Significance
Sea urchins (echinoids) consist of two subclasses, cidaroids and euechinoids. Research on gene regulatory networks (GRNs) in the early development of three euechinoids indicates that little appreciable change has occurred to their linkages since they diverged ∼90 million years ago (mya). I asked whether this conservation extends to all echinoids. I systematically analyzed the spatiotemporal expression and function of regulatory genes segregating euechinoid ectoderm and mesoderm in a cidaroid. I report marked divergence of GRN architecture in early embryonic specification of the oral–aboral axis in echinoids. Although I found evidence for diverged regulation of both mesodermal and ectodermal genes, comparative analyses indicated that, since these two clades diverged 268 mya, mesodermal GRNs have undergone significantly more alterations than ectodermal GRNs.
Keywords: gene regulatory network, echinoderms, sea urchin embryogenesis, embryonic axis specification, echinoids
Abstract
Developmental gene regulatory networks (GRNs) are assemblages of gene regulatory interactions that direct ontogeny of animal body plans. Studies of GRNs operating in the early development of euechinoid sea urchins have revealed that little appreciable change has occurred since their divergence ∼90 million years ago (mya). These observations suggest that strong conservation of GRN architecture was maintained in early development of the sea urchin lineage. Testing whether this holds for all sea urchins necessitates comparative analyses of echinoid taxa that diverged deeper in geological time. Recent studies highlighted extensive divergence of skeletogenic mesoderm specification in the sister clade of euechinoids, the cidaroids, suggesting that comparative analyses of cidaroid GRN architecture may confer a greater understanding of the evolutionary dynamics of developmental GRNs. Here I report spatiotemporal patterning of 55 regulatory genes and perturbation analyses of key regulatory genes involved in euechinoid oral–aboral patterning of nonskeletogenic mesodermal and ectodermal domains in early development of the cidaroid Eucidaris tribuloides. These results indicate that developmental GRNs directing mesodermal and ectodermal specification have undergone marked alterations since the divergence of cidaroids and euechinoids. Notably, statistical and clustering analyses of echinoid temporal gene expression datasets indicate that regulation of mesodermal genes has diverged more markedly than regulation of ectodermal genes. Although research on indirect-developing euechinoid sea urchins suggests strong conservation of GRN circuitry during early embryogenesis, this study indicates that since the divergence of cidaroids and euechinoids, developmental GRNs have undergone significant, cell type–biased alterations.
Integral to early development of a bilaterian is the development of the three embryonic tissue-layer domains: endoderm, ectoderm, and mesoderm. Asymmetrically distributed RNA and proteins in the egg provide the initial inputs into this process and thereby determine the spatial coordinates of domain formation (1, 2). In the context of these maternal factors, zygotic transcription of distinct sets of regulatory genes occurs in specific regions of the embryo, initiating the genomically encoded regulatory program and its output of regulatory genes. In this way, the embryo becomes populated by distinct sets of transcription factors and cell signaling molecules called regulatory states (3). Providing each cell with its molecularly distinct and functional identity, regulatory states are the spatial readout of developmental gene regulatory networks (GRNs) (4, 5).
Sea urchins (class Echinodea) have long served as model systems for studying the mechanisms of early development and, more recently, to study fundamental aspects of developmental GRNs. Mechanisms of early development can be readily studied in sea urchins owing to invariant cleavage patterns that give rise to early specification of blastomeres and cell lineages with relatively clear boundaries of embryonic domains (6, 7). Evolution and mechanisms of developmental programs also lend themselves to study in sea urchins, whose lineages have undergone multiple changes in life history strategies (8). Importantly, an excellent fossil record affords dating of evolutionary events (9), which has established that the sister subclasses of sea urchins—cidaroids and euechinoids—diverged at least 268 million years ago (mya) (10). Differences in the timing of developmental events in embryogenesis of cidaroids and euechinoids have long been a topic of interest, but have become the subject of molecular research only recently (11–18).
Research on the early development of the euechinoid purple sea urchin Strongylocentrotus purpuratus (Sp) has brought into high resolution the players and molecular logic directing developmental GRNs that specify Sp’s early embryonic domains (19–30). Abundant comparative evidence exists for other euechinoid taxa as well, including Lytechinus variegatus (Lv) (31–37) and Paracentrotus lividus (Pl) (38–42). Data from these indirect-developing euechinoids indicate that although these taxa diverged from one another ∼90 mya (9, 43), very little appreciable change to their developmental GRNs has accrued (44–46). Although there is evidence of minor alterations to these GRNs, such as a heterochronic shift in snail expression in Lv and Sp (22), numerous studies have made clear the striking conservation of GRN linkages in these lineages. Recent studies of early development of the distantly related, indirect-developing cidaroid sea urchin Eucidaris tribuloides (Et) have revealed that skeletogenic mesoderm specification in this clade is markedly different from that observed in euechinoids (17, 18, 47). The foregoing observations suggest that comparative analyses of GRN circuitry in early development of cidaroids and euechinoids have the potential to illuminate the tempo and mode of evolution of developmental GRNs.
In the present work, I coupled comparative spatiotemporal gene expression analyses with experimental manipulations of regulatory genes specifying euechinoid nonskeletogenic mesodermal (NSM) and ectodermal domains in the cidaroid Et to reveal how these GRNs have changed since the divergence of echinoids. This study focused on oral–aboral (O-A; or dorsal-ventral) patterning, which has consequences for both the ectoderm and mesoderm and is highly conserved in deuterostomes (46, 48, 49). I present evidence that deployment and interactions of regulatory genes specifying sea urchin ectoderm and mesoderm have diverged substantially in indirect-developing echinoids. Importantly, comparative data and analyses suggest that regulatory linkages occurring in ectodermal GRNs have undergone fewer alterations—and thus are less divergent–than those occurring among regulatory genes in mesodermal domains. The conclusions are supported by comparative spatiotemporal data, statistical analyses of timecourse gene expression data in three taxa of echinoids, and perturbation analyses.
Overall, regulatory genes expressed in ectodermal domains exhibited stronger signals of conservation relative to those expressed in mesodermal domains. These results suggest that embryonic domains and cell types in early development of sea urchins have evolved at different rates since the divergence of the two echinoid sister subclasses. Alterations to GRN architecture have occurred frequently throughout the network since their divergence. In addition, these results offer an in-principle explanation for the rapid changes in developmental processes during the convergent evolution of direct-developing, nonfeeding sea urchins (50–53).
Results
Conserved Deployment of Euechinoid Ectodermal Regulatory Genes in the Cidaroid Et.
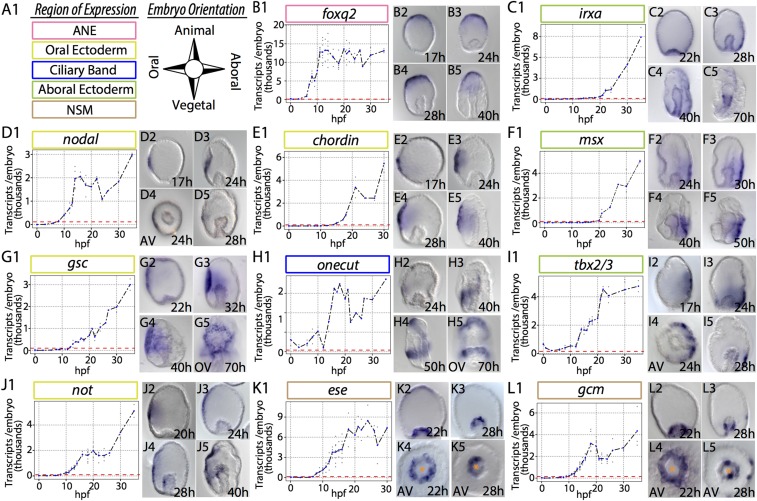
In euechinoids, the ectoderm is segregated into a diverse set of regulatory states defined by the future location of the stomodaeum (46, 54, 55). A critical factor in establishing O-A (47) polarity in sea urchins is Nodal (35, 38). Nodal ligand, via activation by SMAD signaling, is directly upstream of nodal, not, lefty, and chordin in oral ectoderm (OE) (40, 56). In Et, zygotic transcription of nodal, not, and lefty begins by early blastula stage (Fig. 1 D and J and SI Appendix, Fig. S1D). In contrast to euechinoids, transcriptional activation of chordin does not occur until 5 h after this cohort, indicative of an intermediate regulator between nodal and chordin in Et (Fig. 1 D and E). From 17 to 40 h postfertilization (hpf), spatial expression of nodal is observed in a well-defined region in OE that expands slightly as gastrulation proceeds (Fig. 1D, 2–5 and SI Appendix, Fig. S2).
Fig. 1.
Spatiotemporal dynamics of 11 euechinoid ectodermal and mesodermal regulatory genes in the cidaroid Et. mRNA transcripts were visualized by WMISH; mRNA transcript abundance was determined by qPCR. Individual data points are in gray. Blue data points represent the mean at that timepoint. Red dashed lines indicate the onset of zygotic transcription. Orange asterisks denote the position of the archenteron. (A) Key to embryonic domains of expression and orientation of embryos in micrographs. (B) Foxq2 is spatially restricted to ANE by 17 hpf. (C) By 40 hpf, irxa is seen extending anteriorly at the boundary of AE and OE. (D) Nodal is restricted to a few cells in OE up to early to mid-gastrula stage. (E) chordin expands from a few cells early and later from the perianal ectoderm to ANE. (F) msx is expressed exclusively in lateral AE. (G) By 22 hpf, gsc is detected in a broad region surrounding OE and later is observed near the stomodaeum. (H) By 40 hpf, onecut is detected in the future post-oral CB and is initiated in a band moving from the posterior to the anterior. (I) tbx2/3 at 17 hpf is detected broadly in AE and later extends from the perianal ectoderm to the lateral AE. (J) Not is first detected in OE and subsequently expands by 28 hpf to the oral side of the archenteron. (K) By 28 hpf, ese is observed at the tip of the archenteron and is asymmetrically polarized. (L) As gastrulation begins, gcm is expressed broadly in NSM; by 28 hpf, it is restricted to one side of the archenteron and is observed in a few ectodermal cells.
The spatial distributions of nodal and its targets are not restricted solely to a small field of cells in OE. Lefty (also known as Antivin), an antagonist of Nodal, exhibits a broader pattern of expression that, by 50 hpf, expands into the oral side of the archenteron (SI Appendix, Figs. S1D, 2–5 and S2). Similarly, chordin transcripts expand in OE throughout embryogenesis (Fig. 1E, 2–5 and SI Appendix, Fig. S2). The homeobox gene Not, which is known to play a role directly downstream of nodal in euechinoid O-A ectoderm and mesoderm polarity (27, 57), is detected in OE during gastrulation and by 28 hpf is observed in the mesoderm of the archenteron (Fig. 1J, 2–5).
In euechinoids, Bmp2/4 ligand is directly downstream of Nodal and is translocated across the embryo to the aboral side, where it stimulates aboral ectoderm (AE) regulatory genes, such as irxa, msx, and tbx2/3 (38, 58, 59). In Et, tbx2/3 exhibits spatial distribution complementary to OE genes in lateral AE (Fig. 1I, 2–5). By midgastrula stage, tbx2/3 is detected in the archenteron and much later, by 70 hpf, appears in skeletogenic bilateral clusters (SI Appendix, Fig. S2). This pattern is similar to that seen in two euechinoids with notable heterochronic differences (60, 61). Later in development, another Bmp2/4- responsive factor in AE, msx, is spatially distributed similar to tbx2/3 (Fig. 1F, 1–5). The considerable delay between the zygotic activation of msx and its candidate euechinoid upstream genes, including tbx2/3, which is expressed in an overlapping domain, suggest that the regulation of msx in Et may be indirectly under the control of Bmp2/4 and Tbx2/3.
Finally, the Forkhead family transcription factor Foxq2 is sequentially restricted to and specifically expressed in embryonic anterior neural ectoderm (ANE) in deuterostomes (62). In euechinoids, Foxq2 sets the anterior boundary of OE by restricting the transcription of nodal in ANE (23, 55). In Et, foxq2 transcripts exhibit an expression pattern consistent with observations in euechinoids and other deuterostomes, suggesting conserved roles for this gene in ANE and O-A specification (Fig. 1B and SI Appendix, Fig. S2).
Conserved Spatiotemporal Deployment of Ciliated Band Regulatory Genes.
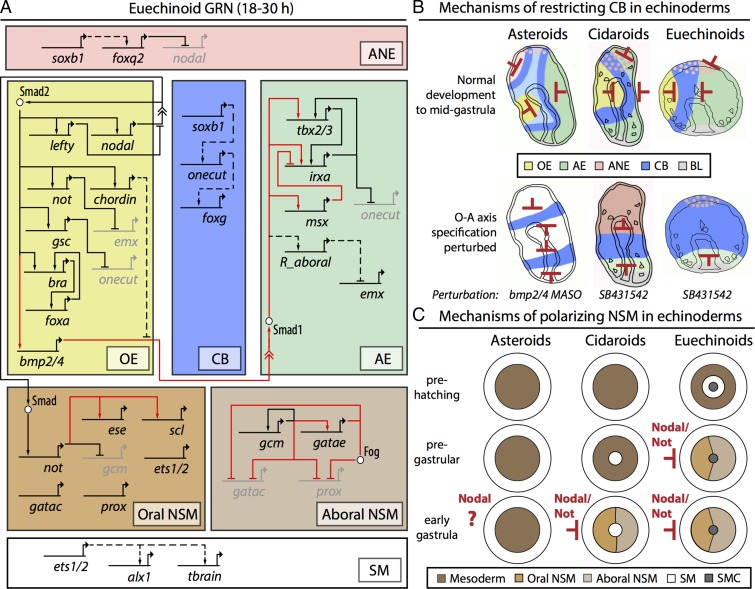
Free-feeding, indirect-developing sea urchins possess a single neurogenic ciliated band (CB) early in development that circumnavigates the larval oral face and facilitates feeding and locomotion (63). This structure has undergone frequent modification in the lineages leading to modern sea urchins (64). In euechinoids, Goosecoid (gsc), Onecut, and Irxa contribute to the geometric patterning of CB formation (28, 40, 65). In euechinoids, gsc is expressed in OE and is directly downstream of Nodal signaling (40), onecut (also known as hnf6) is a maternal factor in euechinoids that is later restricted to a region at the boundaries of OE and AE where progenitor CB territory forms, and irxa is expressed exclusively in AE downstream of Bmp2/4 and Tbx2/3 (40, 66). In the cidaroid Et, gsc is also spatially restricted to OE (Fig. 1G, 1–5). As in euechinoids, onecut is maternally deposited (Fig. 1H, 1), and although its early spatial expression was not obtained, staining was observed in a restricted band of cells encircling the OE by midgastrula stage (Fig. 1H, 4). Spatial deployment of onecut in Et is notable insofar as its spatial distribution in progenitor CB begins in the future post-oral CB and subsequently extends in a narrow band of 4–8 cell diameters toward progenitor pre-oral CB (Fig. 1H, 2–5). This observation starkly contrasts with that in euechinoids, in which onecut is observed ubiquitously and later delimited as a whole to the CB territory by transcriptional repressors in the OE and AE (28, 67). Irxa initiates zygotic expression at midblastula stage (∼14 hpf) in Et, and by 28 hpf is observed broadly in AE (Fig. 1C, 1–5). Unlike in euechinoids, irxa is broadly distributed in Et AE—much more so than tbx2/3—indicating that it is broadly activated in the ectoderm and repressed in OE and ANE. These spatiotemporal observations of key CB regulatory genes suggest that CB patterning mechanisms are likely conserved in echinoids.
NSM in Et Is Polarized by Early Gastrula Stage.
NSM in euechinoids arises at the vegetal pole from early cleavage endomesodermal precursors and gives rise to at least four different cell types (68). Euechinoids rely completely on the presentation of Delta ligand in the adjacent SM to up-regulate NSM regulatory genes in veg2 endomesoderm (32, 69, 70). NSM regulatory genes are subsequently segregated into aboral NSM and oral NSM as a result of repression by Not via Nodal/SMAD in OE (27, 71). The first evidence of NSM polarity in Et is seen in the spatial distribution of ese and gcm transcripts at 4–6 h after the start of gastrulation (Fig. 1 K, 2–5 and L, 2–5). In euechinoids, ese is expressed in oral NSM and gcm is expressed in aboral NSM. In Et, transcription of ese in progenitor NSM commences just before the onset of gastrulation and shortly after archenteron invagination is subsequently restricted to one side of the archenteron (Fig. 1K, 5). Similarly, gcm is expressed transiently in oral and aboral NSM and by 28 hpf is restricted to a cluster of cells just below the tip of the archenteron (Fig. 1L, 3 and 5). Later this expression is seen solely on one side of the archenteron as gcm-positive cells ingress rapidly into the blastocoel at 36 hpf (SI Appendix, Fig. S4). Double-fluorescent whole-mount in situ hybridization (WMISH) revealed that ese and gcm in Et are restricted to opposite sides of the archenteron (SI Appendix, Fig. S4G, 1 and 2), providing the first indication that NSM polarity may be conserved in echinoids.
Nodal signaling in OE plays an important role in segregating NSM regulatory states. To further clarify the extent to which NSM polarity in Et is consistent with euechinoid regulatory states, I conducted additional single- and double-fluorescent WMISH on NSM regulatory genes ets1/2, gatac, gatae, prox, scl, and tbrain. Directly downstream of Gcm in euechinoids is gatae (27). In Sp, gatae is observed in endomesoderm by blastula stage (72). In Et NSM, gatae is expressed throughout the endomesoderm at the time of SM ingression (∼28 hpf), and later is observed restricted to one side near the tip of the archenteron, as well as in the second wave of ingressing mesenchyme (SI Appendix, Figs. S1C, 2–5 and S4). Gatac (gata1/2/3), prox, and scl, all of which are euechinoid oral NSM genes downstream of Gcm repression on the aboral side (27), come off the baseline at similar times in Et and are detected in a few cells in the vegetal pole by 18 hpf (SI Appendix, Figs. S1C, E, and F and S4 B, E, and F). Of these three genes, scl was the first to show O-A NSM polarity, followed shortly by gatac (SI Appendix, Fig. S1 B, 5, E, 5, and F, 5). Surprisingly, by 36 hpf, prox did not exhibit O-A polarity in NSM (SI Appendix, Figs. S1E, 5 and S4E, 6), suggesting that prox is a general mesodermal regulatory factor in Et.
The foregoing observations suggest that NSM polarity is highly divergent in cidaroids and euechinoids. Sequential unfolding of NSM regulatory states in Et has diverged, although evidence presented here indicates that gcm and ese are spatially distributed in a similar fashion to that in euechinoids. In the Et archenteron, previously reported observations suggested that the euechinoid SM-specific regulatory genes ets1/2 and tbrain are distributed broadly in the mesoderm (17). I confirmed that result by double-fluorescent WMISH (SI Appendix, Fig. S4I) and here further clarify the numerous regulatory states at the tip of the Et archenteron. Within this broad ets1/2-tbrain domain lie three regulatory states (SI Appendix, Fig. S4 G–I): (i) orally localized ets1/2, tbrain, and ese; (ii) aborally localized ets1/2, tbrain, and gcm; and (iii) an anteriorly localized micromere-descendant regulatory state at the tip of the archenteron of ets1/2, tbrain, ese, and alx1. The NSM regulatory states described here indicate that since the echinoid divergence there have been numerous changes to spatiotemporal gene expression in echinoid NSM cell types. The spatial dynamics of NSM regulatory factors, coupled with the presence of euechinoid OE regulatory genes in the archenteron, suggest that Et NSM is likely downstream of the Nodal cascade.
Nodal Signaling Is a Highly Conserved Mechanism Patterning Echinoid O-A Ectoderm and Mesoderm.
I next sought to experimentally perturb O-A specification in Et to determine the extent to which this developmental program is conserved in echinoids. In euechinoids, perturbation of animal-vegetal axis polarity by disruption of nuclearization of β-catenin indirectly disrupts O-A axis specification (31, 33). One mechanism underlying the crosstalk of these two deuterostome specification events is the restriction of foxq2 to ANE, with its presence in OE blocking nodal transcription (23). In Et, disruption of nuclearization of β-catenin at the vegetal pole by overexpression of dn-cadherin RNA (MOE) leads to up-regulation of foxq2 and strong down-regulation of nodal and its euechinoid downstream components nodal, not, and tbx2/3 (SI Appendix, Fig. S5A). This result suggests that the molecular crosstalk among β-catenin/TCF, foxq2, and nodal is conserved in echinoids.
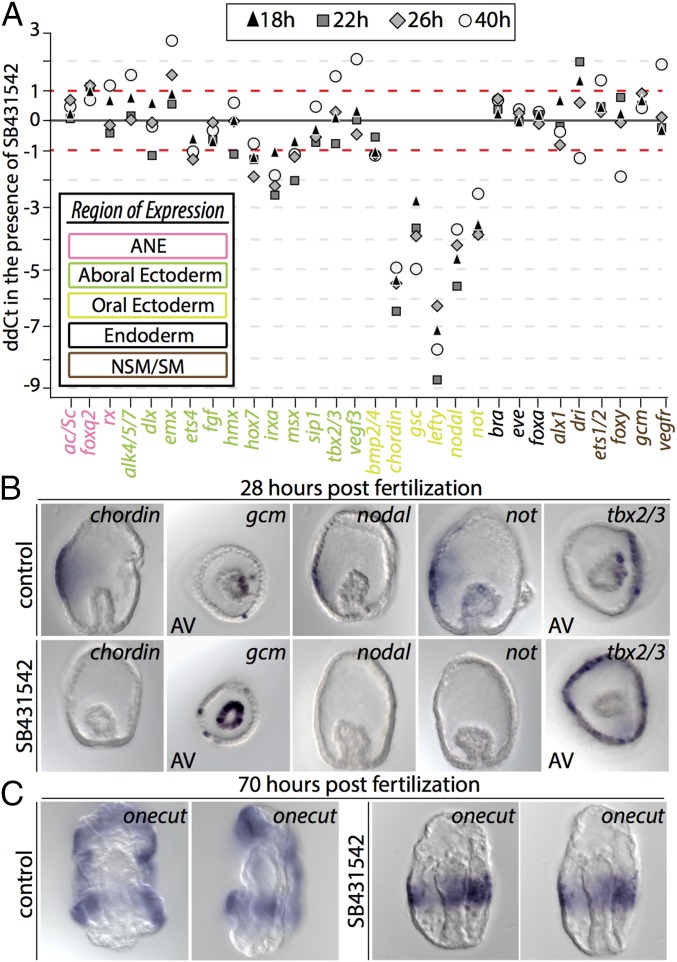
I next aimed to determine the spatiotemporal effects of perturbation of O-A specification by culturing Et embryos in the presence of SB431542, a small-molecule antagonist of the TGF-β (Nodal) receptor Alk4/5/7 (73). At 4 days postfertilization, these embryos exhibited aboralization, archenterons not contacting OE, and supernumerary skeletal elements (SI Appendix, Fig. S5B). Quantitative PCR (qPCR) analyses at four different time points in Et development showed strong down-regulation of OE regulatory factors chordin, gsc, lefty, nodal, and not (Fig. 2A). This result was confirmed spatially by WMISH of chordin, nodal, and not (Fig. 2B and SI Appendix, Fig. S5C). Despite early temporal onset with the Nodal regulatory cohort, the secreted TGF-β ligand bmp2/4 was only modestly affected by SB431542 treatment and cadherin MOE (Fig. 2A and SI Appendix, Fig. S5A). This result is strikingly different from the strong down-regulation of bmp2/4 observed in the euechinoid Pl when it was cultured in the presence of SB431542 or injected with Nodal morpholino (MASO) (38, 40), indicating that regulation of bmp2/4, as well as of its downstream components, may be under alternative control in Et.
Fig. 2.
Perturbation of O-A axis formation in Et reveals alterations to regulatory factor deployment in echinoids. (A) Quantitative effect of SB431542 on expression of 30 Et regulatory genes as revealed by qPCR. Fold change in mRNA transcripts (ddCt) is shown on the y-axis. Two timepoints from two independent replicates are shown. Regulatory factors are listed on the x-axis, and font color designates their embryonic domain: pink, ANE; green, AE; yellow, OE; black, endoderm; brown, mesoderm. (B) WMISH of select ectodermal and mesodermal regulatory genes in embryos cultured in the presence of SB431542. At 28 hpf, expression of chordin, nodal, and not are completely extinguished. Gcm is expressed ectopically throughout the archenteron. In the ectoderm, tbx2/3 expands ectopically into oral ectoderm. (C) The CB marker onecut is normally observed at the boundaries of OE and AE; however, in the presence of SB431542, onecut is expressed ectopically in an equatorial band.
In euechinoids, msx and tbx2/3 are downstream of Bmp2/4 ligand, which diffuses from OE to AE (58, 59, 74). Treatment of Pl embryos with SB431542 inhibitor completely and specifically extinguishes both msx and tbx2/3 in AE, although the latter is still expressed in SM (40). In Et, qPCR data suggest that SB431542 inhibitor has a moderate negative affect on msx and no effect on tbx2/3 regulation (Fig. 2A). However, WMISH of tbx2/3 in the presence of the inhibitor showed the expansion of its domain of expression into an equatorial ring, much like the expansion exhibited by irxa in Pl in a SB431542 background (Fig. 2B and SI Appendix, Fig. S5C). Similarly, whereas in Et qPCR data indicated down-regulation of the AE regulatory factor irxa (Fig. 2A), in Pl its domain of expression expands into OE on SB431542 treatment (40). In Et, however, irxa is down-regulated in the presence of SB431542, providing further evidence suggesting that the GRN circuitry downstream of Nodal has undergone alteration. Taken together, the foregoing results suggest that GRN architecture immediately downstream of the initial nodal-bmp2/4 circuitry has diverged in echinoids and indicate divergence of regulatory linkages immediately upstream of bmp2/4, irxa, msx, and tbx2/3 in echinoids.
In euechinoid CB, perturbation of Nodal signaling results in an expansion of onecut throughout the ectoderm, leading to the suggestion that the default fate of ectoderm is a proneural CB territory (40). When this perturbation was carried out in Et, a very different result was obtained, with onecut restricted to a single equatorial band of 6–10 cell diameters (Fig. 2C). A previous study showed that disruption of Et endomesoderm formation by treatment with zinc results in embryos exhibiting a ring of highly concentrated proneural synaptotagmin-B–positive cells at the equator of the embryo (75). Interestingly, this result is similar to that seen in the sea star Patiria miniata, in which CB-specific foxg, immediately downstream of Onecut in euechinoids, is observed on Bmp2/4 perturbation in an equatorial band that later becomes two (76). These observations indicate that the ancestral state of CB patterning in eleutherozoans is anterior repression of CB. In the absence of Nodal signaling, these taxa show a CB domain repressed from both the anterior and the posterior, which for asteroids later becomes two bands by activity of a laterally positioned repressor. Therefore, it is likely that this anteriorly positioned repression mechanism, which is revealed on disruption of the Nodal signal, has been lost in euechinoids.
Given the presence of transcripts of Nodal responsive regulatory genes in the archenteron shortly after gastrulation, I suspected that Nodal signaling likely plays a role in establishing polarity in Et NSM, as it does in euechinoids studied thus far via the transcription factor Not (27, 47, 71). In Et, qPCR data did not indicate consistent differences in mRNA abundance for NSM regulatory genes (Fig. 2A); however, WMISH assays revealed that embryos treated with SB431542 failed to restrict gcm to the aboral side (Fig. 2B and SI Appendix, Fig. S5C). This finding is consistent with the euechinoid GRN linkage immediately downstream of Nodal signaling via the transcription factor Not, which represses aboral NSM in the oral-facing region of the archenteron (27). Indeed, in Et, not is seen on one side of the archenteron throughout gastrulation (Fig. 1J, 3–5). These observations are consistent with a conserved role for Nodal signaling via Not in establishing polarity in NSM cell types in the archenteron of Et.
Regulatory Linkages Restricting the CB Are Conserved in Echinoids.
The spatial distributions of gsc, onecut, and irxa are highly suggestive of a conserved echinoid regulatory apparatus that spatially restricts CB to the boundary of OE and AE (Fig. 1 and SI Appendix, Fig. S3). I introduced by microinjection into Et mutated and wild-type BACs harboring GFP and encoding the cis-regulatory region of Sp onecut. In Sp, onecut is restricted to the progenitor CB territory by Gsc repression in OE and Irxa repression in AE (Fig. 3A). Recently, Barsi and Davidson (65) experimentally validated four cis-regulatory modules (CRMs) directing the activation of onecut by Soxb1 and the repression of onecut by Gsc in OE and Irxa in AE (Fig. 3B). Remarkably, microinjection of the engineered BACs into Et faithfully recapitulated important regulatory transactions observed in Sp (Fig. 3 C–H and SI Appendix, Fig. S7). For instance, BAC reporter 5 recapitulates endogenous Sp onecut expression, and does so in Et as well, where it is seen in numerous cells of CB (Fig. 3H and SI Appendix, Fig. S7 A and B). BAC reporters with mutated activation of cis-regulatory modules (BACs 1, 2, and 3) exhibited either no expression or GFP in a only few CB cells (Fig. 3 D, E, and G). Surprisingly, a BAC harboring mutated repressor sites for OE repressor Gsc and AE repressor Irxa repeatedly exhibited ectopic expression in OE of Et and elevated GFP reporter in AE (Fig. 3 C and E). These results suggest the existence of conserved GRN circuitry guiding the restriction of onecut to CB territory by Irxa and Gsc in echinoids and provide a striking example of kernel-like subcircuit conservation of a developmental program after 268 million years of evolution.
Fig. 3.
Expression in Et of engineered reporter BACs harboring the regulatory locus of Sp onecut. (A) Schematic of CB restriction in the Sp embryo. Red bars indicate repressor genes restricting CB genes in those domains. (B) Schematic showing the Sp onecut locus and the CRMs responsible for its geometric positioning. (C) Results of analysis of reporter expression of five Sp onecut BACs in Et. The BACs were previously used to analyze the cis-regulatory dynamics of onecut spatial control in Sp (65). (D) BAC reporter 1 harbors mutations to all known CRMs and shows markedly reduced reporter expression. (E) BAC reporter 2 harbors mutations to a zygotic activation CRM. (F) BAC reporter 4 harbors mutations to the AE and OE repression CRMs, is ectopically expressed in Et in OE and AE, and exhibits regular reporter expression in CB. (G) BAC reporter 3 harbors mutated postactivation enhancer CRMs and shows markedly reduced reporter expression. (H) BAC Reporter 5 harbors the unperturbed locus of Sp onecut and repeatedly exhibits reporter GFP in CB of Et.
Echinoid Ectodermal and Mesodermal GRNs Have Diverged at Different Rates.
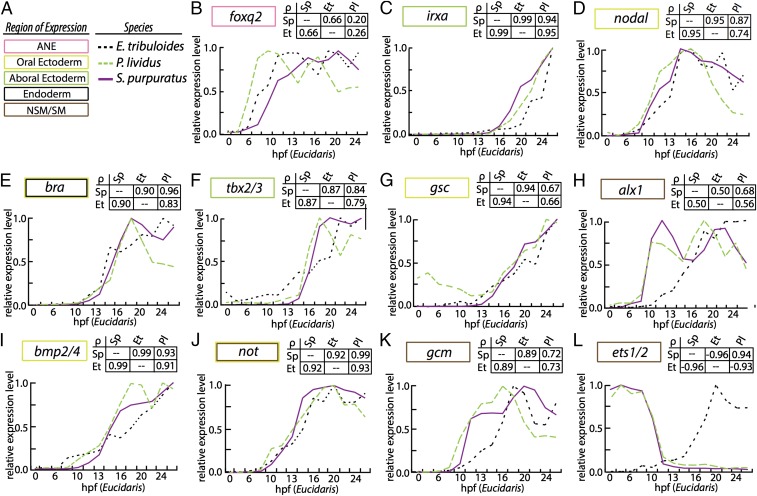
The spatiotemporal data presented thus far are highly suggestive of the hypothesis that O-A axis specification, as well as gastrular CB formation, in Et is consistent with similar processes in euechinoids, and that NSM specification has ostensibly diverged. I sought quantitative analyses that would test this hypothesis and used a comparative gene expression approach to reveal the extent of divergence among these cell types in echinoid clades. I compared the absolute mRNA transcript abundance of 18 regulatory genes in three species in early euechinoid developmental GRNs for 14 early developmental timepoints in Et, and also performed pairwise comparisons of 55 regulatory genes in Sp and Et (77). I scaled timepoints from Et to the euechinoid Pl because both develop at a similar temperature, and scaling of developmental stages between Pl and Sp has been established (SI Appendix, Table S1) (78). I performed pairwise analyses of Spearman’s rank correlation coefficient, ρ, among the three species (SI Appendix, Table S2). Plots of relative mRNA transcriptional dynamics indicate strong correlations for ectodermal regulatory factors and poor correlations for regulatory factors involved in mesoderm specification (Fig. 4 and SI Appendix, Fig. S6).
Fig. 4.
Comparative gene expression analyses of echinoid regulatory orthologs. (A) Domain of expression is indicated by a colored box. Species is indicated by purple line (Sp); green dashed line (Pl); or black dashed line (Et). (B) foxq2. (C) irxa. (D) nodal. (E) bra. (F) tbx2/3. (G) gsc. (H) alx1. (I) bmp2/4. (J) not. (K) gcm. (L) ets1/2. Transcripts per embryo were normalized to its maximal expression over the first 30 h of development and are plotted against Et development on the x-ordinate. Comparative developmental staging for each species is listed in SI Appendix, Table S1. Each analysis is accompanied by a matrix of Spearman correlation coefficients (indicated as ρ).
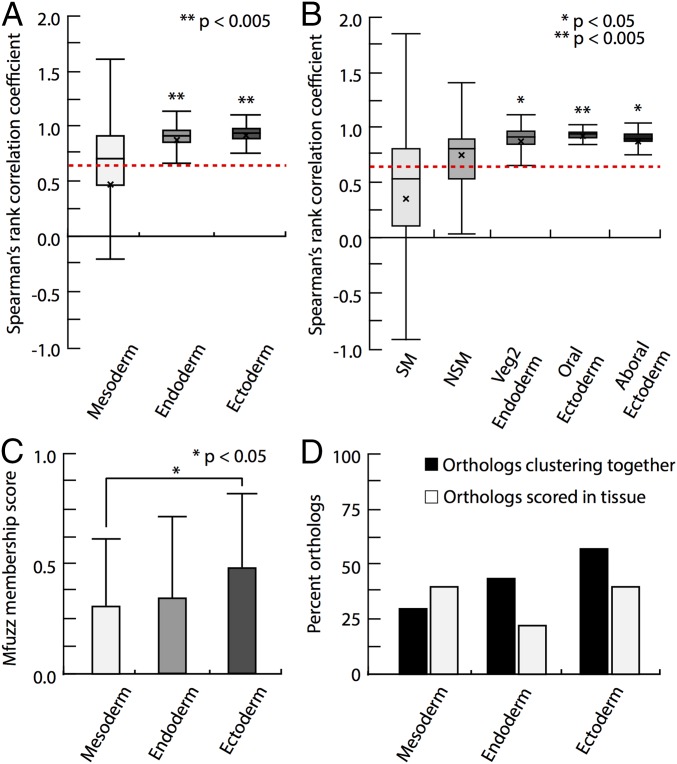
I next considered time course expression data of 55 orthologs of Et and Sp. I binned the orthologs into ectoderm, endoderm, and mesoderm based on their spatial distributions in Sp and compared them against the mean of all ρ values. Regulatory orthologs expressed in ectoderm showed significantly higher ρ values relative to the mean of all ρ values, suggesting strongly conserved transcriptional dynamics in echinoids (Fig. 5A). In contrast, regulatory orthologs expressed in mesodermal cell lineages did not depart significantly from mean ρ, suggesting that the dynamics of mesodermal regulatory factors have changed markedly since the echinoid divergence (Fig. 5 A and B).
Fig. 5.
Statistical analyses of comparative time course data in the euechinoid Sp and the cidaroid Et. (A and B) Distribution plots of Spearman rank correlation coefficients (ρ) of Et and Sp orthologs that were binned by embryonic domain of spatial expression. Boxplot boundaries show interquartile range, mean, and SD. Asterisks mark the statistical significance of a two-tailed t test. (A) Boxplots for statistical distribution of endodermal, ectodermal, and mesodermal regulatory factors in Et and Sp. The mean ρ values for endodermal and ectodermal regulatory factors were significantly higher than the mean ρ value of all orthologs, denoted by a red dashed line. This was not true for mesodermal regulatory factors. (B) Boxplots for the statistical distribution of ectodermal and mesodermal subdomains. (C) Mean membership scores for binned orthologs derived from mfuzz cluster analysis. Et ectodermal orthologs exhibited higher membership scores than mesodermal orthologs (P < 0.05, Mann–Whitney U test). (D) Histogram showing both the distribution of tissue-specific orthologs examined in this study and the percentage of Sp and Et orthologs clustering together.
I conducted comparative clustering analyses to identify statistical differences in timecourse data (79, 80). By clustering Sp regulatory orthologs into six distinct clusters and forcing assignment of each Et ortholog into all clusters, cluster membership scores and clustering similarities for each ortholog were acquired (SI Appendix, Tables S3–S5). Considering Sp orthologs alone revealed regulatory genes clustering together based on their expression maternally, early, or late in development (SI Appendix, Fig. S8). Et orthologs expressed in ectodermal domains showed higher cluster membership scores than orthologs expressed in mesodermal domains (P < 0.05, Mann–Whitney U test) (Fig. 5C). In addition, ectodermal orthologs jumped less frequently between clusters than mesodermal orthologs, suggesting that the time course data for ectodermal genes are more similar than that for mesodermal genes (Fig. 5D and SI Appendix, Fig. S8C). These analyses suggest that deployment and transcriptional dynamics of ectodermal regulatory genes are more similar between euechinoids and cidaroids compared with mesodermal regulatory genes.
Taken together, the foregoing data provide quantitative support for the notion that regulatory linkages occurring in mesodermal developmental GRNs have diverged to a greater degree than those occurring in ectodermal GRNs. These data provide further support for the idea that GRN circuitry evolves at different rates based on the developmental regulatory demands of the network that it inhabits (81).
Discussion
Extensive Divergence of Ectodermal and Mesodermal Regulatory Linkages Has Occurred in Echinoid Developmental GRNs.
In this study, embryonic pattern formation and regulatory interactions of the euechinoid ectodermal and mesodermal GRNs were investigated in a distantly related cidaroid that last shared a common ancestor with euechinoids in the Paleozoic 268 mya (Fig. 6). In contrast to the striking conservation of GRN circuitry functioning in the early development of camaradont euechinoids, numerous regulatory interactions were found to have diverged in these two clades (Fig. 6A). Statistical analyses of comparative time course gene expression data revealed that cidaroid regulatory orthologs expressed in ectodermal cell lineages are more similar to their euechinoid counterparts than those expressed in mesodermal cell lineages. One caveat to these findings is that correlative time course analyses do not necessarily indicate conserved GRN wiring, owing to the pleiotropic nature of gene regulation by transcription factors; nonetheless, they do provide reasonable hypotheses for GRN wiring diagrams. Moreover, these statistical analyses are consistent with the broad differences observed in Et in relation to SM specification, as well as that revealed here in NSM segregation and regulatory states. By comparing these observations with those in other echinoderms, we can begin to appreciate the degree to which embryonic developmental GRNs are constrained or altered over vast evolutionary distances, and can reconstruct the ancestral regulatory states that must have existed in the embryos of echinoderm ancestors (47).
Fig. 6.
Divergent ectodermal and mesodermal GRN architecture in the early development of echinoids. (A) Truncated Sp GRN showing ectodermal and mesodermal regulatory interactions investigated in this study. The solid black lines indicate conserved interactions, the dashed black lines indicate interactions that are likely conserved, and the solid red lines indicate divergent regulatory interactions. Embryonic domains are represented by boxes: pink, ANE; yellow, OE; blue, CB; green, AE; brown, oral NSM; light brown, aboral NSM; white, SM. (B) Restriction of ectodermal CB in three echinoderm taxa. Red bars indicate the presence of gene restriction. Color scheme is the same as in A. The top row of the schema represents the unperturbed state; the bottom row, alternative patterning in perturbation background. (C) Mechanisms of polarization of NSM in the same echinoderm taxa. In both cidaroids and euechinoids, Nodal signaling up-regulates Not, which segregates O-A NSM cell types by restricting genes to the aboral side. Whereas asteroids show no indication of NSM O-A polarity, cidaroid and euechinoid lineages have undergone heterochronic shifts since their divergence.
Divergent Regulatory Linkages in the Evolution of Echinoid Ectodermal and Mesodermal GRNs.
These analyses used the regulatory circuitry of the well-known Sp GRN as a basis for comparisons of the cidaroid Et and euechinoids (Fig. 6A). The comparative approach, in combination with the well-documented fossil record of echinoids, allowed approximation of the magnitude of change incurred by these GRNs since these two clades last shared a common ancestor. Spatiotemporal expression patterns of ectodermal regulatory genes in Et and euechinoids strongly suggest that alteration to this circuitry is nontrivial in early development relative to the circuitry directing the development of mesodermal lineages. However, whereas numerous regulatory linkages are conserved in the ectodermal GRN, deployment and rewiring of circuitry have occurred frequently during the evolution of euechinoid lineages that have direct-developing, nonfeeding larvae (52, 82–84). Moreover, these data indicate that the ectodermal GRN has undergone at least one significant alteration since the cidaroid–euechinoid divergence, viz., to bmp2/4, msx, tbx2/3, and irxa.
These observations support the idea that constraints on alterations of GRN circuitry are not equally weighted throughout the GRN over vast evolutionary timescales. For instance, perturbation of Nodal signaling revealed that although initial specification events are highly similar, alterations to the regulation of bmp2/4 and tbx2/3 likely have occurred. In Et, tbx2/3 is expressed in AE and aboral NSM by mid-gastrula. By late gastrula, it is expressed in the lateral clusters of skeletogenic synthesis, at the tip of the gut, in the gut endoderm, and residually in the ectoderm. This unfolding pattern of tbx2/3 expression in Et has essentially been compressed into the early stages of euechinoid development (60). In Pl, Nodal perturbation with SB431542 extinguishes tbx2/3 specifically in AE while not affecting its expression in SM (40). In Et, expansion of tbx2/3 into OE was observed on perturbation with SB431542 (Fig. 2B and SI Appendix, Fig. S5C). These findings suggest altered GRN circuitry downstream of Nodal at the bmp2/4 GRN node.
Evolution of CB Patterning Mechanisms in Echinoderms.
CB formation and ANE patterning in Et are evolutionarily interesting given that cidaroids lack the pan-deuterostome apical senory organ (12, 13, 85, 86). Here these data provide evidence for a GRN kernel operating in echinoid CB patterning. Such striking conservation is also seen in a subcircuit kernel that runs in both asteroid and euechinoid early development (87). These observations indicate that the CRMs patterning CB in echinoids are conserved over a vast evolutionary distance. These results add significantly to previous studies in Et that suggest that CB and ANE patterning are more similar to outgroup echinoderms than to euechinoids (75). Here patterning and regulatory interactions of CB were observed that are consistent with the hypothesis that this process is conserved in echinoids and markedly consistent with asteroid CB and ANE patterning despite the fact that asteroids exhibit two CBs (Fig. 6B) (88). In addition, ANE patterning is consistent with a pan-bilaterian observation of sequential spatial restriction of foxq2 to the anterior end by endomesodermal wnt factors (23, 89–92), suggesting that specification of the apical sensory organ in Et has been developmentally uncoupled from these events, and also that the loss of this embryonic structure has had little effect on the conserved patterning of CB and anterior localization of foxq2. Furthermore, CB and ANE patterning in echinoderms present an exemplary case study of the evolution of GRN patterning mechanisms in anciently diverged taxa.
Regulatory States and Polarity of NSM in Et.
Examining mesodermal polarity in euechinoids and outgroup echinoderms aids in establishing a timeline of GRN evolution. In euechinoid mesodermal NSM, gcm is directly downstream of Notch signaling and later is restricted before gastrulation to aboral NSM by Not, which is directly downstream of Nodal/SMAD signaling (37, 71, 93, 94). In cidaroids, early expression of gcm likely is not dependent on Delta/Notch (16), and mesodermal polarity is apparent only at 4–6 h after the start of gastrulation (Fig. 1L). Thus, in the early development of echinoids, the regulation of gcm changed markedly and at numerous nodes of the GRN; however, gcm still demarcates NSM polarity, which appears to be an echinoid novelty. This hypothesis is supported by the observation that no significant polarity occurs during embryonic mesodermal specification of asteroids, ophiuroids, and holothuroids (95–97). Thus, polarization of gcm and ese is a derived feature of echinoids that likely first appeared at least 268 mya, and the mechanism of NSM segregation via Nodal/SMAD signaling is likely conserved since that time (Fig. 6C).
At some point after the cidaroid–euechinoid divergence and before the diversification of modern camarodont euechinoid lineages, deployment of GRN circuitry polarizing NSM underwent a heterochronic shift, becoming pregastrular sometime between 170 and 90 mya. The data presented on O-A polarity in the NSM of Et suggest that multiple regulatory domains unfold at and around the tip of the archenteron as gastrulation proceeds, as is also the case in euechinoids (98), and that these embryonic domains have diverged markedly since the divergence of the two modern echinoid clades.
Evolution of Global Embryonic Domains in Early Development of Echinoids.
Previous analyses of embryonic regulatory states in Et surveyed SM regulatory factors (17, 47) and anterior neural ectoderm specification (75). In addition, two previous studies investigated SM and early endomesodermal micromere regulatory factors in the Pacific-dwelling cidaroid Prionocidaris baculosa (16, 99). Integrating these data into the present study affords an analysis of global embryonic regulatory states and GRN linkages in indirect-developing sea urchins. From these studies, numerous alterations to deployment and GRN circuitry at all levels of GRN topology can be enumerated. Here I enumerate 19 changes in spatiotemporal deployment or regulation of ectodermal and mesodermal embryonic regulatory factors since the cidaroid–euechinoid divergence (SI Appendix, Table S6). Prominent among the alterations of regulatory interactions are those that occurred while establishing polarity in mesodermal embryonic domains. Ectodermal specification and regulatory states also have undergone significant changes, but to a lesser degree. One hypothesis that can accommodate these observations is that endodermal and ectodermal developmental programs may be more recalcitrant to change than mesodermal programs because of their more ancient evolutionary origin (5), suggesting that accretion of process over evolutionary time is a mechanism of constraint in developmental programs (5, 100).
Pointedly, it is clear that the regulatory apparatus running in SM was specifically installed into the micromere embryonic address by co-option of the adult GRN skeletogenic program (101). Interestingly, in cidaroids, the micromere embryonic address is the location at which numerous euechinoid SM genes are first activated (17, 47). These observations suggest that in the ancestral echinoid lineage, the adult skeletogenesis program was co-opted to run in the micromeres. Later, in the lineage leading to modern euechinoids, additional layers of GRN topology, which do not exist in modern cidaroids, accrued in euechinoid mesodermal specification, such as the pmar1-hesc double-negative gate novelty (16, 17, 22) and delta-dependent NSM specification (17, 34). The accretion of additional regulatory layers in euechinoid mesodermal GRNs may explain the fact that little to no appreciable change has been observed in the mesodermal developmental programs of Lv, Pl, and Sp, representatives of modern euechinoid lineages that diverged ∼90 mya.
Since the divergence of cidaroids and euechinoids, numerous alterations to developmental GRNs have accrued between these lineages, although not to same degree within camaradont euechinoids. This study revealed that in modern echinoids, changes to mesodermal GRN architecture have occurred more frequently than alterations to ectodermal GRN architecture. These results support the notion that GRN architecture evolves at different rates (81), and provide an in-principle explanation for the rapid evolution observed in both cidaroid and euechinoid sea urchin lineages that have convergently and independently evolved direct-developing larval forms (53). It remains to be determined in future research whether the shared regulatory states between cidaroids and euechinoids elucidated here are the product of conserved stretches of genomic DNA hardwired in the cis-regulatory regions of orthologous regulatory genes or the result of diverged cis-regulatory modules producing similar developmental outcomes.
Materials and Methods
Animal and embryo culture, cloning, and acquisition of spatial and temporal gene expression data and microinjection perturbation data were performed as described previously (17, 47). The dosage of the small-molecule inhibitor SB431542 (15 μM) was determined by dilution series. Comparative gene expression analyses were done using the R application mfuzz. Additional details on previously described methods, statistical analyses, and all other experimental manipulations are provided in SI Appendix.
Supplementary Material
Acknowledgments
I dedicate this paper to my mentor and friend Eric Harris Davidson, who indelibly impacted my life in so many ways and who conceptually influenced the major theme of this paper, as well as provided input on experimental design and assessed portions of the data presented here. I thank Dr. Julius C. Barsi for the Sp onecut engineered BACs; Jennifer Wellman for her insight into statistical analyses; Stefan Materna for the raw data on S. purpuratus mRNA transcript abundance and manuscript critiques; Dr. Matthias Futschik and Cong Liang for mfuzz advice; Drs. Oliver Griffith, David McClay, Tom Stewart, and Jeffrey R. Thompson for their critical commentary on figures and the manuscript; and Prof. Günter P. Wagner for his patience and support. This work was funded by National Science Foundation CREATIV Grant 1240626.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612820113/-/DCSupplemental.
References
- 1.Davidson EH. Gene Activity in Early Development. 3rd Ed Academic; New York: 1986. [Google Scholar]
- 2.Wikramanayake AH, et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426(6965):446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 3.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic; Oxford, UK: 2006. [Google Scholar]
- 4.Peter IS, Davidson EH. Genomic Control Process, Development and Evolution. Academic; Oxford, UK: 2015. [Google Scholar]
- 5.Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519(7542):219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson EH. Spatial mechanisms of gene regulation in metazoan embryos. Development. 1991;113(1):1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Davidson EH, Cameron RA, Ransick A. Specification of cell fate in the sea urchin embryo: Summary and some proposed mechanisms. Development. 1998;125(17):3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- 8.Wray GA, Bely AE. The evolution of echinoderm development is driven by several distinct factors. Dev Suppl. 1994;(Supplement):97–106. [PubMed] [Google Scholar]
- 9.Kroh A, Smith AB. The phylogeny and classification of post-Palaeozoic echinoids. J Syst Palaeontology. 2010;8(2):147–212. [Google Scholar]
- 10.Thompson JR, et al. Reorganization of sea urchin gene regulatory networks at least 268 million years ago as revealed by oldest fossil cidaroid echinoid. Sci Rep. 2015;5:15541. doi: 10.1038/srep15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tennent D. The early influence of the spermatazoan upon the characters of echinoid larvae. Carn Inst Wash Publ. 1914;182:129–138. [Google Scholar]
- 12.Mortensen T. Contributions to the study of the development and larval forms of echinoderms, IV. Danske Vid Selsk Ser. 1938;9(7(3)):1–59. [Google Scholar]
- 13.Schroeder TE. Development of a “primitive” sea urchin (Eucidaris tribuloides): Irregularities in the hyaline layer, micromeres, and primary mesenchyme. Biol Bull. 1981;161(1):141–151. [Google Scholar]
- 14.Wray GA, McClay DR. The origin of spicule-forming cells in a “primitive” sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103(2):305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- 15.Wray GA, McClay DR. Molecular heterochronies and heterotopies in early echinoid development. Evolution. 1989;43(4):803–813. doi: 10.1111/j.1558-5646.1989.tb05178.x. [DOI] [PubMed] [Google Scholar]
- 16.Yamazaki A, Kidachi Y, Yamaguchi M, Minokawa T. Larval mesenchyme cell specification in the primitive echinoid occurs independently of the double-negative gate. Development. 2014;141(13):2669–2679. doi: 10.1242/dev.104331. [DOI] [PubMed] [Google Scholar]
- 17.Erkenbrack EM, Davidson EH. Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proc Natl Acad Sci USA. 2015;112(30):E4075–E4084. doi: 10.1073/pnas.1509845112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao F, et al. Juvenile skeletogenesis in anciently diverged sea urchin clades. Dev Biol. 2015;400(1):148–158. doi: 10.1016/j.ydbio.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Angerer LM, et al. A BMP pathway regulates cell fate allocation along the sea urchin animal-vegetal embryonic axis. Development. 2000;127(5):1105–1114. doi: 10.1242/dev.127.5.1105. [DOI] [PubMed] [Google Scholar]
- 20.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295(5560):1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 21.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104(30):12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105(16):5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaguchi S, Yaguchi J, Angerer RC, Angerer LM. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell. 2008;14(1):97–107. doi: 10.1016/j.devcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Su Y-H, et al. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev Biol. 2009;329(2):410–421. doi: 10.1016/j.ydbio.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340(2):188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter IS, Davidson EH. A gene regulatory network controlling the embryonic specification of endoderm. Nature. 2011;474(7353):635–639. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Materna SC, Ransick A, Li E, Davidson EH. Diversification of oral and aboral mesodermal regulatory states in pregastrular sea urchin embryos. Dev Biol. 2013;375(1):92–104. doi: 10.1016/j.ydbio.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barsi JC, Li E, Davidson EH. Geometric control of ciliated band regulatory states in the sea urchin embryo. Development. 2015;142(5):953–961. doi: 10.1242/dev.117986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam J, et al. cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev Biol. 2007;306(2):860–869. doi: 10.1016/j.ydbio.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui M, Siriwon N, Li E, Davidson EH, Peter IS. Specific functions of the Wnt signaling system in gene regulatory networks throughout the early sea urchin embryo. Proc Natl Acad Sci USA. 2014;111(47):E5029–E5038. doi: 10.1073/pnas.1419141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wikramanayake AH, Huang L, Klein WH. Beta-catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc Natl Acad Sci USA. 1998;95(16):9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126(8):1703–1713. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- 33.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126(2):345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 34.Sweet HC, Gehring M, Ettensohn CA. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development. 2002;129(8):1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 35.Flowers VL, Courteau GR, Poustka AJ, Weng W, Venuti JM. Nodal/activin signaling establishes oral-aboral polarity in the early sea urchin embryo. Dev Dyn. 2004;231(4):727–740. doi: 10.1002/dvdy.20194. [DOI] [PubMed] [Google Scholar]
- 36.Ettensohn CA, Kitazawa C, Cheers MS, Leonard JD, Sharma T. Gene regulatory networks and developmental plasticity in the early sea urchin embryo: Alternative deployment of the skeletogenic gene regulatory network. Development. 2007;134(17):3077–3087. doi: 10.1242/dev.009092. [DOI] [PubMed] [Google Scholar]
- 37.Croce JC, McClay DR. Dynamics of Delta/Notch signaling on endomesoderm segregation in the sea urchin embryo. Development. 2010;137(1):83–91. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duboc V, Röttinger E, Besnardeau L, Lepage T. Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell. 2004;6(3):397–410. doi: 10.1016/s1534-5807(04)00056-5. [DOI] [PubMed] [Google Scholar]
- 39.Duboc V, Lapraz F, Besnardeau L, Lepage T. Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev Biol. 2008;320(1):49–59. doi: 10.1016/j.ydbio.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Saudemont A, et al. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 2010;6(12):e1001259. doi: 10.1371/journal.pgen.1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lhomond G, McClay DR, Gache C, Croce JC. Frizzled1/2/7 signaling directs β-catenin nuclearisation and initiates endoderm specification in macromeres during sea urchin embryogenesis. Development. 2012;139(4):816–825. doi: 10.1242/dev.072215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavalieri V, Spinelli G. Early asymmetric cues triggering the dorsal/ventral gene regulatory network of the sea urchin embryo. eLife. 2014;3:e04664. doi: 10.7554/eLife.04664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith AB, et al. Testing the molecular clock: Molecular and paleontological estimates of divergence times in the Echinoidea (Echinodermata) Mol Biol Evol. 2006;23(10):1832–1851. doi: 10.1093/molbev/msl039. [DOI] [PubMed] [Google Scholar]
- 44.Croce J, et al. Wnt6 activates endoderm in the sea urchin gene regulatory network. Development. 2011;138(15):3297–3306. doi: 10.1242/dev.058792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ettensohn CA. Lessons from a gene regulatory network: Echinoderm skeletogenesis provides insights into evolution, plasticity and morphogenesis. Development. 2009;136(1):11–21. doi: 10.1242/dev.023564. [DOI] [PubMed] [Google Scholar]
- 46.Molina MD, de Crozé N, Haillot E, Lepage T. Nodal: Master and commander of the dorsal-ventral and left-right axes in the sea urchin embryo. Curr Opin Genet Dev. 2013;23(4):445–453. doi: 10.1016/j.gde.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Erkenbrack EM, et al. Ancestral state reconstruction by comparative analysis of a GRN kernel operating in echinoderms. Dev Genes Evol. 2016;226(1):37–45. doi: 10.1007/s00427-015-0527-y. [DOI] [PubMed] [Google Scholar]
- 48.Lapraz F, Haillot E, Lepage T. A deuterostome origin of the Spemann organiser suggested by Nodal and ADMPs functions in Echinoderms. Nat Commun. 2015;6:8927. doi: 10.1038/ncomms9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duboc V, Lepage T. A conserved role for the nodal signaling pathway in the establishment of dorso-ventral and left-right axes in deuterostomes. J Exp Zoolog B Mol Dev Evol. 2008;310(1):41–53. doi: 10.1002/jez.b.21121. [DOI] [PubMed] [Google Scholar]
- 50.Wray GA, Raff RA. The evolution of developmental strategy in marine invertebrates. Trends Ecol Evol. 1991;6(2):45–50. doi: 10.1016/0169-5347(91)90121-D. [DOI] [PubMed] [Google Scholar]
- 51.Smith MS, Collins S, Raff RA. Morphogenetic mechanisms of coelom formation in the direct-developing sea urchin Heliocidaris erythrogramma. Dev Genes Evol. 2009;219(1):21–29. doi: 10.1007/s00427-008-0262-8. [DOI] [PubMed] [Google Scholar]
- 52.Smith MS, Turner FR, Raff RA. Nodal expression and heterochrony in the evolution of dorsal-ventral and left-right axes formation in the direct-developing sea urchin Heliocidaris erythrogramma. J Exp Zoolog B Mol Dev Evol. 2008;310(8):609–622. doi: 10.1002/jez.b.21233. [DOI] [PubMed] [Google Scholar]
- 53.Raff RA. Origins of the other metazoan body plans: The evolution of larval forms. Philos Trans R Soc Lond B Biol Sci. 2008;363(1496):1473–1479. doi: 10.1098/rstb.2007.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su Y-H. Gene regulatory networks for ectoderm specification in sea urchin embryos. Biochim Biophys Acta. 2009;1789(4):261–267. doi: 10.1016/j.bbagrm.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Li E, Cui M, Peter IS, Davidson EH. Encoding regulatory state boundaries in the pregastrular oral ectoderm of the sea urchin embryo. Proc Natl Acad Sci USA. 2014;111(10):E906–E913. doi: 10.1073/pnas.1323105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li E, Materna SC, Davidson EH. Direct and indirect control of oral ectoderm regulatory gene expression by Nodal signaling in the sea urchin embryo. Dev Biol. 2012;369(2):377–385. doi: 10.1016/j.ydbio.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li E, Materna SC, Davidson EH. New regulatory circuit controlling spatial and temporal gene expression in the sea urchin embryo oral ectoderm GRN. Dev Biol. 2013;382(1):268–279. doi: 10.1016/j.ydbio.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lapraz F, Besnardeau L, Lepage T. Patterning of the dorsal-ventral axis in echinoderms: Insights into the evolution of the BMP-chordin signaling network. PLoS Biol. 2009;7(11):e1000248. doi: 10.1371/journal.pbio.1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben-Tabou de-Leon S, Su Y-H, Lin K-T, Li E, Davidson EH. Gene regulatory control in the sea urchin aboral ectoderm: Spatial initiation, signaling inputs, and cell fate lockdown. Dev Biol. 2013;374(1):245–254. doi: 10.1016/j.ydbio.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Croce J, Lhomond G, Gache C. Coquillette, a sea urchin T-box gene of the Tbx2 subfamily, is expressed asymmetrically along the oral-aboral axis of the embryo and is involved in skeletogenesis. Mech Dev. 2003;120(5):561–572. doi: 10.1016/s0925-4773(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 61.Gross JM, Peterson RE, Wu SY, McClay DR. LvTbx2/3: A T-box family transcription factor involved in formation of the oral/aboral axis of the sea urchin embryo. Development. 2003;130(9):1989–1999. doi: 10.1242/dev.00409. [DOI] [PubMed] [Google Scholar]
- 62.Range R. Specification and positioning of the anterior neuroectoderm in deuterostome embryos. Genesis. 2014;52(3):222–234. doi: 10.1002/dvg.22759. [DOI] [PubMed] [Google Scholar]
- 63.Strathmann RR. The feeding behavior of planktotrophic echinoderm larvae: mechanisms, regulation, and rates of suspensionfeeding. J Exp Mar Biol Ecol. 1971;6(2):109–160. [Google Scholar]
- 64.Wray GA. The evolution of larval morphology during the postpaleozoic radiation of echinoids. Paleobiology. 1992;18(3):258–287. [Google Scholar]
- 65.Barsi JC, Davidson EH. cis-regulatory control of the initial neurogenic pattern of onecut gene expression in the sea urchin embryo. Dev Biol. 2016;409(1):310–318. doi: 10.1016/j.ydbio.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 66.Poustka AJ, et al. A global view of gene expression in lithium and zinc treated sea urchin embryos: New components of gene regulatory networks. Genome Biol. 2007;8(5):R85. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otim O, Amore G, Minokawa T, McClay DR, Davidson EH. SpHnf6, a transcription factor that executes multiple functions in sea urchin embryogenesis. Dev Biol. 2004;273(2):226–243. doi: 10.1016/j.ydbio.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 68.Cameron RA, Davidson EH. Cell type specification during sea urchin development. Trends Genet. 1991;7(7):212–218. doi: 10.1016/0168-9525(91)90367-y. [DOI] [PubMed] [Google Scholar]
- 69.Sweet HC, Hodor PG, Ettensohn CA. The role of micromere signaling in Notch activation and mesoderm specification during sea urchin embryogenesis. Development. 1999;126(23):5255–5265. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- 70.Materna SC, Davidson EH. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev Biol. 2012;364(1):77–87. doi: 10.1016/j.ydbio.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duboc V, et al. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development. 2010;137(2):223–235. doi: 10.1242/dev.042531. [DOI] [PubMed] [Google Scholar]
- 72.Lee PY, Davidson EH. Expression of Spgatae, the Strongylocentrotus purpuratus ortholog of vertebrate GATA4/5/6 factors. Gene Expr Patterns. 2004;5(2):161–165. doi: 10.1016/j.modgep.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 73.Duboc V, Röttinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9(1):147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Bradham CA, et al. Chordin is required for neural but not axial development in sea urchin embryos. Dev Biol. 2009;328(2):221–233. doi: 10.1016/j.ydbio.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bishop CD, MacNeil KE, Patel D, Taylor VJ, Burke RD. Neural development in Eucidaris tribuloides and the evolutionary history of the echinoid larval nervous system. Dev Biol. 2013;377(1):236–244. doi: 10.1016/j.ydbio.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 76.Yankura KA, Koechlein CS, Cryan AF, Cheatle A, Hinman VF. Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc Natl Acad Sci USA. 2013;110(21):8591–8596. doi: 10.1073/pnas.1220903110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Materna SC, Nam J, Davidson EH. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr Patterns. 2010;10(4-5):177–184. doi: 10.1016/j.gep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gildor T, Ben-Tabou de-Leon S. Comparative study of regulatory circuits in two sea urchin species reveals tight control of timing and high conservation of expression dynamics. PLoS Genet. 2015;11(7):e1005435. doi: 10.1371/journal.pgen.1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Israel JW, et al. Comparative developmental transcriptomics reveals rewiring of a highly conserved gene regulatory network during a major life history switch in the sea urchin genus Heliocidaris. PLoS Biol. 2016;14(3):e1002391. doi: 10.1371/journal.pbio.1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yanai I, Hunter CP. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009;19(12):2214–2220. doi: 10.1101/gr.093815.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311(5762):796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 82.Wilson KA, Andrews ME, Rudolf Turner F, Raff RA. Major regulatory factors in the evolution of development: The roles of goosecoid and Msx in the evolution of the direct-developing sea urchin Heliocidaris erythrogramma. Evol Dev. 2005;7(5):416–428. doi: 10.1111/j.1525-142X.2005.05046.x. [DOI] [PubMed] [Google Scholar]
- 83.Wilson KA, Andrews ME, Raff RA. Dissociation of expression patterns of homeodomain transcription factors in the evolution of developmental mode in the sea urchins Heliocidaris tuberculata and H. erythrogramma. Evol Dev. 2005;7(5):401–415. doi: 10.1111/j.1525-142X.2005.05045.x. [DOI] [PubMed] [Google Scholar]
- 84.Raff RA, Snoke Smith M. Chapter 7: Axis formation and the rapid evolutionary transformation of larval form. Curr Top Dev Biol. 2009;86(86):163–190. doi: 10.1016/S0070-2153(09)01007-2. [DOI] [PubMed] [Google Scholar]
- 85.Emlet RB. Larval form and metamorphosis of a primitive sea urchin, Eucidaris thouarsi (Echinodermata, Echinoidea, Cidaroida), with implications for developmental and phylogenetic studies. Biol Bull. 1988;174(1):4–19. doi: 10.2307/1541754. [DOI] [PubMed] [Google Scholar]
- 86.Bennett KC, Young CM, Emlet RB. Larval development and metamorphosis of the deep-sea cidaroid urchin Cidaris blakei. Biol Bull. 2012;222(2):105–117. doi: 10.1086/BBLv222n2p105. [DOI] [PubMed] [Google Scholar]
- 87.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100(23):13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yankura KA, Martik ML, Jennings CK, Hinman VF. Uncoupling of complex regulatory patterning during evolution of larval development in echinoderms. BMC Biol. 2010;8:143. doi: 10.1186/1741-7007-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev Biol. 2006;300(1):49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 90.Santagata S, Resh C, Hejnol A, Martindale MQ, Passamaneck YJ. Development of the larval anterior neurogenic domains of Terebratalia transversa (Brachiopoda) provides insights into the diversification of larval apical organs and the spiralian nervous system. Evodevo. 2012;3(1):3. doi: 10.1186/2041-9139-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu JK, et al. The Fox genes of Branchiostoma floridae. Dev Genes Evol. 2008;218(11-12):629–638. doi: 10.1007/s00427-008-0229-9. [DOI] [PubMed] [Google Scholar]
- 92.Fritzenwanker JH, Gerhart J, Freeman RM, Jr, Lowe CJ. The Fox/Forkhead transcription factor family of the hemichordate Saccoglossus kowalevskii. Evodevo. 2014;5:17. doi: 10.1186/2041-9139-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev Biol. 2002;246(1):132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- 94.Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297(2):587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 95.McCauley BS, Wright EP, Exner C, Kitazawa C, Hinman VF. Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. Evodevo. 2012;3(1):17. doi: 10.1186/2041-9139-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dylus DV, et al. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo. 2016;7(1):2. doi: 10.1186/s13227-015-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol. 2010;340(2):200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 98.Andrikou C, Pai CY, Su YH, Arnone MI. Logics and properties of a genetic regulatory program that drives embryonic muscle development in an echinoderm. eLife. 2015;4:4. doi: 10.7554/eLife.07343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamazaki A, Kidachi Y, Minokawa T. “Micromere” formation and expression of endomesoderm regulatory genes during embryogenesis of the primitive echinoid Prionocidaris baculosa. Dev Growth Differ. 2012;54(5):566–578. doi: 10.1111/j.1440-169X.2012.01360.x. [DOI] [PubMed] [Google Scholar]
- 100.Gould SJ. Ontogeny and Phylogeny. Harvard Univ Press; Cambridge, MA: 1977. [Google Scholar]
- 101.Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci USA. 2008;105(16):6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.