Significance
The proteasome holoenzyme is an ATP-dependent protease in eukaryotes that degrades ubiquitylated substrates. It is involved in numerous important biological processes, such as cell division, differentiation, innate immunity, adaptive immunity, regulation of gene expression, and response to proteotoxic stress. Using cryoelectron microscopy, we have examined multiple conformational states of the human proteasome at medium to high resolution. Our results reveal that the substrate-conducting channel in the core particle is transiently opened and accompanied by dynamic changes in structure of the particle. These observations provide new insights into how the proteasome recognizes ubiquitylated substrates and translocates them through a channel and gate to degradation sites in the core particle.
Keywords: ubiquitin-proteasome system, AAA-ATPase, cyroelectron microscopy
Abstract
The proteasome is the major engine of protein degradation in all eukaryotic cells. At the heart of this machine is a heterohexameric ring of AAA (ATPases associated with diverse cellular activities) proteins that unfolds ubiquitylated target proteins that are concurrently translocated into a proteolytic chamber and degraded into peptides. Using cryoelectron microscopy, we determined a near–atomic-resolution structure of the 2.5-MDa human proteasome in its ground state, as well as subnanometer-resolution structures of the holoenzyme in three alternative conformational states. The substrate-unfolding AAA-ATPase channel is narrowed by 10 inward-facing pore loops arranged into two helices that run in parallel with each other, one hydrophobic in character and the other highly charged. The gate of the core particle was unexpectedly found closed in the ground state and open in only one of the alternative states. Coordinated, stepwise conformational changes of the regulatory particle couple ATP hydrolysis to substrate translocation and regulate gating of the core particle, leading to processive degradation.
The amount of each protein in the cell depends on its rates of synthesis and degradation. In eukaryotic cells, selective protein degradation is mostly carried out by a set of pathways of ubiquitylation that terminate in a 2.5-MDa protein proteolytic complex, called the 26S proteasome. The ubiquitin-proteasome pathways are essential parts of important biological processes, such as cell division, differentiation, innate immunity, adaptive immunity, regulation of gene expression, and the response to proteotoxic stress (1–4). The proteasome is also an important therapeutic target in multiple myeloma (5, 6).
The proteasome is composed of a 28-subunit barrel-shaped core particle (CP) in the center capped at the top and bottom by 19-subunit regulatory particles (RPs) (SI Appendix, Fig. S1) (7–10). The CP forming the catalytic chamber contains three proteolytically active threonine residues. Heptameric α-rings, positioned on each side of the catalytic chamber, control substrate entry into this space. Opening of a channel within the α-ring is thought to result from association of the RP and the CP (5, 11–13). However, the mechanism of the core regulatory step of the proteasome channel opening remains mysterious.
The RP is formed from two subcomplexes known as the lid and the base. Recognition of a substrate with the requisite number and configuration of ubiquitin is mediated principally by the base subunit Rpn13 and by another receptor, Rpn10 (4). To allow substrate degradation, ubiquitin is first removed by Rpn11, a metalloprotease subunit in the lid. The globular domains of a substrate are then unfolded mechanically by a ring-like heterohexameric complex consisting of six distinct subunits, Rpt1 to Rpt6, which belong to the ATPases-associated-with-diverse-cellular-activities (AAA) family. Both Rpn11-dependent deubiquitylation of the substrate and unfolding of substrate globular domains require prior engagement with the translocation machinery.
High-resolution structures of the isolated CP have been available for several species (5, 12, 13). The intact proteasome has not been resolved to a level at which a reliable Cα-backbone can be traced with spatial assignment of amino acids, although major advances have been made in recent years (2, 3, 7–11, 14–25). Several RP subunits have been resolved at high resolution by X-ray crystallography (14–16, 18, 21–24). Recent cryoelectron microscopy (cryo-EM) analysis of the complete proteasome at moderate resolution (6 to 10 Å) revealed an overall subunit organization of the RP (7–10, 25–28). The lid subcomplex, which consists of nine Rpn proteins (Rpn3, Rpn5 to Rpn9, Rpn11, Rpn12, and Dss1/Sem1), exhibits a horseshoe-like architecture and is organized around an elaborate bundle assembled from the C-terminal helices of each subunit but Dss1/Sem1 (20, 29). The six Rpt subunits of the base share a general domain organization, starting with a coiled-coil (CC) domain at the N terminus, followed by an oligonucleotide- and oligosaccharide-binding (OB) domain and an AAA domain toward the C terminus (7, 23). The yeast proteasome holoenzyme was previously observed to assume three distinct conformations, hypothetically considered the substrate-accepting (s1), commitment (s2), and translocating (s3) states (25–28). The limited resolution, however, precludes understanding of the critical molecular mechanisms underlying ATP-dependent degradation by the proteasome holoenzyme. The nucleotide-binding states of AAA-ATPases, architecture of the peptide-unfolding channel, and allosteric regulation of the substrate-translocation pathway remain particularly elusive.
We report here structures of the human proteasome in four conformational states by single-particle cryo-EM. These structures offer atomic-level details of key intersubcomplex interfaces in the human proteasome assembly. A detailed dissection of the AAA-ATPase channel provides key insights into the molecular mechanism of ATP-dependent, processive substrate translocation. Importantly, the four conformational states reveal dynamic gating of the CP channel, which is elegantly regulated through highly coordinated conformational changes among distinct subcomplexes.
Results
Cryoelectron Microscopy of the Proteasome.
Cryo-EM data were collected on the proteasome holoenzyme purified from human embryonic kidney (HEK) 293 cells (30) using a 200-kV cryogenic electron microscope equipped with a direct electron detector (Fig. 1 A and B and SI Appendix, Fig. S2 A and B). We reconstructed the complete, doubly capped proteasome complex after imposing C2 symmetry using 237,083 single-particle images. The CP component was refined to a nominal resolution of 3.6 Å (Fig. 1B and SI Appendix, Fig. S2C), whereas the resolutions of its two RPs were significantly lower than that of the CP in this map, suggesting that the RPs fluctuate conformationally (SI Appendix, Fig. S2F). Maximum likelihood-based classification identified a dataset of 85,420 particles of improved structural homogeneity, which improved the reconstruction of AAA-ATPase to around 4-Å resolution (SI Appendix, Fig. S2 C and D). However, the lid subcomplex still demonstrates significantly lower resolution, suggesting it is the most flexible component in the proteasome holoenzyme.
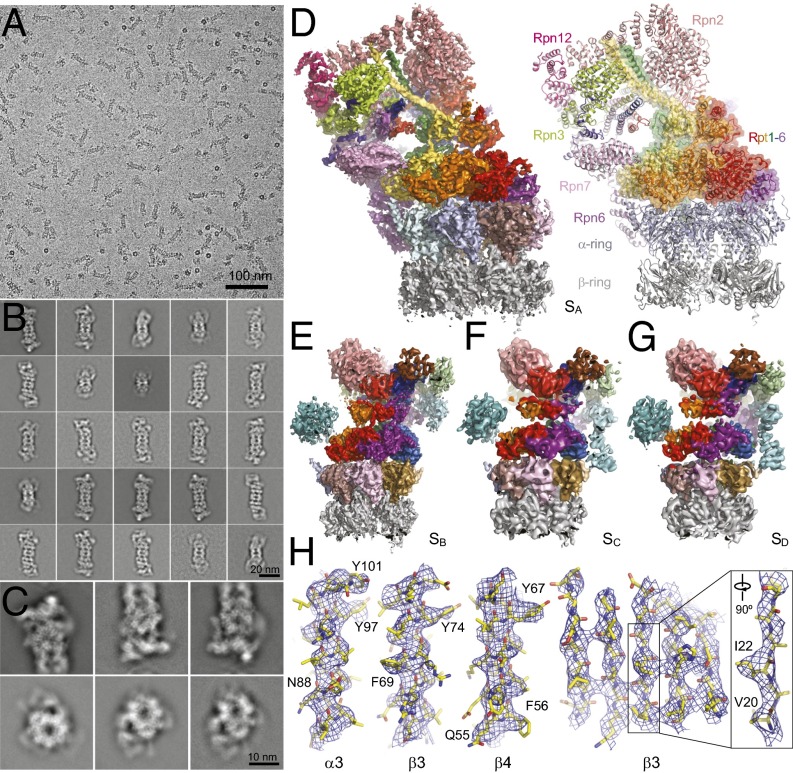
Fig. 1.
Cryo-EM structure determination of the human proteasome in four conformational states. (A) A typical cryo-EM micrograph of the human proteasome imaged with a Tecnai Arctica and a Gatan K2 Summit direct detector camera. (B) Typical reference-free 2D class averages of the doubly capped proteasome computed by the ROME software (49). (C) Typical reference-free 2D class averages of the RP–CP subcomplex, showing great detail corresponding to secondary structures of the complex. (D) The cryo-EM density map of the RP–CP subcomplex in a surface representation (Left) and the atomic model built from the density map (Right). (E–G) The cryo-EM densities are shown as solid surfaces for the SB (E), SC (F), and SD (G) states. (H) Representative cryo-EM densities of secondary structures of α-helices in the α3, β3, and β4 subunits (Left three panels) and β-strands in the β3 subunit (Right two panels) in the SA state are superimposed with the fitted atomic model shown as a stick representation, showing that density quality is sufficient to allow side-chain fitting. The residue numbers of selected bulky side chains are labeled.
To investigate the conformational dynamics, we boxed half of the holoenzyme, including half of the CP, in complex with a complete RP (hereafter referred to as the RP–CP subcomplex) using additional cryo-EM data (Fig. 1C and SI Appendix, Figs. S2 and S3). We conducted iterative maximum likelihood-based classification focusing on the RP structure by using single-particle images of the RP–CP subcomplex with the CP density subtracted (31, 32) (SI Appendix, Fig. S3 and Materials and Methods). After exhaustive computational purification, we obtained structurally homogeneous datasets of the RP–CP subcomplex corresponding to four distinct conformational states, namely a major state (SA) and three alternative states (SB, SC, and SD) (Fig. 1 D–G and SI Appendix, Fig. S3). We refined the overall RP–CP reconstructions of the SA, SB, SC, and SD states to nominal resolutions of 4.4, 6.8, 8.0, and 8.0 Å, respectively (SI Appendix, Fig. S2E).
The high-resolution features of the doubly capped proteasome map are consistent with those of the RP–CP map in the SA state. In both maps, the ATPase subunits are better-resolved than the lid subcomplex (SI Appendix, Fig. S2 F and G). The ATPase density is best-resolved in the doubly capped map in the SA state, whereas the lid subcomplex is best-resolved in the RP–CP map in SA at a nominal resolution of 4.9 Å (SI Appendix, Fig. S2C). The cryo-EM maps of the SA state enabled atomic modeling and refinement (Fig. 1H and SI Appendix, Figs. S4–S7 and Table S1). The final atomic model of the SA state contains all lid and CP subunits, Rpn2, Rpn10, and Rpt1 to Rpt6 (Fig. 1D). The local resolution of Rpn1 in SA is about 8 Å, which nonetheless suffices to build its pseudoatomic model. Based on the atomic model of the SA state, we built pseudoatomic models for SB, SC, and SD (Fig. 1 E–G). As expected, the ubiquitin-interacting motif of the ubiquitin receptor Rpn10 is disordered and missing in the cryo-EM densities of all conformational states (17, 19). The other ubiquitin receptor, Rpn13, was not observed (SI Appendix, Fig. S1).
Overview of Conformational States of the Human Proteasome.
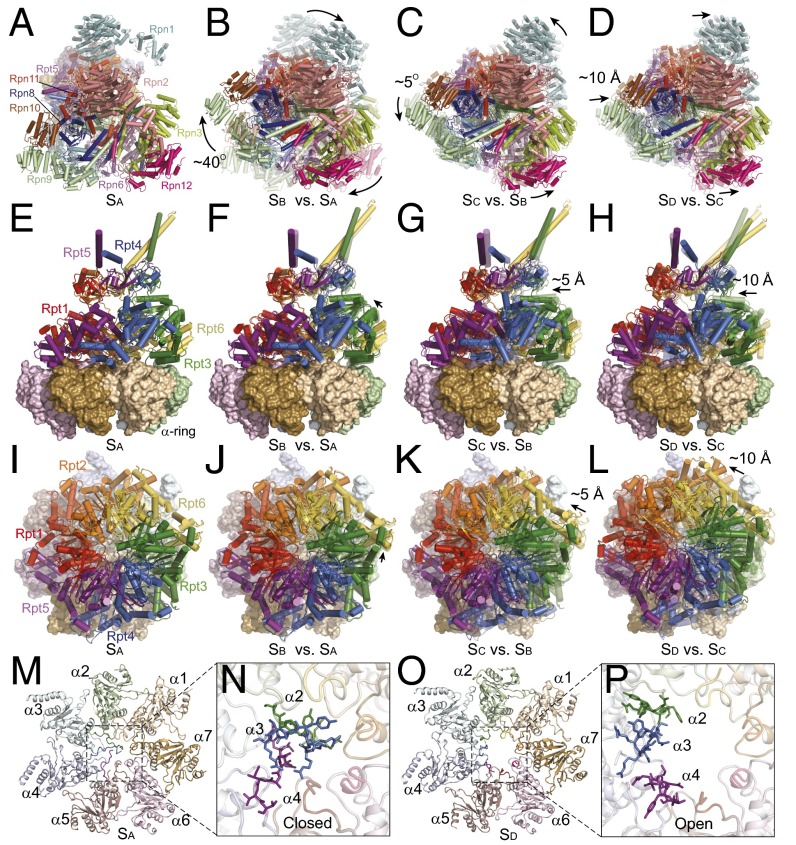
The individual SA, SB, SC, and SD conformational states are represented in the particle populations at 76.2%, 10.1%, 5.8%, and 7.9%, respectively. The lid is rotated ∼40° clockwise in the SB state relative to the SA state but rotated ∼5° counterclockwise in SC relative to SB and translated ∼10 Å in SD relative to SC (Fig. 2 A–D). The AAA-ATPase heterohexamer movements on the α-ring in the SC and SD states exhibit stepwise axial rotation, lateral translation, and vertical rocking, which reduces the tilt of the ATPase ring relative to the α-ring (Fig. 2 E–L).
Fig. 2.
Conformational changes of the human proteasome in four distinct states. (A) Top view of the lid of SA. (B–D) Top views of the lid of SB (B), SC (C), and SD (D) superimposed with transparent cartoons of SA, SB, and SC, respectively. (E) Side view of the ATPase ring above the α-ring in SA. (F–H) Side views of the ATPase ring above the α-ring in SB (F), SC (G), and SD (H) superimposed with transparent cartoons of SA, SB, and SC, respectively. (I) Top view of the ATPase ring in SA. (J–L) Top views of the ATPase ring in SB (J), SC (K), and SD (L) superimposed with transparent cartoons of SA, SB, and SC, respectively. (M) The α-ring in a cartoon representation from the perspective of the AAA-ATPase or the RP–CP interface in SA. (N) Close-up view of the central portion of the α-ring, showing that the CP channel is closed in this conformation. The amino-terminal tails of the α2, α3, and α4 subunits blocking the CP channel are shown in stick representation, whereas the rest of the structure is in cartoon representation. (O) The α-ring in a cartoon representation from the perspective of the AAA-ATPase or the RP–CP interface in SD. (P) Close-up view of the central part of the α-ring, showing that the CP channel is open in this conformation. The amino-terminal tails of the α2, α3, and α4 subunits blocking the CP channel are shown in stick representation, whereas the rest of the structure is in cartoon representation.
The structures of the CP in the SA, SB, and SC states are identical up to their measured resolution. The crystal structure of free human CP with a closed CP channel can be fitted into the cryo-EM maps of these states as a rigid body, including the channel-blocking amino-terminal tails of the α-subunits (5) (Fig. 2 M and N and SI Appendix, Fig. S4 J–M). However, there is considerable structural change in the α-subunits of the SD state, where the CP channel is open (Fig. 2 O and P and SI Appendix, Fig. S4M). A closed channel in the intact holoenzyme structure is unexpected (33), and suggests that association of the RP and CP subcomplexes does not open the CP channel by default in the SA state of the human proteasome. Channel opening in the SD state is accompanied by coordinated conformational changes in the RP (Fig. 2 A–L).
Although multiple conformational states of the yeast proteasome were reported in previous studies (25–28), no human counterparts to these states have been observed before. Comparison between our human holoenzyme in four states and the yeast ones in three states (s1, s2, and s3) suggests considerable structural difference in both the lid and the base (SI Appendix, Fig. S8). However, the SA and SB states of the human holoenzyme generally correspond to the s1 and s2 states of the yeast holoenzyme (28) based on the general features of the Rpn11–OB and RC–CP interfaces (SI Appendix, Fig. S9D). In the human SA and yeast s1 states, Rpn11 is positioned above the Rpt4–Rpt5 interface on the OB ring without blocking the substrate entry port (7, 9, 25, 28). By contrast, Rpn11 is moved over the center of the substrate entry port in both human SB and yeast s2 states (28). In the human SC/SD states, there is prominent translation of the ATPase ring with respect to the CP, a feature shared with the yeast substrate-engaged and s3 states (26, 27).
The Lid–Base Interfaces.
The lid–base interface plays a critical role in coordinating substrate translocation with deubiquitylation (7–10, 26, 28). In the SA state, the interface between the ATPase ring and lid buries ∼3,900 Å2 of interfacial area. Most of these interfaces are contributed by Rpt3 and Rpt6, whose CC–OB domains bury ∼3,100 Å2 of interface area with the lid (SI Appendix, Fig. S9A and Table S2). The CC domain of the Rpt3–Rpt6 heterodimer is encircled by the helical elements of Rpn2, Rpn3, Rpn8, and Rpn11 (SI Appendix, Fig. S9A) (7, 25, 27). This interfacial architecture constitutes the stabilizing core of the lid–base association and is largely invariant in all four conformational states. Using this interface as a pivot, Rpn11 rotates 30° to a position closer to the OB ring of AAA-ATPase in the SB, SC, and SD states (SI Appendix, Fig. S9 D–G).
The lateral lid–base interface exhibits prominent conformational transitions. The AAA domains of Rpt3 and Rpt6 bury ∼1,800 Å2 of interface area with Rpn5 to Rpn7 in the SA state (SI Appendix, Fig. S9B and Table S2). One side of the amino-terminal PCI (proteasome-cyclosome-initiation factor) domain of Rpn7, involving four helix-connecting loops, makes extensive interaction (∼940 Å2) with the Rpt6 AAA domain. This interface is translated clockwise for ∼10 Å in SB relative to SA, and remains nearly invariant in SC and SD. The AAA domains of Rpt3 and Rpt6 make limited contact with Rpn5 and Rpn6, with only ∼340 and ∼480 Å2 buried in the SA state, respectively. Consistent with the small interface area, the cryo-EM densities of the Rpn5/Rpn6 amino-terminal PCI domain are substantially weakened in the SB state, implying local dissociation between Rpn and Rpt subunits at these interfaces. In the SD state, the Rpn5–Rpt3 interface is reestablished. This analysis suggests that the Rpn7–Rpt6 interface may be retained during conformational changes of the lid, whereas the smaller interfaces of Rpn5–Rpt3 and Rpn6–Rpt6 are labile (25–27).
The AAA-ATPase Heterohexamer.
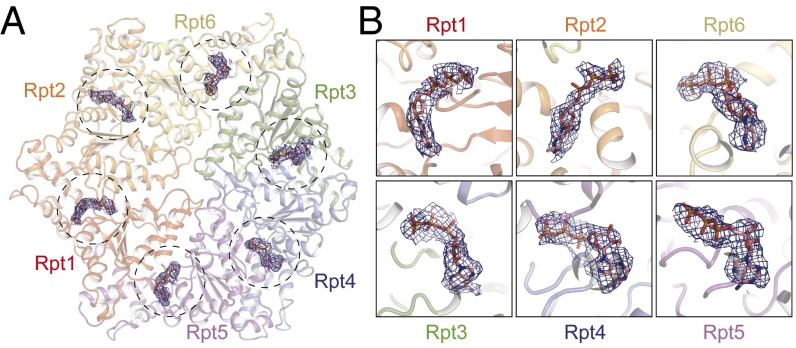
ATP binds the Walker A motif located next to a short linker between the small and large AAA subdomains of the ATPases. The nucleotide-binding state regulates the conformations of the Rpt subunits by modifying the geometric relationship between the small and large AAA subdomains (27, 34, 35). In the density map of the SA state, we identified nucleotide densities in all Rpt subunits (Fig. 3A). We tentatively modeled six ATPs into the atomic structure of the SA state. However, a caveat is noted. Because cryo-EM density is built from averaging many single-particle images of individual molecular copies, it does not sufficiently differentiate potential coexistence of ADP and ATP in certain Rpt subunits in different copies of single particles. Further work to establish the nucleotide composition in different states is needed.
Fig. 3.
Structure of the six nucleotide-binding sites of the proteasome. (A) Overview of six nucleotide-binding sites in the AAA-ATPase heterohexamer of the SA state. Bound nucleotides are shown in stick representation superimposed with cryo-EM densities of the nucleotide that are shown in blue mesh. (B) Close-up views of nucleotide conformations in the six nucleotide-binding sites in SA. ATP is tentatively modeled into the nucleotide density of each Rpt subunit in SA.
The AAA domains of Rpt1 to Rpt6 form a staircase beneath the OB ring, with Rpt3 located at the top position, Rpt2 at the bottom, and Rpt6 bridging the two in the SA state (7–10) (SI Appendix, Figs. S10 and S11A). The large AAA subdomain contacts both the large and small AAA subdomains of the neighboring subunit, forming an L-shaped intersubunit interface with a dihedral angle of about 120° (SI Appendix, Fig. S11B). The interfacial area between the two neighboring large AAA subdomains varies from 0.96 to 1.45 times that between the large and small AAA subdomains (SI Appendix, Fig. S11 D and E). The buried interface (∼1,100 Å2) between the AAA domains of Rpt3 and Rpt6 is half of the interfacial area between the other two adjacent Rpt subunits in SA, suggesting that the former interface may be capable of greater structural rearrangement (26) (SI Appendix, Figs. S11D and S12). Indeed, Rpt3 demonstrates the greatest shift and rotation compared with other Rpt subunits during state transitions from SA to SC (SI Appendix, Fig. S12 and Table S3). In summary, the six AAA-ATPases are not arranged symmetrically; rather, a strong asymmetry in the intersubunit interfacial stability is a hallmark of the proteasomal AAA-ATPase heterohexamer (36).
During transitions from SA to any of the alternative states, the OB ring behaves as a rigid body as its position slides above the AAA domains. By contrast, each Rpt AAA domain exhibits a distinct reconfiguration, manifested most obviously in different extents of hinge-like rotation between the small and large AAA subdomains (SI Appendix, Fig. S12 and Table S3). Both the OB and AAA rings translate and tilt to align axially with the CP channel (25–27). The current resolutions of the SB, SC, and SD states are insufficient to resolve nucleotide densities. This leaves an open question as to whether and how ATPase repositioning is correlated with ATP hydrolysis cycles.
The Substrate-Translocation Pathway.
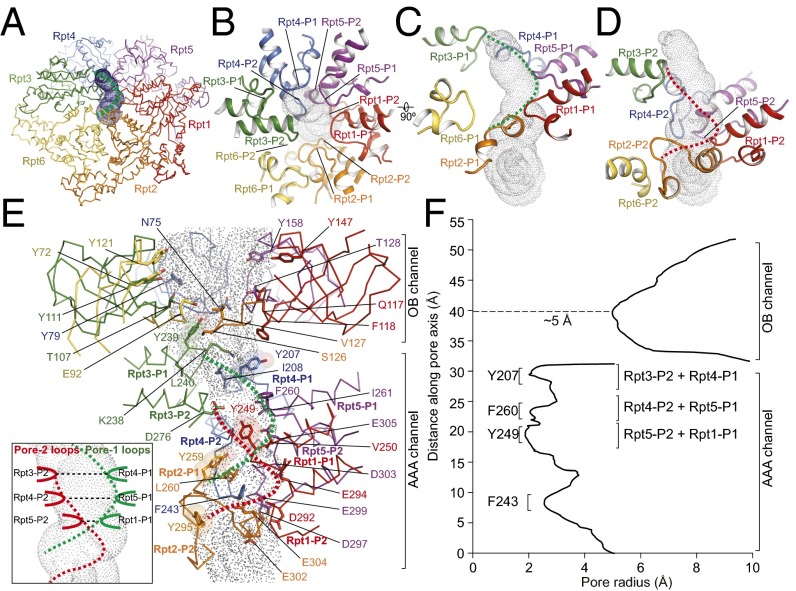
The substrate-translocation channel in the center of the ATPase ring is mainly shaped by inward-facing pore loops (23, 36, 37). Each ATPase contains four pore loops, two on the OB domain and two on the AAA domain (Fig. 4). The right-handed helical architecture in the AAA channel gives rise to a constriction much narrower than the OB channel (Fig. 4 E and F). The interior of the AAA channel is largely negatively charged. By contrast, the interior of the OB channel is positively charged (SI Appendix, Fig. S11C). Both channels are dramatically enriched for tyrosine residues. The OB channel has six tyrosine residues (Tyr147 in Rpt1, Tyr72 and Tyr121 in Rpt6, Tyr111 in Rpt3, Tyr79 in Rpt4, and Tyr158 in Rpt5), with their oxygen atoms pointed toward the substrate-translocation pathway (Fig. 4E). The AAA channel has five tyrosine residues from Rpt1 to Rpt4 (Fig. 4E and SI Appendix, Fig. S13).
Fig. 4.
Architecture of the substrate-translocation channel. (A) Overview of the AAA channel calculated by the HOLE program (50). The channel is rendered by surface dots. The dashed green curve indicates the spiral shape formed by the pore-1 loops. (B) Top view of the pore loops aligning along the channel axis from the perspective of the OB domain. (C) Close-up side views of the pore-1 loops from six Rpt subunits decorating the channel, which align along the channel in a spiral staircase formed from Rpt1 to Rpt5, with a backward recession in the Rpt6 pore-1 loop that is slightly away from the major channel pathway. The pore-1 loops form a helical part of the channel interior as illustrated by the dashed green line. (D) Close-up side view of the pore-2 loops from six Rpt subunits decorating the channel, which form a complete spiral staircase from Rpt1 to Rpt6. These pore-2 loops form another helical part of the channel interior, illustrated by the red dashed line. (E) Side view of the complete ATPase channel, including components from both the OB and AAA domains, calculated by the HOLE program (50). Side-chain patterns observed along the substrate-translocation pathway are highlighted. The five tyrosine residues, highlighted by transparent sphere representation, and a number of hydrophobic and negatively charged residues decorate the AAA channel; color codes for ATPase protomers match those shown in A. (Inset) A schematic cartoon showing that the pore-2 loops of Rpt3, Rpt4, and Rpt5 pair laterally with the pore-1 loops of Rpt4, Rpt5, and Rpt1, respectively, to form the three narrowest constrictions in the AAA channel. (F) The channel radius along the pore axis approximately estimated by HOLE (50), showing the three narrowest constrictions in the AAA channel but only one narrow constriction in the OB channel that is more than twice as wide as those of the AAA channel.
In the SA state, pore-1 loops from Rpt3, Rpt4, Rpt5, Rpt1, and Rpt2 constitute a helical inner surface. These loops feature the conserved hydrophobic [Tyr/Phe]-[Val/Leu/Ile]-Gly sequence pattern that has been previously suggested to drive substrate translocation in many ATP-dependent unfoldases such as HslU, ClpX, ClpA, LonA, FtsH, and PAN (37, 38) (Fig. 4 C and E and SI Appendix, Figs. S13 A and B and S14). The pore-2 loops constitute a second helical inner surface running parallel to the pore-1 loops (Fig. 4 B and D). Note that these pore loops directly extend from the Walker B motif, which is involved in ATP hydrolysis (39). Unlike the hydrophobic pore-1 loops, the pore-2 loops are heavily populated with conserved, charged residues (seven glutamates and two aspartates) (Fig. 4E and SI Appendix, Fig. S13C).
The AAA channel in the SA state has three narrow constrictions, defined by the axial area of overlap between the two pore-loop helices (Fig. 4E, Inset). In this region, Tyr207 and Ile208 from Rpt4, Phe260 and Ile261 from Rpt5, and Tyr249 and Val250 from Rpt1, all pore-1 loop residues, are paired with neighboring charged residues in the pore-2 loops of Rpt3, Rpt4, and Rpt5, respectively (Fig. 4 E and F). Interestingly, the phenotypic effects of pore-loop mutations are similar for the two members of each pore-loop pair. Thus, for the pore-1 loop, Rpt4 has the strongest phenotype, whereas for the pore-2 loop the partner Rpt3 has the strongest phenotype among the paired loops (36). Also, the strength of the phenotype diminishes with distance along the pore axis. As a result of pore-loop pairing, the channel radius is constricted to as little as ∼2 Å (Fig. 4F). Among the pore loops, Rpt6 is uniquely displaced from the channel axis and does not contribute to these constrictions in the SA and SB states (Fig. 4 C and D). Consistent with the observed displacement, mutating the pore-2 loop of Rpt6 had little impact on protein-degradation rates (36).
RP–CP Interactions.
The interface between the RP and CP is dominated by the associations of the hexameric AAA-ATPases and the heptameric α-ring (Fig. 5A). This symmetry-mismatched interface buries ∼3,600 Å2 of surface area in the SA state (SI Appendix, Table S2). All Rpt subunits but Rpt6 are in direct contact with the α-ring (Fig. 5A). A large gap between Rpt6 and the α2-subunit is forced by the amino-terminal PCI domain of Rpn6, which itself contacts α2, preventing Rpt6 from directly contacting the α-ring (Fig. 5A). The amino-terminal PCI domain of Rpn6 extends down to the lateral surface of the α2-subunit, making an ∼620-Å2 contact, which is perhaps critical for stabilizing the lateral position of Rpn6 (SI Appendix, Fig. S15 A and C). The amino-terminal PCI domain of Rpn5 is also found lateral to the CP, although its contact area with α1 is small (∼50 Å2).
Fig. 5.
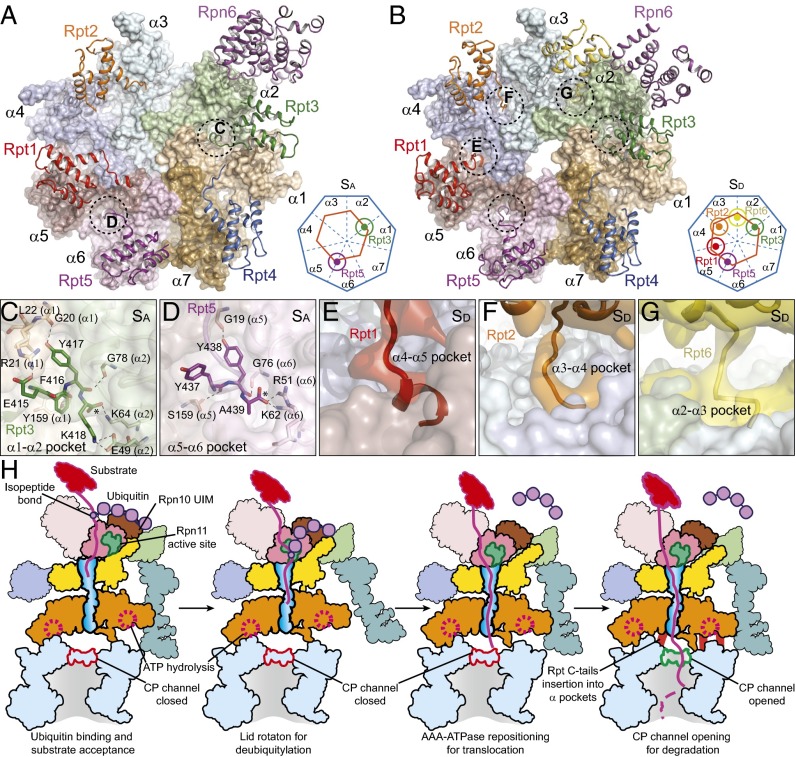
RP–CP interface regulates gating of the CP channel. (A) Overview of the RP–CP interface in which the carboxyl-terminal tails of Rpt3 and Rpt5 are shown to insert into the α-pockets in the SA state. Rpt6 is not shown, for clarity. The dashed circles mark the HbYX motifs of two Rpt subunits that insert into the α-pockets. (B) Overview of the RP–CP interface in which the carboxyl-terminal tails of Rpt1, Rpt2, Rpt6, Rpt3, and Rpt5 are shown to insert into the α-pockets in the SD state. The dashed circles mark the HbYX motifs of five Rpt subunits that insert into the α-pockets. (A and B, Insets) Simplified illustrations of the tail–pocket interactions between the ATPase ring and CP. The orange hexagons and blue heptagons represent the ATPase and the α-ring, respectively. The small circles connected to the orange hexagon represent the carboxyl-terminal tails of the Rpt subunits inserted into the α-pockets. (C) Close-up view of interactions between the C-terminal HbYX motifs of Rpt3 and the α1–α2 pocket in the SA state. (D) Close-up view of interactions between the C-terminal HbYX motifs of Rpt5 and the α5–α6 pocket in the SA state. (E–G) Close-up view of interactions between the Rpt1 (E), Rpt2 (F), and Rpt6 (G) C termini and the α4–α5, α3–α4, and α2–α3 pockets in the SD state, respectively. (H) Hypothetical structure-based model for substrate degradation by the human proteasome. UIM, ubiquitin-interacting motif.
Previous biochemical studies on the yeast proteasome suggested that the C termini of Rpt2, Rpt3, and Rpt5 contain conserved C-terminal HbYX motifs that insert into pockets formed between adjacent α-subunits (40, 41). In the SA state, the HbYX motifs of both Rpt3 and Rpt5 insert into the α1–α2 and α5–α6 pockets, respectively (Fig. 5 C and D and SI Appendix, Fig. S15 G and H). Consistent with previous structural data, Tyr417 in Rpt3 and Tyr438 in Rpt5 interact with Gly20-O of the α1-subunit and Gly19-O of the α5-subunit, respectively, through hydrogen bonding. The terminal carboxyl groups of Lys418 in Rpt3 and Ala438 in Rpt5 form hydrogen bonds with Lys64 of α2 (Fig. 5C) and Lys62 of α6 (Fig. 5D), respectively. These contacts are remarkably conserved, as they have previously been noted for homomeric proteasome precursors from archaea (41). By contrast, experimental density corresponding to the C-terminal HbYX tail (residues 434 to 440) of Rpt2 is missing, whereas there is no additional density in the α3–α4 pocket (SI Appendix, Fig. S15D). Similarly, the carboxyl-terminal tails of Rpt1 and Rpt4 do not insert into any α-pockets (SI Appendix, Fig. S15 E and F). These structural features may explain the previous finding that the extreme C termini of Rpt3 and Rpt5 are essential for assembly of the human proteasome (42, 43).
Gating of the CP Channel.
In the states in which the CP channel is closed (i.e., SA, SB, and SC), only Rpt3 and Rpt5 are found to have their C termini inserted into α-pockets (Fig. 5A and SI Appendix, Fig. S15 G–J). By contrast, in the SD state, the carboxyl-terminal tails of Rpt1, Rpt2, and Rpt6 are inserted into the α4–α5, α3–α4, and α2–α3 pockets, respectively (Fig. 5 E–G). Thus, except for Rpt4, all Rpt carboxyl-terminal tails are inserted into α-pockets in the SD state (SI Appendix, Fig. S15 K–M). The principal channel-blocking tails are from α2 and α4; the α3-tail, which behaves as a lynchpin of the gate (44), is controlled by Rpt2 (Fig. 2N) (40). The reorientation of these tails constitutes gating and controls substrate entry into the CP. Consistently, the AAA-ATPase heterohexamer is translated for ∼5 Å and rotates for ∼2° on the α-ring in the SC state, and is further translated for ∼10 Å and rotated for ∼4° in the SD state. Although the amino-terminal tails of α2, α3, and α4 are rotated over a large angle to roughly align along the heptameric axis to open the CP gate in the SD state (Figs. 2 O and P and 5 A and B), the helical elements connected to the gate-blocking tails in the α-ring are nearly identical to those in other states. This is reminiscent of the observation of the open gate of the yeast CP in complex with the 11S regulator from Trypanosoma brucei (33, 41).
Comparison of the RP–CP interface among different conformational states reveals a structural mechanism for gating of the CP channel. The initial repositioning of the ATPase ring on the α-ring in the SC state follows a large rotation of ∼40° in the lid in the SB state. However, this movement is insufficient to allow the insertion of additional Rpt carboxyl-terminal tails into the α-pockets, leaving the CP channel closed in the SC state. Further movement of the ATPase ring on the α-ring eventually allows the carboxyl-terminal tails of Rpt1, Rpt2, and Rpt6 to reach their nearest α-pockets. The insertion of these Rpt carboxyl-terminal tails into the α-pockets rotates the three gate-blocking tails of α2–α3–α4 to approximately align along the axial direction, which opens the CP channel in the SD state. Importantly, our observations indicate that the open state of the CP channel in the human holoenzyme cannot be immediately achieved through RP–CP association. Instead, opening of the CP channel is primed through a series of coordinated, stepwise remodeling events around the RP–CP interface, including a lid reconfiguration, ATPase repositioning, and insertion of three additional Rpt carboxyl-terminal tails into the α-pockets (Fig. 5H).
Discussion
The coexistence of four conformational states, one of which is open in its CP gate under common solution conditions, provides insights into the dynamic regulation of the proteasome holoenzyme (Fig. 5H). We may assume SA, the most populated state, to represent a ground state, whereas SD, the state most distinct from SA, to represent the translocating or fully engaged state of the proteasome, in accordance with prior studies of the yeast proteasome (7, 25–28) (SI Appendix, Fig. S8 E and F). By structural criteria (SI Appendix, Table S3), the SB and SC states may represent intermediates between SA and SD, and may describe a progression of conformational transitions that is temporally ordered: SA is converted to SB, then to SC and SD (Fig. 5H). These transitions, although apparently inherent to the proteasome, are expected to be guided by interactions with substrate, particularly the engagement of substrate by the AAA pore loops. The transition from SA to SB involves local dissociation of the Rpn5–Rpt6 and Rpn6–Rpt6 interfaces. This may topologically destabilize the lid conformation in the SA state, allowing it to rotate around ATPase with a large angle (∼40°). The key consequence of this lid rotation is the repositioning of Rpn11 above the OB ring, which was thought to free the Rpn11 catalytic loop and mobilize its Ins-1 region (15, 18, 25). This suggests that Rpn11 may be enhanced in its deubiquitinating activity in the SB, SC, and SD states (25).
Rearrangement of the lid may not only reposition Rpn11 but also perturb the RP–CP interface, including the lateral contact of Rpn6 to the CP. The dissolution of this contact may release a constraint on movement of the ATPase heterohexamer on the α-ring in the SC and SD states. The stepwise repositioning of the ATPases on the α-ring aligns axially the OB ring and the AAA channel with the CP channel (25–27), allowing a major change in the ATPase–CP interface, where additional Rpt carboxyl-terminal tails are inserted into the α-pockets, to eventually open the CP gate in the SD state.
In both the SC and SD states, a major rearrangement of the AAA channel would allow the pore loops from Rpt3 and Rpt4, the first Rpt pair interacting with an incoming substrate, to move toward the CP (SI Appendix, Fig. S12). The contrasting side-chain patterns of the pore loops—the hydrophobic pore-1 versus the highly charged pore-2 loops—suggest that they might play distinct roles in processive substrate translocation (36, 45). We speculate that the pore-1 loops may propel substrates forward along the AAA channel through hydrophobic interactions, whereas the pore-2 loops translate their interactions with substrates into enhancement of ATP hydrolysis (46), possibly by controlling the configuration of the conserved glutamate residue in the Walker B motif (39) (SI Appendix, Fig. S16).
Taken together, our structural data lead to a model where stepwise, distributed conformational changes within the holoenzyme are coordinated so as to generate a series of enzymatic regulatory events: The deubiquitinating activity of Rpn11 is enhanced in an SB-like state; the ATPase activity is likely heightened in an SC-like state, which can initiate substrate translocation; and eventually the CP gate is opened for degradation in a translocation-competent SD-like state. In the presence of a substrate in the translocation channel, we would imagine that the open gate would be stabilized until completion of substrate degradation.
Confirmatory evidence of our ground-state structure is as follows: Coincident with this study, two other groups determined the high-resolution cryo-EM structures of the human proteasome holoenzyme in the ground state (47, 48). Their structures are largely consistent with our proteasome structure in the SA state. Key structural features of their ground-state structures are virtually identical to our SA state, including (i) the closed CP gate, (ii) the insertion of the carboxyl-terminal tails of Rpt3 and Rpt5 into the α-pockets, and (iii) occupancy of all six nucleotide-binding sites.
Materials and Methods
Human proteasomes were affinity-purified on a large scale from a stable HEK293 cell line (30). Cryo-EM data were collected with the supercounting mode of a Gatan K2 Summit direct electron detector installed on an FEI Tecnai Arctica microscope operating at a nominal magnification of 21,000× and an acceleration voltage of 200 kV. Details of experimental procedures and cryo-EM data processing are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded in part by a grant from the NIGMS (GM026875; to M.W.K.), a grant of the Thousand Talents Plan of China (to Y.M.), an Intel academic grant (to Y.M.), a grant from the National Natural Science Foundation of China (91530321; to Y.M. and Q.O.), and an NIH grant (GM43601; to D.J.F.). The cryo-EM experiments were performed in part at the Center for Nanoscale Systems at Harvard University, which is supported by the National Science Foundation under NSF Award 1541959. The cryo-EM facility was funded through NIH Grant AI100645 and the Center for HIV/AIDS Vaccine Immunology and Immunogen Design.
Footnotes
The authors declare no conflict of interest.
Data deposition: The single-particle reconstructions and atomic coordinates reported in this paper have been deposited in the Electron Microscopy Data Bank, www.emdatabank.org (accession nos. EMD-8332 to EMD-8337) and Protein Data Bank, www.wwpdb.org [PDB ID codes 5T0C (the doubly capped proteasome in the SA state), 5T0G, 5T0H, 5T0I, and 5T0J (the half proteasome in the SA, SB, SC, SD states, respectively)]. The raw micrographs and particle data have been deposited in the Electron Microscopy Pilot Image Archive, www.ebi.ac.uk/pdbe/emdb/empiar (accession no. EMPIAR-10072).
See Commentary on page 12896.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614614113/-/DCSupplemental.
References
- 1.Finley D, Chen X, Walters KJ. Gates, channels, and switches: Elements of the proteasome machine. Trends Biochem Sci. 2016;41(1):77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kish-Trier E, Hill CP. Structural biology of the proteasome. Annu Rev Biophys. 2013;42:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harshbarger W, Miller C, Diedrich C, Sacchettini J. Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure. 2015;23(2):418–424. doi: 10.1016/j.str.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Huber EM, Groll M. Inhibitors for the immuno- and constitutive proteasome: Current and future trends in drug development. Angew Chem Int Ed Engl. 2012;51(35):8708–8720. doi: 10.1002/anie.201201616. [DOI] [PubMed] [Google Scholar]
- 7.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109(37):14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109(5):1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46(1):54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Löwe J, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 12.Groll M, et al. Structure of 20S proteasome from yeast at 2.4Å resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 13.Unno M, et al. The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure. 2002;10(5):609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 14.Pathare GR, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA. 2012;109(1):149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathare GR, et al. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci USA. 2014;111(8):2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riedinger C, et al. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem. 2010;285(44):33992–34003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Young P, Walters KJ. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol. 2005;348(3):727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol. 2014;21(3):220–227. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 19.Zhang N, et al. Structure of the S5a:K48-linked diubiquitin complex and its interactions with Rpn13. Mol Cell. 2009;35(3):280–290. doi: 10.1016/j.molcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dambacher CM, Worden EJ, Herzik MA, Martin A, Lander GC. Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition. eLife. 2016;5:e13027. doi: 10.7554/eLife.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehringer J, et al. Structural and functional characterization of Rpn12 identifies residues required for Rpn10 proteasome incorporation. Biochem J. 2012;448(1):55–65. doi: 10.1042/BJ20120542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, et al. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric α-helical rings. Structure. 2012;20(3):513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34(4):473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, et al. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34(4):485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luan B, et al. Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc Natl Acad Sci USA. 2016;113(10):2642–2647. doi: 10.1073/pnas.1601561113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20(7):781–788. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Śledź P, et al. Structure of the 26S proteasome with ATP-γS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci USA. 2013;110(18):7264–7269. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unverdorben P, et al. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci USA. 2014;111(15):5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estrin E, Lopez-Blanco JR, Chacón P, Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure. 2013;21(9):1624–1635. doi: 10.1016/j.str.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, et al. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46(11):3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 31.Scheres SH, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4(1):27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 32.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408(6808):115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 34.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 35.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: Common structure–diverse function. Genes Cells. 2001;6(7):575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 36.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nat Struct Mol Biol. 2013;20(10):1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139(4):744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iosefson O, Nager AR, Baker TA, Sauer RT. Coordinated gripping of substrate by subunits of a AAA+ proteolytic machine. Nat Chem Biol. 2015;11(3):201–206. doi: 10.1038/nchembio.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Wigley DB. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat Struct Mol Biol. 2008;15(11):1223–1227. doi: 10.1038/nsmb.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian G, et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011;18(11):1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadtmueller BM, et al. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J Biol Chem. 2010;285(1):13–17. doi: 10.1074/jbc.C109.070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YC, DeMartino GN. C termini of proteasomal ATPases play nonequivalent roles in cellular assembly of mammalian 26 S proteasome. J Biol Chem. 2011;286(30):26652–26666. doi: 10.1074/jbc.M111.246793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SH, Moon JH, Yoon SK, Yoon JB. Stable incorporation of ATPase subunits into 19 S regulatory particle of human proteasome requires nucleotide binding and C-terminal tails. J Biol Chem. 2012;287(12):9269–9279. doi: 10.1074/jbc.M111.316208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7(11):1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 45.Erales J, Hoyt MA, Troll F, Coffino P. Functional asymmetries of proteasome translocase pore. J Biol Chem. 2012;287(22):18535–18543. doi: 10.1074/jbc.M112.357327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peth A, Kukushkin N, Bossé M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. J Biol Chem. 2013;288(11):7781–7790. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweitzer A, et al. Structure of the human 26S proteasome at a resolution of 3.9 Å. Proc Natl Acad Sci USA. 2016;113(28):7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang X, Luan B, Wu J, Shi Y. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol. 2016;23(9):778–785. doi: 10.1038/nsmb.3273. [DOI] [PubMed] [Google Scholar]
- 49.Wu J, et al. 2016. Unsupervised single-particle deep classification via statistical manifold learning. arXiv:1604.04539.
- 50.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14(6):354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.