Significance
This study shows successful experimental phasing methods (single-wavelength anomalous diffraction, single isomorphous replacement, and single isomorphous replacement with anomalous scattering) for crystal structure determination of a membrane protein by serial femtosecond crystallography with X-ray free electron lasers. Our iodine-containing detergent provided strong anomalous and isomorphous difference signals, which enabled experimental phasing using lower-resolution reflections (worse than 3 Å) from fewer indexed images than phasing attempts reported previously. The findings of this study will be applicable to a wide range of target proteins in structural biology, especially membrane proteins that often diffract to low resolution.
Keywords: phase problem, de novo phasing, heavy atom, detergent, G protein-coupled receptor
Abstract
The 3D structure determination of biological macromolecules by X-ray crystallography suffers from a phase problem: to perform Fourier transformation to calculate real space density maps, both intensities and phases of structure factors are necessary; however, measured diffraction patterns give only intensities. Although serial femtosecond crystallography (SFX) using X-ray free electron lasers (XFELs) has been steadily developed since 2009, experimental phasing still remains challenging. Here, using 7.0-keV (1.771 Å) X-ray pulses from the SPring-8 Angstrom Compact Free Electron Laser (SACLA), iodine single-wavelength anomalous diffraction (SAD), single isomorphous replacement (SIR), and single isomorphous replacement with anomalous scattering (SIRAS) phasing were performed in an SFX regime for a model membrane protein bacteriorhodopsin (bR). The crystals grown in bicelles were derivatized with an iodine-labeled detergent heavy-atom additive 13a (HAD13a), which contains the magic triangle, I3C head group with three iodine atoms. The alkyl tail was essential for binding of the detergent to the surface of bR. Strong anomalous and isomorphous difference signals from HAD13a enabled successful phasing using reflections up to 2.1-Å resolution from only 3,000 and 4,000 indexed images from native and derivative crystals, respectively. When more images were merged, structure solution was possible with data truncated at 3.3-Å resolution, which is the lowest resolution among the reported cases of SFX phasing. Moreover, preliminary SFX experiment showed that HAD13a successfully derivatized the G protein-coupled A2a adenosine receptor crystallized in lipidic cubic phases. These results pave the way for de novo structure determination of membrane proteins, which often diffract poorly, even with the brightest XFEL beams.
Serial femtosecond crystallography (SFX) using ultrabright femtosecond pulses from X-ray free electron lasers (XFELs) has opened a new area of structural biology (1–3). Using high photon density in the focused XFEL beam, which achieves single-pulse diffraction within femtosecond exposure time, SFX enables protein structure determination from micrometer- to submicrometer-sized crystals at ambient temperature. The femtosecond pulse duration allows the “diffraction-before-destruction” approach by circumventing radiation damage of the sample, because the diffraction process can be terminated on a timescale shorter than that of the damage process (4–6). Moreover, SFX has become a powerful tool for time-resolved analyses of light-driven structural changes and chemical dynamics (7–9).
Phase determination has been a central problem in protein crystallography. In synchrotron radiation crystallography (SRX), single-wavelength anomalous diffraction (SAD) or multiwavelength anomalous diffraction (MAD) is currently the most commonly used de novo phasing method (10). Single isomorphous replacement (SIR) or multiple isomorphous replacement and methods with single isomorphous replacement with anomalous scattering (SIRAS) or multiple isomorphous replacement with anomalous scattering can be used for diffraction data collected at relatively low resolution compared with SAD. However, in SFX, most structures have been phased by the molecular replacement method using homologous structures as the search model (11–14). Only four de novo phasing cases have been published so far for SFX structures. In 2014, Gd-SAD phasing of lysozyme was shown for the first time in SFX with an 8.5-keV (1.459-Å) XFEL beam (15). Last year, we reported Hg-SIRAS phasing for luciferin-regenerating enzyme with a 12.6-keV (0.984-Å) beam (16) and native S/Cl-SAD phasing for lysozyme with a 7.0-keV (1.771-Å) beam (17), and this year, we reported Cu-SAD phasing for nitrite reductase with a 10.8-keV (1.418-Å) beam (18). Although successful, these phasing attempts required tens of thousands of indexed images and high-resolution reflections (better than 2.5 Å). This point is especially important for membrane proteins, which attract much interest from both basic and applied scientists, because their crystals often diffract poorly to low resolution, even with the brightest XFEL sources. During the review of this manuscript, S-SAD phasing for thaumatin with a 6.0-keV (2.066-Å) beam and reanalysis of the Gd-SAD dataset for lysozyme were reported (19), marking important steps by showing phasing at lower resolution (2.9 Å for thaumatin) and using fewer indexed images (7,000 for lysozyme). Progress since 2014 has been supported by improvements in data processing (for example, scaling of intensities and detector metrology refinement).
To use SFX techniques for a variety of protein targets and promote SFX-based structural biology, it is very important to explore de novo phasing methodology and accumulate examples using diverse heavy atoms. In SRX, heavy atoms, such as Se, Hg, Au, and Pt, which have absorption edges near 12.4 keV (1.000 Å), are frequently used for de novo phasing with large crystals, because most macromolecular crystallography beamlines at synchrotron facilities are optimized at this wavelength. However, in SFX, X-ray beams at lower energies may be used, because the smaller crystal size often used mitigates absorption effects that would otherwise adversely impact data quality. Therefore, other heavy atoms possessing higher anomalous scattering factors at lower energies can be used for efficient phasing in XFEL [e.g., iodine (I) atoms, which have f″ = 8.6e at 7.0 keV (1.771 Å) (20, 21)]. Although I-containing compounds [for example, magic triangle, I3C (22, 23) and 4-iodopyrazole (24)] have been used to phase proteins, their use was limited to SRX and soluble proteins.
Given that de novo phasing of membrane proteins has not been reported so far for SFX (25, 26), we addressed this issue by synthesizing a heavy-atom additive 13a (HAD13a) comprised of I3C and a hydrophobic alkyl chain to improve its affinity for membrane proteins (Fig. 1). The HAD13a showed detergent properties and successfully bound in crystallo to model membrane proteins, the light-driven proton pump bacteriorhodopsin (bR), and the G protein-coupled A2a adenosine receptor containing a thermostabilized apocytochrome b562RIL (A2a-BRIL) (27). We, furthermore, show I-SAD, -SIR, and -SIRAS phasing in SFX for bR, which was derivatized with HAD13a.
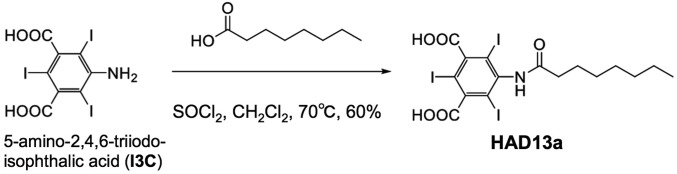
Fig. 1.
Synthetic reaction scheme for HAD13a (molecular weight = 685.8), which can be readily produced via a single-step synthesis from I3C.
Results
Synthesis and Analysis of HAD13a.
HAD13a (molecular weight = 685.8) was synthesized by substituting an octanoyl tail onto an aromatic head group known as I3C, which bears three I atoms (Fig. 1) (22). The compound I3C, called magic triangle, has previously been used for experimental phasing (23). We expected that the hydrophobic tail would increase the affinity of HAD13a for bicelles and lipidic cubic phases (LCPs) and facilitate its binding to the surface of membrane proteins during crystallization. Because the structure of HAD13a implies that it will act as a detergent, we measured the critical micelle concentration (CMC). The CMC of HAD13a was estimated at 4.6 mM, approximately 30 times as large as the 0.14 mM CMC of n-dodecyl-β-d-maltopyranoside (DDM), and the foaming of HAD13a was less than that of DDM (SI Appendix, Fig. S1).
To evaluate the effect of HAD13a on dispersity and stability of membrane proteins solubilized in detergent solution, we conducted size exclusion chromatography-based thermostability (SEC-TS) assays (28, 29) using bR and A2a-BRIL as model proteins. For bR, the addition of 4 mM HAD13a resulted in broad, decreased peak heights in size exclusion chromatography (SEC) compared with controls lacking HAD13a (SI Appendix, Fig. S2 A and B). In contrast, addition of 4 mM n-nonyl-β-d-maltopyranoside (NM), which has a similar CMC (∼6 mM) and molecular weight (468.5) to HAD13a, slightly improved the recovery of bR as evidenced by the sharp, increased peak heights (SI Appendix, Fig. S2C) compared with the control. A melting curve of bR in the presence of NM shifted to a higher temperature than that in the control or the presence of HAD13a (SI Appendix, Fig. S2D). The difference between HAD13a and NM in their effects on dispersity and stability of bR is interesting. The ionic carboxylates and I atoms on the amphiphilic head of HAD13a may interact with bR differently from nonionic detergent NM, although the two detergents have similar CMC and molecular weight values. For A2a-BRIL, the addition of 4 mM HAD13a did not affect the peak heights and shapes in SEC or the resulting melting curve compared with controls without HAD13a (SI Appendix, Fig. S3), indicating that HAD13a did not impair the monodispersity/stability of A2a-BRIL in detergent solution.
Experimental Phasing of bR.
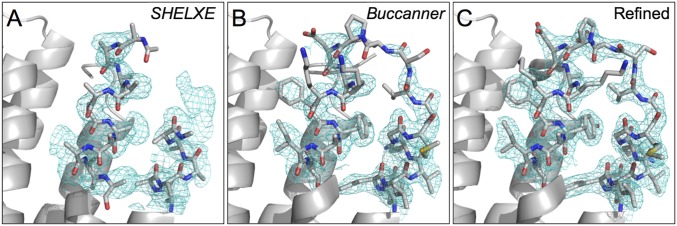
Microcrystals of native bR obtained by bicelle crystallization were mixed with HAD13a. Then, SFX datasets for the native and the HAD13a-derivative crystals were collected at the SPring-8 Angstrom Compact Free Electron Laser (SACLA) (Table 1). We tried SAD phasing with the HAD13a dataset alone. Using reflections up to 2.1-Å d spacing from all 23,347 indexed images, substructure solution and phase determination were successful (Fig. 2). SHELXD (30) located six I atoms in the asymmetric unit, which corresponded to two HAD13a molecules (Fig. 3). Six I atoms provide an expected anomalous difference <|ΔFano|>/<|F|> ∼ 10.5% according to the formula in the work by Hendrickson and Teeter (31). SHELXE traced 204 residues in five chains, and Buccaneer (32) almost completed the model; 238 residues were built in four fragments, 217 of which could be assigned to protein sequences (Fig. 4). The Rwork and Rfree factors after automatic modeling were 29.3% and 33.6%, respectively. When the correct substructure was supplied, SHELXE was able to trace the protein using only 16,000, but not 14,000, indexed images. However, substructure determination was impossible when the dataset was truncated to 22,000 indexed images. Thus, substructure determination, not phasing, was the limiting factor.
Table 1.
Data collection and refinement statistics for bR calculated for datasets merged with Friedel’s law, except for CCano
| Dataset name (PDB ID code) | Native (5B35) | HAD13a (5B34) | I3C | BaCl2 |
| Data collection | ||||
| XFEL wavelength (Å) | 1.771 | 1.771 | 1.771 | 1.771 |
| Space group | C2221 | C2221 | C2221 | C2221 |
| Unit cell parameters a, b, c (Å) | 46.2, 103.0, 128.7 | 46.2, 103.0, 128.7 | 46.2, 103.0, 128.7 | 46.2, 103.0, 128.7 |
| Beam time used (min) | 120 | 90 | 80 | 60 |
| No. of collected images | 206,280 | 142,272 | 130,942 | 175,550 |
| No. of hits | 9,807 | 47,946 | 27,485 | 37,335 |
| No. of indexed patterns | 6,817 | 23,347 | 8,524 | 12,419 |
| Indexing rate from hits (%) | 69.5 | 48.7 | 31.0 | 33.3 |
| Indexed pattern nos. used | 6,817 | 23,347 | 8,524 | 12,419 |
| No. of total reflections | 1,068,219 | 3,730,121 | 1,340,525 | 1,700,316 |
| No. of unique reflections | 13,212 | 18,388 | 13,209 | 11,691 |
| Multiplicity | 80.9 (56.5) | 202.9 (78.9) | 101.5 (92.3) | 145.4 (130.3) |
| Resolution range (Å; outer shell) | 47.8–2.35 (2.42–2.35) | 40.1–2.10 (2.17–2.10) | 42.9–2.35 (2.43–2.35) | 42.9–2.45 (2.54–2.45) |
| Completeness (%) | 100 | 100 | 100 | 100 |
| Rsplit (%) | 13.4 (111.7) | 8.14 (120.5) | 15.9 (72.9) | 12.2 (112.1) |
| CC1/2 (%) | 98.4 (43.1) | 99.1 (43.7) | 96.0 (48.2) | 97.7 (38.4) |
| CCano (%) | n/a | 5.7 (−1.5) | −3.8 (−6.4) | 2.9 (2.1) |
| <I/σ(I)> | 4.83 (0.94) | 7.73 (0.88) | 5.31 (1.43) | 6.52 (0.86) |
| Refinement | ||||
| No. of reflections | 12,535 | 17,471 | ||
| Rwork/Rfree (%) | 16.6/22.0 | 16.3/21.6 | ||
| Mean B factor (Å2) | 66.3 | 68.5 | ||
| No. of non-H atoms | 1,986 | 1,997 | ||
| rmsd from Ideal | ||||
| Bond lengths (Å)/angles (°) | 0.015/1.945 | 0.016/1.901 |
Because per-pattern resolution cutoff was not used, completeness and multiplicity for higher-resolution shells must be evaluated with care; they reflect the numbers of observations merged into the final datasets, but signals may or may not be present under the integration mask. n/a, Not applicable.
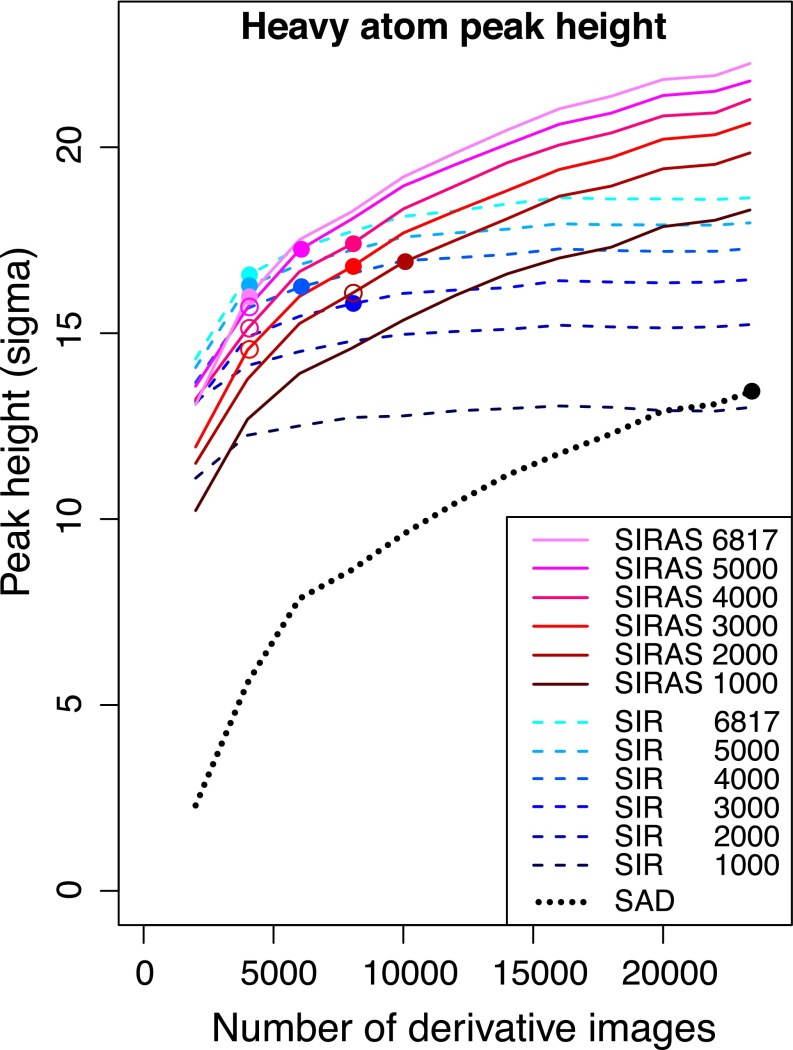
Fig. 2.
Improvement in anomalous difference peak heights with the number of derivative images of bR crystals. SAD, SIR, and SIRAS phasing are shown using black, blue, and red lines, respectively. For SIR and SIRAS phasing, the number of native images was also varied from 1,000 to 6,817. Colored circles indicate the minimum number of derivative images necessary for each phasing method and the given number of native images. Using substructures determined by the SIR method, the number of images necessary in SIRAS could be reduced (outlined circles).
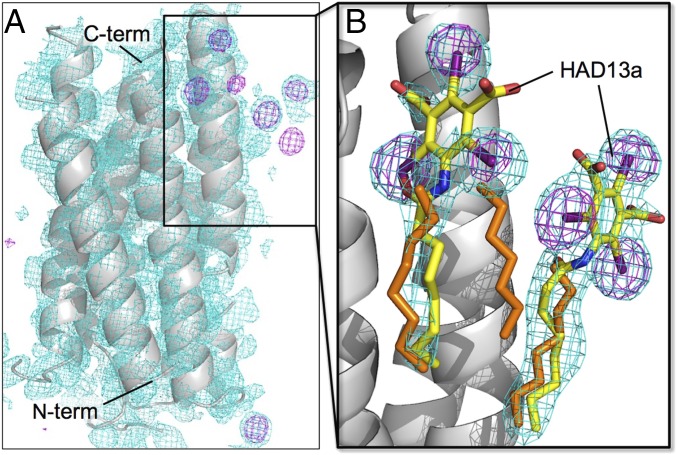
Fig. 3.
I-SAD phasing of bR with HAD13a. (A) Initial overall structure (gray ribbon model) with experimental map (cyan mesh; contoured at 1.0 σ) after SHELXE with density modification and polyalanine autotracing. Anomalous difference maps (purple mesh; 4.0 σ) are also shown. (B) Close-up view of the HAD13a binding site in the final refined structure with 2mFo–DFc maps (cyan mesh; 1.0 σ). The C, N, O, and I atoms of HAD13a (stick models) are colored yellow, blue, red, and purple, respectively. Alkyl chains derived from native lipid or DMPC/CHAPSO bicelles are depicted as orange stick models.
Fig. 4.
Improvement of electron density maps (cyan mesh; contoured at 1.0 σ) and models around residues 26–46 of bR from the various steps of the I-SAD phasing process: (A) SHELXE model and experimental map after density modification and polyalanine autotracing, (B) Buccaneer model and 2mFo–DFc map after automatic model building, and (C) the final refined model and 2mFo–DFc map. The C, N, O, and S atoms in stick models are colored gray, blue, red, and yellow, respectively.
We next carried out SIR and SIRAS phasing. Phasing was successful with fewer images than SAD phasing (Fig. 2). In SIRAS phasing, about 12,000 indexed images (native/derivative: 5,000/6,000, 4,000/8,000, 3,000/8,000, or 2,000/10,000) were sufficient for substructure determination and phasing. However, substructure determination became impossible with fewer images. In contrast, substructure solution was successful in the SIR regime with only 5,000/4,000, 4,000/4,000, 3,000/4,000, or 2,000/8,000 indexed images. However, the datasets from 4,000/4,000, 3,000/4,000, or 2,000/8,000 indexed images did not lead to interpretable maps by SIR. At least 5,000/4,000, 4,000/6,000, or 3,000/8,000 indexed images were necessary for successful SIR phasing (chain tracing). Thus, we combined SIR and SIRAS using SIR for substructure solution and SIRAS for phasing. With this method, phasing was successful with 5,000/4,000, 4,000/4,000, or 3,000/4,000 indexed images. These numbers indicate that SIR was more effective for substructure determination than SIRAS, whereas SIRAS was more powerful in phasing.
We also evaluated the low-resolution limit of our experimental phasing by the SIRAS method using all 6,817 native and 23,347 derivative indexed images. At progressively lower resolutions, SHELXD and SHELXE were executed with the same high-resolution cutoff (SHEL keyword and -d option). The I atoms could be located even when the dataset was truncated at 4.0-Å resolution (SI Appendix, Fig. S4). The correctness of the top solution was confirmed by phenix.emma. However, density modification and automatic chain tracing became increasingly difficult. At 3.3-Å resolution, SHELXE produced an interpretable map, where seven helices were clearly visible. Compared with phasing at 2.2-Å resolution, Buccaneer’s output at 3.3-Å resolution was more fragmented (215 residues in 13 fragments, the longest having 69 residues), and more than one-half of the residues had their sequence unassigned. Maps became less continuous at 3.4- and 3.5-Å resolutions and uninterpretable at 3.6-Å or lower resolution. The real space cross-correlation (CCmap) of the autotraced map from the Buccaneer pipeline to the final refined model was calculated to 3.5-Å resolution. The CCmap result from phasing up to 2.2-Å resolution was 0.692, whereas maps from phasing at lower resolution (3.2, 3.3, 3.4, and 3.5 Å) gave lower CCmap values of 0.548, 0.543, 0.524, and 0.480, respectively.
Binding Sites of HAD13a on bR.
Two HAD13a molecules bind to the outer surface of bR, where the alkyl tail of HAD13a positions at the hydrophobic transmembrane surface and the head group interacts with a hydrophilic intracellular surface (Fig. 3). The two HAD13a molecules locate side by side at a distance of 8 Å via a lipid/detergent molecule present between them (Fig. 3B). One HAD13a (occupancy: 0.6) interacts with residues V173, N176, V177, and V180 from a bR molecule and I198 from a neighboring symmetry-related bR molecule within 4.3-Å distance. The other HAD13a (occupancy: 0.5) interacts with E166, T170, G218, I222, R225, S226, and R227 from a single bR molecule.
When HAD13a was first assigned to full occupancy during the structure refinement, negative peaks appeared on I atoms in the Fo–Fc maps. Therefore, the occupancy of HAD13a was decreased until the negative peaks disappeared. However, positive Fo–Fc peaks appeared instead around the alkyl chain of HAD13a. Because a lipid or detergent molecule is located in the corresponding site of the native bR structure, we concluded that HAD13a bound to the same site, partially replacing the lipid/detergent. Thus, we modeled an alkyl chain derived from the lipid/detergent with partial occupancy in the site together with HAD13a (Fig. 3B).
To confirm the importance of the alkyl tail of HAD13a for binding to bR, we collected a dataset from microcrystals soaked in I3C lacking the tail (Table 1). Anomalous difference maps calculated with refined phases showed no significant peaks corresponding to I3C. Thus, the alkyl chain was essential for the binding of HAD13a to bR. Although we collected another dataset from BaCl2-mixed microcrystals to test the potential of inorganic heavy atoms for phasing (Table 1), no binding of Ba2+ was detected in the anomalous difference map.
Compatibility of HAD13a with LCP.
To test whether HAD13a is compatible with the LCP crystallization method, microcrystals of A2a-BRIL grown in LCP were soaked in 12 mM HAD13a, and an SFX dataset was collected (SI Appendix, Table S1). Of 15,518 images, we successfully indexed 14,877. Unfortunately, we could not solve the substructure of this dataset by SAD phasing because of the limited number of images. However, the anomalous difference map calculated from the refined model clearly showed binding of an HAD13a molecule and the positions of several S atoms (SI Appendix, Fig. S6). The HAD13a (occupancy: 0.4) was located at an interface region between two A2a-BRIL molecules and partially replaced an oleic acid seen in the synchrotron structure [Protein Data Bank (PDB) ID code 4EIY]. HAD13a interacted with residues F201, A204, R205, L208, Q226, V229, and K233 from an A2a-BRIL molecule and W32 from a neighboring molecule within 4.0-Å distance. These results confirmed that HAD13a can bind to A2a-BRIL in LCP crystals and contribute to anomalous signals. Unfortunately, our limited beam time did not allow us to optimize soaking procedures to increase the occupancy and collect more diffraction images to achieve SAD phasing.
Discussion
As indicated by the number of indexed images necessary for substructure determination and phasing in our SIRAS and SAD attempts for bR, substructure determination was more difficult than phasing. This difficulty is in contrast to previous Hg-SIRAS phasing at SACLA (16), which required more than 18,000 indexed images in total for structure solution, whereas only 1,000/2,000 indexed images were sufficient for localization of Hg atoms. SIR enabled substructure solution with the fewest images. This success is probably due to the strong isomorphous difference signal arising from the high atomic number of the I atom (Z = 53; <Fa>/<Fp> ∼ 32%). When the number of images is very small, anomalous signals are weak and noisy, which might make substructure solution difficult. In contrast, weaker and noisier signals can still positively contribute to phase calculation and density modification. Thus, SIRAS was more powerful than SIR and SAD in phasing. This trend parallels standard practice in macromolecular crystallography, where one uses only low-resolution, highly accurate data for substructure solution (33) and uses more reflections for density modification and refinement (34).
The total number of images necessary for success in SIR/SIRAS phasing (7,000) was less than one-third of that in the SAD method (23,347). For efficient use of precious XFEL beam time, it would be desirable to collect both native and derivative datasets rather than dedicating all of the beam time to derivative crystals. It is interesting that 3,000/4,000 indexed images were sufficient for successful phasing, because both the native and derivative datasets are not fully converged at this number. For example, the CC1/2 value of the 4,000-image derivative dataset was only 95.0% (12.3% for 2.17−2.10 Å), whereas that of the full dataset was 99.1% (43.7% for 2.17−2.10 Å) (SI Appendix, Fig. S5). This observation suggests that accuracy, but not precision, of the Monte Carlo integrated intensities improves more quickly than previously thought. The fact that the CCano was still low when SAD phasing was successful at 23,347 indexed images also supports this suggestion.
Microcrystals might be more suitable for heavy-atom derivatization by soaking than large crystals used in conventional crystallography. A higher surface to volume ratio enables rapid diffusion of compounds. A smaller volume means less crystal strain during soaking; even when crystals are damaged (for example, cracked), they do not diffract well and can simply be ignored in subsequent processing. In contrast, harvesting, cryoprotecting, mounting, and screening hundreds of crystals to find good crystals at synchrotron-based beamlines would be tedious and time-consuming. The HAD13a soak method compatible with SFX at room temperature can be chosen as an alternative protocol for membrane protein structure determination.
In previous SRX experiments, MAD phasing with an Se-labeled detergent was reported with a membrane protein, leukotriene C4 synthase, in complex with dodecyl-β-d-selenomaltoside (35). In another study, although phasing was not successful, binding sites of Br- or Hg-labeled phospholipids or detergents were identified at membrane protein reaction centers (36, 37). These results encouraged us to prepare an I-containing detergent for efficient experimental phasing in SFX. As we have shown, the alkyl chain is essential for HAD13a to bind to the hydrophobic, membrane-facing surface of the transmembrane region of bR. This binding specificity is different from those of traditional heavy-atom reagents, such as inorganic cations and organomercury compounds. Thus, HAD13a and traditional phasing reagents can be used in a complementary manner for experimental phasing of membrane proteins. In particular, HAD13a would be useful for membrane proteins lacking exposed cysteine residues, in which the thiol group often gives strong anomalous and/or isomorphous signals by forming mercaptides with heavy metals. HAD13a was readily prepared via a single-step reaction by amido coupling of I3C. The ease of preparation should expand its application range.
Strong anomalous and isomorphous difference signals from HAD13a enabled structure solution of bR from data truncated at 3.3-Å resolution. Given that most membrane protein crystals diffract poorly, our demonstration of low-resolution phasing by SFX is an important development. Because actual low-resolution datasets from poorly diffracting crystals are weaker and noisier than truncated datasets from well-diffracting crystals, it is certainly possible that experimental phasing of actual low-resolution datasets is more difficult than that from truncated datasets. However, we note that, in previous phasing attempts by SFX, higher resolution was used for phasing [1.8 Å for Gd-SAD (15), 1.7 Å for Hg-SIRAS (16), 2.3 Å for S/Cl-SAD (17), 1.9 Å for Cu-SAD (18), and 2.9 Å for S-SAD (19)] and necessary at least for the S/Cl-SAD and Cu-SAD cases. Recently, a 2.5-Å structure of the A2a adenosine receptor solved by S-SAD phasing using XFEL was deposited in the PDB (PDB ID code 5K2A). Although the number of images that they used has not yet been disclosed, using HAD13a will give stronger anomalous signals than S atoms alone and probably reduce the number of images necessary for phasing.
To conclude, we have shown experimental phasing of a membrane protein by means of SFX. Strong isomorphous and anomalous signals from HAD13a enabled phasing with small numbers of indexed images and at low resolution. Of course, improvements in CrystFEL (38) also contributed to the data quality. More sophisticated merging methods (39–43) might further improve the data quality attainable from a given number of images. We could not explore this possibility, because the multiport charge-coupled device (MPCCD) detector (44) in SACLA was not yet fully supported by programs cctbx.xfel and cppxfel. It is noteworthy that we took only 120 and 90 min to complete the native and HAD13a-derivative datasets (6,817 + 23,347 indexed images), respectively (Table 1), and that less than one-quarter of the images (3,000 + 4,000) were necessary for successful phasing. With more beam time, we can probably solve new structures of more difficult membrane protein targets by de novo phasing using HAD13a or its analogs. We also note that HAD13a might be useful at synchrotron sources, although most beamlines are not optimized for longer wavelengths. In this regard, bromine derivatives of HAD13a might be more attractive. Variants with different lengths and saturation of alkyl tails will increase the scope of the targets. Applications of these methods to crystals grown in LCP and optimization of the alkyl tail length of HAD13a are underway for both SFX and SRX experiments.
Materials and Methods
Preparation of 5-Capryloylamino-2,4,6-Triiodo-Isophthalic Acid (HAD13a).
The heavy atom-substituted aromatic core 5-amino-2,4,6-triiodo-isophthalic acid (250 mg, 0.45 mmol), which is known as magic triangle, I3C (22, 23), and n-octanoic acid (1.0 g, 6.9 mmol) were mixed, and thionyl chloride (0.1 mL, 1.4 mmol) was added. The mixture was stirred for 15 h at room temperature without solvent. Then, an additional portion of thionyl chloride (0.1 mL, 1.4 mmol) was added and stirred for 2 h at 70 °C. The mixture was then cooled to room temperature and quenched with H2O (20 mL). The precipitate was collected by filtration, and the residue was purified by SiO2 chromatography (CHCl3:MeOH = 1:1) and SepPak C18 (MeOH:H2O = 1:10) to produce HAD13a (184 mg, 0.27 mmol, 60% yield) as a white solid. 1H NMR (400 MHz CD3OD) δ 2.40 (2H, triplet, J = 7.2 Hz), 1.75 (2H, quintet, J = 7.2 Hz), 1.35 (8H, multiplet), 0.89 (3H, triplet, J = 6.7 Hz); High-resolution electrospray ionization mass spectrometry calculated C16H19I3NO5 685.8397 [M + H]+, found 685.8391.
Crystallization and SFX Data Collection for bR.
Detailed methods are described in SI Appendix. Briefly, purple membranes from Halobacterium salinarum strain R1M1 were prepared according to a standard method (45). Microcrystals of bR were grown by the bicelle method at 20 °C. For preparation of heavy-atom derivative crystals, HAD13a, I3C, or BaCl2 was added to the microcrystals at a final concentration of 4 mM and incubated for 3 d. The microcrystals of bR were mixed with a grease matrix, Synthetic Grease Super Lube (21030; Synco Chemical), and packed in an injector syringe as described previously (46). Diffraction patterns were collected at beamline BL3(EH4) of SACLA (47). The injector was installed in a diffraction chamber enclosure DAPHNIS (48). Data collection was guided by real time analysis by the Cheetah pipeline adapted for SACLA (49).
Phasing and Structure Determination of bR.
Using the Cheetah pipeline, raw images were retrieved from SACLA API (50) and filtered by Cheetah (51). Algorithm 6 in Cheetah was used for hit finding. The relevant parameters are hitfinderADC = 40, hitfinderMinSNR = 5, hitfinderMinPixCount = 2, hitfinderMaxPixCount = 40, and hitfinderMinPeakSeparation = 3. Images with more than 20 spots were retained as hits and converted to the HDF5 format for subsequent processing in CrystFEL (version 0.6.1) (52). Detector metrology was refined by geoptimizer and detector shift in the CrystFEL suite. Indexing was performed by DirAx (53). Spot finding was performed by the built-in spot finder (54) in CrystFEL with threshold of 400, minimum gradient of 1,500,000, and minimum signal-to-noise ratio of 3. The radii of the integration and background masks were three and seven, respectively. Intensities were merged by process_hkl in the CrystFEL suite with linear scaling (–scale option). No per-pattern resolution cutoff and no saturation cutoff were used. The structure of the HAD13a derivative was determined by SAD, SIR, or SIRAS phasing. For SAD and SIRAS phasing, intensities of the derivative dataset were merged in the 222-point group. The native dataset for SIRAS or SIR phasing and the derivative dataset for SIR phasing were merged in the mmm point group. The phasing was performed using SHELX (30). Intensities were processed by SHELXC. The heavy-atom substructure was solved by SHELXD (2013/2) using reflections up to 2.5 Å, except for low-resolution phasing attempts. Up to 10,000 trials were performed. Phase calculation and density improvement were carried out in SHELXE (2016/1) by 25 iterations (-a25) of 20 cycles of density modification (-m20) and automatic chain tracing with α-helix search (-q option). Different combinations of parameters were tested, varying the high-resolution limit (-d option; 2.1 or 2.2 Å unless otherwise stated), number of sites to be used (-z option for SIR and SIRAS and -h option for SAD; four to seven), and solvent content (-s option; 50 or 52%). The polyalanine model generated by SHELXE was completed by Buccaneer (32). Manual model building was carried out in Coot (55). Refmac5 (56) in the CCP4 suite (6.5.016) (57) was used for structure refinement. Anomalous difference maps were calculated with ANODE (58) to 2.3 Å using final refined models as the phase source. The peak heights for six I atom sites were averaged in Fig. 2. CCmaps to the final model were calculated by the phenix.get_cc_mtz_pdb command (59). The correctness of the substructure was tested by the phenix.emma command (59), but the result was not used for de novo phasing attempts. Protein structures were drawn with PyMOL (1.5.0.4; https://www.pymol.org). Data statistics are summarized in Table 1. We note that the CCano of the Monte Carlo integrated intensities tends to be lower than those of synchrotron datasets [e.g., our lysozyme S/Cl-SAD (17) and AfNiR Cu-SAD (18) cases], which results from higher variances of each observation (without partiality correction) than the size of the anomalous signals [the work by Assmann et al. (60)].
CMC Measurement, SEC-TS Assay, and SFX Experiments of A2a-BRIL.
These methods are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank the beamline staff at the SACLA for technical assistance and Keitaro Yamashita for discussion of the manuscript. We also thank the SACLA High Performance Computing system and the Mini-K Super Computer System for support. SFX experiments were performed at the SACLA with the approval of Japan Synchrotron Radiation Research Institute Proposals 2013B8045, 2014A8031, 2015B8042, 2015B8047, and 2016A8041. This work was supported by the X-ray Free Electron Laser Priority Strategy Program of the Ministry of Education, Culture, Sports, Science and Technology in Japan; Exploratory Research for Advanced Technology of the Japan Science and Technology Agency (JST); the Research Acceleration Program of the JST; and Japan Society for the Promotion of Science KAKENHI Grants 15K18487 and 16H06315.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.B. is a Guest Editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 5B34 (heavy-atom additive 13a) and 5B35 (native)]. The serial femtosecond crystallography diffraction images have been deposited in the Coherent X-Ray Imaging Data Bank (CXIDB ID code 43).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602531113/-/DCSupplemental.
References
- 1.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470(7332):73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlichting I, Miao J. Emerging opportunities in structural biology with X-ray free-electron lasers. Curr Opin Struct Biol. 2012;22(5):613–626. doi: 10.1016/j.sbi.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutet S, et al. High-resolution protein structure determination by serial femtosecond crystallography. Science. 2012;337(6092):362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neutze R, Wouts R, van der Spoel D, Weckert E, Hajdu J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406(6797):752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 5.Barty A, et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nat Photonics. 2012;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda Y, et al. Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography. J Biochem. 2016;159(5):527–538. doi: 10.1093/jb/mvv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenboer J, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346(6214):1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kupitz C, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513(7517):261–265. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barends TR, et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015;350(6259):445–450. doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Hendrickson WA. Crystallographic phasing from weak anomalous signals. Curr Opin Struct Biol. 2015;34:99–107. doi: 10.1016/j.sbi.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson LC, et al. Lipidic phase membrane protein serial femtosecond crystallography. Nat Methods. 2012;9(3):263–265. doi: 10.1038/nmeth.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339(6116):227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, et al. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342(6165):1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barends TR, et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature. 2014;505(7482):244–247. doi: 10.1038/nature12773. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita K, et al. An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography. Sci Rep. 2015;5:14017. doi: 10.1038/srep14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakane T, et al. Native sulfur/chlorine SAD phasing for serial femtosecond crystallography. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 12):2519–2525. doi: 10.1107/S139900471501857X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda Y, et al. Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. Proc Natl Acad Sci USA. 2016;113(11):2928–2933. doi: 10.1073/pnas.1517770113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nass K, et al. Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data. IUCrJ. 2016;3(Pt 3):180–191. doi: 10.1107/S2052252516002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yogavel M, Gill J, Sharma A. Iodide-SAD, SIR and SIRAS phasing for structure solution of a nucleosome assembly protein. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):618–622. doi: 10.1107/S0907444909013171. [DOI] [PubMed] [Google Scholar]
- 21.Abendroth J, et al. SAD phasing using iodide ions in a high-throughput structural genomics environment. J Struct Funct Genomics. 2011;12(2):83–95. doi: 10.1007/s10969-011-9101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck T, Sheldrick GM. 5-Amino-2,4,6-triiodo-isophthalic acid monohydrate. Acta Crystallogr Sect E Struct Rep Online. 2008;64(Pt 7):o1286. doi: 10.1107/S1600536808017741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck T, Krasauskas A, Gruene T, Sheldrick GM. A magic triangle for experimental phasing of macromolecules. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 11):1179–1182. doi: 10.1107/S0907444908030266. [DOI] [PubMed] [Google Scholar]
- 24.Bauman JD, Harrison JJ, Arnold E. Rapid experimental SAD phasing and hot-spot identification with halogenated fragments. IUCrJ. 2016;3(Pt 1):51–60. doi: 10.1107/S2052252515021259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlichting I. Serial femtosecond crystallography: The first five years. IUCrJ. 2015;2(Pt 2):246–255. doi: 10.1107/S205225251402702X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neutze R, Brändén G, Schertler GF. Membrane protein structural biology using X-ray free electron lasers. Curr Opin Struct Biol. 2015;33:115–125. doi: 10.1016/j.sbi.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337(6091):232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancusso R, Karpowich NK, Czyzewski BK, Wang DN. Simple screening method for improving membrane protein thermostability. Methods. 2011;55(4):324–329. doi: 10.1016/j.ymeth.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20(8):1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrickson WA, Teeter MM. Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulfur. Nature. 1981;290(5802):107–113. doi: 10.1038/290107a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 33.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 10 Pt 2):1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 34.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saino H, Ago H, Ukita Y, Miyano M. Seleno-detergent MAD phasing of leukotriene C4 synthase in complex with dodecyl-β-D-selenomaltoside. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 12):1666–1673. doi: 10.1107/S1744309111042345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cogdell RJ, et al. Mapping lipid and detergent molecules at the surface of membrane proteins. Biochem Soc Trans. 2011;39(3):775–779. doi: 10.1042/BST0390775. [DOI] [PubMed] [Google Scholar]
- 37.Roszak AW, Gardiner AT, Isaacs NW, Cogdell RJ. Brominated lipids identify lipid binding sites on the surface of the reaction center from Rhodobacter sphaeroides. Biochemistry. 2007;46(11):2909–2916. doi: 10.1021/bi062154i. [DOI] [PubMed] [Google Scholar]
- 38.White TA, et al. Recent developments in CrystFEL. J Appl Cryst. 2016;49(Pt 2):680–689. doi: 10.1107/S1600576716004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uervirojnangkoorn M, et al. Enabling X-ray free electron laser crystallography for challenging biological systems from a limited number of crystals. eLife. 2015;4:4. doi: 10.7554/eLife.05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White TA. Post-refinement method for snapshot serial crystallography. Philos Trans R Soc Lond B Biol Sci. 2014;369(1647):20130330. doi: 10.1098/rstb.2013.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauter NK. XFEL diffraction: Developing processing methods to optimize data quality. J Synchrotron Radiat. 2015;22(2):239–248. doi: 10.1107/S1600577514028203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginn HM, Evans G, Sauter NK, Stuart DI. On the release of cppxfel for processing X-ray free-electron laser images. J Appl Cryst. 2016;49(Pt 3):1065–1072. doi: 10.1107/S1600576716006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginn HM, et al. A revised partiality model and post-refinement algorithm for X-ray free-electron laser data. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 6):1400–1410. doi: 10.1107/S1399004715006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kameshima T, et al. Development of an X-ray pixel detector with multi-port charge-coupled device for X-ray free-electron laser experiments. Rev Sci Instrum. 2014;85(3):033110. doi: 10.1063/1.4867668. [DOI] [PubMed] [Google Scholar]
- 45.Oesterhelt D, Stoeckenius W. Isolation of the cell membrane of Halobacterium halobium and its fractionation into red and purple membrane. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 46.Sugahara M, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat Methods. 2015;12(1):61–63. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 47.Ishikawa T, et al. A compact X-ray free-electron laser emitting in the sub-angstrom region. Nat Photonics. 2012;6(8):540–544. [Google Scholar]
- 48.Tono K, et al. Diverse application platform for hard X-ray diffraction in SACLA (DAPHNIS): Application to serial protein crystallography using an X-ray free-electron laser. J Synchrotron Radiat. 2015;22(3):532–537. doi: 10.1107/S1600577515004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakane T, et al. Data processing pipeline for serial femtosecond crystallography at SACLA. J Appl Cryst. 2016;49(Pt 3):1035–1041. doi: 10.1107/S1600576716005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joti Y, et al. Data acquisition system for X-ray free-electron laser experiments at SACLA. J Synchrotron Radiat. 2015;22(3):571–576. doi: 10.1107/S1600577515004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barty A, et al. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J Appl Cryst. 2014;47(Pt 3):1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White TA, et al. Crystallographic data processing for free-electron laser sources. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 7):1231–1240. doi: 10.1107/S0907444913013620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duisenberg AJM. Indexing in single-crystal diffractometry with an obstinate list of reflections. J Appl Crystallogr. 1992;25:92–96. [Google Scholar]
- 54.Zaefferer S. New developments of computer-aided crystallographic analysis in transmission electron microscopy. J Appl Crystallogr. 2000;33:10–25. [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorn A, Sheldrick GM. ANODE: Anomalous and heavy-atom density calculation. J Appl Cryst. 2011;44(Pt 6):1285–1287. doi: 10.1107/S0021889811041768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Assmann G, Brehm W, Diederichs K. Identification of rogue datasets in serial crystallography. J Appl Cryst. 2016;49(Pt 3):1021–1028. doi: 10.1107/S1600576716005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.