Significance
A nucleotide-specific 2H-edited NMR approach was used to determine the nature of the intermolecular interface in the intact, dimeric HIV type-1 (HIV-1) 5′-leader RNA (230 kD). The studies distinguish between previously proposed extended duplex and kissing hairpin models and identify additional intermolecular interaction sites. A one-dimensional 2H-edited NMR method that allows temporal monitoring of intermolecular base-pair formation revealed that the observed “extended dimer interface” forms rapidly, even in the absence of RNA chaperones. In addition to addressing long-standing questions about retroviral genome dimerization, these studies illustrate the utility of 2H-edited NMR for determining the structures and folding kinetics of relatively large RNAs.
Keywords: HIV-1, 5′-leader, RNA, 5′-untranslated region, structure
Abstract
HIV type-1 (HIV-1) contains a pseudodiploid RNA genome that is selected for packaging and maintained in virions as a noncovalently linked dimer. Genome dimerization is mediated by conserved elements within the 5′-leader of the RNA, including a palindromic dimer initiation signal (DIS) that has been proposed to form kissing hairpin and/or extended duplex intermolecular contacts. Here, we have applied a 2H-edited NMR approach to directly probe for intermolecular interactions in the full-length, dimeric HIV-1 5′-leader (688 nucleotides; 230 kDa). The interface is extensive and includes DIS:DIS base pairing in an extended duplex state as well as intermolecular pairing between elements of the upstream Unique-5′ (U5) sequence and those near the gag start site (AUG). Other pseudopalindromic regions of the leader, including the transcription activation (TAR), polyadenylation (PolyA), and primer binding (PBS) elements, do not participate in intermolecular base pairing. Using a 2H-edited one-dimensional NMR approach, we also show that the extended interface structure forms on a time scale similar to that of overall RNA dimerization. Our studies indicate that a kissing dimer-mediated structure, if formed, exists only transiently and readily converts to the extended interface structure, even in the absence of the HIV-1 nucleocapsid protein or other RNA chaperones.
Like all retroviruses, HIV type-1 (HIV-1) selectively packages two copies of its unspliced, positive-sense RNA genome (1). Both strands are used for strand transfer-mediated recombination during reverse transcription (2–4), a process that promotes antiviral resistance by enhancing genetic diversity (5) and enables transcription at RNA breaks that occur naturally or are induced by restriction nucleases (6). Genomes are selected for packaging as dimers (7, 8), and studies indicate that dimerization and packaging are mechanistically coupled (9–11). Dimerization is promoted by a conserved “dimer initiation signal” (DIS) located within in the 5′-leader of the viral RNA (7, 12–19), the most conserved region of the viral genome (20). Elements that promote transcriptional activation (TAR) and splicing (SD) are also located within the 5′-leader, and these functions similarly appear to be modulated by dimerization (21–24) [although the role of dimerization on translation has been questioned (25)].
The DIS is a pseudopalindrome that contains a central, 4–6-nucleotide GC-rich element. Structural studies with oligoribonucleotides have shown that the DIS can adopt a dimeric kissing loop structure, in which residues of the GC-rich loops participate in intermolecular base pairs and the flanking residues form intramolecular base pairs within the stem of the hairpin (26–30). The DIS is also capable of adopting an extended duplex conformer in which all base pairing is intermolecular, and structures of DIS oligonucleotides in this extended duplex state have also been determined (29, 31, 32). Notably, DIS oligonucleotides with relatively long stems that include nonpaired bulges are capable of forming kissing dimers that can be converted to thermodynamically more stable extended duplex dimers (33), a process that may be facilitated by conformational fluctuations in the stem (34, 35). In addition, genomes isolated from immature virions can be readily dissociated by mild heating (36), whereas genomes from mature virions are more heat stable, with stability that increases with the age of the virus (37). The combined data are consistent with a model in which genomes are selected for packaging as less stable kissing dimers that convert to the more stable extended dimer conformers during viral maturation (13, 38–41). Other elements within the 5′-leader, including the pseudopalindromic TAR hairpin, the SD element, and residues overlapping the Unique-5′ (U5) and gag start codon regions (AUG; Fig. 1), have also been implicated in dimerization (42–44).
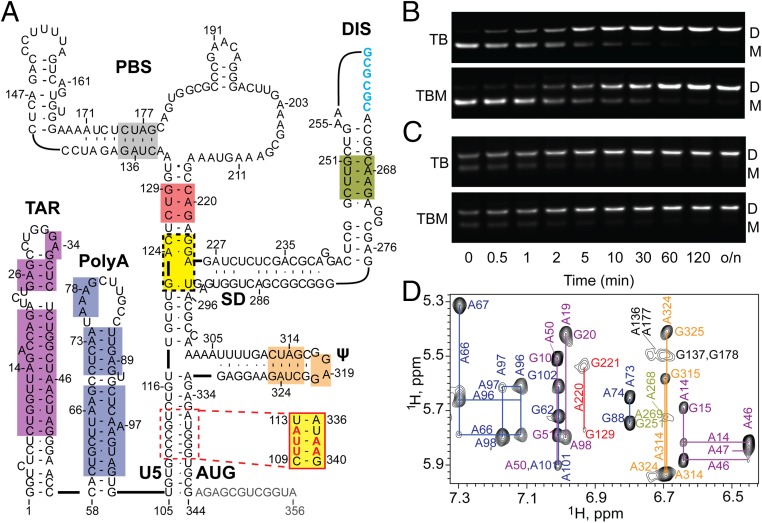
Fig. 1.
(A) Secondary structure of the HIV-1 5′-L356 in its DIS-exposed, dimer-promoting state (adapted from ref. 57; residues truncated in 5′-L344 colored gray). The 5′-L344 NMR signals with resolved and assigned 2D 1H–1H NOESY cross-peaks are denoted by color shaded boxes; yellow denotes sites that were only assignable in mutant constructs that either lacked the upper PBS loop (5′-L344ΔPBS) (57) or contained an lrAID substitution in the U5:AUG stem (5′-L344-UUA) (48). Intact (B) and truncated (C) 5′-leader constructs rapidly dimerize when incubated in PI buffer at 37 °C. Dimerization profiles on TB and TBM gels are indistinguishable, indicating that the dimers are nonlabile. o/n, overnight. (D) Region of the 2D 1H–1H NOESY spectrum of A2rGrCr-labeled 5′-L344 RNA. Assignments (adenosine-H2 and ribose-H1′ assignments labeled vertically and horizontally, respectively) are color-coded to match the highlighted elements within the secondary structure in A.
In this study, we developed a 2H-edited NMR strategy to identify the intermolecular interface of the HIV-1 dimeric 5′-leader RNA (5′-L). The approach is conceptually similar to an 15N-edited NMR line-shape method used to distinguish between inter- and intramolecular base pairing in a 39-nucleotide DIS oligonucleotide (45), but provides additional 1H NMR chemical shift and NOE information and affords sensitivity and resolution sufficient to allow probing of the intact, dimeric HIV-1 5′-leader (NL4-3 strain; 688-nucleotide dimer; 230 kDa). Our studies reveal that, under physiological-like conditions and in the absence of nucleocapsid or other RNA chaperones, the DIS element adopts an extended duplex structure and U5 participates in intermolecular base pairing with AUG. The time scale for formation of this extended interface is similar to that of overall RNA dimerization, suggesting that kissing structures, if they exist, occur transiently along the dimerization pathway.
Results
Sample Preparation and Characterization.
The structural and dimerization properties of retroviral 5′-leader RNAs have been studied under a range of experimental conditions using RNAs prepared and purified by different methods. Previous studies in our laboratory used plasmids that had been linearized with restriction endonucleases as templates for RNA transcription, producing RNAs that contained several nonnative nucleotides at the 3′-end. We recently found that the inclusion of nonnative nucleotides at the 3′-end of 5′-leader samples alters the dimerization properties of those RNAs (46), consistent with earlier observations (42). Here, we used a method for preparation of HIV-1NL4-3 5′-leader RNA samples that precluded the use of nonnative 3′-residues (47). This approach also inhibited self-templated run-on during transcription, which was a readily detectable and significant problem for one of the constructs used in the current study (see SI Materials and Methods and Fig. S1). Samples prepared using this approach afforded 1H NMR spectra with narrower signals, consistent with improved sample homogeneity. The full-length 5′-leader prepared by this method (residues 1–356, 5′-L356; Fig. 1A) readily formed dimers when incubated under conditions of physiological-like ionic strength (PI) (PI buffer = 140 mM K+, 10 mM Na+, and 1 mM Mg+2) and detected by native agarose gel electrophoresis (Fig. 1B). Equilibrium was reached ∼50-fold faster compared with constructs containing three nonnative 3′-cytidines (48).
Fig. S1.
Incorporation of DMSO and 2´ O-methyl–modified templates eliminates self-templated run-on transcription by T7 RNAP. (A) Secondary structure of the 5′-L344-UUG RNA construct (lr-AID mutations are shown in red) including the proposed self-coded run-off transcription product (blue). One-dimensional proton spectra of U5:AUG control oligo-RNAs containing either the UUA (B) or UUG (C) lrAID motif. (D) One-dimensional proton spectrum of 5′-L344-UUG RNA prepared without addition of DMSO or a 2´ O-methyl–modified template. The signal at 6.48 ppm (highlighted with yellow box) is inconsistent with the expected UUG-UAA base pairing engineered at U5:AUG but rather is consistent with UUA–UAA base pairing that would result from self-coded 3′ extension of the RNA. (E) Run-off transcription is prevented by the inclusion of DMSO and 2´ O-methyl–modified template in the transcription reaction. The absence of a signal at 6.48 ppm in the 1D proton spectrum of the 5′-L344-UUG RNA prepared by this method is consistent with the elimination of run-off transcription at the 3′ terminus of the RNA.
Previous studies have shown that HIV-1LAI leader constructs are capable of forming a “labile” dimer that can only be distinguished from monomers by electrophoresis when Mg+2 is included in the Tris–borate gel and running buffers [Tris-borate magnesium (TBM)] (49) and that the labile dimer can be converted to a nonlabile dimer (detectable in agarose gels using Tris–borate buffers that lack magnesium; TB) by incubation at 60 °C (50). The labile dimer was proposed to contain a kissing DIS interface and the nonlabile dimer an extended duplex DIS interface (50). Some fragments of the HIV-1MAL leader (51), as well as intact leaders of other lentiviruses including HIV-2 and SIV (46), also form labile dimers that can only be detected by including Mg+2 in the electrophoresis running buffer. In contrast, we did not observe significant differences in the dimerization rate of the HIV-1NL4-3 5′-leader when measured using TB or TBM running buffers (Fig. 1B), consistent with reports that the leaders of some strains of HIV-1 do not form labile dimers (51).
The 3′-end of the HIV-1 5′-leader contains a conserved stretch of residues that are capable of either forming a local hairpin structure or base pairing with U5 (43, 48, 52). NMR studies indicate that the hairpin species is adopted in the monomeric form of the RNA and that U5:AUG pairing occurs in the dimer (48). Deletions or mutations that prevent formation of the AUG hairpin but do not affect U5:AUG pairing promote dimerization (48). The present studies were therefore performed with 3′-truncated leader constructs that include all residues required for U5:AUG base pairing but lack the 3′-residues necessary for formation of the competing AUG hairpin structure (residues 1–344, 5′-L344; Fig. 1A) (48). The 5′-L344 formed nonlabile dimers, and the rate of dimerization exceeded that of the intact leader (time to reach equilibrium, ∼10 min and ∼30 min, respectively; Fig. 1 B and C).
The improved NMR spectral quality obtained with the present samples enabled us to extend assignments of adenosine-H2 NMR signals observed in 2H-edited 2D NOESY NMR spectra (53). A portion of the 2D NOESY spectrum obtained for a dimeric 5′-L344 sample containing nonexchangeable protons on the adenosine C2 and ribose (A2r), guanosine ribose (Gr), and cytosine ribose (Cr) carbons (all other nonexchangeable sites deuterated; 5′-L344-A2rGrCr) is shown in Fig. 1D, and assigned residues are labeled in Fig. 1A. Some regions that could not be directly examined due to severe signal overlap were observable in constructs that either lacked the upper PBS loop (5′-L344ΔPBS) or contained conservative base-pair substitutions in the U5:AUG region that resulted in resolved adenosine-H2 signals (long-range Adenosine Interaction Detection, lrAID) (48) (Fig. 1A).
DIS Adopts an Extended Duplex Structure in the Dimeric HIV-1 5′-Leader.
Although the above NMR approach enabled direct detection of secondary structure elements in the dimeric leader, it did not distinguish between inter- and intramolecular interactions. To directly probe for intermolecular base pairing, we used a modified 2H-edited NMR method that involved 2D 1H–1H NOESY studies of differentially labeled heterodimers. With this approach, two RNA samples with identical sequences were prepared separately using different nucleotide-specific 2H labeling schemes and purified by denaturing gel electrophoresis, then mixed, washed with salt, and incubated under conditions that promote refolding (PI buffer; 37 °C; 1 h). Samples prepared in this manner contain a statistical 50:50 distribution of heterodimers and homodimers, and labeling schemes are chosen such that cross-helix NOEs would only be observable for the heterodimeric interaction being probed.
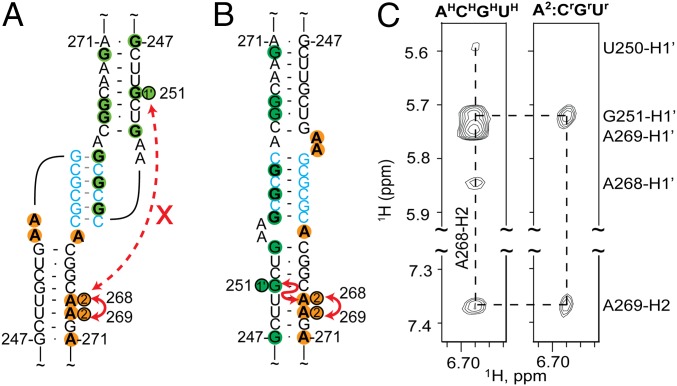
As a proof of concept, this strategy was applied to a 47-nucleotide oligo-RNA corresponding to the isolated HIV-1NL4-3 DIS element [DIS237–281; residues G237-C281, plus a nonnative 5′-G to improve transcription yield (54) and 3′-C to close the helix]. The 2D NOESY spectrum of the fully protonated DIS237–281 dimer exhibited NOEs between H2 protons of adenosines on one side of the helix minor groove and H1′ protons of residues across the minor groove, which are characteristic of A-form helices and indicate that the proton pairs are separated by ∼5 Å or less. To determine if these are intramolecular NOEs indicative of a kissing dimer or intermolecular NOEs indicative of an extended duplex dimer, a mixture was prepared in which one RNA contained protons only on the adenosine–C2 carbons (DIS237–281-A2) and the other only on the guanosine, cytosine, and uridine ribose carbons (DIS237–281-GrCrUr), with all remaining nonexchangeable sites of both RNAs being deuterated (Fig. 2A). The A268-H2 and G251-H1′ protons are separated by more than 40 Å in the kissing dimer and would not give rise to an A268-H2 to G251-H1′ intermolecular NOE, whereas these protons would be in close proximity (<4 Å) and give rise to a significant NOE signal in the extended duplex conformer (Fig. 2 A and B). The observation of a well-resolved A268-H2 to G251-H1′ intermolecular NOE in the spectrum obtained for the mixed sample provides strong direct evidence that DIS237–281 adopts an extended duplex structure under the conditions used (Fig. 2C).
Fig. 2.
DIS-containing RNAs prepared with different deuterium labeling schemes can form heterodimers with kissing (A) or extended duplex (B) interfaces. The A268-H2 to G251-H1′ separation is too great for NOE detection in the kissing conformer but short enough to be readily detected in extended duplex conformer. (C) Portions of the 2D 1H–1H NOESY spectra obtained for uniformly protonated DIS237–281 oligo-RNA (AHCHGHUH) (Left) and an equimolar mixture of A2 and CrGrUr-labeled DIS237–281 (Right).
To investigate the nature of the base pairing within the DIS element in the HIV-1 5′-leader, two HIV-1 5′-L344 RNAs were prepared with different 2H labeling strategies. An A2-labeled 5′-L344 RNA was mixed with a Gr-labeled 5′-L344 RNA. The 2D 1H–1H NOESY spectrum collected on the mixed sample was compared with that collected on a uniformly A2Gr-labeled sample. Consistent with our results from the isolated DIS237–281 RNA, an NOE from A268-H2 to G251-H1′ is observed (Fig. 3A). The combined NOE and chemical shift information provide direct evidence that DIS adopts an extended intermolecular duplex structure in the context of the dimeric 5′-leader.
Fig. 3.
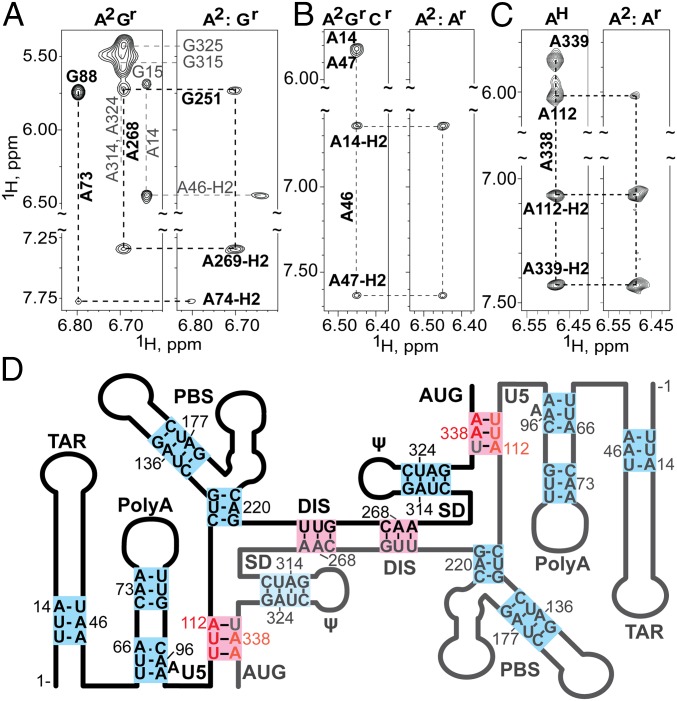
(A–C) Portions of 2D NOESY spectra obtained for mixed, differentially 2H-labeled 5′-L344 RNAs that probe the DIS and PolyA helices (NOEs that differentiate inter- or intramolecular base pairing are denoted with black labels and thick lines; noninformative NOEs are labeled gray). (A) Spectra obtained for A2Gr (Left) and mixed A2:Gr 5′-L344 (Right) samples. The A268-H2 to G251-H1′ NOE observed for the mixed sample is indicative of intermolecular base pairing at DIS (see also Fig. 2C). In contrast, the absence of an A73-H2 to G88-H1′ in the mixed sample indicates that base pairing in the PolyA helix is intramolecular. (B) Regions of spectra obtained for A2GrCr and A2:Ar 5′-L344 samples that probe TAR. Absence of an A46-H2 to A14-H1′ cross-strand NOE for the mixed sample is indicative of intramolecular base pairing. (C) Spectra obtained for AH and mixed A2:Ar-labeled 5′-L344-UUA samples that probe the U5:AUG helix. The cross-strand NOE from A338-H2 to A112-H1′ observed for the mixed sample reveals that U5:AUG base pairing is intermolecular. (D) Secondary structure consistent with the combined NMR probing data, in which the TAR, PolyA, PBS, and ψ helices are formed by intramolecular base pairs (blue) and the U5:AUG and DIS helices comprise intermolecular base pairs (pink).
Identification of Other Intermolecular Interaction Sites.
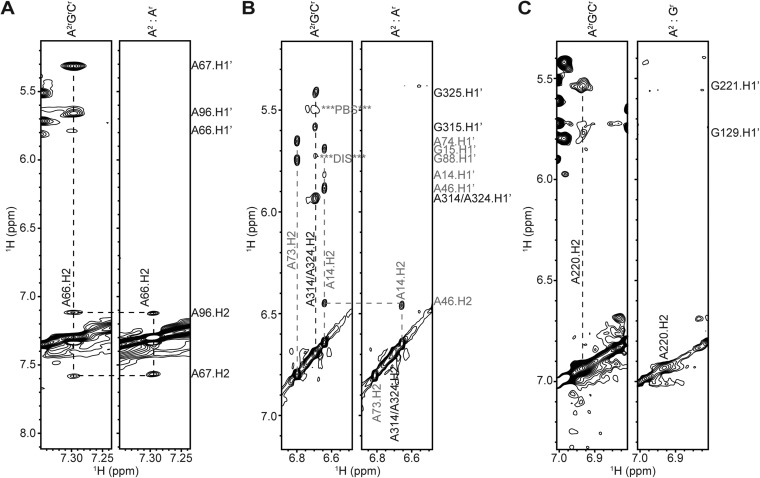
By using different labeling combinations, it was possible to probe for intermolecular base pairings for all adenosines that gave rise to resolved 1H NMR signals. The TAR hairpin contains an unusual triplet of base pairs, [5′-12UUA14]:[5′-45UAA47], that gives rise to a well-resolved A46-H2 NMR signal (∼6.5 ppm) (48). In a homodimeric 5′-L344 sample with A2rGrCr labeling, A46-H2 exhibits a typical cross-strand NOE with the H1′ proton of A14, as well as a sequential NOE with A47-H2 (Fig. 3B). Spectra obtained for an A2:Ar mixed 5′-L344 sample (Fig. 3B) exhibited the sequential A46-H2 to A47-H2 NOE but not the sequential A46-H2 to A47-H1′ NOE, as neither RNA molecule contains protons at both the aromatic C2 and ribose carbons. Importantly, the cross-strand A46-H2 to A14-H1′ signal was also absent in the mixed sample, indicating that base pairing in the helical region of TAR is intramolecular. Similar results were obtained for samples that probed the PolyA, ψ, and PBS helices, indicating that these helical substructures also contain intramolecular base pairs (Fig. 3A and Fig. S2).
Fig. S2.
Base pairing at PolyA, ψ, and lower PBS is intramolecular. (A) NOE strips of A2rGrCr and A2:Ar 5′-L344 samples focusing on A66-H2 proton (located in the base of the PolyA hairpin). In the mixed sample, A66-H2 does not give an NOE to the cross-strand A96-H1′, providing evidence that the base pairing at the base of PolyA is intramolecular. (B) NOE strips of A2rGrCr and A2:Ar 5′-L344 samples focusing on A314 and A324-H2 protons. In the mixed sample, A314-H2 does not give an NOE to the cross-strand A324-H1′. Likewise, A324-H2 does not give an NOE to the cross-strand A314-H1′. These data are consistent with intramolecular base pairing in the ψ hairpin. (C) NOE strips of A2rGrCr and A2:Gr 5′-L344 samples focusing on A220-H2 proton. In the mixed sample, A220-H2 does not give an NOE to the cross-strand G129-H1′, providing evidence that the base pairing at the lower stem of PBS is intramolecular.
Several helical regions of the native 5′-leader could not be probed by this approach due to severe signal overlap. However, it was possible to probe for intermolecular interactions in the U5:AUG helix using an lrAID modified leader, in which three sequential base pairs ([5′-109CCG112]:[5′-337UGG339]) are conservatively substituted by a triplet of adenosine-containing base pairs ([5′-UUA]:[5′-UAA]) that gives rise to a readily detectable adenosine-H2 signal. This approach was previously used to probe for U5:AUG interactions in the context of the HIV-1 5′-L (48) and was used again to probe for analogous interactions in the dimeric HIV-2ROD 5′-leader (46). The [5′-UUA]:[5′-UAA] triplet is native in the TAR hairpin; therefore, an additional mutation (A46G) was incorporated to avoid spectral overlap with the TAR signal. As a control, an lrAID-modified HIV-1 5′-L334 sample (5′-L334-UUA) was prepared with only adenosines protonated (AH). The 2D NOESY spectrum obtained for this sample exhibited sequential A338-H2 to A339-H1′ and -H2 and cross-strand A338-H2 to A112-H1′ NOEs, as expected (48). Importantly, the A338-H2 to A112-H1′ NOE was also observed in a mixed A2:Ar 5′-L344-UUA sample, demonstrating that the U5:AUG helix comprises intermolecular base pairs (Fig. 3C). Collectively, these data provide a detailed picture of the intermolecular interface of the HIV-1 5′-L RNA (Fig. 3D).
Extended Dimer Interface Forms Rapidly.
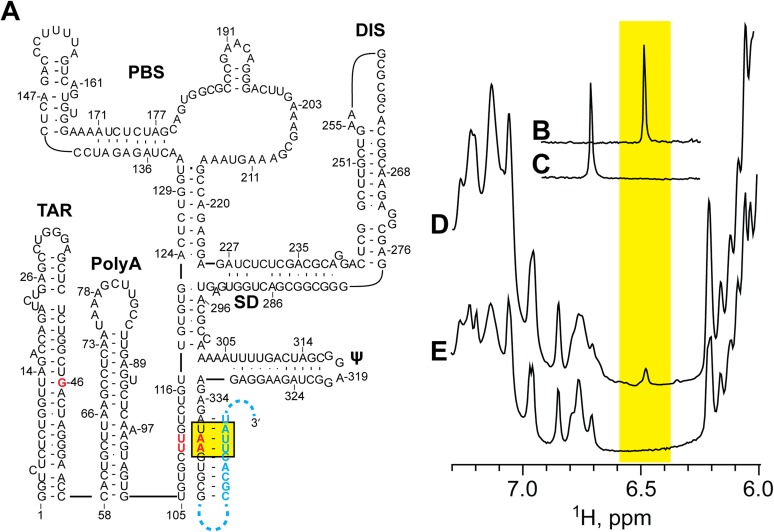
The above gel-based assays indicated that HIV-1 5′-L RNA dimerization occurs rapidly and reaches equilibrium at ∼30 min. However, the 2D NMR experiments that probed for intermolecular interactions required at least 72 h (but typically 144 h) of data collection, and we thus could not rule out the possibility that 5′-L might form a kissing dimer that slowly converts to the extended interface structure.
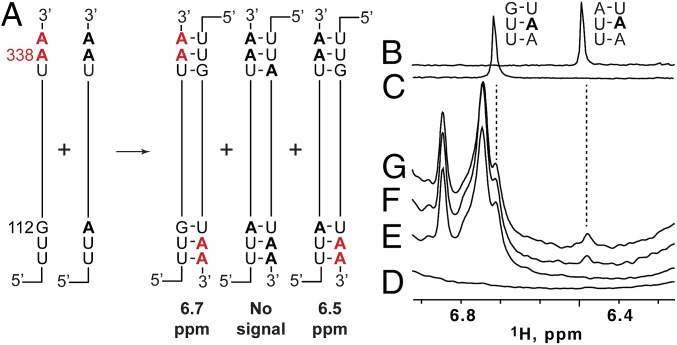
To probe the nature of the intermolecular interface at shorter time intervals, we developed a 1D 1H NMR strategy that involves mixing two mutant 5′-L RNAs that differ in both sequence and 2H-isotopic labeling (Fig. 4A). The strategy is based on the fact that the central adenosine in [UUA]:[UAA] base-pair triplets gives rise to an A-H2 signal at 6.48 ppm (Fig. 4B), whereas the same adenosine in the closely related [UUG]:[UAA] triplet gives rise to an A-H2 signal at 6.7 ppm (Fig. 4C). The U5:AUG helix was selected as the site of mutagenesis and NMR probing, as it is involved in the intermolecular interface yet is well removed from DIS, the site where dimerization is believed to nucleate. In both 5′-L344 RNAs, the native 337UGG triplet was substituted by UAA. One RNA contained an additional 110UUG substitution (5′-L344-UUG) and in the other a 110UUA (5′-L344-UUA) substitution (Fig. 4A). The latter RNA corresponds precisely to the lrAID 5′-L344 construct used in the 2D NOESY experiments to probe for intermolecular U5:AUG base pairing. The 5′-LUUG construct was prepared to be fully protonated (AHGHCHUH-5′-L344-UUG), and the 5′-L344-UUA sample contained fully protonated G, C, and U residues and perdeuterated adenosines (GHCHUH-5′-L344-UUA).
Fig. 4.
Extended dimer forms rapidly, as measured using a 2H-edited 1D NMR approach. (A) Two RNAs with different sequences and 2H-labeling schemes (5′-AHGHCHUH-L344-UUG and GHCHUH-5′-L344-UUA) are mixed under low-salt conditions that favor the monomer. Dimerization, induced by addition of PI buffer, results in statistical mixture of [AHGHCHUH-L344-UUG]2 and [GHCHUH-L344-UUA]2 homodimers and a [GHCHUH-L344-UUA]:[AHGHCHUH-L344-UUG] heterodimer. The [GHCHUH-L344-UUA]:[AHGHCHUH-L344-UUG] heterodimer is the only species with an adenosine-protonated [UUA]:[UAA] base-pair triplet that could give rise to a detectable adenosine-H2 NMR signal at 6.48 ppm. (B and C) Regions of 1H NMR spectra obtained for control oligo-RNAs showing that the H2 signal of the central adenosine in [UUG]:[UAA] and [UUA]:[UAA] triplets occurs at significantly different frequencies (6.72 and 6.48 ppm, respectively). (D and E) Regions of 1D 1H NMR spectra obtained for dimeric forms of the isolated GHCHUH 5′-L344-UUA (D) and AHGHCHUH-L344-UUG (E) RNAs (samples incubated in PI buffer for 1 h at 37 °C). (F and G) Portions of 1H NMR spectra obtained for a mixed GHCHUH 5′-L344-UUA + AHGHCHUH-L344-UUG sample upon addition of dimer-promoting PI buffer. The 6.48 ppm signal appears within 20 min (F), with intensity similar to that observed after 12 h of incubation (G).
The isolated GHCHUH-5′-L344-UUA construct did not give rise to 1H NMR signals in the range of 6.3–6.9 ppm, as expected, as all of the adenosines were deuterated (Fig. 4D). The isolated AHGHCHUH-5′-L344-UUG RNA gave rise to a number of adenosine H2 NMR signals in the range of 6.7–6.9 ppm, including a signal at 6.7 ppm diagnostic of a UUG:UAA triplet (Fig. 4E). Note that neither isolated sample gave rise to a signal at 6.48 ppm diagnostic of the UUA:UAA triplet because (i) the 5′-L344-UUA sample lacks protonated adenosines and (ii) the 5′-L344-UUG construct can only form [UUG]:[UAA] base-pair triplets (Fig. 4A). However, after mixing the two samples and acquiring a 1D 1H NMR spectrum (20 min total time), a well-resolved signal at 6.48 ppm was observed indicating that (i) heterodimerization has occurred and (ii) the U5:AUG helix is comprised of strands from both RNAs (Fig. 4 A and F). The intensity of this signal did not increase significantly in subsequently obtained NMR spectra (Fig. 4G). Thus, the intermolecular U5:AUG interface forms on the same time scale as that of overall RNA dimerization.
SI Materials and Methods
Preparation of DNA Template.
DNA oligonucleotides that served as templates for the synthesis of DIS237–281, U5:AUGUUA, and U5:AUGUUG RNAs (once annealed with the T7 promoter) were purchased from IDT DNA Technology. HIV-1 5′-L DNA templates were generated by PCR amplification (EconoTaq PLUS 2X Master Mix, Lucigen) on appropriate plasmids. Reverse primers contained 2′ O-methyl modifications on the first two nucleotides (65). A complete list of plasmids and primers is shown in Tables S1 and S2.
Table S1.
Plasmids used in this study
| Plasmid | Description |
| T-SL4_native_blunt | WT HIV-1NL4-3 5′-L sequence (nt 1–356) |
| T-SL4-345 | WT HIV-1NL4-3 5′-L sequence (nt 1–345) |
| T-SL4-345-dPBS | HIV-1NL4-3 5′-L sequence with a GAGA tetraloop capping the lower PBS hairpin |
| T-SL4-345_UUA_lrAID | HIV-1NL4-3 5′-L sequence (nt 1–345), A46G, C110T, C111T, G112A, G338A, G339A |
| T-SL4-345_UUG_lrAID | HIV-1NL4-3 5′-L sequence (nt 1–345), A46G, C110T, C111T, G338A, G339A |
Table S2.
Primers used for the PCR-based preparation of DNA templates for RNA transcription
| Primer | Description | Sequence, 5′–3′* |
| 5′-primer_80 | Forward amplification primer for all 5′-L samples | GGGATGTGCTGCAAGGCGATTAAGTTGGG |
| R_TSL4nativeBlunt | Reverse amplification primer for 5′-L356 template | mUmACCGACGCTCTCGCACCCATCTCTC |
| R_TSL4-344 | Reverse amplification primer for 5′-L344 and 5′-L344ΔPBS template | mCmGCACCCATCTCTCTCCTTCTAGCCTCC |
| R_TmSL4-344-lrAID | Reverse amplification primer for 5′-L344-UUA lrAID and 5′-L344-UUG lrAID template | mCmGCACTTATCTCTCTCCTTCTAGCCTCCGC |
| Top17 | T7 promoter | TAATACGACTCACTATA |
| DIS_237-281 | Template for transcription of DIS237–281 RNA | mGmGCCCCTCGCCTCTTGCCGTGCGCGCTTCAGCAAGCCGAGTCCTGCCTATAGTGAGTCGTATTA |
| U5-gaga-AUG-UUAlrAID | Template for transcription of U5:AUGUUA lrAID RNA | mGmGCGCACTTATCTTCTCAGATAAGCACACCTATAGTGAGTCGTATTA |
| U5-gaga-AUG-UUGlrAID | Template for transcription of U5:AUGUUG lrAID RNA | mGmGCGCACTTATCTTCTCAGACAAGCACACCTATAGTGAGTCGTATTA |
m denotes the incorporation of 2′ O-methyl RNA bases.
Selective C8 Protonation and Deuteration of ATP and GTP.
Perdeuterated and partially deuterated rNTP reagents used for in vitro transcription were obtained from Cambridge Isotope Laboratories (CIL). Deuteration of the C8 position of fully protonated GTP and ATP was achieved by incubation with triethylamine (TEA, 5 equiv) in D2O (99.8% deuteration; CIL) at 60 °C for 24 h and for 5 d, respectively (66). Similarly, deuteration of the C8 position of rATP (ribose-1′, 2′, 3′, 4′, 5′, 5′′-D6) was achieved by analogous treatment. TEA was subsequently removed by lyophylization.
In Vitro RNA Transcription.
RNAs were synthesized by in vitro transcription using T7 RNA polymerase in 10–30 mL reactions, each containing ∼1.5 mg of PCR-amplified DNA template (for 5′-L constructs) or ∼8 pmol of annealed DNA template (for oligo-RNA constructs), 2 mM spermidine, 80 mM Tris·HCl (pH 8.5), 2 mM DTT, 20% (vol/vol) DMSO, 0.3 mg T7 RNA polymerase, 10–20 mM MgCl2, and 3–6 mM NTPs. Selectively protonated RNA samples were prepared using appropriate combinations of deuterated and fully or partially protonated NTPs. After a 3-h incubation at 37 °C, the reaction was quenched by addition of an EDTA–Urea mixture (250 mM EDTA, pH 8.0, 7 M Urea), and the RNA was purified by electrophoresis on urea-containing polyacrylamide denaturing gels (SequaGel, National Diagnostics) using FisherBiotech DNA sequencing system at 20 W overnight. The gel bands were visualized by UV-shadowing, excised, and eluted using the Elutrap electroelution system (Whatman) at 120 V overnight. The eluted RNAs were washed with 2 M NaCl and then desalted using a 30-kDa (5′-L RNAs) or 3-kDa (oligo-RNAs) MWCO Amicon Ultra-4 Centrifugal Filter Device (Millipore). The concentration of each sample was determined by measuring the optical absorbance at 260 nm.
Native Agarose Gel Electrophoresis.
RNA (20 μL, 0.9 μM) was incubated at 37 °C in PI buffer (20 mM Tris, pH 7.5, 1 mM MgCl2, 140 mM KCl, 10 mM NaCl) for the time indicated such that all samples would be finished at the same time. Samples were removed from the incubator and immediately placed on ice. Native agarose gel loading solution [40% (vol/vol) Sucrose, 0.17% Bromophenol Blue] was added to each sample mixed on ice and briefly centrifuged. We loaded 10 μL of each sample onto a 1% (vol/vol) agarose TB gel and loaded 10 μL onto a 1% (vol/vol) agarose TBM gel (agarose gel containing 0.2 mM MgCl2 in both the gel and running buffer), each containing ethidium bromide at a final concentration of 0.5 μg/mL TB and TBM agarose gels, and buffers were refrigerated before use. Gels were run on ice at 115 V for 65 min and visualized by UV illumination. The percentages of monomer and dimer were quantified using the ImageJ software (67). To address the concern that lowering the temperature to 4 °C might not inhibit dimerization, we conducted an experiment in which one RNA sample was incubated at 4 °C with PI buffer, and a second sample was incubated at 37 °C in PI. The RNA maintained at 4 °C remained monomeric, whereas the 37 °C-incubated RNA formed a dimer.
NMR Sample Preparation.
DIS237–281 NMR samples [550 μL of ∼200 μM RNA in in D2O (99.96%; CIL)] were prepared in a standard 5 mm NMR tube with 20 mM Tris–d11 buffer (pD = 7.5). The 5′-L344 NMR samples (340 μL of ∼150 μM RNA in D2O in a Bruker Shaped sample tube) were prepared in 20 mM Tris–d11 buffer (pD = 7.5), 140 mM KCl, and 1 mM MgCl2. Mixed samples were transcribed and purified independently. After elution from the denaturing gel, equal molar samples were pooled together, washed, lyophilized, and then prepared for NMR as described above.
NMR Spectroscopy.
All NMR data were processed with NMRFx (68) and analyzed with NMRViewJ (69). NMR data were collected with a Bruker AVANCE spectrometer (800 MHz, 1H, 37 °C). Signals in 2D NOESY spectra were assigned by comparisons with subspectra obtained for isolated oligonucleotide fragments (e.g., TAR, PolyA, and U5:AUG oligo-RNAs) and validated by both NOE analyses and comparisons with database and predicted 1H NMR chemical shifts (53, 70, 71). This approach (53) enabled direct detection of a number of secondary structure elements in the dimeric 5′-L344 RNA (Fig. 1A).
Extended Dimer Formation Monitored by NMR.
The 5′-LUUA and 5′-LUUG RNAs were independently transcribed and purified as described above. Equal quantities (∼17 nmol) of GHCHUH-5′-LUUA and AHGHCHUH-5′-LUUG RNA were lyophilized from water. In addition, 340 μL of NMR buffer (see above) was lyophilized. Each RNA was reconstituted in 170 μL D2O (99.96%, CIL), mixed together, and then added to the tube containing lyophilized buffer, at which point timing began. The sample was mixed, briefly centrifuged, and loaded into a Bruker Shaped tube. The sample was immediately placed into the spectrometer, where the experiment parameters were checked and acquisition started. A series of identical 1D proton experiments were queued to run sequentially.
Discussion
The importance of the DIS element in promoting HIV genome dimerization has been well documented (12–19, 55), and there is now strong genetic evidence that the central six-nucleotide palindrome is the primary determinant of RNA partner selection (7, 56). Studies with short oligoribonucleotides indicate that DIS is capable of adopting both kissing (26–30) and extended duplex (29, 31, 32) conformers, and both types of interfaces have been proposed to play roles in replication. Although the secondary structures of recombinant HIV-1 5′-leader RNAs have been extensively studied, the nature of the dimer interface has not been probed. The present studies reveal that the HIV-1NL4-3 5′-leader adopts an extended duplex DIS conformation and that formation of the extended dimer interface occurs on a similar time scale as that of overall RNA dimerization. Furthermore, formation of the extended duplex DIS structure in the context of the intact 5′-leader does not require the presence of nucleocapsid or other RNA chaperones and occurs without heating above physiological temperatures (37 °C).
Previous studies with truncated HIV-1NL4-3 5′-leader RNAs and antisense oligonucleotides indicate that SD also plays a role in dimerization (42), and this finding is consistent with the intermolecular base pairing of SD residues shown in Fig. 1A (57). Unfortunately, we were unable to directly detect NMR signals for these or other residues of the central helix in the intact dimeric leader due to overlap with numerous and sometimes broad adenosine-H2 signals associated with the upper PBS loop. It was, however, possible to directly detect cross-helix NOEs involving A124 in a mutant 5′-L344 construct that lacked the upper PBS loop (57) as well as in a 155-nucleotide “core encapsidation signal” RNA (57). Thus, the only segments of the native dimeric leader that we were unable to probe by this NMR approach, due to issues of signal overlap, were the central helix of the tandem three-way junction ([U118-G123]:[U294-A300]) and the lower region of the extended DIS stem ([A227-A242]:[G278-A292]).
Other regions of the HIV-1 5′-leader have also been proposed to play roles in dimerization. For example, deletion of the TAR hairpin has been shown to inhibit dimerization of the HIV-1HXB2 5′-leader (43). Consistent with this hypothesis, antisense oligonucleotides that target the apical loop of TAR have been shown to promote dimerization (58). However, mutagenesis studies indicated that it is the upper Tat-binding bulge and not the apical loop residues that promote dimerization (59). Other studies indicated that TAR is not essential for genome dimerization and packaging and that mutations that disrupt base pairing in the stem of TAR can lead to aberrant dimerization and packaging (which was attributed to misfolding) (60). On the other hand, chemical probing studies suggested that the base of the TAR hairpin, rather than the Tat-binding bulge or apical loop, may be involved in regulating a monomer–dimer structural switch (61). The present studies provide clear evidence that residues of TAR adopt an intramolecular hairpin structure in the context of the intact 5′-leader. However, we cannot rule out the possibility that residues in the loops or lower base of TAR, which we did not observe in our NMR experiments, might participate in dimer-stabilizing interactions with undefined downstream elements (59).
Our data are incompatible with an early proposal that residues of PolyA form an intermolecular duplex (62). The binding of tRNALys3 (and an 18-nucleotide DNA analog) to the PBS was also reported to promote dimerization, suggesting that the PBS could play a direct or indirect role in dimerization (63). The present studies show that the lower stem of PBS comprises intramolecular base pairs in the dimeric 5′-leader. Although we cannot rule out the possibility that other regions of the PBS make intermolecular contacts in the dimer, the fact that 5′-leader–containing vector RNAs that lack the tRNA-binding site within PBS are packaged with avidity similar to that of the intact 5′-leader (53, 57) is consistent with studies indicating that the PBS does not contribute to dimerization (53).
Studies also indicate that U5:AUG interactions promote genome dimerization. Helper RNAs are unable to rescue packaging of RNAs with AUG mutations (44), and this is likely due to a defect in U5:AUG-dependent exposure of the DIS (48). Mutations in U5 and/or AUG that would disrupt U5:AUG pairing inhibited dimerization (43, 64), and it was suggested that this is most likely due to an indirect effect and not intermolecular base pairing (43). Consistent with this hypothesis, addition of short oligonucleotides complementary to the U5 region of the HIV-1NL4-3 5′-leader promoted 5′-leader dimerization, and this was attributed to displacement of monomer-stabilizing intramolecular U5:DIS base pairing (48). However, the present study shows that, at equilibrium, U5:AUG base pairing is intermolecular in the dimeric 5′-leader.
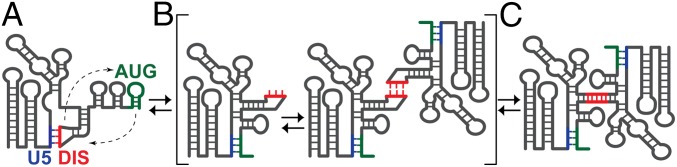
The collective data are consistent with a dimerization model in which the DIS is originally sequestered by base pairing with U5 in the monomeric form of the 5′-leader (Fig. 5A). Subsequent intramolecular base pairing of the downstream AUG element with U5 would displace and expose the DIS, thus leading to a monomeric RNA that is poised to form DIS-mediated kissing interactions with another RNA molecule (Fig. 5B). The next species formed along this dimerization trajectory would be the labile kissing dimer species, in which the U5:AUG helix comprises intramolecular base pairs and only the DIS:DIS interface contains intermolecular base pairs (Fig. 5B). Finally, the labile kissing dimer undergoes rearrangement to the thermodynamically nonlabile extended dimer that includes intermolecular DIS and U5:AUG helices (Fig. 5C). In the case of the HIV-1NL4-3 leader, the DIS-exposed monomer and kissing dimer species are undetected by the methods used here and therefore, if they exist, must only be transiently formed along the dimerization pathway. We now plan to apply this approach to the 5′-leader RNAs of retroviruses that form labile dimers to determine if they adopt kissing or “labile-extended” species.
Fig. 5.
Mechanism of HIV-1 5′-leader dimerization. (A) In the monomeric conformer, the DIS hairpin is sequestered by intramolecular base pairing with U5 (48). (B) AUG base pairs with U5, displacing the DIS and enabling formation of a labile kissing intermolecular interface, which rapidly transitions to the extended dimer conformer (C). NMR signatures for the kissing species were not observed in the present studies, indicating that it represents a transient species along the dimerization pathway.
The present studies illustrate both the potential and weaknesses of the 2H-edited adenosine-detected 1H NMR method for differentiating inter- and intramolecular interactions in relatively large RNAs. The sensitivity and chemical shift dispersion was sufficient for detection of 28 out of 85 total adenosine residues, which enabled probing of all of the known helices of the native, dimeric 5′-leader except the lower helix of the extended DIS stem, the central helix of the tandem three-way junction, and the native U5:AUG helix. The majority of unassigned adenosines (50%) reside within the upper PBS loop, a region for which we do not have appropriate control oligo-RNAs to aid in assignment and which may be conformationally heterogeneous in the absence of the primer tRNA binding partner. The lrAID approach enabled probing of U5:AUG; however, we were unable to use this method to probe the short, central helix of the tandem three-way junction due to apparent RNA misfolding caused by the lrAID mutations. It thus appears that the lrAID approach may only be readily applicable if the helical elements being probed are sufficiently stable to tolerate incorporation of a short stretch of sequential adenosines. Finally, the combination of mixed 2H-labeling with mixed lrAID mutations provides a powerful approach for temporal monitoring of intermolecular helix formation in dimeric RNAs as large as 230 kDa.
Materials and Methods
RNAs were synthesized by in vitro transcription using T7 RNA polymerase with slight modifications to published procedures (57) (plasmids and primers are summarized in Tables S1 and S2, respectively). NMR data were collected with a Bruker AVANCE spectrometer (800 MHz, 1H, 37 °C). For studies involving mixed RNAs with different labeling schemes, RNAs were transcribed and purified independently. After elution from the denaturing gel, equal molar amounts were pooled, washed, and lyophilized before addition of dimer-promoting PI buffer. Samples for time-dependent NMR studies were prepared as follows: Equal quantities (∼17 nmol) of GHCHUH-5′-LUUA and AHGHCHUH-5′-LUUG RNA were lyophilized from water and independently reconstituted in 170 μL D2O. The RNAs were pooled together, added to a tube containing lyophilized NMR buffer, and immediately placed into the spectrometer. A series of identical 1D proton experiments were queued to run sequentially. For details, see SI Materials and Methods.
Acknowledgments
We thank the Howard Hughes Medical Institute (HHMI) staff at University of Maryland Baltimore County for technical assistance. This research was supported by National Institute of General Medical Sciences (NIGMS) Grants R01 GM42561 and P50 GM 103297 (to M.F.S.). S.M. and J.S. were supported by NIGMS Grant MARC U*STAR 2T34 GM008663 for enhancing minority access to research careers and by an HHMI undergraduate education grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614785113/-/DCSupplemental.
References
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Lab Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- 2.Coffin JM. Structure, replication, and recombination of retrovirus genomes: Some unifying hypotheses. J Gen Virol. 1979;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Hu W-S, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87(4):1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 5.Nora T, et al. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81(14):7620–7628. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73(3):451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore MD, et al. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81(8):4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikolaitchik OA, et al. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9(3):e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3(8):643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 10.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410(4):609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzembayeva M, Dilley K, Sardo L, Hu W-S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454-455:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H-J, Lee K, O’Rear JJ. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198(1):336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 13.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91(11):4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7(8):1097–1114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clever JL, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70(9):5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paillart J-C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93(11):5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paillart JC, et al. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70(12):8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70(5):2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70(10):6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foley B, et al. 2013. HIV Sequence Compendium (Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM)
- 21.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70(2):944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greatorex J. The retroviral RNA dimer linkage: Different structures may reflect different roles. Retrovirology. 2004;1:22. doi: 10.1186/1742-4690-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbink TEM, Berkhout B. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J Virol. 2008;82(6):3090–3098. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jablonski JA, Buratti E, Stuani C, Caputi M. The secondary structure of the human immunodeficiency virus type 1 transcript modulates viral splicing and infectivity. J Virol. 2008;82(16):8038–8050. doi: 10.1128/JVI.00721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44(25):9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 26.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5(6):432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 27.Dardel F, Marquet R, Ehresmann C, Ehresmann B, Blanquet S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998;26(15):3567–3571. doi: 10.1093/nar/26.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol. 2001;8(12):1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 29.Baba S, et al. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J Biochem. 2005;138(5):583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 30.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34(1):343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mujeeb A, Parslow TG, Zarrinpar A, Das C, James TL. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 1999;458(3):387–392. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 32.Ulyanov NB, et al. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J Biol Chem. 2006;281(23):16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi K-I, et al. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6(1):96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mihailescu M-R, Marino JP. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc Natl Acad Sci USA. 2004;101(5):1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Zhang Q, Al-Hashimi HM. Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: Role in the kissing-duplex structural transition. Nucleic Acids Res. 2007;35(5):1698–1713. doi: 10.1093/nar/gkm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68(8):5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J Mol Biol. 2007;371(4):1084–1098. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Paillart JC, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269(44):27486–27493. [PubMed] [Google Scholar]
- 39.Muriaux D, Girard P-M, Bonnet-Mathonière B, Paoletti J. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270(14):8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 40.Muriaux D, Fossé P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35(15):5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 41.Muriaux D, De Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271(52):33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 42.Deforges J, Chamond N, Sargueil B. Structural investigation of HIV-1 genomic RNA dimerization process reveals a role for the Major Splice-site Donor stem loop. Biochimie. 2012;94(7):1481–1489. doi: 10.1016/j.biochi.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Song R, Kafaie J, Laughrea M. Role of the 5′ TAR stem-loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47(10):3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- 44.Nikolaitchik O, Rhodes TD, Ott D, Hu W-S. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J Virol. 2006;80(10):4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi K, et al. NMR analysis of intra- and inter-molecular stems in the dimerization initiation site of the HIV-1 genome. J Biochem. 2000;127(4):681–686. doi: 10.1093/oxfordjournals.jbchem.a022657. [DOI] [PubMed] [Google Scholar]
- 46.Tran T, et al. Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12:83. doi: 10.1186/s12977-015-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helmling C, et al. Rapid NMR screening of RNA secondary structure and binding. J Biomol NMR. 2015;63(1):67–76. doi: 10.1007/s10858-015-9967-y. [DOI] [PubMed] [Google Scholar]
- 48.Lu K, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334(6053):242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laughrea M, Jetté L. HIV-1 genome dimerization: Kissing-loop hairpin dictates whether nucleotides downstream of the 5′ splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry. 1997;36(31):9501–9508. doi: 10.1021/bi970862l. [DOI] [PubMed] [Google Scholar]
- 50.Laughrea M, Jetté L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35(5):1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 51.Marquet R, Paillart J-C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22(2):145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem. 2003;278(13):11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 53.Heng X, et al. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417(3):224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 55.Laughrea M, Shen N, Jetté L, Wainberg MA. Variant effects of non-native kissing-loop hairpin palindromes on HIV replication and HIV RNA dimerization: Role of stem-loop B in HIV replication and HIV RNA dimerization. Biochemistry. 1999;38(1):226–234. doi: 10.1021/bi981728j. [DOI] [PubMed] [Google Scholar]
- 56.Moore MD, et al. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5(10):e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keane SC, et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348(6237):917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reyes-Darias JA, Sánchez-Luque FJ, Berzal-Herranz A. HIV RNA dimerisation interference by antisense oligonucleotides targeted to the 5′ UTR structural elements. Virus Res. 2012;169(1):63–71. doi: 10.1016/j.virusres.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Jalalirad M, Saadatmand J, Laughrea M. Dominant role of the 5′ TAR bulge in dimerization of HIV-1 genomic RNA, but no evidence of TAR-TAR kissing during in vivo virus assembly. Biochemistry. 2012;51(18):3744–3758. doi: 10.1021/bi300111p. [DOI] [PubMed] [Google Scholar]
- 60.Das AT, Vrolijk MM, Harwig A, Berkhout B. Opening of the TAR hairpin in the HIV-1 genome causes aberrant RNA dimerization and packaging. Retrovirology. 2012;9:59. doi: 10.1186/1742-4690-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenyon JC, Prestwood LJ, Le Grice SF, Lever AM. In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res. 2013;41(18):e174. doi: 10.1093/nar/gkt690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Höglund S, Öhagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233(2):271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 63.Seif E, Niu M, Kleiman L. Annealing to sequences within the primer binding site loop promotes an HIV-1 RNA conformation favoring RNA dimerization and packaging. RNA. 2013;19(10):1384–1393. doi: 10.1261/rna.038497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology. 2002;303(1):152–163. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 65.Kao C, Zheng M, Rüdisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5(9):1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang X, Yu P, LeProust E, Gao X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25(23):4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Norris M, Fetler B, Marchant J, Johnson BA. NMRFx Processor: A cross-platform NMR data processing program. J Biomol NMR. 2016;65(3-4):205–216. doi: 10.1007/s10858-016-0049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 70.Barton S, Heng X, Johnson BA, Summers MF. Database proton NMR chemical shifts for RNA signal assignment and validation. J Biomol NMR. 2013;55(1):33–46. doi: 10.1007/s10858-012-9683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown JD, Summers MF, Johnson BA. Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. J Biomol NMR. 2015;63(1):39–52. doi: 10.1007/s10858-015-9961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]