Significance
Copper is central to the metabolism of methanotrophs, methane-oxidizing bacteria that are of interest because of their potential applications in bioremediation and bioconversion processes. Methanotrophs convert methane to methanol using particulate methane monooxygenase (pMMO), a copper-dependent, membrane-bound enzyme. To fulfill pMMO’s high requirement for copper, methanotrophs secrete and re-internalize methanobactin, a peptide-derived, copper-chelating natural product (CuMbn). Here we characterize the protein machinery used for import and periplasmic transport of CuMbn. These data provide important insights into the molecular mechanisms of CuMbn recognition and handling.
Keywords: methanobactin, chalkophore, copper transport, methane monooxygenase, metal homeostasis
Abstract
Methanotrophic bacteria use methane, a potent greenhouse gas, as their primary source of carbon and energy. The first step in methane metabolism is its oxidation to methanol. In almost all methanotrophs, this chemically challenging reaction is catalyzed by particulate methane monooxygenase (pMMO), a copper-dependent integral membrane enzyme. Methanotrophs acquire copper (Cu) for pMMO by secreting a small ribosomally produced, posttranslationally modified natural product called methanobactin (Mbn). Mbn chelates Cu with high affinity, and the Cu-loaded form (CuMbn) is reinternalized into the cell via an active transport process. Bioinformatic and gene regulation studies suggest that two proteins might play a role in CuMbn handling: the TonB-dependent transporter MbnT and the periplasmic binding protein MbnE. Disruption of the gene that encodes MbnT abolishes CuMbn uptake, as reported previously, and expression of MbnT in Escherichia coli confers the ability to take up CuMbn. Biophysical studies of MbnT and MbnE reveal specific interactions with CuMbn, and a crystal structure of apo MbnE is consistent with MbnE's proposed role as a periplasmic CuMbn transporter. Notably, MbnT and MbnE exhibit different levels of discrimination between cognate and noncognate CuMbns. These findings provide evidence for CuMbn–protein interactions and begin to elucidate the molecular mechanisms of its recognition and transport.
Methanotrophs, bacteria that use methane as their sole carbon source (1), have attracted intense interest because of their potential uses in methane mitigation, as well as bioremediation and bioconversion processes (2). In the first step of methanotroph metabolism, methane is oxidized to methanol by metalloenzymes known as methane monooxygenases (MMOs). Two MMOs have been characterized: a soluble cytoplasmic enzyme (sMMO) and a membrane-bound, particulate enzyme (pMMO). sMMO belongs to a large family of bacterial multicomponent monooxygenases and requires iron for activity (3–5) whereas pMMO requires copper (Cu) for activity (5–8). In most methanotroph species, pMMO catalyzes methane oxidation, but some species have genes for both sMMO and pMMO. In these methanotrophs, MMO expression is regulated by the metal availability: under Cu-limiting conditions, sMMO is expressed, and under Cu-replete conditions, pMMO is expressed (9–11).
In many methanotrophs, pMMO accounts for up to 20% of the proteome (12), and its requirement for Cu is met by a specialized system for Cu acquisition (13). When grown under conditions of limiting Cu, some methanotrophs secrete small molecules that bind Cu with high affinity [equilibrium binding constant, Kb, of 1011–14 M−1 for Cu(II) and Kb of 1019–21 M−1 for Cu(I)] (14). Originally known as Cu-binding compounds (15), these compounds are now called methanobactins (Mbns). Mbns are ribosomally synthesized posttranslationally modified natural products (16, 17), produced by some methanotrophic and possibly also nonmethanotrophic bacteria species (17). Mbn was first isolated from the growth medium of Methylosinus (Ms.) trichosporium OB3b (15, 18, 19). The Ms. trichosporium OB3b Mbn chelates a single Cu(I) ion in a distorted tetrahedral geometry using two oxazolone and neighboring thioamide groups; Mbns isolated from other methanotrophs have similar N-containing heterocycles and Cu coordination environments (SI Appendix, Fig. S1) (14, 16, 19–21). Mbn can bind both Cu(I) and Cu(II), but Cu(II) is rapidly reduced to Cu(I) via an unknown mechanism (22, 23). The Cu(I)-loaded Mbn (CuMbn) is stabilized, and this form is reinternalized to fulfill the bacterium’s Cu requirements (24).
Direct evidence for import of intact CuMbn came from our studies of Ms. trichosporium OB3b (24). This methanotroph takes up both soluble Cu and CuMbn, but according to isotopic and fluorescent labeling studies, CuMbn is imported intact. Moreover, CuMbn uptake is an active transport process (24), relying on energy transduced across the inner membrane. Likely candidates for CuMbn transport include TonB-dependent transporters (TBDTs), outer membrane receptors that interact with the inner membrane TonB complex to transport substrates (25, 26), including siderophores, sugars, metal chelates, and large protein cofactors like cobalamin (27), all of which bind with relatively high affinity. TBDTs also exhibit substrate specificity, binding even very similar substrates with different affinities (28).
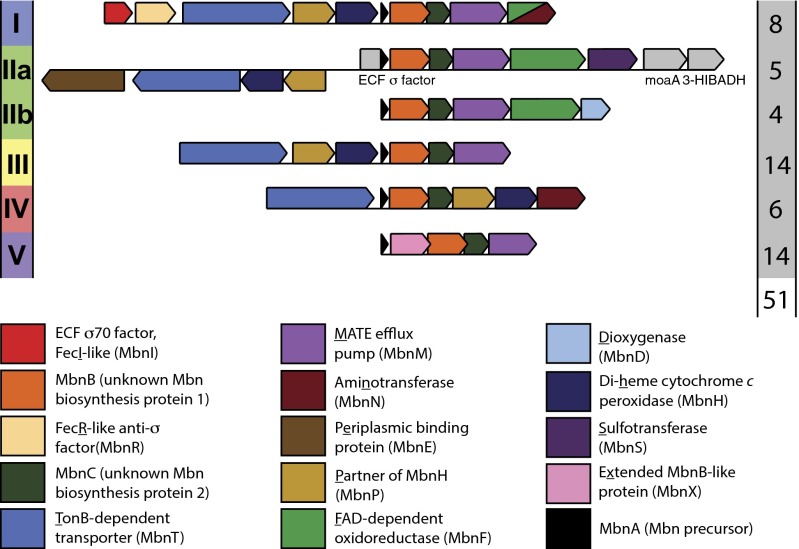
Extensive bioinformatics analyses revealed that in addition to the gene encoding the Mbn precursor peptide, mbnA, there is an entire operon, termed mbn, of surrounding genes that encode putative Mbn biosynthetic, transport, and regulatory proteins (Fig. 1 and SI Appendix, Fig. S2). These operons were divided into families (groups I–V) on the basis of operon content and sequence analysis (17); analysis of additional Mbn operons identified in the last 2 years suggests that the operons previously categorized as group II might better be split into two subgroups, IIA and IIB. In some genomes, multiple distinct Mbn operons are present. One of the genes identified in most mbn operons is mbnT, which encodes a TBDT. Disruption of this gene abolishes Mbn uptake, indicating that it is the Mbn importer (29). Also present in most of these operons is mbnM, which encodes a member of the multidrug and toxic compound extrusion protein family (16, 17, 30, 31). As such, MbnT and MbnM were proposed in 2013 to be the prime candidates for the CuMbn importer and exporter, respectively (17). In addition, 9 kb upstream of mbnT in the Ms. trichosporium OB3b genome is the previously unidentified mbnE, which encodes a periplasmic binding protein (PBP) that may transport periplasmic CuMbn. Recent quantitative RT-PCR (qRT-PCR) studies of Ms. trichosporium OB3b (32) revealed that these genes, including mbnE, are coregulated with mbnA and are all down-regulated in response to Cu addition. Taken together with the cessation of Mbn production in an mbnA deletion mutant (33) and the reported mbnT knockout strain (29), there is strong genetic evidence for the involvement of mbnT and mbnE in Mbn transport. However, the interaction of these proteins with Mbn has not been investigated biochemically. Here we have probed the roles of MbnT and MbnE using bioinformatic, genetic, biochemical, and biophysical approaches. Taken together, our results identify MbnT as the CuMbn importer and MbnE as a CuMbn PBP.
Fig. 1.
Composition of Mbn operons. Mbn operons can be subdivided into six groups on the basis of operon content, precursor peptide (MbnA) sequence, and phylogenetic analysis of MbnA, MbnB, and MbnC, which are present in all operons. The number of complete operons of a given type is presented to the right of the operon. Ms. trichosporium OB3b belongs to group I, Mc. parvus OBBP has both a group I and a group IIb operon, and Mc. hirsuta CSC1 has a group IIa operon. Although genes related to Mbn biosynthesis, regulation, and transport are present in all operons, genes for the putative Mbn exporter, MbnM, and importer, MbnT, are notably widespread.
Results and Discussion
Bioinformatics Analysis of the Transport Proteins.
The Mbn operons and their component genes were originally identified via bioinformatics (17) (Fig. 1). We have now extended and updated this analysis with a focus on transport-related genes, resulting in a more complete model for Mbn transport. TBDTs are present in operons for four of five Mbn groups (17) (Fig. 1), consistent with the need for active transport of a large metal-bound natural product. However, the TBDTs present in Mbn operons are more heterogeneous than might be expected, given the high conservation of other Mbn-related genes. As noted previously (17), MbnT genes that neighbor genes encoding sigma and anti-sigma factor pairs also encode the N-terminal extensions necessary for signal transduction between TBDTs and anti-sigma factors (34–36). As additional Mbn operons have been identified, it has become increasingly clear that these fecIRA-like mbnIRT triads are present only in group I Mbn operons, suggesting that only those MbnTs play a role in signal transduction. A larger comparison of putative MbnTs with extant TBDTs in the Joint Genome Institute/Integrated Microbial Genomes & Microbiomes and UniProt databases indicates that the differences between MbnTs extend beyond the presence or absence of an N-terminal extension. MbnTs from group I Mbn operons (MbnT1s) are distinct in sequence and domain content from group II MbnTs (MbnT2s), and both are distinct from MbnTs in group III and IV (MbnT3s); all MbnTs appear to be distinct from previously categorized TBDT groups (SI Appendix, Fig. S3 A and B and SI Results and Discussion and Datasets S1–S3). Notably, many methanotrophs contain multiple genes encoding MbnTs, sometimes from multiple MbnT groups.
Although not described in the initial bioinformatics-based characterization of Mbn operons (17), there is some evidence that PBPs may also play a role in Mbn uptake. These proteins are periplasmic (in Gram-negative bacteria) or membrane-anchored (in Gram-positive bacteria) and are associated with ATP-binding cassette (ABC) importers (37, 38). The only Mbn operons containing PBPs are group IIA operons, which include the MbnE gene, but close homologs of the Methylocystis (Mc.) and the Ms. trichosporium OB3b mbnE genes can be found elsewhere in the genomes of other Mbn-producing methanotrophs. In some cases, multiple potential homologs are present. All these PBPs are members of solute binding protein (SBP) Family 5 (39), which comprises primarily oligopeptide-binding proteins; in a sequence similarity network of this family, MbnEs cluster separately from all characterized PBPs except a microcin C7 transporter, YejA (SI Appendix, Fig. S4A and Dataset S4). Potential homologs are observed in the genomes of most species producing group III–V Mbns, but they are also similar to YejA (40). The similarities between the two PBP subgroups (SI Appendix, Fig. S4B) complicate any definitive assignment without evidence of coregulation with Mbn operons or biochemical interactions with Mbns. However, it is notable that yejA encodes a microcin C7 transporter; microcin C7 is a ribosomally produced, posttranslationally modified natural product with a heptapeptide backbone and a final mass within the range observed for Mbns (40).
Gene Regulation Patterns of Transport Proteins in Ms. trichosporium OB3b.
A previous set of gene regulation studies confirmed that the entirety of the mbn operon in Ms. trichosporium OB3b [mbn(E)IRTABCMNPH] (SI Appendix, Fig. S2) is coregulated in a Cu-dependent manner (32). Many of the genes in the operon lack close homologs elsewhere in the genome. However, there are several TBDTs that are closely related to mbnT as well as two other PBPs classified via bioinformatics as oligopeptide-binding proteins and possible mbnE homologs. In total, there are 45 TBDTs and 3 Family 5 PBPs in the Ms. trichosporium OB3b genome (41). Thus, to ensure that we were pursuing the candidates most likely to be related to Mbn transport, we monitored Cu-dependent gene regulation of the closest mbnE and mbnT homologs.
qRT-PCR results confirm that, although mbnE is down-regulated by Cu in concert with the Mbn operon, the two most closely related PBPs, a hypothetical protein (MettrDRAFT_1622) and yejA (MettrDRAFT_3640) (SI Appendix, Fig. S2), are not (SI Appendix, Fig. S5 and Table S1 and Dataset S5). The TBDT results are more complicated. Two mbnIRTPH operons are present elsewhere in the genome in close proximity to one another (SI Appendix, Fig. S2). Both operons (containing the mbnT1 homologs MettrDRAFT_1229 and MettrDRAFT_1241) are Cu-regulated, but down-regulation levels differ (particularly notably for the nonregulatory genes mbnPH, which are 5- to 10-fold down-regulated, compared with the 25- to 50-fold down-regulation of their mbn operon counterparts) (Dataset S5). Differences in regulation patterns after Cu addition are also observed, particularly in the operon containing the mbnT1 gene MettrDRAFT_1241: unlike the regulatory genes in the main mbn operon or the other mbnIRTPH operon, which respond to Cu addition swiftly, the mbnIR homologs in this operon are not down-regulated completely within the first 15 min, but continue decreasing at the 5-h and 24-h time points. These regulatory differences are statistically significant. Whether analyzed via hierarchical clustering or calculation of gene-to-gene Spearman correlation values, expression patterns of genes within the mbn operon correlate significantly more closely to other genes within that operon than they do to genes outside of that operon (SI Appendix, Fig. S5 and Table S1 and Dataset S5). These differing responses to Cu suggest that, even if the MbnT1s encoded by these operons are capable of binding Mbn, the systems are not fully interchangeable with the native mbn operon. Further work will be necessary to identify the extent of regulatory crosstalk between the three systems and native substrate-triggered regulation and/or uptake. However, these gene-regulation results provide support for the preliminary assignment of the originally identified mbnE and mbnT genes (MettrDRAFT_3413 and MettrDRAFT_3421) as the PBP and TBDT most likely to function in import of the native Mbn. A possible function of the close MbnT1 homologs could be uptake of nonnative CuMbns produced by other methanotrophs in the environment.
In Vivo Functional Characterization of MbnT.
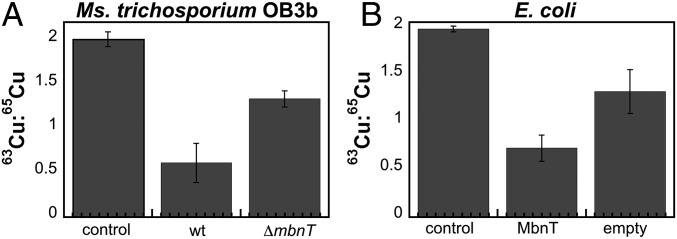
We took a two-pronged approach to definitively assign MbnT’s function: loss-of-function experiments using a gene disruption mutant of Ms. trichosporium OB3b and gain-of-function experiments using a heterologous host. For the first approach, we used marker exchange mutagenesis to prepare a strain of Ms. trichosporium OB3b in which mbnT was replaced with a gene conferring gentamicin resistance (SI Appendix, Fig. S6). Both WT and ΔmbnT cells were incubated with 65CuMbn (final concentration of 5 μM) for 2 h followed by washing and inductively coupled plasma mass spectrometry (ICP-MS) analysis. Natural Cu has two stable isotopes, 63Cu and 65Cu, in a 2.3:1 ratio. By adding 65CuMbn to cells, we can monitor the levels of 65Cu directly and correlate the values to the amount of 65CuMbn in the cells. Our results show a clear difference in the levels of 65Cu in WT Ms. trichosporium OB3b and ΔmbnT Ms. trichosporium OB3b (Fig. 2A), with the ΔmbnT strain exhibiting much lower levels of 65Cu. This result supports the experiments reported recently by Gu and coworkers (29).
Fig. 2.
In vivo characterization of CuMbn import. 65CuMbn uptake in (A) WT Ms. trichosporium OB3b and the mutant lacking MbnT (ΔmbnT) and (B) E. coli cells expressing MbnT or containing the empty pET-20b vector. Columns represent three averaged biological replicates, and error bars represent SD. Buffers used to wash the cells were used as controls. Uptake of 65CuMbn results in a decreased 63Cu:65Cu ratio.
We then expressed MbnT in E. coli (SI Appendix, Fig. S7) and tested for uptake of CuMbn. E. coli was previously shown to be incapable of CuMbn uptake (24). In this experiment, MbnT expression was induced in E. coli cells that were then incubated for 3 h before 65CuMbn addition (final concentration of 5 μM), followed by incubation for 30 min, washing, and ICP-MS analysis. The data reveal a clear difference in the 65Cu content of cells expressing MbnT and cells containing only the empty vector (Fig. 2B). These combined data provide strong in vivo evidence for the identification of MbnT as the CuMbn importer.
Interaction of CuMbn with Purified MbnT.
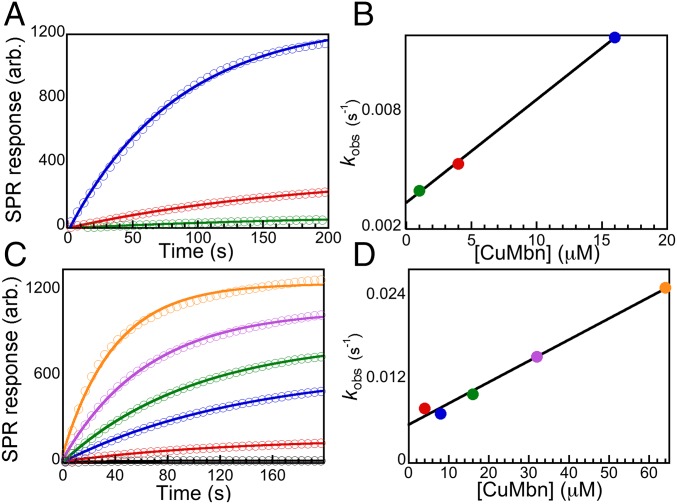
We then investigated the interaction of MbnT with CuMbn in vitro. MbnT was expressed in E. coli, solubilized in n-dodecyl-N,N-dimethylamine-N-oxide and purified via Ni-affinity chromatography (SI Appendix, Fig. S8). Size-exclusion chromatography suggests that the purified protein exists in multiple oligomeric states. Binding of Ms. trichosporium OB3b CuMbn to MbnT was assessed using surface plasmon resonance (SPR) (42). SPR is a detergent-free means to characterize ligand binding that has been used previously to characterize TBDTs (43). Purified MbnT was immobilized on a sensor chip via amine coupling, and solutions of CuMbn were applied. Binding of CuMbn to the immobilized MbnT elicits a response that is not observed with the application of buffer solutions (Fig. 3A). The response curves were fit with an equation describing the approach to equilibrium with pseudo-first-order kinetics (44); the pseudo-first-order rate constants, kobs, obtained in these traces directly correlate to the concentration of CuMbn. A plot of kobs versus the concentration of CuMbn can be fit to a line (Fig. 3B) where the slope is the rate constant for association (kon) and the intercept is the rate constant for dissociation (koff) (44). An equilibrium constant for dissociation, Kd, of 6.22 μM, was obtained. The determined Kd value is higher than those reported for other TBDTs binding to their cognate substrates (45–48). However, most of those studies were performed in vivo using native TBDTs whereas MbnT was isolated from a heterologous host, and in vitro binding experiments were conducted in the absence of membranes or detergents.
Fig. 3.
Binding of CuMbn to MbnTs. (A) Application of 1 μM (green), 4 μM (red), and 16 μM (blue) CuMbn to Ms. trichosporium OB3b MbnT elicits SPR responses in a concentration-dependent fashion, yielding apparent rate constants of association, kobs. (B). Plot of kobs versus the concentration of CuMbn fit to a line with slope (kon) of 5.3 × 10−4 μM−1⋅s−1 and intercept (koff) of 3.3 × 10−3 s−1, giving a Kd value of 6.22 μM. (C and D) Binding of 4 μM (red), 8 μM (blue), 16 μM (green), 32 μM (purple) and 64 μM (orange) Ms. trichosporium OB3b CuMbn to the MbnT from Mc. rosea SV97T with values for kon of 3.0 × 10−4 μM−1·s−1, koff of 5.4 × 10−3 s−1, and Kd of 17.6 μM.
The specificity of Ms. trichosporium OB3b MbnT for its cognate CuMbn was investigated via SPR experiments in which cobalamin (a proxy for a large, metal-containing compound), ferric pyoverdine (a proxy for a peptide-derived, metal-containing natural product), and ferric citrate (a small, metal-containing compound) were applied to immobilized MbnT. None of these molecules gave an appreciable SPR response (SI Appendix, Fig. S9A), indicating that they do not bind to MbnT. To further ensure that observed interactions result from specific recognition of CuMbn by MbnT, we immobilized myoglobin (a protein that should not bind CuMbn or Cu) to the SPR sensor chip and applied CuMbn. No SPR response was observed for any concentration of CuMbn (SI Appendix, Fig. S9B). To probe the specificity of a given MbnT for its cognate CuMbn, we also cloned, expressed, and purified the MbnT from Mc. rosea SV97. Although Mc. rosea SV97 CuMbn has two nitrogen-containing heterocycles and two thioamide groups like Ms. trichosporium OB3b CuMbn, there is no N-terminal transamination or disulfide formation, the first oxazolone is replaced by a pyrazinedione, and a sulfonated threonine is present (SI Appendix, Fig. S1B) (14, 16). Interestingly, the addition of Ms. trichosporium OB3b CuMbn to Mc. rosea SV97 MbnT does elicit an SPR response (Fig. 3C), yielding a Kd value of 17.6 μM (Fig. 3D). These data suggest a complex mechanism for CuMbn recognition and binding, wherein MbnT binds only structurally similar substrates, and individual MbnTs may bind cognate CuMbns with higher affinity than nonnative CuMbns. Similar trends are observed for the ferric pyoverdine receptor FpvA from Pseudomonas aeruginosa ATCC 15692, which, unlike other Pseudomonas TBDTs, binds noncognate pyoverdines, albeit with different affinities (28).
Interaction of CuMbn with Purified MbnE.
To investigate whether MbnE might play a role in binding CuMbn in the periplasm, perhaps for delivery to periplasmic enzymes or, alternatively, to an inner membrane ABC transporter (37), we cloned, expressed, and purified Ms. trichosporium OB3b MbnE. Most Ms. trichosporium OB3b MbnE is expressed in inclusion bodies, but a small amount remained soluble and was purified via affinity chromatography. Size-exclusion chromatography revealed the protein to be a single oligomer (SI Appendix, Fig. S10), and circular dichroism spectroscopy suggests that it is mostly composed of α-helices and unordered loops (SI Appendix, Fig. S11).
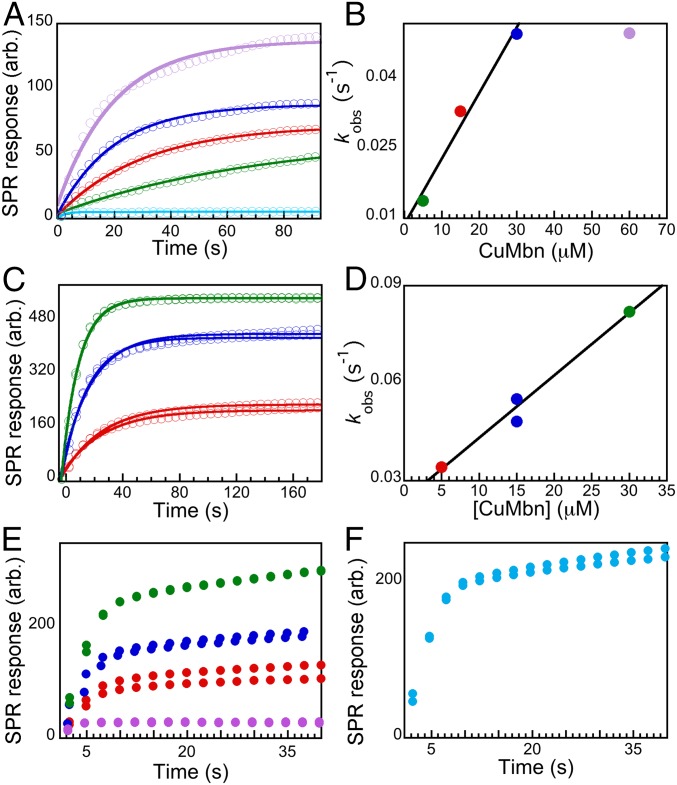
Binding of Ms. trichosporium OB3b CuMbn to its cognate purified MbnE was then tested via SPR. The application of CuMbn elicits an SPR response with kobs directly proportional to the concentration of CuMbn (Fig. 4A), up to 30 μM. At 60 μM CuMbn, the kobs diverges from linearity (Fig. 4B), perhaps due to inactivation that reduces the protein’s ability to bind substrate. The plot of kobs vs. [CuMbn] fits to a line that yields kon of 1.4 × 10−3 μM−1⋅s−1, koff of 8.5 × 10−3 s−1, and a Kd value of 6.07 μM. This value is within the range that has been observed for binding of oligopeptides to PBPs (Kd values of 0.3 nM–50 μM) (49, 50). The specificity of MbnE for CuMbn was investigated by adding cobalamin, ferric pyoverdine, and CuSO4 to the immobilized protein. In these experiments, we observed rapid association followed by an even faster dissociation (SI Appendix, Fig. S12A), indicating that these molecules do not bind to an appreciable extent. Therefore, MbnE, like MbnT, is specific for CuMbn and does not bind soluble Cu, siderophores, or metal chelates.
Fig. 4.
Binding of CuMbn to MbnEs. (A) SPR response curves (average of three injections) showing the binding of 0 μM (teal), 5 μM (green), 15 μM (red), 30 μM (blue), and 60 μM (purple) CuMbn to the MbnE from Ms. trichosporium OB3b. (B) Plot of kobs values versus the concentration of CuMbn fit to a line that yields a Kd value of 6.07 μM. (C) Binding of 5 μM (red), 15 μM (blue), and 30 μM (green) CuMbn isolated from Mc. hirsuta CSC1 to the MbnE from that organism. (D) A plot of kobs vs. concentration of CuMbn fit to a line that yields a Kd value of 12.1 μM for CuMbn binding. (E) SPR response curves showing the binding of 0 μL (pink), 5 μL (red), 15 μL (blue), and 30 μL (green) of Cu-supplemented spent medium isolated from Mc. parvus OBBP to the MbnE from that organism. (F) SPR response showing the binding a low-molecular-weight CuMbn candidate compound isolated from Mc. parvus OBBP to the MbnE from that organism.
To investigate whether MbnE recognizes and binds noncognate CuMbns, we cloned, expressed, and purified MbnEs from Mc. hirsuta CSC1 and Mc. parvus OBBP. The Mc. hirsuta CSC1 mbnE gene is found within the Mbn operon (SI Appendix, Fig. S2 and Dataset S6); there are two close nonoperon mbnE homologs in Mc. parvus OBBP, but the one used in this study, O5ODRAFT_02185, is more similar to group II mbnEs (SI Appendix, Fig. S2). Both MbnEs are monomeric (SI Appendix, Fig. S13), and the Mc. hirsuta CSC1 MbnE comprises primarily α-helices and unordered loops (SI Appendix, Fig. S14). Binding of cognate CuMbn to the Mc. hirsuta CSC1 MbnE was demonstrated by SPR experiments in which semipurified CuMbn (spent medium from the Cu-starved cultures supplemented with CuSO4 and purified on a Diaion HP-20 column) was applied to the immobilized protein. SPR responses were observed (Fig. 4C), and the rates and amplitudes of the response curves were directly proportional to the amount of semipurified CuMbn applied. Using the absorbance at 322 nm and the reported extinction coefficient at that wavelength (14), we estimate the amounts of applied CuMbn to be 5, 15, and 30 μM each. A plot of the kobs vs. these concentrations of CuMbn can be fit to a line with a kon of 1.9 × 10−3 μM−1⋅s−1 and koff of 2.3 × 10−2 s−1 determined, yielding a Kd of 12.1 μM (Fig. 4D). Like Ms. trichosporium OB3b MbnE, Mc. hirsuta CSC1 MbnE does not bind cobalamin, ferric pyoverdine, and CuSO4, but surprisingly, it also does not bind the semipurified CuMbn from Ms. trichosporium OB3b (SI Appendix, Fig. S12B). These data suggest that MbnEs may be less promiscuous than MbnTs in their ability to recognize and bind noncognate CuMbns.
We also investigated the interactions between Mc. parvus OBBP MbnE and CuMbn by SPR. There are two distinct Mbn operons and MbnEs in Mc. parvus OBBP (SI Appendix, Fig. S2). The first operon is a group I Mbn operon, containing a 16-amino-acid-long precursor core peptide. The second is a group IIb operon that has a shorter, eight-residue core peptide. There is at least one Cu-binding compound that can be isolated from the spent medium of Cu-starved Mc. parvus OBBP cells via a Diaion HP-20 column (SI Appendix, Fig. S15): a low molecular weight species that is likely to be the group IIb CuMbn. We obtained Diaion- and HPLC-purified (SI Appendix, Fig. S15A) samples of this low-molecular-weight compound in the presence and absence of Cu. It has an Mbn-like, Cu-responsive, UV-visible spectrum (SI Appendix, Fig. S15B), and a Cu-bound form of this species appears after Cu addition (SI Appendix, Fig. S15 C and D). A second higher-molecular-weight species was observed in some samples (SI Appendix, Fig. S15C), but low yields and apparent instability render it difficult to ascertain if this is the group I Mbn. Application of fractions containing the HPLC-purified Cu-binding compound to immobilized MbnE results in an SPR response, and the response is dependent on the amount of the samples added (Fig. 4E). Application of the semipurified CuMbn from Ms. trichosporium OB3b did not give an appreciable response, nor did cobalamin, ferric pyoverdine, and CuSO4 (SI Appendix, Fig. S12C). The fact that a response is also observed with the low-molecular-weight Cu-binding compound (Fig. 4F) strongly suggests that this compound binds specifically to MbnE. Thus, we have demonstrated specific interactions between three different MbnEs and their cognate CuMbns and no apparent cross-recognition by MbnE of CuMbns from different species.
Crystal Structure of Mc. parvus OBBP MbnE.
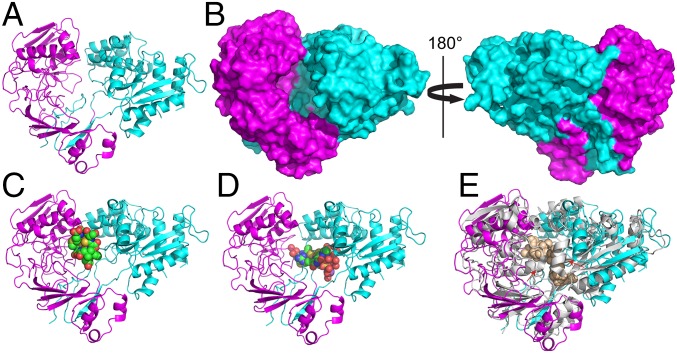
As a first step toward defining the molecular basis for CuMbn recognition by MbnE, we crystallized and determined the 1.9-Å resolution structure of Mc. parvus OBBP MbnE (SI Appendix, Table S2) in the absence of CuMbn. This apo structure (residues 35–609) reveals two major domains (Fig. 5A). The N-terminal domain includes residues 35–313, and the second domain includes residues 314–575. The C-terminal residues 576–609 form a β-strand and a loop that are part of the N-terminal domain. The domain–domain interface includes several extended loops that likely rearrange during the binding of substrate. There is a cavity at the domain–domain interface that measures ∼15 × 30 Å (Fig. 5B) with solvent access from one side only. To determine whether this cavity is large enough to accommodate CuMbn or CuMbn-like molecules, we used the GRAMMX server to generate docking models with the crystal structures of the structurally distinct CuMbns from Ms. trichosporium OB3b and Mc. sp. M. These models indicate that the interdomain cavity is indeed of sufficient size to bind either CuMbn (Fig. 5 C and D).
Fig. 5.
The 1.9-Å resolution crystal structure of Mc. parvus OBBP apo MbnE. (A) Cartoon representation with the two domains colored in magenta and cyan. (B) Surface representations of the structure show a cavity at the domain–domain interface that measures ∼15 Å across and 30 Å high. The cavity is open only to one side, consistent with it being the only entry point for the substrate. Docking models generated with the structures of CuMbns from Ms. trichosporium OB3b (C) and Mc. sp. M (D) reveal that the cavity can accommodate both forms of CuMbns. (E) Superposition of the Mc. parvus OBBP MbnE structure with that of nonapeptide-bound AppA from B. subtilis (PDB 1XOC). Major structural rearrangements include the ordering of a loop in the N-terminal domain and the movement of a α-helix closer to the domain–domain interface (red arrows).
A Dali search (51) with the apo Mc. parvus OBBP MbnE structure revealed several PBPs and SBPs structurally similar to Mc. parvus OBBP MbnE. Of particular interest is the structure of the SBP AppA from Bacillus subtilis (PBD 1XOC, rmsd 1.151 Å for 122 Cα atoms). AppA was copurified and crystallized with a nonapeptide (52). Surface representations of both structures reveal that MbnE is in an open conformation, whereas AppA is in a closed conformation with its nonapeptide substrate inaccessible to solvent (SI Appendix, Fig. S16). There are two major structural differences between the proteins: a loop in the N-terminal domain of the Mc. parvus MbnE is ordered into an α-helix in AppA, and an α-helix in the C-terminal domain is shifted significantly toward the cavity in AppA (Fig. 5E). These regions of AppA contact the bound nonapeptide and shield it from solvent, and this structural comparison provides significant insight into how MbnEs might bind CuMbn.
Conclusions.
Since the discovery of Mbns over a decade ago, these natural products have been postulated to play a central role in methanotroph Cu acquisition (19, 53, 54). Although uptake of intact CuMbn by methanotrophs has been demonstrated (24), the detailed molecular mechanisms of its recognition, import, and export have not been elucidated. Here we show that both the MbnT TBDT and the MbnE PBP encoded by Mbn operons are involved in Mbn transport. Disruption of the gene encoding MbnT in Ms. trichosporium OB3b impairs Mbn import, and expression of MbnT in E. coli confers the ability to import CuMbn. In addition, purified MbnT and MbnE interact with their cognate CuMbns with similar affinities. Notably, MbnT can recognize noncognate CuMbns whereas MbnE is not similarly promiscuous. The crystal structure of apo MbnE provides further insight into how MbnEs might bind their cognate CuMbns. These studies provide a framework for understanding not only Cu acquistion by methanotrophs, but also the transport of Mbn and related molecules by a range of bacteria.
Materials and Methods
Detailed procedures for bioinformatics analyses, qRT-PCR assay preparation, validation, interpretation, methanotroph growth and mutant generation, isolation of Mbns, and 65CuMbn uptake assays are included in the SI Appendix, SI Materials and Methods. Cloning, expression, and purification of MbnT and MbnE were accomplished by standard procedures. Also provided are protocols for SPR studies of CuMbn binding to MbnE and MbnT as well as details of MbnE crystallization and structure determination.
Supplementary Material
Acknowledgments
Funding for this work was provided by National Science Foundation Grant MCB0842366 (to A.C.R.) and National Institutes of Health Grant GM118035 (to A.C.R.) and Grant F32GM110934 (to L.M.K.D.). L.M.K.D. is the recipient of a Postdoctoral Enrichment Program grant from the Burroughs Wellcome Fund. G.E.K. was supported by American Heart Association Predoctoral Fellowship 14PRE20460104. Constructs of the Ms. trichosporium OB3b and Mc. parvus OBBP MbnEs were provided by Anthony S. Gizzi and Steven C. Almo (Albert Einstein College of Medicine), who are supported by the Price Family Foundation and New York Structural Genomics Research Center (National Institute of General Medical Sciences Grant U54-GM094663). Additional institutional and core facility acknowledgments are in the SI Appendix.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Methylocystis hirsuta CSC1 preliminary draft genome has been deposited and made publicly available in the Joint Genome Institute/Integrated Microbial Genomes & Microbiomes database (genome ID 2698536815), and the coordinates of apo Mc. parvus OBBP MbnE have been deposited in the Protein Data Bank (PDB ID code 5ICQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603578113/-/DCSupplemental.
References
- 1.Sirajuddin S, Rosenzweig AC. Enzymatic oxidation of methane. Biochemistry. 2015;54(14):2283–2294. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strong PJ, Xie S, Clarke WP. Methane as a resource: Can the methanotrophs add value? Environ Sci Technol. 2015;49(7):4001–4018. doi: 10.1021/es504242n. [DOI] [PubMed] [Google Scholar]
- 3.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366(6455):537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 4.Lipscomb JD. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48(1):371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 5.Sazinsky MH, Lippard SJ. Methane monooxygenase: Functionalizing methane at iron and copper. Met Ions Life Sci. 2015;15:205–256. doi: 10.1007/978-3-319-12415-5_6. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434(7030):177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 7.Balasubramanian R, et al. Oxidation of methane by a biological dicopper centre. Nature. 2010;465(7294):115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Culpepper MA, Rosenzweig AC. Architecture and active site of particulate methane monooxygenase. Crit Rev Biochem Mol Biol. 2012;47(6):483–492. doi: 10.3109/10409238.2012.697865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prior SD, Dalton H. The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath) J Gen Microbiol. 1985;131(1):155–163. [Google Scholar]
- 10.Murrell JC, McDonald IR, Gilbert B. Regulation of expression of methane monooxygenases by copper ions. Trends Microbiol. 2000;8(5):221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 11.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annu Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 12.Martinho M, et al. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus bath: Evidence for a Diiron center. J Am Chem Soc. 2007;129(51):15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34(4):496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 14.El Ghazouani A, et al. Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc Natl Acad Sci USA. 2012;109(22):8400–8404. doi: 10.1073/pnas.1112921109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiSpirito AA, et al. Copper-binding compounds from Methylosinus trichosporium OB3b. J Bacteriol. 1998;180(14):3606–3613. doi: 10.1128/jb.180.14.3606-3613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krentz BD, et al. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry. 2010;49(47):10117–10130. doi: 10.1021/bi1014375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney GE, Rosenzweig AC. Genome mining for methanobactins. BMC Biol. 2013;11(1):17–35. doi: 10.1186/1741-7007-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Téllez CM, Gaus KP, Graham DW, Arnold RG, Guzman RZ. Isolation of copper biochelates from Methylosinus trichosporium OB3b and soluble methane monooxygenase mutants. Appl Environ Microbiol. 1998;64(3):1115–1122. doi: 10.1128/aem.64.3.1115-1122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HJ, et al. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305(5690):1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 20.Behling LA, et al. NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J Am Chem Soc. 2008;130(38):12604–12605. doi: 10.1021/ja804747d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandow N, et al. Spectral and copper binding properties of methanobactin from the facultative methanotroph Methylocystis strain SB2. J Inorg Biochem. 2012;110:72–82. doi: 10.1016/j.jinorgbio.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Hakemian AS, et al. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds copper(I) J Am Chem Soc. 2005;127(49):17142–17143. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi DW, et al. Spectral, kinetic, and thermodynamic properties of Cu(I) and Cu(II) binding by methanobactin from Methylosinus trichosporium OB3b. Biochemistry. 2006;45(5):1442–1453. doi: 10.1021/bi051815t. [DOI] [PubMed] [Google Scholar]
- 24.Balasubramanian R, Kenney GE, Rosenzweig AC. Dual pathways for copper uptake by methanotrophic bacteria. J Biol Chem. 2011;286(43):37313–37319. doi: 10.1074/jbc.M111.284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: Regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schalk IJ, Yue WW, Buchanan SK. Recognition of iron-free siderophores by TonB-dependent iron transporters. Mol Microbiol. 2004;54(1):14–22. doi: 10.1111/j.1365-2958.2004.04241.x. [DOI] [PubMed] [Google Scholar]
- 27.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: Do we only see the ‘tip of the iceberg’? Trends Biochem Sci. 2008;33(7):330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Greenwald J, et al. FpvA bound to non-cognate pyoverdines: Molecular basis of siderophore recognition by an iron transporter. Mol Microbiol. 2009;72(5):1246–1259. doi: 10.1111/j.1365-2958.2009.06721.x. [DOI] [PubMed] [Google Scholar]
- 29.Gu W, et al. A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2016;82(6):1917–1923. doi: 10.1128/AEM.03884-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda T, Tsuchiya T. Multidrug efflux transporters in the MATE family. Biochim Biophys Acta. 2009;1794(5):763–768. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci. 2006;27(11):587–593. doi: 10.1016/j.tips.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Kenney GE, Sadek M, Rosenzweig AC. Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b. Metallomics. 2016;8(9):931–940. doi: 10.1039/c5mt00289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semrau JD, et al. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ Microbiol. 2013;15(11):3077–3086. doi: 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 34.Koebnik R. TonB-dependent trans-envelope signalling: The exception or the rule? Trends Microbiol. 2005;13(8):343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Braun V, Endriss F. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals. 2007;20(3-4):219–231. doi: 10.1007/s10534-006-9072-5. [DOI] [PubMed] [Google Scholar]
- 36.Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr Opin Microbiol. 2003;6(2):173–180. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 37.Linton KJ. Structure and function of ABC transporters. Physiology (Bethesda) 2007;22(2):122–130. doi: 10.1152/physiol.00046.2006. [DOI] [PubMed] [Google Scholar]
- 38.Wichelecki DJ, et al. ATP-binding cassette (ABC) transport system solute-binding protein-guided Identification of novel d-altritol and galactitol catabolic pathways in Agrobacterium tumefaciens C58. J Biol Chem. 2015;290(48):28963–28976. doi: 10.1074/jbc.M115.686857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berntsson RPA, Smits SHJ, Schmitt L, Slotboom D-J, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584(12):2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 40.Severinov K, Nair SK. Microcin C: Biosynthesis and mechanisms of bacterial resistance. Future Microbiol. 2012;7(2):281–289. doi: 10.2217/fmb.11.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein LY, et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J Bacteriol. 2010;192(24):6497–6498. doi: 10.1128/JB.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pattnaik P. Surface plasmon resonance: applications in understanding receptor-ligand interaction. Appl Biochem Biotechnol. 2005;126(2):79–92. doi: 10.1385/abab:126:2:079. [DOI] [PubMed] [Google Scholar]
- 43.James KJ, Hancock MA, Moreau V, Molina F, Coulton JW. TonB induces conformational changes in surface-exposed loops of FhuA, outer membrane receptor of Escherichia coli. Protein Sci. 2008;17(10):1679–1688. doi: 10.1110/ps.036244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Copeland RA. Protein–Ligand Binding Equilibria. Enzymes. John Wiley & Sons, Inc.; New York: 2002. pp. 76–108. [Google Scholar]
- 45.Newton SM, Igo JD, Scott DC, Klebba PE. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol Microbiol. 1999;32(6):1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 46.Locher KP, Rosenbusch JP. Oligomeric states and siderophore binding of the ligand-gated FhuA protein that forms channels across Escherichia coli outer membranes. Eur J Biochem. 1997;247(3):770–775. doi: 10.1111/j.1432-1033.1997.t01-1-00770.x. [DOI] [PubMed] [Google Scholar]
- 47.Kenley JS, Leighton M, Bradbeer C. Transport of vitamin B12 in Escherichia coli. Corrinoid specificity of the outer membrane receptor. J Biol Chem. 1978;253(5):1341–1346. [PubMed] [Google Scholar]
- 48.Clément E, Mesini PJ, Pattus F, Schalk IJ. The binding mechanism of pyoverdin with the outer membrane receptor FpvA in Pseudomonas aeruginosa is dependent on its iron-loaded status. Biochemistry. 2004;43(24):7954–7965. doi: 10.1021/bi049768c. [DOI] [PubMed] [Google Scholar]
- 49.Picon A, van Wely KHM. Peptide binding to the Bacillus subtilis oligopeptide-binding proteins OppA and AppA. Mol Biol Today. 2001;2(2):21–25. [Google Scholar]
- 50.Klepsch MM, et al. Escherichia coli peptide binding protein OppA has a preference for positively charged peptides. J Mol Biol. 2011;414(1):75–85. doi: 10.1016/j.jmb.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 51.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levdikov VM, et al. The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J Mol Biol. 2005;345(4):879–892. doi: 10.1016/j.jmb.2004.10.089. [DOI] [PubMed] [Google Scholar]
- 53.Balasubramanian R, Rosenzweig AC. Copper methanobactin: A molecule whose time has come. Curr Opin Chem Biol. 2008;12(2):245–249. doi: 10.1016/j.cbpa.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kenney GE, Rosenzweig AC. Chemistry and biology of the copper chelator methanobactin. ACS Chem Biol. 2012;7(2):260–268. doi: 10.1021/cb2003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.