT hosts.
Fig. 5.
PTPN22 deficiency controls CD4 T-cell number and function in a T-cell–extrinsic manner. (A) CD45.1+ SMARTA cells (104) were purified by MACS and transferred into CD45.2+ WT or PTPN22−/− T-cell–sufficient hosts. The following day these mice were infected with 106 pfu LCMV cl13. (B) Spleens were restimulated with GP61–80 peptide on day 8 postinfection, and intracellular staining for IFNγ, TNFα, and IL-2 was performed. (C) CD45.1+ PTPN22−/− SMARTA and WT SMARTA cells (104) were purified by MACS and transferred into CD45.2+ WT or PTPN22−/− (TCRβ−/−δ−/−) hosts. The following day the four groups of mice were infected with 106 pfu LCMV cl13. (D) The percentage of SMARTA cells in the lymphocyte gate on day 8 postinfection in the spleen. (E–G) Spleens were restimulated with GP61–80 peptide on day 8 postinfection, and intracellular staining for TNFα (E), IL-2 (F), and IFNγ (G) was performed. Experiments were performed twice and pooled. Each symbol represents one mouse. The data in the graphs are shown as the mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
We repeated this experiment using TCRβ−/−δ−/− hosts to avoid rejection of PTPN22−/− SMARTA cells and also to avoid competition with endogenous viral-specific CD4 cells. Hosts received either WT or PTPN22−/− SMARTA cells and were infected with LCMV cl13 on the following day; the spleens were analyzed on day 8 postinfection (Fig. 5C). There was a significant host effect on the percentage of SMARTA cells recovered; the accumulation of both PTPN22-deficient and WT cells was greater in the PTPN22−/− host than in the WT host (Fig. 5D and Fig. S5A). There also was a slight increase in the percentage of PTPN22−/− SMARTA cells compared with WT SMARTA cells in the PTPN22−/− host, although this increase did not achieve significance. PTPN22 deficiency in the host results in a significant increase in the production of TNFα and IL-2 (Fig. 5 E and F). PTPN22 deficiency in either the host or T cells results in increased production of IFNγ (Fig. 5G and Fig. S5 B–D). Taken together, these data suggest that PTPN22 affects CD4 T-cell function in a predominantly cell-extrinsic manner during chronic infection.
Fig. S5.
PTPN22 deficiency controls CD4 T-cell number and function in a T-cell–extrinsic manner (related to Fig. 5). CD45.1+ PTPN22−/− SMARTA and WT SMARTA cells (104) were purified by MACS and transferred into CD45.2+ WT or PTPN22−/− (TCRβ−/−δ−/−) hosts. The following day the four groups of mice were infected with 106 pfu LCMV cl13. (A) Representative flow cytometry plots showing the percentage of SMARTA cells in the lymphocyte gate on day 8 postinfection in the spleen. (B–D) Spleens were restimulated with GP61–80 peptide on day 8 postinfection, and intracellular staining for TNFα (B), IL-2 (C), and IFNγ (D) was performed. Each graph is a representative flow cytometry plot gated on SMARTA cells. (E) CD45.1+ WT SMARTA cells (104) were purified by MACS and transferred into CD45.2+ WT or PTPN22−/− (TCRβ−/−δ−/−) uninfected hosts. The spleens were analyzed at day 8 posttransfer for SMARTA cells. The data in graphs are shown as the mean ± SEM.
To assess the potential contribution of differences in homeostatic proliferation leading to the differences between host PTPN22 genotypes in the expansion of SMARTA cells, 104 carboxyfluorescein succinimidyl ester (CFSE)-labeled SMARTA cells were transferred into WT and PTPN22−/− TCRβ−/−δ−/− hosts without LCMV cl13 infection. Eight days later no detectable SMARTA cells were recovered in the spleens of either host type, indicating that lymphopenic-driven expansion of SMARTA cells is minimal in this model (Fig. S5E). This result is consistent with reports showing that naive SMARTA cells do not proliferate in various lymphopenic hosts (40, 41). Furthermore, production of IL-7, which is important for the homeostatic proliferation and survival of naive CD4 T cells, is produced mainly by cell types that do not express PTPN22 and would not be expected to differ in these hosts (42).
cAMP Responsive Element Modulator Is Involved in IL-2 Repression During LCMV cl13 Infection.
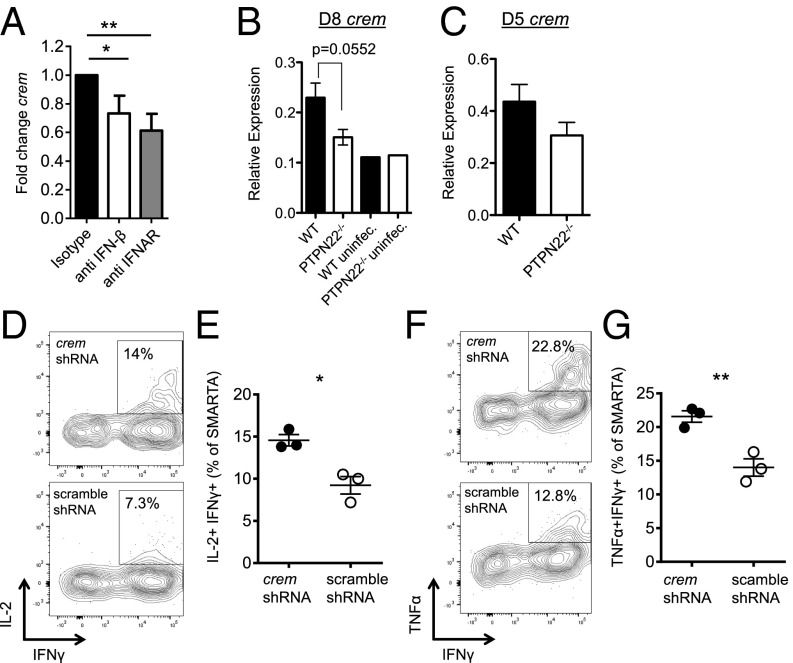
To explore the T-cell–extrinsic mechanism by which CD4 function is restored in PTPN22−/− mice, we investigated expression of an IFN-I–inducible gene, crem (cAMP response element modulator), that is known to inhibit IL-2 transcription (43, 44). Recently it was reported that CREM expression, which is induced by IFNβ, is up-regulated in T cells following LCMV cl13 infection (45). To confirm this IFN-dependent up-regulation of CREM during LCMV c13 infection, we transferred SMARTA cells into WT B6 mice along with blocking antibodies to IFNβ or IFN-1 receptor (IFNAR) 1 d before infection (Fig. 6A). SMARTA cells were sorted from the spleens of mice 7 d postinfection, and crem transcript was measured by quantitative RT-PCR. Blocking either IFNβ or IFNAR significantly reduce the crem transcript in CD4 T cells following LCMV cl13 infection.
Fig. 6.
PTPN22−/− CD4 T cells do not up-regulate CREM, which can lead to enhanced antibody production. (A) CD45.1+ SMARTA cells (103) were adoptively transferred into WT B6 mice along with blocking antibodies against IFNβ, IFNAR, or isotype control. Twenty-four hours later, the mice were infected i.v. with LCMV cl13 (106 pfu). SMARTA cells were sorted from spleens 7 d postinfection, and the crem transcript was measured by quantitative RT-PCR. (B) PTPN22−/− and WT mice were infected i.v. with LCMV cl13 (106 pfu), and CD4/CD44hi cells were sorted by FACS on day 8. Quantitative RT-PCR was performed to measure crem transcript levels. (C) SMARTA cells or PTPN22−/− SMARTA cells were adoptively transferred into WT or PTPN22−/− mice, respectively, and mice were infected with LCMV cl13. Transferred cells were sorted on day 5, and mRNA was isolated. Quantitative RT-PCR was used to measure crem transcript. A–C show data points; each point was pooled from two mouse spleens from two independent experiments. (D–G) SMARTA cells were transduced with a retrovirus expressing shRNA targeting CREM or control (scramble) and were reinjected into WT mice. After at least 5 d of rest, mice were infected with LCMV cl13 (106 pfu i.v.), and spleens were collected on day 8. SMARTA T cells were restimulated with GP61–80 peptide, and TNFα+ IFNγ+ (D and E) and IL-2+ IFNγ+ (F and G) cells were counted by flow cytometry. D and F are representative flow cytometric plots gated on SMARTA cells. E and G are representative of two independent experiments; each symbol represents one mouse. The data in graphs are shown as the mean ± SEM; *P < 0.05; **P < 0.01.
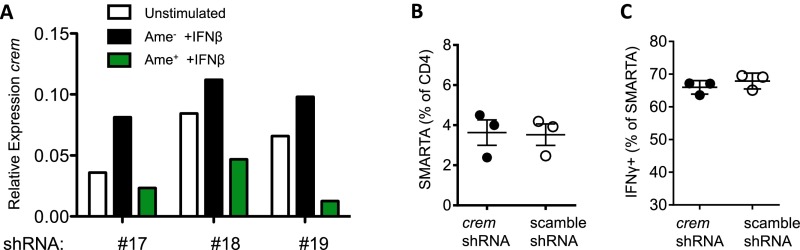
To determine whether the enhanced IL-2 production observed in PTPN22−/− mice may be caused by the reduced expression of CREM, quantitative RT-PCR was performed using mRNA from sorted CD4+/CD44hi T cells from spleens harvested at day 8 postinfection. Crem transcript levels were lower in PTPN22−/− CD4/CD44hi T cells than in WT T cells (Fig. 6B). To measure crem transcript levels at day 5 postinfection, SMARTA adoptive transfer was used to increase antiviral CD4 numbers. PTPN22−/− or WT SMARTA cells were transferred into PTPN22−/− or WT hosts, respectively. Five days after infection with LCMV cl13, SMARTA cells were sorted by FACS, and crem mRNA was measured. As found at day 8 postinfection, the crem transcript at day 5 postinfection is higher in WT CD4 T cells than in PTPN22−/− T cells (Fig. 6C). To determine whether CREM expression directly regulated cytokine production in antiviral CD4 T cells, we used shRNA to knock down CREM in SMARTA CD4 T cells. Retroviral transduction of anti-crem shRNA led to efficient knockdown of crem transcripts (Fig. S6A). Transduced SMARTA cells were adoptively transferred into WT B6 hosts and allowed to rest for at least 5 d before infection with LCMV cl13. Recovery of SMARTA cells at 8 d postinfection was similar in the scramble and CREM knockdown groups (Fig. S6B). Eight days later SMARTA cells were restimulated with GP61–80 peptide, and IL-2 (Fig. 6 D and E) and TNFα (Fig. 6 F and G) production were measured by intracellular cytokine staining. Knockdown of CREM led to significantly increased IL-2 and TNFα production compared with SMARTA cells transduced with scramble shRNA. Of interest, IFNγ production was not affected by CREM knockdown (Fig. S6C). Overall these data show that CREM knockdown phenocopies the increase in IL-2 and TNFα production observed in PTPN22−/− CD4 T cells and is sufficient to prevent the loss of CD4 T-cell function following LCMV cl13 infection.
Fig. S6.
CREM knockdown by shRNA (related to Fig. 6). (A) SMARTA cells were transduced with retroviral vectors expressing three different shRNA sequences (nos. 17, 18, and 19). Transduced cells were sorted by FACS using the fluorescent reporter protein Ametrine (Ame). Transduced (Ame+) and untransduced (Ame−) cells were stimulated overnight with IFNβ, and mRNA was isolated. CREM transcript levels were measured by quantitative RT-PCR. shRNA no.17 was chosen for all subsequent experiments. (B and C) SMARTA CD4 T cells were transduced with a retrovirus expressing shRNA targeting CREM or control and were reinjected into WT B6 mice. After at least 5 d rest, mice were infected i.v. with LCMV cl13 (106 pfu), and spleens were collected on day 8. (B) The percentage of SMARTA cells in the CD4 gate. (C) SMARTA T cells were restimulated with GP61–80 peptide, and IFNγ+ cells were counted by flow cytometry.
PTPN22 Deficiency Prevents Exhaustion of Viral-Specific CD8 T Cells.
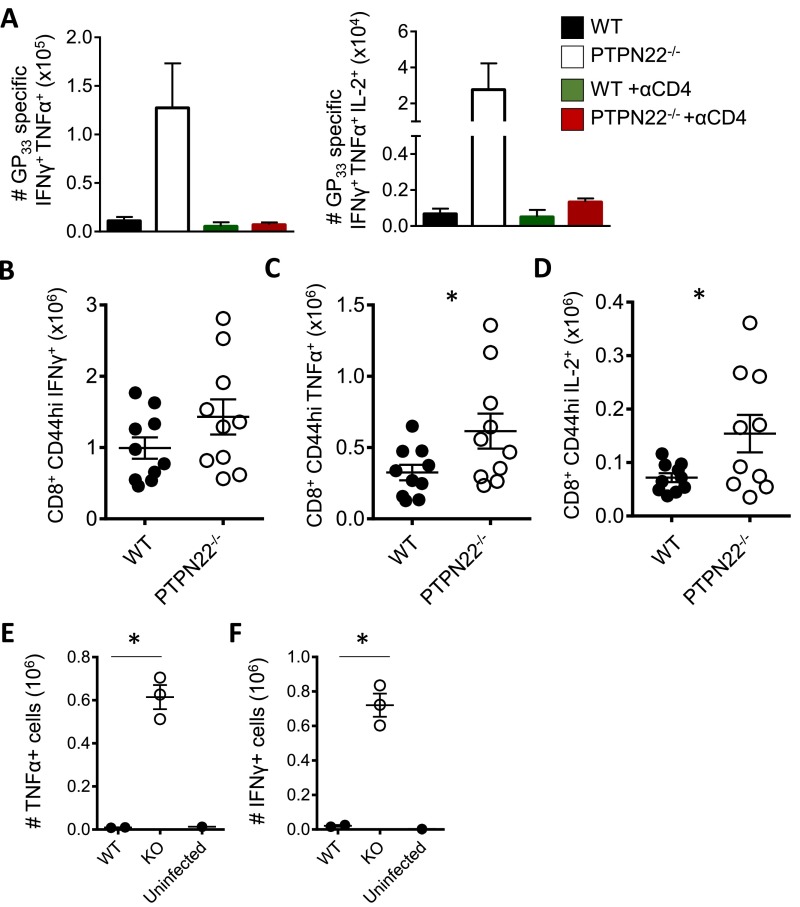
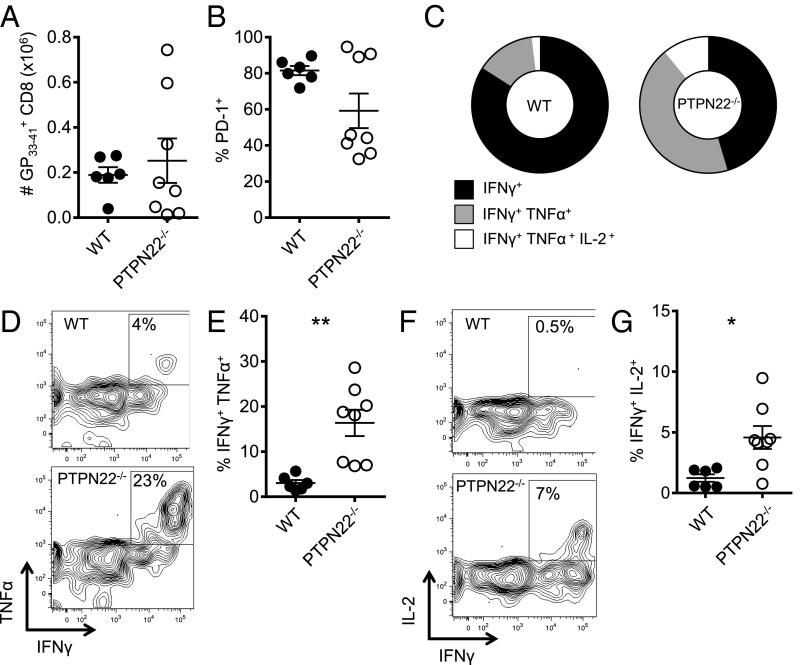
CD4 help, especially by IL-2, affects the number and function of CD8 T cells during chronic infection (16, 19, 38). Because CD4 T-cell function was increased in PTPN22−/− mice after infection with LCMV cl13, we hypothesized that antiviral CD8 T cells would also retain function. Depletion of CD4 T cells in PTPN22−/− mice leads to viral persistence (Fig. 1C) and exhaustion of CD8 T cells (Fig. S7A), further substantiating the importance of CD4 cells in viral elimination. PTPN22−/− and WT mice were infected with LCMV cl13, and spleens were collected and analyzed on day 14. Similar numbers of GP33–41 tetramer-positive CD8 T cells were found in the spleens of PTPN22−/− and WT mice (Fig. 7A). PD-1 expression on tetramer-positive cells was measured, and the percentage of PD-1+ CD8 T cells was lower in the majority of PTPN22−/− mice than in WT mice by day 14, as is consistent with less T-cell exhaustion (Fig. 7B). To assess the function of the CD8 T cells, splenocytes were restimulated with GP33–41 peptide, and intracellular staining for cytokines was performed on GP33–41 tetramer-positive cells. The GP33–41 tetramer-positive cells from PTPN22−/− mice showed broader polyfunctionality than the cells from WT mice, with a larger pool of triple- and double-cytokine–positive cells (Fig. 7C). Although the percentage of GP33–41 tetramer-positive cells that can produce TNFα or IL-2 is very low in WT mice, a significantly higher percentage of GP33–41 tetramer-positive CD8 T cells from PTPN22−/− mice produce both cytokines (Fig. 7 D–F).
Fig. S7.
PTPN22−/− mice have increased CD8 T-cell function (related to Fig. 7). (A) PTPN22−/− and WT mice were injected with CD4-depleting antibody or isotype control and then were infected with 106 pfu of LCMV cl13. At 40 d postinfection spleens were harvested and restimulated with GP33–41 peptide. Intracellular cytokine staining was performed to measure IFNγ, TNFα, and IL-2. (B–D) PTPN22−/− and WT mice were infected with 106 pfu of LCMV cl13. Eight days later spleens were removed and restimulated with GP33–41 peptide. Intracellular cytokine staining for IFNγ (B), TNFα (C), and IL-2 (D) was performed. (E and F) Intracellular cytokine staining following GP33–41 peptide stimulation on day 25 postinfection with low-dose LCMV cl13 (gated on CD8/CD44hi cells). The data in graphs are shown as the mean ± SEM; *P < 0.05.
Fig. 7.
Virus-specific CD8 T cells are not exhausted in PTPN22−/− mice. PTPN22−/− and WT mice were infected i.v. with LCMV cl13 (106 pfu). (A) GP33–41 tetramer-positive CD8 T-cell numbers in the spleen at day 14. (B) PD-1 expression on GP33–41 tetramer-positive CD8 T cells. Spleens were collected on day 14 and restimulated with GP33–41 peptide. TNFα, IFNγ, and IL-2 were measured by intracellular FACS. (C) Piecharts showing polyfunctionality of GP33–41 tetramer-positive cells with respect to cytokine production. (D) Representative FACS plots of TNFα and IFNγ staining on GP33–41 tetramer-positive cells. (E) Combined intracellular cytokine staining data from two independent experiments. (F) Representative FACS plots of IL-2 and IFNγ staining on GP33–41 tetramer-positive cells. (G) Combined intracellular cytokine staining data from two independent experiments. Experiments were performed twice and pooled. Each symbol represents one mouse. In A, B, E, and G data are shown as the mean ± SEM. In C data represents the mean (WT n = 6, KO n = 8). *P < 0.05; **P < 0.01.
Viral titers were found to be lower in the spleens of PTPN22−/− mice than in spleens from WT mice at day 14 postinfection. To be certain that reduced virus load was not responsible for enhanced T-cell function, we analyzed the spleen for CD8 function at day 8 postinfection, a time when viral titer was high (Fig. S7). Following GP33–41 peptide restimulation, PTPN22−/− mice had increased numbers of IFNγ-producing CD8 T cells as well as significantly increased numbers of TNF and IL-2–positive cells (Fig. S7 B–D).
Similarly, at the low dose of LCMV cl13 CD8 T cells are more functional at 25 d postinfection in PTPN22−/− mice than in WT mice (Fig. S7 E and F).
Discussion
PTPN22 encodes a tyrosine phosphatase that is expressed in multiple immune cell types and has numerous reported functions. In particular, mice deficient in PTPN22 have increased accumulation of effector and memory T cells and impaired production of IFN-I by myeloid cells (27, 34). In light of these phenotypes, we hypothesized that PTPN22 deficiency would benefit the clearance of LCMV cl13 under conditions that normally result in the exhaustion of antiviral T-cell responses and in chronic infection. In contrast to WT mice, PTPN22−/− mice retained T-lymphocyte function and cleared the LCMV cl13 infection. Because PTPN22 is present in numerous immune cell types that could promote T-cell activity, we sought to characterize the immune response at various times postinfection.
DCs from PTPN22−/− mice produce less IFN-I in response to LCMV cl13 infection, in keeping with recent reports that PTPN22 is involved downstream of TLR- and nucleic acid-sensing pathways in myeloid cells (Fig. 2 A–C) (34, 35). Furthermore DCs and T cells in PTPN22−/− mice exhibit a reduced IFN-I signature at 8 d postinfection (Fig. 2 D and E). IFN-I is strongly induced immediately after LCMV cl13 infection and is induced in much larger amounts by LCMV cl13 than by the rapidly cleared Armstrong strain of LCMV (21). Recent reports demonstrate that overproduction of IFN-I during LCMV cl13 infection is detrimental to the host immune system and contributes to T-cell exhaustion and viral persistence (20–23). Blockade of the IFNAR, genetic ablation of this receptor, or blocking IFNβ leads directly to improved clearance of LCMV cl13 that is associated with increased antiviral CD4 function. A similar outcome was seen PTPN22−/− mice infected with LCMV cl13, suggesting that the reduction in IFNβ production in these mice could contribute to viral clearance.
Following LCMV cl13 infection, PTPN22−/− mice have increased percentages of viral-specific CD4 T cells and significantly higher percentages of CD4 T cells that produce IL-2, TNFα, and IFNγ (Fig. 4). The expansion of the GP66–77 tetramer-positive population in PTPN22−/− mice appears to be the result of both increased proliferation and reduced cell death (Fig. 4). CD4 responses during LCMV cl13 infection contribute to the clearance of the virus, as demonstrated the rapid exhaustion of antiviral CD8 T cells and the persistence of LCMV cl13 following the depletion of CD4 T cells (Fig. 1C) (16, 38). In addition, adoptive transfer of SMARTA CD4 T cells into mice before LCMV cl13 infection results in lower viral titers, which are associated with increased function of GP33–41-specific CD8 T cells and antibody production (18).
The blockade of IFN-I signaling also correlates with reduced PD-L1 expression on DCs and lower serum levels of IL-10 (21, 22). Both of these molecules are known to suppress T-cell responses to LCMV cl13; however, the expression of PD-L1 was not altered significantly at early time points in PTPN22−/− mice (Fig. S2), but the IL-10 concentration in serum was increased significantly in these mice (Fig. 3). An increase in CD4 numbers and function in PTPN22-deficient mice despite unaltered PD-L1 expression on DCs is perhaps not surprising, given that blockade of PD-1 does not affect the number or function of LCMV-specific CD4 T cells (19). Many cell types, including CD4 T cells, produce IL-10 during chronic LCMV infection (2). Stimulation of PTPN22-deficient splenocytes from mice at day 8 postinfection with the CD4 epitope GP61–80 resulted in a significantly increased IL-10 concentration in the culture medium compared with results from WT mice (Fig. 4B). CD4 T-cell IL-10 production has been attributed to the BLIMP+ Th1 subset during chronic LCMV infection, and increased production of IL-10 by CD4 T cells from PTPN22−/− mice may reflect the larger GP66–77 tetramer-positive Th1 pool in these mice compared with WT mice rather than a per-cell increase in IL-10 production (Fig. S4) (46). Overall, despite unaltered PD-L1 expression and increased IL-10 production, PTPN22 deficiency enhances T-cell function during chronic infection. Our results are consistent with a recent report showing that partial blockade of IFN-I signaling (by blocking IFN-β as opposed to the IFNAR) can lead to enhanced LCMV cl13 clearance and increased CD4 numbers and cytokine production despite unaltered levels of IL-10 and PD-L1 on DCs (23). We hypothesize that CD4 function during LCMV cl13 infection may be determined by a balance between suppressive IL-10 signaling and IL-2/TNFα/IFNγ signaling, resulting in exhaustion or retention of function. In the case of PTPN22−/− mice, the production of IL-2, TNFα, and IFNγ by CD4 T cells is significantly enhanced to the point that IL-10 signaling may not be sufficient to impose an exhausted phenotype.
Because PTPN22 affects TCR signaling, we performed experiments aimed at discovering whether the increased expansion and function of antiviral CD4 T cells in PTPN22-deficient mice is intrinsic to the T cell or is modulated through extrinsic factors. To do so, we used an adoptive transfer model in which LCMV-specific CD4 T cells (SMARTA) are transferred into mice deficient in T cells (Fig. 5 and Fig. S5). We observed only a small increase in the numbers of PTPN22-deficient SMARTA cells compared with WT SMARTA cells. In contrast, PTPN22 deficiency in the host was found to increase the number and function of both WT and KO SMARTA CD4 T cells. As a caveat, these experiments were performed in T-cell–deficient mice (Fig. 5 C–G), and although similar results were observed when SMARTA cells were transferred into either WT or PTPN22−/− T-cell–sufficient hosts (Fig. 5 A and B), the reciprocal transfer of PTPN22−/− SMARTA cells into WT hosts could not be performed because these cells were rejected in T-cell–sufficient mice. Nevertheless, these results suggest that deficiency of PTPN22 in a non-T cell influences the CD4 response following viral infection. A recent report characterizes the early cytokine production after LCMV cl13 infection and after Armstrong infection and shows that a number of cytokines, including IFN-I, are differentially expressed (20). It is not known whether any of these cytokines could contribute to the T-cell–extrinsic effect we observed, but it is known that PTPN22 is not involved in the expression of NF-κB–dependent cytokines by myeloid cells (34). Based on our current understanding of PTPN22 in production of IFN-I, IFN-I is a major candidate for the host factor responsible for this phenotype.
To expand on the mechanism by which IFN-I may mediate its effects on T-cell exhaustion, we explored the role of CREM in T cells. CREM is part of the cAMP responsive element modulator family responsible for regulation of gene expression (47). The crem gene encodes numerous proteins produced through alternative splicing and alternative promoter use (48). CREM has been shown to inhibit IL-2 expression by recruiting histone deacetylases that close the chromatin structure in the IL-2 promoter and prevent transcription following TCR stimulation (43–45). Importantly for our studies, CREM expression is induced by IFN-I, and the protein can be detected in T cells following LCMV cl13 infection (45). Our data show that we can reduce CREM expression in CD4 T cells significantly by blocking IFN-I signaling during LCMV cl13 infection (Fig. 6A). We hypothesized that because of the reduced IFN-I production by PTPN22-deficient mice following LCMV cl13 infection, CREM levels in T cells would be lower than in WT mice, thus allowing more efficient IL-2 transcription. In support of this hypothesis, we found that CD4 T cells in PTPN22−/− mice express lower levels of CREM than do CD4 T cells in WT mice, as determined by quantitative RT-PCR (Fig. 6 B and C). To establish a direct link between increased CREM expression following cl13 infection and the loss of cytokine production by virus-specific CD4 cells, shRNA was used to knock down CREM expression in WT SMARTA CD4 T cells. CREM reduction resulted in increased IL-2 and TNFα following LCMV cl13 infection, thus phenocopying PTPN22−/− mice (Fig. 6 D–G). These results implicate the regulation of CREM as a potential factor responsible for the loss of IL-2 and TNFα production during LCMV cl13 infection. We hypothesize that increasing the production of Crem in PTPN22-deficient CD4 cells following viral infection may reestablish exhaustion, although it may be difficult to achieve the physiological levels of Crem that normally occur during infection.
Alternative splicing products from the crem gene include the inducible cAMP repressor (ICER) protein, which acts as a strong transcriptional repressor and is transcribed from the alternative P2 promoter (49). In human medullary thymocytes ICER has been shown to inhibit transcription at NFAT/AP-1 composite DNA sites that are essential for the expression of both IL-2 and TNFα, suggesting that CREM and its alternative protein products such as ICER have wide-ranging suppressive effects on cytokine production in immune cells (50). CREM and its related protein products are poorly understood in the context of immune cell function and represent important targets for preventing the induction and maintenance of T-cell exhaustion.
Chronic TCR signaling in LCMV cl13 coupled with the loss of CD4 help underlies the exhaustion of CD8 T cells (1, 51). PTPN22-KO mice have increased numbers of functional antiviral CD8 T cells at day 8 postinfection, when serum and splenic viral titers are equally high in WT mice, suggesting that there is resistance to exhaustion in these mice (Fig. S7 B–D). Following the increase in CD8 function at day 8, the splenic viral titer in PTPN22−/− mice is reduced significantly by day 14, with most mice clearing the infection (Fig. S1 A and B). This course correlates with the enhanced function of PTPN22−/− CD8 T cells compared with the exhaustion of CD8 T cells in WT mice (Fig. 7). Depletion of CD4 T cells in PTPN22−/− mice results in persistence of virus (Fig. 1 C and D) and CD8 exhaustion (Fig. S7A), suggesting that CD8 function and viral clearance are controlled by the increased CD4 function in these mice. These results are consistent with the hypothesis that the early restoration of CD4 function, in particular IL-2 production, may inhibit exhaustion of CD8 T cells at later times during infection. In keeping with our observations, the Ahmed laboratory has reported that low-dose IL-2 therapy enhances antiviral CD8 responses in LCMV cl13-infected mice, increasing both the number and the cytokine production of CD8 T cells as well as lowering PD-1 expression (19).
Because the loss of PTPN22 leads to the clearance of chronic viral infection, it is interesting to speculate that the presence of the autoimmune-predisposing allele in a high percentage of European populations has been evolutionarily retained for this reason. Although the data generated in this report used mice deficient in PTPN22 rather than mice deficient in the minor allele, many of the effects observed in PTPN22−/− mice, including reduced production of IFN-I, are mirrored in the PTPN22 R619W mutant mice (52, 53). In addition there are reports that the minor allele of PTPN22 is protective against tuberculosis infections (54). We hypothesize that, despite the increased susceptibility to autoimmunity caused by PTPN22 R620W in humans, the resistance conferred by this allele to both bacterial and viral infections may have resulted in a selective advantage. Furthermore healthy carriers of the PTPN22 R620W variant have exaggerated Th1 responses with increased IFNγ, TNFα, and IL-2 production, similar to the data presented in this report (55). It will be interesting to extend this genotypic analysis to other infectious diseases in the future.
Materials and Methods
Mice.
Experimental procedures were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (56). PTPN22−/− mice were obtained from Andrew Chan. Genentech, San Francisco, and have been described previously (27). SMARTA CD45.1+ mice were bred to PTPN22−/− mice.
Virus Infection and Plaque Assays.
LCMV cl13 strains were grown, stored, and quantified according to published methods (57). For determination and quantification of viremia, 10 μL of serum was used to perform 10-fold serial dilutions for plaque assays on VERO cells (58). Tissues were harvested and frozen at −80 °C. At the time of the assay, tissues were thawed, weighed, and homogenized in medium to 10% (wt/vol) and were clarified by low-speed centrifugation; 10 μL of the supernatant was used in plaque assays.
Antibody Treatments.
For CD4 T-cell depletion, mice were treated with 500 µg of the monoclonal GK1.5 antibody or isotype control on days −1 and 0 and with 250 µg on days 3 and 5 postinfection. One milligram of anti-IFNAR (clone MAR-5A3; Leinco Technologies), 250 µg of anti-IFNβ (clone HDβ-4A7) (23), or 1 mg of mouse IgG1 isotype control (Bio X Cell) was injected i.p. at day −1.
Flow Cytometry.
Cells were resuspended in HBSS containing 1% FCS and were incubated with the indicated antibodies for 15 min on ice. Cells then were washed before acquisition on an LSR-II flow cytometer (BD Biosciences), and analysis was performed using FlowJo (TreeStar). Antibodies (all from BioLegend unless otherwise stated) used were anti-mouse CD4 PerCP-Cy5.5, CD45.1 FITC/Pacific blue, SLAM PE, CD8 PerCP-Cy5.5/APC Cy7, PD-1 FITC, CXCR5-biotin (BD Bioscience), CD44 Pacific Blue/PE-Cy7, streptavidin APC, CD11c APC, PDCA-1 Pacific Blue, CD19 APC-Cy7, MHC class II Pe-Cy7, PD-L1 PE, and CD86 FITC. For intracellular staining of markers, an intracellular staining kit (Fix/Perm; eBioscience) was used together with anti-mouse Ki-67 APC (BioLegend). Staining of cleaved caspase 3 used the Cytofix/Cytoperm Kit (BD Bioscience) with anti-cleaved caspase 3 (Cell Signaling Technology) and anti-rabbit Alexa Fluor 647 (Life Technologies).
Tetramers were obtained from the NIH tetramer core facility. Phycoerythrin (PE)-conjugated H2Db GP33–41 tetramer was used to stain CD8 T cells for 30 min on ice. BV421-conjugated I-Ab GP66–77 tetramer was used to stain CD4 T cells for 1 h at 37 °C.
For FACS, cell suspensions were labeled with antibody as described above, and the desired populations were sorted using FACSAria (BD Bioscience) by the Scripps Research Institute flow cytometry core facility.
Intracellular Cytokine Staining.
Splenocyte populations were plated at 2 × 105 cells per well in a 96-well plate and were stimulated with LCMV peptides (GP61–80 and GP33–41) for 4 h in the presence of Brefeldin A (Sigma Aldrich). For pDC intracellular cytokine staining, splenocytes were cultured as above in the presence of Brefeldin A for 3 h. Cells were stained for surface markers and then were fixed in Cytofix/Cytoperm buffer (BD Bioscience). Cells then were permeabilized in Perm buffer (BD Bioscience), and cytokines were stained using antibodies against IL-2 BV421/PE, TNFα PerCP-Cy5.5/PE, IFNγ APC (all from BioLegend), IFNα FITC, and IFNβ FITC (PBL Bioassay).
Adoptive T-Cell Transfer.
Magnetic-activated cell sorting (MACS) of CD4+ T cells was carried out using biotinylated antibodies to deplete unwanted cells types as described in ref. 59. Streptavidin-coated IMag beads (BD Bioscience) were used to deplete labeled cells. SMARTA+ CD45.1+ CD4 T cells were transferred into recipients by i.v. injection. The exact numbers vary by experiment and are noted in the figure legends.
Quantitative RT-PCR.
FACS or MACS was used to purify desired cell populations, and then mRNA was extracted using the RNeasy RNA extraction kit according to the manufacturer’s instructions (Qiagen). cDNA was produced using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. Quantitative RT-PCR was performed to measure levels of crem and irf7 (sequences are available on request) using a Bio-Rad CFX96 machine (Bio-Rad). Cycle threshold (Ct) values were normalized to β-actin using the 2-ΔΔCt method.
Retroviral Transduction of SMARTA T Cells.
shRNAs (shERWOOD-UltraMar set) targeting the crem gene [Gene ID 12916, National Center for Biotechnology Information (NCBI)] were cloned into the pLMP-d Ametrine vector (TransOMIC Technologies) and packaged into retroviral vectors using the Plat-E packaging cell line (Cell Biolabs, Inc.). Transduction of CD3/CD28-activated MACS-purified SMARTA cells was carried out as described in ref. 60 using two 90-min spinfections on consecutive days. FACS-sorted Ametrine-positive cells were adoptively transferred into B6 hosts and rested for at least 5 d before LCMV cl13 infection.
IL-10 ELISA.
Quantitation of IL-10 in the serum and cell-culture supernatants was carried out using the Quantikine IL-10 ELISA kit according to the manufacturer’s instructions (R&D Systems).
Statistical Analysis.
Graphs were assembled and analyzed using Prism 5 software (GraphPad). For multiple group analyses, one-way ANOVA with Tukey’s posttest was carried out. For comparison of two-group datasets Student’s t test was used.
Acknowledgments
We thank The Scripps Research Institute flow cytometry core for assistance with cell sorting and Shane Crotty and Simon Belanger of the La Jolla Institute of Allergy and Immunology for advice and protocols for retroviral transduction. This work was supported by National Institute of Allergy and Infectious Diseases (NIAID) Grant 5R21AI119353 (to L.A.S.) and NIAID Grant 1R01AI123210-01 (to J.R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603738113/-/DCSupplemental.
References
- 1.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 2.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandele A, Mukerjee P, Das G, Ahmed R, Chauhan VS. Phenotypic and functional profiling of malaria-induced CD8 and CD4 T cells during blood-stage infection with Plasmodium yoelii. Immunology. 2011;132(2):273–286. doi: 10.1111/j.1365-2567.2010.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esch KJ, Juelsgaard R, Martinez PA, Jones DE, Petersen CA. Programmed death 1-mediated T cell exhaustion during visceral leishmaniasis impairs phagocyte function. J Immunol. 2013;191(11):5542–5550. doi: 10.4049/jimmunol.1301810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautam S, et al. CD8 T cell exhaustion in human visceral leishmaniasis. J Infect Dis. 2014;209(2):290–299. doi: 10.1093/infdis/jit401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmadzadeh M, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114(8):1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zippelius A, et al. Effector function of human tumor-specific CD8 T cells in melanoma lesions: A state of local functional tolerance. Cancer Res. 2004;64(8):2865–2873. doi: 10.1158/0008-5472.can-03-3066. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed R, Jamieson BD, Porter DD. Immune therapy of a persistent and disseminated viral infection. J Virol. 1987;61(12):3920–3929. doi: 10.1128/jvi.61.12.3920-3929.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford A, Wherry EJ. The diversity of costimulatory and inhibitory receptor pathways and the regulation of antiviral T cell responses. Curr Opin Immunol. 2009;21(2):179–186. doi: 10.1016/j.coi.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 12.Overwijk WW, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198(4):569–580. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks DG, Teyton L, Oldstone MB, McGavern DB. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J Virol. 2005;79(16):10514–10527. doi: 10.1128/JVI.79.16.10514-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyasere C, et al. Diminished proliferation of human immunodeficiency virus-specific CD4+ T cells is associated with diminished interleukin-2 (IL-2) production and is recovered by exogenous IL-2. J Virol. 2003;77(20):10900–10909. doi: 10.1128/JVI.77.20.10900-10909.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lechner F, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191(9):1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68(12):8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze Zur Wiesch J, et al. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med. 2012;209(1):61–75. doi: 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aubert RD, et al. Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proc Natl Acad Sci USA. 2011;108(52):21182–21187. doi: 10.1073/pnas.1118450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West EE, et al. PD-L1 blockade synergizes with IL-2 therapy in reinvigorating exhausted T cells. J Clin Invest. 2013;123(6):2604–2615. doi: 10.1172/JCI67008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan BM, Teijaro JR, de la Torre JC, Oldstone MB. Early virus-host interactions dictate the course of a persistent infection. PLoS Pathog. 2015;11(1):e1004588. doi: 10.1371/journal.ppat.1004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340(6129):207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson EB, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340(6129):202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng CT, et al. Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe. 2015;17(5):653–661. doi: 10.1016/j.chom.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bottini N, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36(4):337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 25.Criswell LA, et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: The PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76(4):561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Todd JA, et al. Genetics of Type 1 Diabetes in Finland Wellcome Trust Case Control Consortium Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasegawa K, et al. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science. 2004;303(5658):685–689. doi: 10.1126/science.1092138. [DOI] [PubMed] [Google Scholar]
- 28.Brownlie RJ, et al. Lack of the phosphatase PTPN22 increases adhesion of murine regulatory T cells to improve their immunosuppressive function. Sci Signal. 2012;5(252):ra87. doi: 10.1126/scisignal.2003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maine CJ, et al. PTPN22 alters the development of regulatory T cells in the thymus. J Immunol. 2012;188(11):5267–5275. doi: 10.4049/jimmunol.1200150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maine CJ, Marquardt K, Cheung J, Sherman LA. PTPN22 controls the germinal center by influencing the numbers and activity of T follicular helper cells. J Immunol. 2014;192(4):1415–1424. doi: 10.4049/jimmunol.1302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmond RJ, Brownlie RJ, Morrison VL, Zamoyska R. The tyrosine phosphatase PTPN22 discriminates weak self peptides from strong agonist TCR signals. Nat Immunol. 2014;15(9):875–883. doi: 10.1038/ni.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zikherman J, et al. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol. 2009;182(7):4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, et al. Identification of substrates of human protein-tyrosine phosphatase PTPN22. J Biol Chem. 2006;281(16):11002–11010. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity. 2013;39(1):111–122. doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maine CJ, et al. The effect of the autoimmunity-associated gene, PTPN22, on a BXSB-derived model of lupus. Clin Immunol. 2015;156(1):65–73. doi: 10.1016/j.clim.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12(11):1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ejrnaes M, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203(11):2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Battegay M, et al. Enhanced establishment of a virus carrier state in adult CD4+ T-cell-deficient mice. J Virol. 1994;68(7):4700–4704. doi: 10.1128/jvi.68.7.4700-4704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: Effects on humoral and cellular immune responses after viral infection. Eur J Immunol. 1998;28(1):390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 40.Martin CE, Kim DM, Sprent J, Surh CD. Is IL-7 from dendritic cells essential for the homeostasis of CD4+ T cells? Nat Immunol. 2010;11(7):547–548, author reply 548. doi: 10.1038/ni0710-547. [DOI] [PubMed] [Google Scholar]
- 41.Ramsey C, et al. The lymphopenic environment of CD132 (common gamma-chain)-deficient hosts elicits rapid homeostatic proliferation of naive T cells via IL-15. J Immunol. 2008;180(8):5320–5326. doi: 10.4049/jimmunol.180.8.5320. [DOI] [PubMed] [Google Scholar]
- 42.Hara T, et al. Identification of IL-7-producing cells in primary and secondary lymphoid organs using IL-7-GFP knock-in mice. J Immunol. 2012;189(4):1577–1584. doi: 10.4049/jimmunol.1200586. [DOI] [PubMed] [Google Scholar]
- 43.Powell JD, Lerner CG, Ewoldt GR, Schwartz RH. The -180 site of the IL-2 promoter is the target of CREB/CREM binding in T cell anergy. J Immunol. 1999;163(12):6631–6639. [PubMed] [Google Scholar]
- 44.Solomou EE, Juang YT, Gourley MF, Kammer GM, Tsokos GC. Molecular basis of deficient IL-2 production in T cells from patients with systemic lupus erythematosus. J Immunol. 2001;166(6):4216–4222. doi: 10.4049/jimmunol.166.6.4216. [DOI] [PubMed] [Google Scholar]
- 45.Otero DC, Fares-Frederickson NJ, Xiao M, Baker DP, David M. IFN-β Selectively Inhibits IL-2 Production through CREM-Mediated Chromatin Remodeling. J Immunol. 2015;194(11):5120–5128. doi: 10.4049/jimmunol.1403181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parish IA, et al. Chronic viral infection promotes sustained Th1-derived immunoregulatory IL-10 via BLIMP-1. J Clin Invest. 2014;124(8):3455–3468. doi: 10.1172/JCI66108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamas M, et al. CREM: A master-switch in the transcriptional response to cAMP. Philos Trans R Soc Lond B Biol Sci. 1996;351(1339):561–567. doi: 10.1098/rstb.1996.0055. [DOI] [PubMed] [Google Scholar]
- 48.Foulkes NS, Sassone-Corsi P. More is better: Activators and repressors from the same gene. Cell. 1992;68(3):411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- 49.Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: An alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75(5):875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 50.Bodor J, Habener JF. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem. 1998;273(16):9544–9551. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
- 51.Fahey LM, et al. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med. 2011;208(5):987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai X, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 2013;123(5):2024–2036. doi: 10.1172/JCI66963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43(9):902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 54.Gomez LM, Anaya JM, Martin J. Genetic influence of PTPN22 R620W polymorphism in tuberculosis. Hum Immunol. 2005;66(12):1242–1247. doi: 10.1016/j.humimm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Vang T, et al. The autoimmune-predisposing variant of lymphoid tyrosine phosphatase favors T helper 1 responses. Hum Immunol. 2013;74(5):574–585. doi: 10.1016/j.humimm.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 57.Borrow P, Evans CF, Oldstone MB. Virus-induced immunosuppression: Immune system-mediated destruction of virus-infected dendritic cells results in generalized immune suppression. J Virol. 1995;69(2):1059–1070. doi: 10.1128/jvi.69.2.1059-1070.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160(2):521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin CE, Frimpong-Boateng K, Spasova DS, Stone JC, Surh CD. Homeostatic proliferation of mature T cells. Methods Mol Biol. 2013;979:81–106. doi: 10.1007/978-1-62703-290-2_9. [DOI] [PubMed] [Google Scholar]
- 60.Choi YS, Crotty S. Retroviral vector expression in TCR transgenic CD4⁺ T cells. Methods Mol Biol. 2015;1291:49–61. doi: 10.1007/978-1-4939-2498-1_5. [DOI] [PubMed] [Google Scholar]