Significance
Autocrine cytokine signaling in cancer can activate members of the Janus kinase (JAK) family, which are generally thought to act by phosphorylating STAT family transcription factors. We report here that JAK1 mediates autocrine IL-6 and IL-10 cytokine signaling in activated B-cell–like (ABC) diffuse large B-cell lymphoma (DLBCL) by a noncanonical epigenetic regulatory mechanism involving phosphorylation of histone H3 on tyrosine 41. We have identified target genes that are activated in ABC DLBCL by this epigenetic mechanism. Knowledge of these epigenetic targets led to our demonstration that JAK1 inhibitors synergize with inhibitors of active B cell receptor signaling in ABC DLBCL, suggesting a new therapeutic strategy for this subtype of DLBCL, which is the most difficult to cure with current therapy.
Keywords: JAK1, epigenetics, histone modification, lymphoma, oncogene
Abstract
Janus kinases (JAKs) classically signal by activating STAT transcription factors but can also regulate gene expression by epigenetically phosphorylating histone H3 on tyrosine 41 (H3Y41-P). In diffuse large B-cell lymphomas (DLBCLs), JAK signaling is a feature of the activated B-cell (ABC) subtype and is triggered by autocrine production of IL-6 and IL-10. Whether this signaling involves STAT activation, epigenetic modification of chromatin, or both mechanisms is unknown. Here we use genetic and pharmacological inhibition to show that JAK1 signaling sustains the survival of ABC DLBCL cells. Whereas STAT3 contributed to the survival of ABC DLBCL cell lines, forced STAT3 activity could not protect these cells from death following JAK1 inhibition, suggesting epigenetic JAK1 action. JAK1 regulated the expression of nearly 3,000 genes in ABC DLBCL cells, and the chromatin surrounding many of these genes was modified by H3Y41-P marks that were diminished by JAK1 inhibition. These JAK1 epigenetic target genes encode important regulators of ABC DLBCL proliferation and survival, including IRF4, MYD88, and MYC. A small molecule JAK1 inhibitor cooperated with the BTK inhibitor ibrutinib in reducing IRF4 levels and acted synergistically to kill ABC DLBCL cells, suggesting that this combination should be evaluated in clinical trials.
Diffuse large B-cell lymphoma (DLBCL), a B-cell non-Hodgkin lymphoma, represents 30–40% of newly diagnosed lymphomas and includes two main molecular subtypes termed activated B-cell like (ABC) and germinal center B-cell like (GCB) (1, 2). ABC DLBCL is more aggressive, with a current cure rate of only ∼40% (3). One distinct feature of this lymphoma is autocrine signaling mediated by the cytokines IL-6 and IL-10, which constitutively activates the transcription factor STAT3 (4–6). Autocrine production of these cytokines results from high NF-κB activity (7), which is a hallmark of ABC DLBCL (4, 8–10). The NF-κB pathway can be engaged by a variety of genetic alterations in ABC DLBCL (9, 11, 12), including somatic mutations targeting components of the B-cell receptor (BCR) and Toll-like receptor (TLR) signaling pathways. For example, 39% of ABC DLBCL tumors have mutations in the gene encoding MYD88, a key signaling adapter in TLR signaling (9). The most prevalent MYD88 mutation (L265P), which is present in 29% of ABC DLBCL tumors, promotes cell survival by engaging the NF-κB pathway and inducing production of IL-6 and/or IL-10. NF-κB is also activated in ABC DLBCL by gain-of-function mutations in the BCR pathway, including mutations targeting the BCR components CD79A and CD79B (11) and the downstream signaling adapter CARD11 (13). Targeting BCR signaling with ibrutinib, a specific inhibitor for Bruton tyrosine kinase (BTK), has emerged as a promising therapy for ABC DLBCL (14).
IL-6 and IL-10 function through their respective receptors and activate members of the Janus kinase (JAK) family (15, 16). In mammals, the JAK family has four members: JAK1, JAK2, JAK3, and TYK2 (15). JAKs phosphorylate STAT family members to promote STAT dimerization, nuclear translocation, and binding to cis-regulatory elements to regulate transcription (15, 17). This canonical JAK/STAT pathway is deregulated in several hematologic malignancies (16). In DLBCL, STAT3 is activated in the ABC subtype and regulates gene expression to promote the survival of the malignant cells (6, 18).
A separate, noncanonical mechanism by which JAK activity perturbs gene expression is by directly phosphorylating chromatin, which was first discovered in Drosophila (19) and subsequently confirmed in human cells (20–22). In leukemia cells, a mutant JAK2 isoform phosphorylates the tail of histone H3 tyrosine 41 (H3Y41), which displaces the inhibitory heterochromatin protein HP1α from chromatin to augment gene transcription (20, 23). We previously reported a similar function of JAK2 in primary mediastinal B-cell lymphoma (PMBL) and Hodgkin lymphoma (HL), in which JAK2 kinase is activated by autocrine IL-13 signaling (21, 24). Through this noncanonical pathway, JAK2 induces expression of more than 2,000 genes, including genes that control the growth and proliferation of the malignant cell such as MYC, JMJD2C, and JAK2 itself, as well as the genes encoding PD-L1 and PD-L2, which inhibit tumor immunity through the T-cell inhibitory receptor PD1 (21, 24, 25).
Here, we demonstrate that JAK1 promotes the malignant phenotype of ABC DLBCL cells by phosphorylating and activating STAT3 and also epigentically by phosphorylating chromatin on H3Y41. We demonstrate that some epigenetic JAK1 target genes are also induced by the BCR/NF-κB signaling pathway and that cotargeting of BCR and JAK signaling with small molecule inhibitors kills ABC DLBCL cells synergistically.
Results
JAK1 Is Required for the Survival of ABC DLBCL Cells.
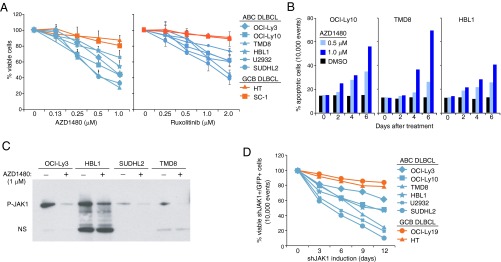
The essential role of autocrine IL-6 or IL-10 signaling in the survival of ABC DLBCL cells has been demonstrated (4, 5), but the molecular mechanisms by which these cytokines promote lymphomagenesis are largely unknown. As a first step, we examined the viability of DLBCL cell lines treated with AZD1480, an inhibitor of JAK1 and JAK2 (26). AZD1480 potently decreased cell viability in ABC but not GDC DLBCL lines (Fig. 1A), and induced apoptosis in ABC DLBCL lines, as indicated by active caspase 3 and cleaved PARP (Fig. 1B). Selective toxicity to ABC DLBCL cell lines was also observed with another inhibitor of JAK1 and JAK2, ruxolitinib (Fig. 1A) (27). Because JAK2 is not activated in ABC DLBCL cells (21), we tested whether JAK1 is the target of AZD1480 action in these lymphomas. Due to the difficulty in detecting JAK1 phosphorylation by a direct immunoblotting assay, we first immunoprecipitated tyrosine-phosphorylated proteins and then immunoblotted with an antibody that detects JAK1 phosphorylation. By this method, JAK1 phosphorylation was evident in all ABC DLBCL cell lines tested and was reduced when cells were treated with AZD1480 (Fig. 1C).
Fig. 1.
JAK1 is required for the survival of ABC DLBCL cells. (A) MTT viability assay conducted in the indicated cell lines after 6 d of treatment with the indicated concentrations of AZD1480 or ruxolitinib. The drugs were replenished every other day. Error bars represent mean ± SD of triplicates. (B) Flow cytometric analysis of apoptosis in the indicated cell lines after a 3-d treatment of AZD1480 or DMSO as measured by cells with active caspase 3 and cleaved PARP. (C) Immunoblot analysis of JAK phosphorylation in ABC DLBCL cells. Ten million cells were treated with AZD1480 (2 μM) or DMSO for 2 h before immunoprecipitation with an antiphosphotyrosine antibody and subsequent immunoblotting using an antiphospho-JAK1 antibody. NS denotes nonspecific bands. (D) Flow cytometric analysis of cell viability in the indicated cell lines after inducing expression of shJAK1#5 or a control shRNA for the indicated times. The viability of shJAK1#5-expressing/GFP+ cells was normalized to that of the cells transduced with control shRNA for each time point and then normalized to day 0 values. Data are representative of three independent experiments.
To investigate the consequence of JAK1 activity in ABC DLBCL, we developed JAK1-specific small hairpin RNAs (shRNAs) that mediate RNA interference and expressed them using a retroviral vector that coexpresses GFP. We identified two shRNAs that reduced JAK1 expression by 50–70% (Fig. S1 A and B). Following retroviral infection, the percentage of viable shRNA-expressing (GFP+) cells relative to day 0 was monitored over time by FACS. Over the course of 12 d, JAK1 shRNA expression decreased the viability of all six ABC DLBCL lines examined, whereas two GCB DLBCL lines were insensitive (Fig. 1D). By contrast, the viability of ABC DLBCL cells was not affected by a JAK2 shRNA (Fig. S1C), which is toxic for JAK2-dependent PMBL and HL cell lines (21). Together, these data demonstrate that JAK1 promotes ABC DLBCL cell survival.
Fig. S1.
Verification of JAK shRNAs. (A) Immunoblotting with JAK1 antibody for JAK1 shRNA-expressing cells. QPCR analysis of JAK1 mRNA in cells expressing JAK1 shRNAs (normalized to β-2-microglobulin). Error bars represent mean ± SD of triplicates. (B) Flow cytometric analysis of the percentage of transduced TMD8 cells with JAK1 shRNAs with the GFP marker. (C) Flow cytometric analysis of the percentage of transduced cell lines with JAK2 shRNA.
JAK1-Mediated H3Y41 Phosphorylation and Epigenetic Gene Regulation in ABC DLBCL.
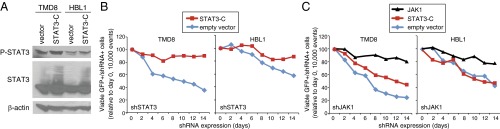
We and others have demonstrated that ABC DLBCL cell survival is promoted by overexpression and activation of STAT3 by autocrine IL-6 and IL-10 signaling (4–6). We therefore investigated whether a constitutively activated form of STAT3 (STAT3-C) (28) would prevent the apoptotic cell death induced by JAK1 inhibition. STAT3-C forms a dimer that is driven by oxidation of cysteines rather than by JAK-mediated phosphorylation of tyrosine 705 and can transform fibroblasts (28, 29). We used a retrovirus to express STAT3-C in two ABC DLBCL lines, TMD8 and HBL1, and immunoblot analysis demonstrated that STAT3-C increased the levels of active, phosphorylated STAT3 (Fig. 2A). To test the ability of STAT3-C to promote ABC DLBCL survival, we knocked down endogenous STAT3 expression using an shRNA targeting the STAT3 3′-untranslated region in two ABC DLBCL lines that had been transduced with either STAT3-C or an empty vector. In cells transduced with empty vector, STAT3 knockdown caused a time-dependent loss of viable cells, but this was reversed by expression of STAT3-C (Fig. 2B). By contrast, in two ABC DLBCL lines induced to express a JAK1 shRNA, expression of STAT3-C produced partial or no rescue of viability, whereas ectopic expression of JAK1 itself was able to rescue the cells (Fig. 2C).
Fig. 2.
STAT3-C does not rescue viability of ABC DLBCL cells following JAK1 knockdown. (A) Immunoblot analysis of the indicated proteins in TMD8 and HBL1 ABC DLBCL cells transduced with STAT3-C or empty vector, after 2 d of doxycycline induction of STAT3-C expression. (B) Flow cytometric analysis of TMD8 and HBL1 ABC DLBCL cells transduced with STAT3-C or empty vector following induction of shSTAT3. Shown are the percentages of shSTAT3+/GFP+ cells over time of shSTAT3 induction. (C) Flow cytometric analysis of TMD8 and HBL1 ABC DLBCL cells transduced with STAT3-C, JAK1, or empty vector following induction of shJAK1. Shown are the percentages of shJAK1+/GFP+ cells over time of shJAK1 induction. Data are representative of three independent experiments.
These findings suggested that STAT3 mediates some but not all of the prosurvival function of JAK1 in ABC DLBCL cells, prompting us to investigate the role of noncanonical JAK signaling in these cells. First, we investigated the subcellular distribution of JAK1 in ABC DLBCL cell lines. JAK1 was detected in cytoplasmic fractions enriched for tubulin but also in nuclear extracts enriched for histone H3, consistent with the possibility of noncanonical JAK1 signaling in the nucleus (Fig. 3A). Because noncanonical JAK2 signaling in PMBL and HL promotes expression of MYC (21), we investigated whether this is also true for JAK1 in ABC DLBCL. Immunoblot analysis revealed that MYC expression was strongly reduced following knockdown of JAK1 in ABC DLBCL cells, but was not affected by knockdown of JAK2 (Fig. 3B). The small molecule JAK1 inhibitors AZD1480 and ruxolitinib also significantly reduced MYC expression in a dose-dependent manner (Fig. 3C). Moreover, these inhibitors decreased global H3Y41 phosphorylation, consistent with noncanonical JAK1 signaling (Fig. 3C).
Fig. 3.
Nuclear JAK1 phosphorylates H3Y41 and up-regulates MYC expression in ABC DLBCL. (A) Immunoblot analysis of the indicated proteins in cytoplasmic (C) and nuclear (N) fractions of TMD8 and HBL1 ABC DLBCL cells. (B) Immunoblot analysis of the indicated proteins in TMD8 cells after 2 d of shRNA doxycycline-induced expression of shJAK1#5, shJAK2, or a control shRNA. (C) Immunoblot analysis of the indicated proteins in TMD8 cells after treatment with the JAK inhibitors AZD1480 (2 μM) or ruxolitinib (4 μM) for the indicated times. (D) ChIP analysis of H3Y41-P at the MYC locus in TMD8 cells with and without treatment with AZD1480 (2 μM) for 4 h. Quantitative PCR was performed using the primers targeting the indicated regions of the MYC locus and negative control primers targeting the ubiquitin B promoter. The mean values of H3Y41-P signals were normalized to the input DNA signal. ChIP using IgG is shown as a negative control. Error bars represent SD (n = 3).
We next investigated H3Y41 phosphorylation at the MYC locus by chromatin immunoprecipitation (ChIP) and quantitative PCR analysis using primers spanning several regulatory regions of the MYC locus, as described (21). We performed this analysis in TMD8 ABC DLBCL cells treated with the JAK1 inhibitor AZD1480 or with DMSO as a control. H3Y41 phosphorylation was evident at several MYC regions, and AZD1480 reduced these ChIP signals. The largest effect was observed at a regulatory region in intron 1 (Fig. 3D) that is also phosphorylated by JAK2 in PMBL and HL cells (21). These data suggest that the potent up-regulation of MYC by JAK1 is a consequence of H3Y41 phosphorylation of the MYC locus.
Identification of JAK1 Target Genes by H3Y41-P ChIP Sequencing in ABC DLBCL.
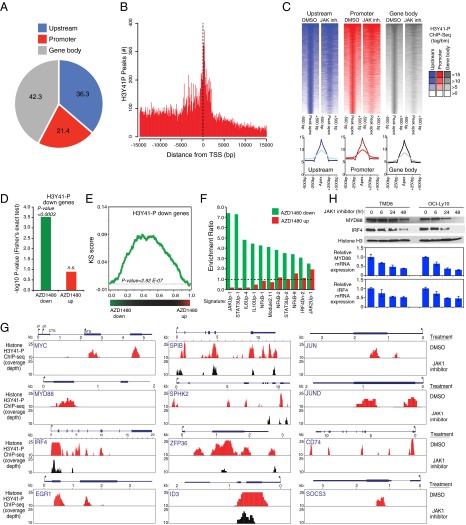
To identify the targets of noncanonical JAK1 signaling genome-wide, we performed H3Y41-P ChIP coupled with next-generation sequencing (ChIP-Seq) in the ABC DLBCL cell line TMD8. Using a stringent filter for peak calling, we identified a total of 36,634 H3Y41-P peaks (Dataset S1), with the vast majority (70.3%) mapping near a protein-coding gene within a window extending from −15 kb 5′ of the transcriptional start site (TSS) to the 3′ end of any annotated transcript associated with the gene. Of those peaks, 36.3% were located upstream of the proximal promoter (−15 kb to −2 kb relative to the TSS), 21.4% were within the proximal promoter region (−2 kb to +2 kb relative to the TSS), and the remaining 42.3% mapped within the gene body (+2 kb to the 3′ end of annotated transcripts) (Fig. 4 A and B). Similar results were obtained when peak calling was performed at a different stringency, confirming that the observed genomic distribution reflects an intrinsic characteristic of H3Y41-P and does not depend on the height of the peaks considered for the analysis. Notably, treatment of TMD8 cells with the JAK inhibitor AZD1480 reduced H3Y41-P modification at the majority of these genomic locations (Fig. 4C). Peaks in the upstream, promoter, and gene body regions were similarly affected by AZD1480 treatment (Fig. 4C, Bottom), consistent with a global impairment of noncanonical JAK1 signaling. Notably, less than 1% of H3Y41-P bound sites (269/36,634) have a STAT binding motif.
Fig. 4.
H3Y41-P ChIP-Seq in the ABC DLBCL TMD8 cell line. (A) Distribution of H3Y41-P peaks relative to protein-coding genes in the TMD8 ABC DLBCL cell line. TMD8 cells were treated with AZD1480 (2 μM) or DMSO for 4 h before ChIP-Seq (see text for details). (B) Frequency histogram of H3Y41-P ChIP-Seq tags mapping in the proximity of an annotated TSS of a protein-coding gene in TMD8 cells. (C) Density heatmaps of H3Y41-P ChIP-Seq sequence tags displayed relative to ChIP-Seq peak apex. Peak calling was performed in DMSO-treated TMD8 cells and peaks were assigned to locations relative to protein-coding genes as in A. Genes were ranked by H3Y41-P density in DMSO-treated cells (−500 bp to +500 bp from peak apex) and the heatmaps for each gene in cells treated with the JAK inhibitor AZD1480 (2 μM) are displayed accordingly. (D) Fisher’s exact test of the overlap between the set of genes with H3Y41-P peaks and the sets of genes that were either down-regulated or up-regulated by AZD1480 treatment (2 μM) of TMD8 cells. The −log10 enrichment P value is shown. (E) GSEA comparing H3Y41-P-containing genes and genes down-regulated by AZD1480 in TMD8 cells. Genes were ranked based on the changes induced by AZD1480 treatment, and H3Y41-P genes were displayed accordingly (black lines). The corresponding KS score is shown in green (P value = 2.92E-07, see Materials and Methods for detail). (F) Overlap between the sets of genes down-regulated or up-regulated by AZD1480 treatment of TMD8 cells and gene expression signatures described in ref. 30. Shown is the enrichment ratio relative to the overlap expected by chance. (G) The H3Y41-P ChIP-Seq tracks of DMSO-treated (red) or AZD1480-treated (black) TMD8 cells are shown for the indicated genes. The coverage depth is displayed and depths greater than 25 are truncated. (H) TMD8 or OCI-Ly10 ABC DLBCL cells were treated with 2 μM AZD1480 before immunoblot analysis of the indicated proteins. Below shows quantitative PCR analysis of MYD88 and IRF4 mRNA levels (normalized to β-2-microglobulin mRNA levels). Error bars represent mean ± SD of triplicates.
We identified a total of 4,366 H3Y41-P peaks (associated with 2,956 genes) that were significantly reduced by AZD1480 treatment (P < 0.01, see Materials and Methods for detail) (Dataset S1). This gene regulation mechanism by JAK1 is distinct from the canonical pathway because there is no statistical enrichment of the STAT motif within H3Y41-P peaks and more than 90% (2,686/2,956) of corresponding genes do not bear a STAT motif in their promoter region (Dataset S1). To functionally validate the role of H3Y41-P in the expression of these genes, we performed a time course analysis of gene expression changes resulting from AZD1480 treatment of TMD8 cells. Notably, genes with H3Y41-P peaks were enriched among genes that were down-regulated by AZD1480 in TMD8 cells (P < 0.0003, Fisher’s exact test) but not among genes up-regulated by AZD1480 treatment (Fig. 4D). Gene set enrichment analysis (GSEA) confirmed a significant enrichment of H3Y41-P genes among those whose transcription was blocked following JAK1 inhibition (P = 2.92 × 10−7, Kolmogorov–Smirnov (KS) test; Fig. 4E). In parallel, we performed GSEA using a database of signatures that reflect signaling and regulatory processes in normal and malignant hematopoietic cells (30). As expected, genes down-regulated by AZD1480 treatment were strongly enriched for JAK/STAT-dependent genes, as well as for NF-κB target genes (Fig. 4F). Altogether, these findings suggest that JAK1-dependent H3Y41 phosphorylation contributes to the transcriptional output of JAK1 signaling.
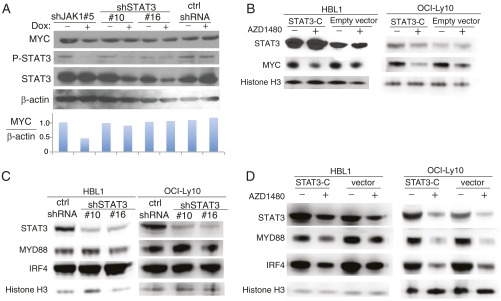
Representative genes with JAK1-dependent H3Y41-P peaks are shown in Fig. 4G, including some that are known to be central to ABC DLBCL biology, such as MYC, IRF4, SPIB, and MYD88. Reduced IRF4 and MYD88 protein and mRNA expression after JAK1 inhibition was confirmed by immunoblot analysis and quantitative PCR in two ABC DLBCL cell lines (Fig. 4H). To test whether MYC, IRF4, and MYD88 are regulated by conventional, JAK1-induced STAT3 activation, we knocked down expression of STAT3 with an shRNA or expressed the constitutively active STAT3-C isoform in ABC DLBCL cells and then analyzed expression of MYC, IRF4, or MYD88 by immunoblotting. MYC expression was not altered in cells expressing either the STAT3 shRNA or STAT3-C, but was down-regulated by a JAK1 shRNA or by AZD1480 treatment (Fig. S2 A and B). Similar results were obtained for IRF4 and MYD88 (Fig. S2 C and D). Thus, these JAK1 target genes are regulated in a STAT3-independent fashion, consistent with noncanonical epigenetic regulation by JAK1. Other genes of note with H3Y41-P peaks that were down-regulated by JAK1 inhibition (Fig. 4G) encode the transcription factors JUN and JUND, which may contribute to ABC DLBCL survival (31); EGR1, an early growth response gene that is induced by BCR stimulation and promotes B-cell proliferation and differentiation (32, 33); SPHK2, which is a molecular target in primary effusion lymphoma (34); ZFP36, an RNA binding protein that regulates mRNA stability (35); and SOCS3, a negative regulator of JAK1 signaling (36).
Fig. S2.
STAT3-independent expression of MYC, MYD88, and IRF4 in ABC DLBCL. (A) Immunoblotting with the indicated antibodies in TMD8 cells after 3 d of induction of JAK1#5, ShSTAT3#10, and #16, and control shRNAs. MYC protein levels relative to β-actin were quantitated by densitometry. (B) After 3 d of STAT3-C expression, immunoblotting was conducted with the indicated antibodies after treatment with AZD1480 (2 μM) in HBL1 cells for 2 h or in OCI-Ly10 cells for 8 h. (C) Immunoblotting with the indicated antibodies in HBL1 cells after 3 d of induction or in OCI-Ly10 cells after 5 d of induction of STAT3 and control shRNAs. (D) After 3 d of STAT3-C expression, immunoblotting was conducted with the indicated antibodies after treatment with AZD1480 (2 μM) in HBL1 cells for another 24 h or OCI-Ly10 cells for another 48 h.
Cotargeting JAK1 and BTK in ABC DLBCL.
The transcription factor IRF4 is a master regulator of the ABC DLBCL phenotype and is a target of both JAK1 signaling (see above) and chronic active BCR signaling in ABC DLBCL (10). We therefore hypothesized that combined inhibition of JAK1 and BCR signaling would have synergistic toxicity in ABC DLBCL. To test this hypothesis, we measured viability of two ABC DLBCL lines, TMD8 and OCI-Ly10, and a control GCB DLBCL line OCI-Ly19, following treatment with AZD1480 and/or ibrutinib, a BTK inhibitor that blocks BCR-dependent NF-κB activation. As expected, we observed a dose-dependent toxicity of ibrutinib treatment in the TMD8 and OCI-Ly10 lines but not in OCI-Ly19 (Fig. S3). Data in Fig. 5A were normalized both to the viability of cells at time 0 and to the viability of cells treated with the indicated concentrations of ibrutinib alone. Addition of ibrutinib to AZD1480 shifted the viability curves for the two ABC DLBCL lines to the left, indicating more than additive killing by the combination, whereas these drugs had no effect on the viability of the GCB DLBCL lines (Fig. 5A). Similarly, when JAK1 was knocked down using an shRNA in ABC DLBCL lines, the toxicity was increased by the addition of ibrutinib, but no toxicity of this combination was observed in a GCB DLBCL line (Fig. 5B).
Fig. S3.
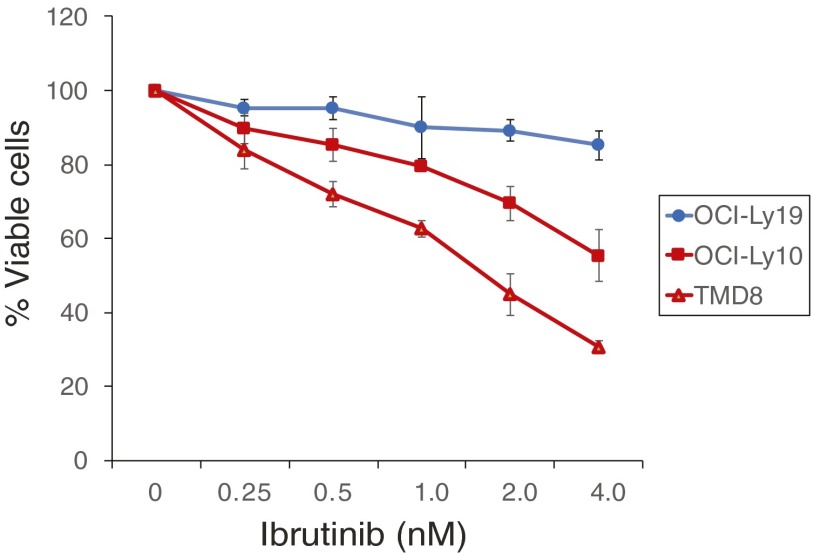
Ibrutinib is toxic to ABC DLBCL cells. The indicated DLBCL lines were treated with the indicated concentrations of ibrutinib for 6 d and evaluated for viability by trypan blue dye exclusion. The drug was replenished on day 3. Error bars represent mean ± SD of triplicates. The GCB DLBCL cell line OCI-Ly19 served as a negative control cell line.
Fig. 5.
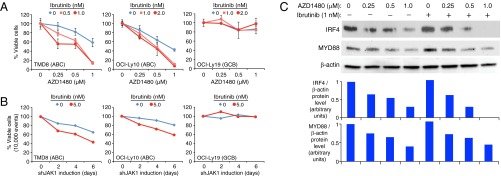
Synergy of JAK1 inhibition and ibrutinib in killing of ABC DLBCL cells. (A) The indicated DLBCL lines were treated with the indicated concentrations of AZD1480 and/or ibrutinib for 6 d and evaluated for viability by trypan blue dye exclusion. Both drugs were replenished on day 3. Error bars represent mean ± SD of triplicates. The GCB DLBCL cell line OCI-Ly19 served as a negative control cell line. Data are normalized to toxicity of treatment with the indicated concentrations of ibrutinib alone. A shift of the AZD1480 toxicity curves to the left indicates a greater than additive effect of the two drugs. (B) Ibrutinib increases JAK1 shRNA toxicity in TMD8 and OCI-Ly10 cells. Flow cytometric analysis of the percentage of viable, shJAK1-expressing cells in the presence of 5 nM ibrutinib or DMSO. (C) Immunoblot analysis of the indicated proteins in OCI-Ly10 ABC DLBCL cells after 48 h of treatment with the indicated concentrations of AZD1480 ± 1 nM ibrutinib. Shown below are IRF4 and MYD88 protein levels relative to β-actin levels as quantified by densitometry.
We next investigated whether decreased IRF4 levels were related to the combined toxicity of the JAK1 and BCR signaling inhibitors. Immunoblot analysis revealed a dose-dependent reduction in IRF4 levels with AZD1480 treatment, and the addition of ibrutinib decreased IRF4 levels further (Fig. 5C). By contrast, another JAK1 target, MYD88, was decreased in expression by AZD1480, as expected, but the addition of ibrutinib did not decrease MYD88 levels further, in keeping with the fact that MYD88 levels are not regulated by BCR signaling (Fig. 5C). Thus, decreased levels of the master regulatory transcription factor IRF4 could explain, at least in part, the observed synergistic killing of ABC DLBCL cells by JAK1 and BCR signaling inhibitors.
Discussion
Here we report a molecular mechanism by which JAK1 contributes to the malignant phenotype of ABC DLBCL. Although STAT3 is activated in ABC DLBCL cells by JAK1, STAT3 does not mediate all of the downstream consequences of JAK1 activity because a constitutively active STAT3 isoform was unable to sustain ABC DLBCL viability following inhibition of JAK1. Rather, epigenetic regulation by JAK1 plays a prominent role in the gene expression program of ABC DLBCL cells by phosphorylating chromatin on H3Y41. The chromatin of nearly 3,000 genes had JAK1-dependent H3Y41 phosphorylation marks and required JAK1 for their expression. Two of these direct JAK1 epigenetic target genes, IRF4 and MYD88, play pivotal roles in ABC DLBCL biology. MYD88 is activated by recurrent somatic mutations in ABC DLBCL, and the ABC DLBCL lines tested harbor the MYD88 L265P allele (9). MYD88 signaling activates the NF-κB and p38 MAP kinase pathways, both of which promote the expression of IL-6 and IL-10, which initiate autocrine signaling and activate JAK1. Thus, epigenetic regulation of MYD88 by JAK1 initiates a positive feedback loop. In addition to IL-6 and IL-10, MYD88 induces many other NF-κB target genes, which most likely contribute to survival of ABC DLBCL cells (9). Conversely, MYD88 signaling in ABC DLBCL also activates type I IFN expression, which is toxic for these cells (10). IRF4 suppresses this type I IFN axis by repressing IRF7, a key transcriptional activator of type I IFN genes (10). Of note, this function of IRF4 requires its dimerization with SPIB, which is also an epigenetic target of JAK1 in ABC DLBCL cells (10). An additional important action of IRF4/SPIB is to up-regulate CARD11, a key signaling adapter that is responsible for NF-κB activation downstream of BCR signaling (10). Thus, the epigenetic regulation of IRF4 and SPIB by JAK1 in ABC DLBCL accentuates the prosurvival NF-κB pathway while dampening the prodeath type I IFN pathway.
Our study adds to a growing literature demonstrating epigenetic regulation of gene expression by JAK kinases, which followed the initial discovery that the Drosophila JAK homolog Hopscotch regulates development by preventing heterochromatin formation (19, 37). This noncanonical mechanism of JAK signaling was further characterized in human leukemia and lymphoma cells in which nuclear JAK2 directly phosphorylates H3Y41, thereby blocking recruitment of the heterochromatin protein HP1α (20, 21). In embryonic stem cells, H3Y41 is phosphorylated by both JAK1 and JAK2 and this is essential for self-renewal (22). In ABC DLBCL, JAK1 epigenetic targets are largely nonoverlapping with a set of STAT3 target genes defined in ABC DLBCL by integrated STAT3 ChIP-Seq with RNA-Seq analysis (18). Of these STAT3 target genes in ABC DLBCL, most (1,241/1,540, 80.5%) do not have H3Y31-P peaks. Conversely, of the genes that have H3Y41-P peaks in ABC DLBCL and were down-regulated by AZD1480 treatment, most (79/113, 70%) were not identified as STAT3 target genes. For example, the JAK1 epigenetic target gene IRF4 does not appear to be a target of STAT3 because its expression remains unchanged when STAT3 is knocked down or overexpressed (Fig. S2). STAT3 transactivates the genes encoding IL-6 and IL-10, thereby augmenting both the canonical JAK1/STAT3 pathway as well as the noncanonical, JAK1 epigenetic pathway. In addition, STAT3 reinforces the canonical pathway by positively autoregulating its own expression (18). STAT3 also regulates a number of genes that are also regulated by NF-κB, including NFKBIA, NFKBIZ, CXCR5, CD44, and PIM2 (18), which could be due to the ability of STAT3 to form a complex with NF-κB transcription factors (38). We conclude that JAK1 uses STAT3-dependent and STAT3-independent regulatory mechanisms to influence the malignant phenotype of ABC DLBCL cells.
Our study supports the development of JAK1 inhibitors for the treatment of ABC DLBCL. We demonstrated that JAK1, not JAK2, is required for ABC DLBCL survival, which suggests that JAK1-selective inhibitors could prove effective in ABC DLBCL. The JAK inhibitors approved thus far for clinical use target multiple JAK family members (see ref. 39 for review). The drug ruxolitinib, approved for treatment of myelofibrosis and polycythemia vera, inhibits both JAK1 and JAK2. As a consequence of JAK2 inhibition, on-target side effects of ruxolitinib include thrombocytopenia and anemia. Tofacitinib is an inhibitor of JAK1, JAK2, and JAK3 that is approved for the treatment of rheumatoid arthritis. JAK3 inhibition blocks T-cell responses by preventing signaling by cytokines that act through the common γ-chain receptor, including IL-2, IL-7, IL-9, IL-15, and IL-21. Selective JAK1 inhibitors are in development that may avoid some side effects associated with inhibition of other JAK family members (39) and should be evaluated in ABC DLBCL.
Although recent clinical investigation of ibrutinib in DLBCL revealed frequent responses in ABC DLBCL (14), most responses were not long lasting, suggesting that combinations of targeted agents that inhibit distinct survival pathways will be necessary. Here we provide a rational basis for combining JAK1 inhibitors with inhibitors of BCR signaling that block NF-κB activation, such as ibrutinib. We identified IRF4 as a direct target of JAK1 epigenetic regulation, and IRF4 is also a target of NF-κB downstream of BCR signaling in ABC DLBCL. As a consequence, IRF4 levels are reduced in ABC DLBCL cells by treatment with ibrutinib (10). The combination of the JAK1 inhibitor AZD1480 and ibrutinib cooperated to reduce IRF4 to undetectable levels, which most likely contributes to their synergy in killing ABC DLBCL cells. Our findings support the development of JAK1 inhibitors in conjunction with ibrutinib for the treatment of ABC DLBCL.
Materials and Methods
Full details of the methods used and data analysis are presented in SI Materials and Methods.
Cell Lines and Culture.
All doxycycline-inducible human DLBCL cell lines that express the bacterial tetracycline repressor were engineered as described previously (40). Doxycycline (20 ng/mL) was used to induce the expression of genes or shRNAs of interest. All cultures were routinely tested for mycoplasma contamination.
ChIP-Seq Analysis of H3Y41 Phosphorylation.
ChIP-enriched DNA samples were used to create adapter-ligated libraries for massively parallel sequencing with the ChIP-Seq DNA Sample Prep Kit (Illumina) following the manufacturer’s protocol, as described previously (21). All ChIP-Seq data are available at www.ncbi.nlm.nih.gov/geo/ (accession GSE86264).
Gene Expression.
Gene expression profiling was performed using Agilent 4 × 44K whole genome arrays, as described previously (21). Signal from DMSO control cells (Cy3) was compared with JAK inhibitor-treated cells (Cy5). Array data are available at www.ncbi.nlm.nih.gov/geo/ (accession GSE86265).
SI Materials and Methods
Cell Lines and Culture.
All doxycycline-inducible human diffuse large B-cell lymphoma cell lines that express the bacterial tetracycline repressor were engineered as described previously (40). Doxycycline (20 ng/mL) was used for inducing the expression of genes of interest. The cell lines were grown in RPMI 1640 media (HyClone) supplemented with 20% (vol/vol) FBS (Atlanta Biologicals), 100 units/mL penicillin, 100 μg/mL streptomycin (Corning Cellgro), 2 mM GlutaGRO (Corning Cellgro), 1× MEM-NEAA (Quality Biological), and 1 mM sodium pyruvate solution (HyClone). All cultures were routinely tested for mycoplasma contamination. Human embryonic kidney cell line 293T was cultured in DMEM (HyClone) with 10% (vol/vol) FBS. All cell lines were cultured at 37 °C in a 5% CO2 atmosphere.
Plasmid Construction and Retroviral Transduction.
The STAT3-C (www.addgene.org/24983/) was verified with the A661C and N663C mutations and subcloned into the pRetroCMV/TO vector, which is an inducible CMV/TO retroviral vector (eGFP−) with a selection marker puromycin (40). shRNAs were prepared in a doxycycline-inducible retroviral vector with the marker eGFP and a selection marker puromycin as described previously (21), with sequences of shJAK1#2 (forward: 5′-GCGATATATTCCAGAAACATT, reverse: 5′-AATGTTTCTGGAATATATCGC), shJAK1#5 (forward: 5′-GACAGTCACAAGACTTGTGAA, reverse: 5′-TTCACAAGTCTTGTGACTGTC), shSTAT3#10 (forward: 5′-GGCAAAGGCTTACTGATAAAC, reverse: 5′-GTTTATCAGTAAGCCTTTGCC), and shSTAT3#16 (forward: 5′-GGGCTTACCATTGGGTTTAAA, reverse: 5′-TTTAAACCCAATGGTAAGCCC). These STAT3 shRNAs only target total endogenous STAT3 expression, and have no effect on exogenous STAT3.
The STAT3-C or shRNA constructs, the mutant ecotropic envelope-expressing plasmid pHIT/EA6 × 3* and gag-pol expressing plasmid pHIT60 were used to transfect the 293T cells using the Genejuice transfection reagent (Novagen) as described previously (40). The doxycycline-inducible lymphoma cell lines were infected by retroviral supernatants in the presence of 8 μg/mL polybrene.
Apoptosis Assays.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) assays were performed as described previously (21). In brief, cells grown in 96-well plates were treated with JAK inhibitors for 72 h. MTT was added to the cells 2 h before harvesting. Cells were lysed completely in isopropanol with 1% hydrochloric acid. The plate was read with a 96-well spectrometer using a 570-nm filter. The background was subtracted using a dual-wavelength setting of 570 and 630 nm.
Apoptosis was also analyzed by FACS for intracellular cleaved PARP and active caspase-3. Cells were fixed in PBS containing 2% (vol/vol) paraformaldehyde, and then permeabilized and costained with anti-PARP (BD Pharmingen, clone F21-852) and anti-caspase-3 (BD Pharmingen 51–68655X) antibodies diluted in FACS buffer containing 0.25% saponin (Sigma).
Immunoblotting and Immunoprecipitation.
Cell pellets were lysed in the lysis buffer (Cell Signaling Technology). Total proteins were separated on 4–12% (vol/vol) SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The following antibodies were used: JAK1 (Millipore/Upstate 06–272), phospho-JAK1 (Y1022/1023) (Millipore/Upstate 07–849), IRF4 (Santa Cruz, sc-6059), MYD88, STAT3, phospho-STAT3 (Y705), and β-actin (Cell Signaling Technology 4283, 9139, 9145, and 4967), MYC (Epitomics 1472–1), and H3 (Abcam, ab1791). Phosphotyrosine 4G10 antibody (Millipore/Upstate 05–3210) was used for immunoprecipitation. Ten million cells per sample were collected and cell lysates were precipitated with the antiphosphotyrosine antibody 4G10 before immunoblotting with the phospho-JAK1 antibody. The NIH ImageJ 142 software was used for densitometry.
Gene Expression.
Quantitative RT-PCR (QPCR) was performed using ABI 7700 Taqman machine for 40 cycles with annealing temperature of 60 °C. Gene expression was normalized to the expression of β-2-microglobulin (B2M) for all samples. The Taqman gene expression probes for IRF4 (Hs01056533_m1), MYD88 (Hs01573837_g1), and B2M (Hs00187842_m1) were all from Applied Biosystems. Signal values were calculated using ΔΔCt method (ABI).
Gene expression profiling was performed using Agilent 4 × 44K whole genome arrays, as described previously (21). Signal from DMSO control cells (Cy3) was compared with JAK inhibitor-treated cells (Cy5). The JAK1-dependent signature was defined as the set of genes (n = 854) down-regulated by AZD1480 treatment in TMD8 cell lines with a fold change < −0.75 log2 in at least two of the four time points (1, 3, 6, and 24 h) considered during the experiments.
Signature Enrichment/Overrepresentation Analysis.
To perform signature enrichment analysis we used a database of gene expression signatures that reflect signaling and regulatory processes in normal and malignant hematopoietic cells (30). For each signature, the gene overlap with a given AZD gene set was identified and a two-by-two contingency table with observed values for each association was generated. P values for signature enrichments were calculated using a Fisher’s exact test. The 95% confidence intervals for enrichment were estimated by calculating the enrichment scores for all possible two by two tables with fixed marginals, and then assuming a conditional multinomial distribution for the cell contents with probabilities equal to the cell proportions observed in the true data. Only confidence intervals >2 were considered significant. The AZD DW gene set was defined as the set of genes down-regulated by AZD1480 treatment in TMD8 cell line with a fold change of less than −0.75 log2 in at least two of the four time points considered during the experiments. The AZD UP gene set was defined as the set of genes up-regulated by AZD1480 treatment in TMD8 cell line with a fold change greater than +0.75 log2 in at least two of the four time points considered during the experiments.
Chromatin Immunoprecipitation (ChIP).
ChIP experiments were performed essentially as previously described (21). Briefly, 10 to 20 million cells were treated for cross-linking with formaldehyde. Chromatin shearing was performed by sonication in a Bioruptor (Diagenode). Immunoprecipitations were performed with protein G-coupled magnetic beads (Dynal/Invitrogen) according to the manufacturer’s recommendations. The antibody was prebound to washed magnetic beads by incubating 45 min at room temperature in the Bind Buffer [0.1 M sodium acetate; 0.2% Tween-20, 0.2% (wt/vol) BSA]. Unprocessed lysates were processed in parallel as “input” material along with washed beads. ChIP samples and corresponding input DNA thus prepared were analyzed by real-time PCR after 30-fold dilution in triplicates using 2× SYBR green master mix (ABI) on an ABI7500 Taqman machine (40–45 cycles, annealing at 60 °C) using self-designed primers, previously screened for lack of primer–dimer artifacts and for single species amplification. Signal values were calculated using ΔΔCt method (ABI). The negative control primer pairs were derived from the ubiquitin B promoter. The phospho-H3Y41 (Abcam, ab26310) and normal rabbit IgG (Santa Cruz, sc-2027) antibodies were used. ChIP primers for the MYC locus were described previously (21).
ChIP-Seq Analysis of H3Y41 Phosphorylation.
ChIP-enriched DNA samples were used to create adapter-ligated libraries for massively parallel sequencing with the ChIP-Seq DNA Sample Prep Kit (Illumina) following the manufacturer’s protocol with the following modifications: adapters were diluted 1:20; 200- to 600-bp fragment size range was selected with agarose gel electrophoresis; libraries were PCR enriched for 21 cycles with 45-second extension steps. Standard Solexa/Illumina kits, reagents, protocols, and instruments were used for generating clusters on flowcells and sequencing the two lanes of each library on a Genome Analyzer GAII (Illumina). Images from Illumina sequencer GAII were processed with Illumina GOAT pipeline for final base calling and quality score at each base. Resulting sequences were then aligned to human genome build 36 (hg18) with ELAND_EXTENDED software. Reads with 36 bp in length from same library in multiple lanes of same or different flowcell were pooled by concatenating their alignment files before subjected to summarization of read alignment. Only reads with up to two mismatches but uniquely mapping to reference genome were counted.
To analyze the H3Y41-P ChIP-Seq data, redundant tags were discarded and only unique tags within each given experiment (DMSO or AZD1480) were used to perform peak calling. The 36-bp sequence tags were aligned to human genome build 36 (hg18) with Bowtie software. A maximum of two mismatches was allowed for each read. To perform “peak calling,” the genome was divided into “bins” of 25 bp. Each sequencing tag was associated with the bin of its start site and an additional extension of seven bins (for a total of 200 bp) along the direction of its read to match the average library size. For a given experiment, the number of “hits” in a bin was defined as the number of extended tags associated with that bin. Peaks were defined by merging consecutive significant bins. The height of the peak was defined as the largest number of hits in any of the bins forming the peak. The “apex” of the peak was defined as the bin that had this largest number of hits. In the case of ties, the earliest such peak was chosen as the apex.
A threshold of apex >10 tags/bin was used to define high-stringency H3Y41-P binding peaks. To compare the results of two experiments (DMSO vs. AZD1480), ChIP-Seq data were normalized by total tags count. A binomial P value was calculated for the degree to which the ratio between the tag counts of the two experiments in a given bin differs from the ratio of the total number of tags in the two experiments. Bins for which this resulted in a (one-sided) P value <0.01 were deemed significant. The P value of the peak was set to be the most significant binomial P value for the bins that made up that peak.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH National Cancer Institute, the University of Wisconsin–Madison (UW–Madison) start-up funds, KL2 Scholar Awards UL1TR0000427 and KL2TR000428, the National Cancer Institute Grant 1R01 CA187299 (to L.R.), and the UW–Madison T32 Hematology Training Award T32 HL07899 (to A.C.D.).
Footnotes
Conflict of interest statement: Dr. Levine and Dr. Staudt separately collaborated with the same laboratory and have therefore appeared as coauthors on a paper published as Sanda et al. (2013) TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov 3(5):564–577, although they did so independently and without knowledge of each other’s participation in this study.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE86264 and GSE86265).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610970113/-/DCSupplemental.
References
- 1.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362(15):1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenz G, et al. Lymphoma/Leukemia Molecular Profiling Project Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam LT, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-kappaB pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111(7):3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding BB, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008;111(3):1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scuto A, et al. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 2011;71(9):3182–3188. doi: 10.1158/0008-5472.CAN-10-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194(12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21(6):723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compagno M, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 14.Wilson WH, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36(4):503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012;36(4):529–541. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368(2):161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardee J, et al. STAT3 targets suggest mechanisms of aggressive tumorigenesis in diffuse large B-cell lymphoma. G3 (Bethesda) 2013;3(12):2173–2185. doi: 10.1534/g3.113.007674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi S, et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38(9):1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson MA, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461(7265):819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rui L, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths DS, et al. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol. 2011;13(1):13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Rui L, Schmitz R, Ceribelli M, Staudt LM. Malignant pirates of the immune system. Nat Immunol. 2011;12(10):933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 25.Steidl C, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471(7338):377–381. doi: 10.1038/nature09754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedvat M, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16(6):487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mesa RA, Yasothan U, Kirkpatrick P. Ruxolitinib. Nat Rev Drug Discov. 2012;11(2):103–104. doi: 10.1038/nrd3652. [DOI] [PubMed] [Google Scholar]
- 28.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Devgan G, Darnell JE, Jr, Bromberg JF. Constitutively activated Stat3 protects fibroblasts from serum withdrawal and UV-induced apoptosis and antagonizes the proapoptotic effects of activated Stat1. Proc Natl Acad Sci USA. 2001;98(4):1543–1548. doi: 10.1073/pnas.041588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaffer AL, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 31.Juilland M, et al. CARMA1- and MyD88-dependent activation of Jun/ATF-type AP-1 complexes is a hallmark of ABC diffuse large B-cell lymphomas. Blood. 2016;127(14):1780–1789. doi: 10.1182/blood-2015-07-655647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glynne R, et al. How self-tolerance and the immunosuppressive drug FK506 prevent B-cell mitogenesis. Nature. 2000;403(6770):672–676. doi: 10.1038/35001102. [DOI] [PubMed] [Google Scholar]
- 33.Dinkel A, et al. The transcription factor early growth response 1 (Egr-1) advances differentiation of pre-B and immature B cells. J Exp Med. 1998;188(12):2215–2224. doi: 10.1084/jem.188.12.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin Z, et al. Targeting sphingosine kinase induces apoptosis and tumor regression for KSHV-associated primary effusion lymphoma. Mol Cancer Ther. 2014;13(1):154–164. doi: 10.1158/1535-7163.MCT-13-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee N, et al. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014;15(1):R12. doi: 10.1186/gb-2014-15-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krebs DL, Hilton DJ. SOCS: Physiological suppressors of cytokine signaling. J Cell Sci. 2000;113(Pt 16):2813–2819. doi: 10.1242/jcs.113.16.2813. [DOI] [PubMed] [Google Scholar]
- 37.Shi S, et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10(4):489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, et al. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21(11):1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz DM, Bonelli M, Gadina M, O’Shea JJ. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nat Rev Rheumatol. 2016;12(1):25–36. doi: 10.1038/nrrheum.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441(7089):106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.