Significance
Contrary to the paradigm, the signal peptide of CD18, an adhesive protein expressed on the surface of leukocytes of cattle and other ruminants, is not cleaved. Intriguingly, leukotoxin produced by Mannheimia haemolytica binds to the intact signal peptide and causes lysis of ruminant leukocytes, resulting in acute pneumonia. In this proof-of-principle study, we used precise gene editing to induce a single amino acid substitution in CD18, which resulted in the cleavage of the signal peptide and abrogation of leukotoxin-induced cytolysis of leukocytes of the gene-edited bovine fetus. This report demonstrates the feasibility of developing lines of cattle genetically resistant to M. haemolytica-caused pneumonia, which will have significant impact on the sustainability of food-animal production in the United States and elsewhere.
Keywords: Mannheimia haemolytica, leukotoxin-resistant, cattle, cloning
Abstract
Signal peptides of membrane proteins are cleaved by signal peptidase once the nascent proteins reach the endoplasmic reticulum. Previously, we reported that, contrary to the paradigm, the signal peptide of ruminant CD18, the β subunit of β2 integrins, is not cleaved and hence remains intact on mature CD18 molecules expressed on the surface of ruminant leukocytes. Leukotoxin secreted by Mannheimia (Pasteurella) haemolytica binds to the intact signal peptide and causes cytolysis of ruminant leukocytes, resulting in acute inflammation and lung tissue damage. We also demonstrated that site-directed mutagenesis leading to substitution of cleavage-inhibiting glutamine (Q), at amino acid position 5 upstream of the signal peptide cleavage site, with cleavage-inducing glycine (G) results in the cleavage of the signal peptide and abrogation of leukotoxin-induced cytolysis of target cells. In this proof-of-principle study, we used precise gene editing to induce Q(‒5)G substitution in both alleles of CD18 in bovine fetal fibroblast cells. The gene-edited fibroblasts were used for somatic nuclear transfer and cloning to produce a bovine fetus homozygous for the Q(‒5)G substitution. The leukocyte population of this engineered ruminant expressed CD18 without the signal peptide. More importantly, these leukocytes were absolutely resistant to leukotoxin-induced cytolysis. This report demonstrates the feasibility of developing lines of cattle genetically resistant to M. haemolytica-caused pneumonia, which inflicts an economic loss of over $1 billion to the US cattle industry alone.
Mannheimia (Pasteurella) haemolytica is a Gram-negative coccobacillus commonly found as a commensal organism in the nasopharynx of healthy cattle and other ruminants (1). In conjunction with active viral infection and stress factors, it reaches the lungs, multiplies, and causes an acute fibrinonecrotic pleuropneumonia (2). It produces several virulence factors including the capsule, outer membrane proteins, lipopolysaccharide and leukotoxin (3). Leukotoxin-induced polymorphonuclear leukocyte (PMN) lysis and degranulation is the primary cause of the acute inflammation and lung injury characteristic of this disease (4, 5). Furthermore, leukotoxin-deletion mutants of M. haemolytica cause reduced mortality and much milder lung lesions than the wild-type organisms in calves (6–8) and other ruminants (9). Therefore, leukotoxin is recognized as the most critical virulence factor of this organism that is primarily responsible for the induction of respiratory disease and death in calves.
Previously, we showed that leukotoxin binds to amino acids 5–17 of the signal peptide of ruminant CD18, the β subunit of β2 integrins, which, contrary to its paradigmatic cleavage in nonruminants, remains uncleaved and intact on mature CD18 molecules expressed on the cell surface of ruminant leukocytes (10). Furthermore, our studies revealed that the signal peptide sequence of ruminant CD18 contains cleavage-inhibiting glutamine (Q), whereas that of nonruminants contains cleavage-inducing glycine (G) at amino acid position 18, which is position 5 upstream of the cleavage site. Site-directed mutagenesis leading to the substitution of Q with G at amino acid position ‒5 resulted in the cleavage of the signal peptide and abrogation of leukotoxin-induced cytolysis of target cells (10), revealing an approach to genetically engineer cattle that will produce leukocytes resistant to leukotoxin via the expression of signal peptide-less CD18 on their surface. In this proof-of-principle study demonstrating the utility of this approach, we used precise gene editing to modify both alleles of CD18 to create bovine fetal fibroblasts bearing the Q(‒5)G substitution and somatic nuclear transfer to clone a bovine fetus producing leukocytes that express signal peptide-less CD18 on their surface.
Results
Cotransfection of Fetal Fibroblasts with Custom-Made Zinc Finger Nuclease and CD18 Oligonucleotide Carrying the Q(‒5)G Substitution Results in Biallelic Modification of CD18.
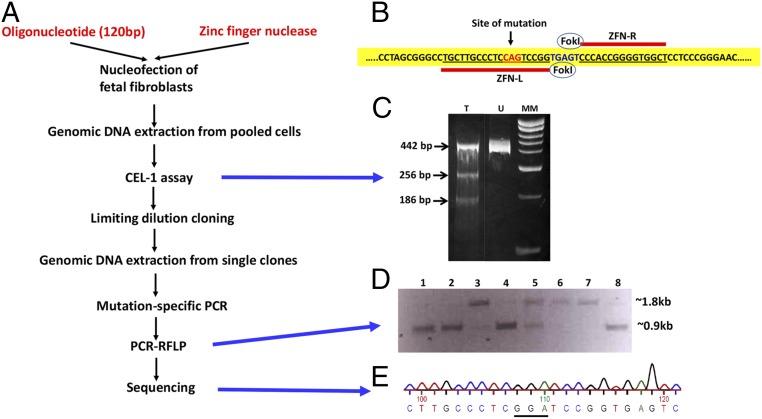
A primary fibroblast cell line was established from a 42-d-old male Holstein fetus. Three pairs of zinc-finger nucleases (ZFN) were custom-made (Sigma‒Aldrich) to target exon 2 of bovine ITGB2 (integrin subunit beta 2) that encodes the signal peptide of CD18. The ZFN pair with the highest activity in the MEL-1 assay (Sigma) was used for gene targeting. Fetal fibroblasts were subjected to nucleofection (Lonza) using the pair of ZFN mRNAs along with a single-stranded oligonucleotide (120 nt, Integrated DNA Technologies) carrying the glycine-encoding GGA codon instead of glutamine-encoding CAG codon as a template for homology-directed repair (HDR) (Fig. 1 A and B). Transfected cells were subjected to transient hypothermia to increase the level of stable ZFN-induced gene disruption and thereby enhance the efficacy of ZFNs (11). ZFN-induced gene disruption was confirmed by the surveyor nuclease (Cel-1) assay. Although a small number of double-strand breaks are fixed by HDR, the majority are repaired by nonhomologous end joining (NHEJ). Quantification of repairs made by NHEJ by the Surveyor Nuclease (Cel-1) assay provides an indication of the cleavage efficiency of the ZFNs. In this assay, a 442-bp region encompassing the FokI-nuclease cleavage site was PCR-amplified. The amplicons were digested with Surveyor Nuclease S enzyme, and the products were visualized after electrophoresis. The lower intensity of the 442-bp band and the presence of the 256-bp and 186-bp bands in the digest from the ZFN-transfected cells, in comparison with the digest from the untransfected cells, indicated a relatively high rate of ZFN-mediated double-strand breaks (Fig. 1C).
Fig. 1.
Development of bovine fetal fibroblasts containing the Q(‒5)G substitution in both alleles of CD18. (A) Schematic representation of ZFN-mediated gene editing of bovine ITGB2. (B) ZFN-mediated induction of the Q(‒5)G substitution. The pair of ZFNs were designed to cleave bovine ITGB2 in the vicinity of the codon targeted for editing (red fonts). Sequences bound by each ZFN are underlined in red. ITGB2 domain targeted for FokI-cleavage is in blue fonts. (C) Cel-1 assay. Lanes T and U show the digestion products from the ZFN-transfected and ZFN-untransfected cells, respectively. Lane MM represents molecular weight markers. (D) PCR-RFLP assay. The ∼1.8-kb and ∼0.9-kb bands represent the undigested and digested products, respectively. In this figure, lanes 1, 2, 4, and 8 show clones that are homozygous, whereas lane 5 shows a clone that is heterozygous for the mutation. Lanes 3, 6, and 7 show clones without any modification. (E) Sequencing analysis. Sequencing data shows the presence of glycine-encoding GGA codon instead of the glutamine-encoding CAG codon.
Limiting dilution cloning of the transfectants yielded single colonies, 500 of which were screened for presence of Q(‒5)G substitution in CD18 by a mutation-specific PCR assay. Twenty-seven colonies were found to be positive, suggesting that these colonies may contain the Q(‒5)G substitution in one or both alleles. Colonies containing the biallelic modification were further identified by a PCR-restriction fragment length polymorphism (RFLP)-based assay (Fig. 1D). The replacement of glutamine-encoding CAG codon by glycine-encoding GGA codon introduces a BamHI restriction site at the point of mutation. The PCR-RFLP assay was performed by amplifying a 1,820-bp fragment of bovine ITGB2 encompassing the targeted sequence followed by digestion with BamHI. Amplicons from clonal lines that do not contain the mutation are not digested by BamHI and hence yield only the undigested fragment of 1,820 bp. Amplicons from cells that are heterozygous for the edited allele yield both undigested (1,820 bp) and digested fragments (910 bp). Amplicons from cells that are homozygous for the mutation yield only the digested fragments (910 bp). This assay revealed nine colonies that were homozygous for the intended mutation. A fragment of CD18 containing the mutation site was PCR-amplified from the genomic DNA isolated from these nine colonies. Sequencing of the amplicons further confirmed biallelic Q(‒5)G editing events (Fig. 1C).
Somatic Nuclear Transfer and Cloning Results in a Bovine Fetus Carrying the Q(‒5)G Substitution in CD18.
A clonal cell line was selected for somatic cell nuclear transfer, embryo development, and implantation, using standard procedures (12, 13). At 125-d gestational age, a fetus was harvested. Physical examination of the fetus was conducted by a clinical veterinarian. Based on the gross appearance, weight, and length (crown/rump) of the fetus, color and amount of amniotic and allantoic fluid, and appearance of the placenta and cotyledons, the fetus was confirmed to be healthy. Blood was collected from the fetus, and leukocytes were isolated.
CD18 of the Gene-Edited Fetus Carries the Q(‒5)G Substitution.
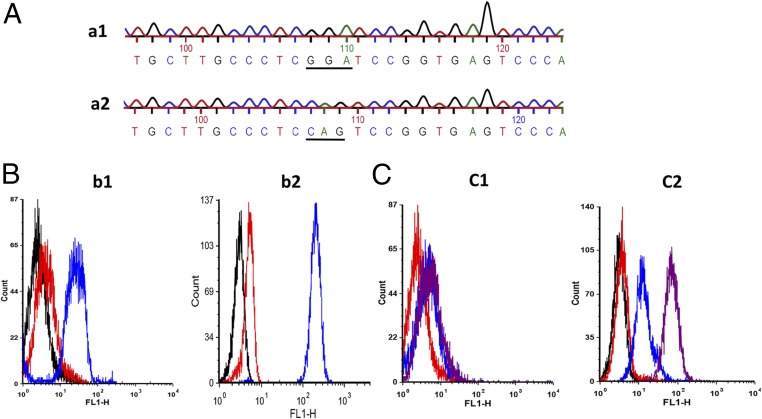
Genomic DNA was isolated from the leukocytes of the gene-edited fetus and a wild-type calf. A fragment of CD18 containing the mutation site was PCR-amplified from the genomic DNA, and the amplicons were sequenced. DNA from the gene-edited fetus contained glycine-encoding GGA codon instead of glutamine-encoding CAG codon at the targeted position demonstrating successful allele replacement by HDR (Fig. 2A).
Fig. 2.
Leukocytes from the gene-edited fetus express CD18 without the signal peptide. (A) Leukocytes from the gene-edited fetus contain the Q(‒5)G substitution in both alleles of CD18. Sequencing data shows the presence of glycine-encoding GGA codon in the CD18 DNA from the gene-edited fetus (a1) and glutamine-encoding CAG codon in the CD18 DNA from the wild-type CD18 DNA (a2). (B) CD18 expression on leukocytes from the gene-edited fetus is comparable to that on leukocytes from a wild-type calf. CD18 expression on the leukocytes from the gene-edited fetus (b1) or a wild-type calf (b2) was determined by flow cytometry. The blue and red histograms represent binding of anti-CD18 antibodies and isotype-matched control antibodies, respectively. The black histogram represents unstained cells. (C) CD18 expressed on leukocytes from the gene-edited fetus lacks the signal peptide. The presence of signal peptide on CD18 expressed on the leukocytes from the gene-edited fetus (c1) or a wild-type calf (c2) was determined by flow cytometry. The magenta and blue histograms represent binding of chicken anti-signal peptide serum and unimmunized chicken serum, respectively. The red histogram represents unstained cells. Leukocytes from the wild-type calf expressed CD18 containing the signal peptide, whereas the leukocytes from the gene-edited fetus expressed CD18 without the signal peptide. Results of one representative experiment of two are shown.
Leukocytes from the Gene-Edited Fetus Exhibit Normal Expression of CD18.
Comparative flow cytometric analysis of leukocytes from the gene-edited fetus and a wild-type calf using a mouse monoclonal anti-bovine CD18 antibody (BAQ30A, IgG1) and isotype-matched control antibody (MM601, IgG1) followed by FITC-conjugated goat-anti-mouse Ig antibodies revealed comparable surface expression of bovine CD18 on the leukocytes of the gene-edited fetus and the wild-type calf (Fig. 2B).
Leukocytes from the Gene-Edited Fetus Express Signal Peptide-Less CD18.
To determine whether the signal peptide was cleaved from CD18 expressed on leukocytes of the gene-edited fetus, flow cytometric analysis was performed on leukocytes from the gene-edited fetus and a wild-type calf, with chicken anti-signal peptide serum (10) followed by FITC-conjugated rabbit anti-chicken Ig antibodies. As expected, leukocytes from the wild-type calf were stained by the anti-signal peptide serum, whereas leukocytes from the gene-edited fetus were not, confirming the cleavage of signal peptide from CD18 (Fig. 2C).
Leukocytes from the Gene-Edited Fetus Are Resistant to Leukotoxin-Induced Cytolysis.
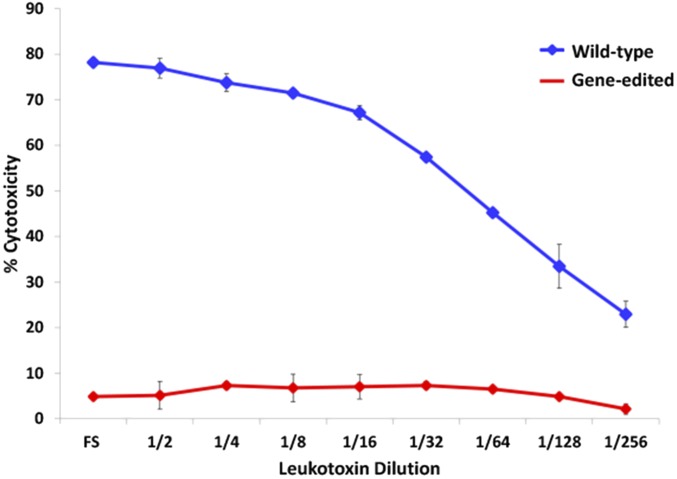
To confirm whether the cleavage of the signal peptide from CD18 results in the abrogation of leukotoxin-induced cytolysis of leukocytes, the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] dye-reduction cytotoxicity assay (14) with leukotoxin was performed on leukocytes from the gene-edited fetus and a wild-type calf. Leukocytes from the wild-type calf were effectively lysed by leukotoxin in a concentration-dependent manner, whereas the leukocytes from the gene-edited fetus were not (Fig. 3). It is noteworthy that the leukotoxin caused 50% cytotoxicity to wild-type leukocytes up to a dilution of 1:32, whereas the leukocytes from the gene-edited fetus were not lysed by leukotoxin even at full strength, indicating the absolute resistance of leukocytes from the gene-edited fetus. Taken together these results clearly indicate that the Q(‒5)G substitution in CD18 indeed confers complete resistance to leukotoxin-induced cytolysis on bovine leukocytes.
Fig. 3.
Leukocytes from the gene-edited fetus are resistant to M. haemolytica leukotoxin-induced cytolysis. Leukocytes from the gene-edited fetus and a wild-type calf were subjected to the MTT dye-reduction cytotoxicity assay with serial dilutions of M. haemolytica leukotoxin. Leukocytes from the wild-type calf (blue line) were lysed in a concentration-dependent manner, whereas the leukocytes from the gene-edited fetus (red line) were not lysed even by the undiluted leukotoxin. All data are expressed as mean ± SD (n = 3).
Discussion
Studies on interactions between human β2 integrins and their ligand ICAM-1 have revealed that the amino acid motifs of CD18 that interact with ICAM-1 do not lie at the N-terminal end of CD18 (15, 16). The modification that we have introduced is directed to the N-terminal end of CD18. More importantly, we are not altering the N-terminal end of the mature CD18 protein, but reintroducing a cleavage site to induce cleavage of the signal peptide after it serves its designated function of leading the nascent CD18 molecule to the endoplasmic reticulum. Hence, the cleavage of the signal peptide, which consistently occurs in nonruminant CD18, is very unlikely to interfere with the interaction between β2 integrins and ICAM-1 and the corresponding orchestration of immune responses. Furthermore, this genetically engineered fetus contains only native DNA because the gene-editing technology used does not introduce any foreign DNA into the genome.
The development of a technology with the potential to develop cattle that are naturally resistant to M. haemolytica leukotoxin represents a very significant advancement toward control of this economically devastating disease. Although respiratory viruses, including bovine herpesvirus 1, bovine respiratory syncytial virus, and bovine viral diarrhea virus, predispose cattle to bovine respiratory disease complex, commonly known as shipping fever, acting alone, these viruses typically do not cause the death of the animals. Severe pneumonia and death of cattle are caused by secondary bacterial infection, particularly by M. haemolytica. Even partial reduction of the economic loss due to bovine respiratory disease will result in significant savings with clear impact on the long-term enhancement of sustainability of food-animal production in the United States and elsewhere. Furthermore, production of calves that are inherently resistant to M. haemolytica leukotoxin will also result in a commensurate reduction in the need for prophylactic and therapeutic antibiotics.
Methods
This study was carried out in conformance with US Department of Agriculture animal research guidelines, under protocol #4211 approved by the Washington State University Institutional Animal Care and Use Committee.
Establishment of a Fetal Bovine Fibroblast Cell Line.
A bovine fibroblast cell line was derived from a fetus using a previously published method with minor modification (12). A 42-d-old bovine fetus was removed by hysterotomy. The fetus was rinsed three times in Dulbecco’s PBS (DPBS) containing antibiotic-antimycotic (16 μL/mL; Cellgro) and fungizone (8 μL/mL; Gibco). The head and internal organs were removed, and the remaining tissues were chopped into very small pieces. The fibroblasts were released from the tissue by a standard trypsinization procedure using 0.05% trypsin-EDTA (17) (Gibco). The cells were seeded into 100-mm tissue culture plates in complete growth medium [α-MEM (α-MEM); HyClone] supplemented with 15% (vol/vol) FBS (Atlanta Biologicals), 2 mM l-glutamine (Gibco), 0.003% β-mercaptoethanol (Sigma), and 20 μg/mL of gentamicin (Invitrogen). Cells were frozen in freezing medium containing FBS [90% (vol/vol)] and dimethyl sulfoxide [10% (vol/vol); ATCC] until they were used for transfection.
Zinc Finger Nucleases and Oligonucleotide.
ZFNs targeting the CD18 double-stranded DNA in the vicinity of the site of mutation were engineered by Sigma-Aldrich. A 120-bp single-stranded CD18 oligonucleotide donor template carrying the Q to G substitution was synthesized and purified by PAGE (Integrated DNA Technologies). The oligonucleotide sequence: CAGGACATGCTGCGCCAGCGCCCCCAGCTGCTGCTCCTAGCGGGCCTGCTTGCCCTCGGATCCGGTGAGTCCCACCGGGGTGGCTCCTCCCGGGAACAGCGGGACCCACCCTCACACCCT. The oligonucleotide was diluted with RNase-free water to 100 μM and stored at ‒20 °C.
Genetic Modification of Bovine Fibroblasts.
Bovine fetal fibroblasts were cultured in complete growth medium at 37 °C and 5% (vol/vol) CO2. The cells subcultured 1:2 the previous day were harvested using 0.05% trypsin‒EDTA on the day of transfection. One million cells were cotransfected with 2 µg ZFN mRNA and 1 pmol oligonucleotide using nucleofection (Lonza) and incubated at 30 °C and 5% (vol/vol) CO2. After 2 d, the incubation temperature was raised to 37 °C (11). On the fourth day of transfection, cells were transferred to replica plates, one for genomic DNA extraction and the other for limiting dilution cloning. Genomic DNA was extracted from one of the replica plates of transfected and nontransfected cells using QIAamp DNA Mini Kit (Qiagen) following the protocol for DNA purification from cultured cells.
Surveyor Nuclease (Cel-I) Assay.
This assay was carried out as described previously (18). The target region (ZFN binding site) of 442 bp was PCR-amplified using genomic DNA from transfected and untransfected fibroblast cells as the template and the following primers: Cel1-F 5′GGAGTGGAGCACATGACCA-3′and Cel1-R 5′ATCAGGAAGCAGGGAGGAAA-3′. The Surveyor nuclease (Surveyor mutation detection kit; Transgenomic) was used according to the manufacturer’s instructions to determine the cleavage efficiency of ZFN. Briefly, the PCR amplicon was denatured under high temperature. The temperature was gradually lowered to allow some wild-type and nonhomologous end joining (NHEJ) products to hybridize to form double-stranded DNA with mismatches around the cleavage site, which were then cleaved by the enzyme nuclease S. The products were resolved by PAGE.
Selection of Colonies Containing the Q to G Substitution in both Alleles of CD18.
Transfected cells were subjected to limiting dilution cloning to obtain single-cell colonies. Genomic DNA extracted from single colonies (QuickExtract DNA extraction solution, Epicenter) 1 wk after cloning was screened for the integration of the Q(‒5)G substitution by performing the following assays: (i) mutation–specific PCR assay; (ii) PCR-based RFLP; and (iii) sequencing.
Mutation-Specific PCR Assay.
A 1,119-bp CD18 fragment was amplified using genomic DNA from single colonies of transfected fibroblasts as the template and the mutation-specific forward primer, 3′F 5′-GGCCTGCTTGCCCTCGGA-3′ and the reverse primer, which was positioned away from the oligonucleotide site, 3′R 5′-GGCTCGTGTCTGCAACCTAC-3′. The amplification was carried out with GoTaq master mix (Promega), using the following cycling conditions: 95 °C for 5 min for initial denaturation, 32 cycles at 95 °C for 30 s, 54 °C for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min.
PCR-Restriction Fragment Length Polymorphism-Based Assay.
Genomic DNA was PCR-amplified with primers flanking the single-stranded oligonucleotide target region. The primer pair, 5UTR-F 5′-ATCAGTTTGTGTCCAGTTCTCTGATTG-3′ and 3UTR-R 5′-AGAGGGTCTTGCAAGTAAAACATGAAG-3′, was used to generate a 1,822-bp fragment. The amplification was carried out with GoTaq master mix (Promega), using the following cycling conditions: 95 °C for 5 min for initial denaturation, 32 cycles at 95 °C for 30 s, 57.4 °C for 30 s and 72 °C for 2 min, and a final extension at 72 °C for 10 min. All of the PCR amplicons were digested with 10 U of BamHI at 37 °C overnight and resolved on agarose gel.
Sequencing.
To confirm whether homologous recombination and replacement of Q to G have occurred as expected, PCR amplicons of all of the colonies, which were positive on PCR-based RFLP, were sequenced. A DNA fragment containing the CD18 mutation site was generated by PCR using genomic DNA from leukocytes of the gene-edited fetus and a wild-type calf as template and sequenced to confirm the replacement of Q to G in both alleles of CD18.
Embryonic Cloning.
The cloned fetus was produced by TransOva Genetics using somatic nuclear transfer procedure, as described previously (12, 13).
Fetus Removal and Isolation of Leukocytes.
The 125-d-old fetus was removed by hysterotomy. Peripheral blood was collected from the gene-edited fetus and a wild-type calf by jugular venipuncture into heparinized tubes. Leukocytes were isolated from heparinized blood by osmotic lysis of RBCs using deionized water and centrifugation.
Flow Cytometry.
Leukocytes from the fetus and wild-type calf were examined for cell surface expression of bovine CD18 by using an anti-CD18 monoclonal antibody (mAb), BAQ30A (IgG1; VMRD), by flow cytometry, as previously described (19). Briefly, 5 × 106 cells mL−1 in 50 µL of FACS buffer [3% (vol/vol) horse serum and 0.01% sodium azide in PBS] were incubated with 50 µL of mAb BAQ30A (15 µg/mL) at 4 °C for 20 min. As an isotype control antibody, anti-M. haemolytica leukotoxin-specific mAb MM601 (IgG1) was used. After three washes in FACS buffer, the cells were incubated with 50 µL of FITC-conjugated goat anti-murine Ig Ab (1:200 dilution; Caltech Laboratories) at 4 °C for 20 min. The cells were washed three times with FACS buffer, resuspended in PBS, and analyzed by a flow cytometer (FACSort; Becton‒Dickinson Immunocytometry Systems).
Presence of the signal peptide on CD18 of leukocytes from the gene-edited fetus and wild-type calf was examined by flow cytometric analysis with the chicken antiserum developed against the synthetic peptide spanning amino acids 5–19 of bovine CD18 signal peptide (1) (Sigma‒Genosys). Unimmunized chicken serum was used as the negative control. The procedure was similar to the one described above for the detection of CD18 expression.
MTT dye-reduction cytotoxicity assay for leukotoxin-induced cytolysis of leukocytes.
Production of M. haemolytica leukotoxin and detection of leukotoxin-induced cytolysis of leukocytes were performed as previously described by us (14). This assay measures the ability of the ER-resident enzymes in viable cells to convert a tetrazolium dye [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; Sigma] into a purple formazan precipitate, which is later dissolved in acid isopropanol. The OD of the end product, representing the intensity of the purple color developed, is directly proportional to the viability of the cells. Briefly, the target cells were resuspended in colorless RPMI-1640 medium (without neutral red) at a concentration of 5 × 106 cells mL−1 and seeded into 96-well round-bottom microtiter plates (50 µL per well) containing the serially diluted leukotoxin in duplicates and incubated at 37 °C for 1 h. After incubation, cells were centrifuged at 600 × g for 5 min, and the supernatant fluid was discarded. The cell pellets were resuspended in 100 µL of colorless RPMI-1640 medium, and 20 µL of 0.5% MTT dye was added to each well. After 4 h of incubation at 37 °C, the plates were centrifuged at 600 × g for 5 min, and the supernatant fluid was removed. The formazan precipitate was thoroughly dissolved in 100 µL of acid isopropanol, and the OD of the samples was measured by using an ELISA reader at 540 nm. The percentage cytotoxicity was calculated as follows: percentage cytotoxicity = [1 ‒ (OD of toxin-treated cells/OD of toxin-untreated cells)] × 100.
Acknowledgments
We thank Yoshimi Kuroiwa from Kyowa Hakko Kirin, California, for technical advice on ZFN-mediated mutation in CD18. This study was funded by Grant 2011-03230 from the US Department of Agriculture.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Frank GH. In: Pasteurellosis of cattle. Pasturella and Pasteurellosis. Adlam C, Rutter JM, editors. Academic; New York: 1989. pp. 197–222. [Google Scholar]
- 2.Rehmtulla AJ, Thomson RG. A review of the lesions in shipping fever of cattle. Can Vet J. 1981;22(1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 3.Confer AW, Panciera RJ, Clinkenbeard KD, Mosier DA. Molecular aspects of virulence of Pasteurella haemolytica. Can J Vet Res. 1990;54(Suppl):S48–S52. [PubMed] [Google Scholar]
- 4.Slocombe RF, Malark J, Ingersoll R, Derksen FJ, Robinson NE. Importance of neutrophils in the pathogenesis of acute pneumonic pasteurellosis in calves. Am J Vet Res. 1985;46(11):2253–2258. [PubMed] [Google Scholar]
- 5.Breider MA, Walker RD, Hopkins FM, Schultz TW, Bowersock TL. Pulmonary lesions induced by Pasteurella haemolytica in neutrophil sufficient and neutrophil deficient calves. Can J Vet Res. 1988;52(2):205–209. [PMC free article] [PubMed] [Google Scholar]
- 6.Petras SF, et al. Antigenic and virulence properties of Pasteurella haemolytica leukotoxin mutants. Infect Immun. 1995;63(3):1033–1039. doi: 10.1128/iai.63.3.1033-1039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatum FM, et al. Construction of an isogenic leukotoxin deletion mutant of Pasteurella haemolytica serotype 1: Characterization and virulence. Microb Pathog. 1998;24(1):37–46. doi: 10.1006/mpat.1997.0181. [DOI] [PubMed] [Google Scholar]
- 8.Highlander SK, et al. Inactivation of Pasteurella (Mannheimia) haemolytica leukotoxin causes partial attenuation of virulence in a calf challenge model. Infect Immun. 2000;68(7):3916–3922. doi: 10.1128/iai.68.7.3916-3922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dassanayake RP, et al. Mannheimia haemolytica serotype A1 exhibits differential pathogenicity in two related species, Ovis canadensis and Ovis aries. Vet Microbiol. 2009;133(4):366–371. doi: 10.1016/j.vetmic.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 10.Shanthalingam S, Srikumaran S. Intact signal peptide of CD18, the β-subunit of β2-integrins, renders ruminants susceptible to Mannheimia haemolytica leukotoxin. Proc Natl Acad Sci USA. 2009;106(36):15448–15453. doi: 10.1073/pnas.0906775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyon Y, et al. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7(6):459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 12.Kasinathan P, et al. Effect of fibroblast donor cell age and cell cycle on development of bovine nuclear transfer embryos in vitro. Biol Reprod. 2001;64(5):1487–1493. doi: 10.1095/biolreprod64.5.1487. [DOI] [PubMed] [Google Scholar]
- 13.Kuroiwa Y, et al. Cloned transchromosomic calves producing human immunoglobulin. Nat Biotechnol. 2002;20(9):889–894. doi: 10.1038/nbt727. [DOI] [PubMed] [Google Scholar]
- 14.Gentry MJ, Srikumaran S. Neutralizing monoclonal antibodies to Pasteurella haemolytica leukotoxin affinity-purify the toxin from crude culture supernatants. Microb Pathog. 1991;10(5):411–417. doi: 10.1016/0882-4010(91)90086-p. [DOI] [PubMed] [Google Scholar]
- 15.Tibbetts SA, et al. Peptides derived from ICAM-1 and LFA-1 modulate T cell adhesion and immune function in a mixed lymphocyte culture. Transplantation. 1999;68(5):685–692. doi: 10.1097/00007890-199909150-00015. [DOI] [PubMed] [Google Scholar]
- 16.Tibbetts SA, Seetharama Jois D, Siahaan TJ, Benedict SH, Chan MA. Linear and cyclic LFA-1 and ICAM-1 peptides inhibit T cell adhesion and function. Peptides. 2000;21(8):1161–1167. doi: 10.1016/s0196-9781(00)00255-2. [DOI] [PubMed] [Google Scholar]
- 17.Freshney IR. Culture of Animal Cells. 3rd Ed. Wiley-Liss, Inc.; New York: 1994. pp. 310–348. [Google Scholar]
- 18.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 19.Deshpande MS, Ambagala TC, Ambagala AP, Kehrli ME, Jr, Srikumaran S. Bovine CD18 is necessary and sufficient to mediate Mannheimia (Pasteurella) haemolytica leukotoxin-induced cytolysis. Infect Immun. 2002;70(9):5058–5064. doi: 10.1128/IAI.70.9.5058-5068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]