Significance
Because protein folding is crucial to proper cellular function, there must be evolutionary pressures on how a protein achieves and maintains its folded structure. The outcome of these pressures on a folding pathway should be reflected in trends and patterns over a protein’s evolutionary history. To understand how folding pathways evolve, we characterized how reconstructed ancestral proteins of the ribonuclease H family fold. The deepest ancestors fold and unfold faster than their modern descendants, and kinetic stability evolved along both mesophilic and thermophilic lineages. This trend is possible because of a conserved partially folded intermediate state, which uncouples thermodynamic and kinetic stability to allow each parameter to evolve independently.
Keywords: protein folding, energy landscape, protein evolution, ancestral sequence reconstruction
Abstract
Proper folding of proteins is critical to producing the biological machinery essential for cellular function. The rates and energetics of a protein’s folding process, which is described by its energy landscape, are encoded in the amino acid sequence. Over the course of evolution, this landscape must be maintained such that the protein folds and remains folded over a biologically relevant time scale. How exactly a protein’s energy landscape is maintained or altered throughout evolution is unclear. To study how a protein’s energy landscape changed over time, we characterized the folding trajectories of ancestral proteins of the ribonuclease H (RNase H) family using ancestral sequence reconstruction to access the evolutionary history between RNases H from mesophilic and thermophilic bacteria. We found that despite large sequence divergence, the overall folding pathway is conserved over billions of years of evolution. There are robust trends in the rates of protein folding and unfolding; both modern RNases H evolved to be more kinetically stable than their most recent common ancestor. Finally, our study demonstrates how a partially folded intermediate provides a readily adaptable folding landscape by allowing the independent tuning of kinetics and thermodynamics.
Because most proteins need to fold to function, there must be selective pressures for efficient folding and maintenance of structure. Although many studies have investigated the evolution of protein function, a detailed understanding of the evolutionary constraints on protein folding is lacking.
The amino acid sequence of a protein encodes its energy landscape, which determines its folding trajectory—its mechanism, rates, and energetics (1). Thus, the energy landscape defines a protein’s ability to fold in a biologically reasonable time scale, avoid misfolding, and prevent frequent unfolding—all of which are likely important for the overall fitness of an organism. Indeed, numerous studies have proposed how protein folding might affect molecular evolution (2–8), and there is growing evidence that folding pathways and their associated kinetics and energetics are evolutionarily constrained. To date, however, there is little experimental evidence detailing how folding properties have changed throughout evolution. Are there trends in the folding mechanism or rates? Which aspects of the folding mechanism have been conserved, and how have they played a role in the evolution of modern-day homologs? We might naively expect folding to optimize, so that modern proteins generally fold faster than their ancestors. Or perhaps the energetics of the folding landscape evolved to avoid kinetic traps or misfolded states. If maintaining the folded state is important for fitness, then we might expect an improvement in kinetic stability over time, although a folded state that is too stable might be detrimental for conformational dynamics. These insights, and others, are critical for our understanding of how evolutionary forces affect the properties of proteins, including folding.
Evaluating these hypotheses and others requires direct access to the evolutionary history of a protein. Although comparing sequences and properties of extant homologs has enriched our understanding of protein folding (9–11), it does not directly access the evolutionary history that generated these homologs. In addition, such comparative studies are often limited by epistatic effects, which complicate the identification of sequence determinants and mechanisms (12, 13). More importantly, these extant homologs can reveal only the outcomes of an evolutionary process, and not the mechanism by which different properties arose from a common ancestral state (14).
In contrast, ancestral sequence reconstruction (ASR) allows direct interrogation of a protein’s history by using evolutionary relationships and sequences of modern homologs to statistically infer ancestral states within a family’s phylogenetic tree (12–14). Characterizing these ancestral proteins will reveal trends in folding properties and also help identify the evolutionary demands on folding. In addition, ASR provides a constellation of proteins that differ systematically in sequence and folding properties, which can shed light on sequence determinants of the folding landscape. This information then can be used to engineer proteins with desired folding properties. ASR has already provided important insights into evolutionary trends in stability, specificity, and other biophysical properties (12–20).
Previously, we used ASR to study the thermostability of the ribonuclease H (RNase H) family and demonstrated divergent trends in thermostability along mesophilic and thermophilic lineages (16). Here we examined the folding mechanism and associated rates of these reconstructed ancestral RNases H. The folding pathway of the two extant homologs, Escherichia coli RNase H (ecRNH) and Thermus thermophilus RNase H (ttRNH) are well characterized; the observed kinetics for both can be modeled using a three-state folding pathway with an on-pathway partially folded intermediate () (21–23). We now find that this overall three-state folding pathway is preserved over billions of years of evolution. Surprisingly, the last common ancestor of ecRNH and ttRNH folds and unfolds faster than the modern descendants, with decreasing trends in both the folding and unfolding rates along mesophilic and thermophilic lineages. Thus, although the thermodynamic stabilities diverge between the two lineages, kinetic stability increases along both. The presence of the folding intermediate permits this paradoxical uncoupling of thermodynamics and kinetics, allowing for independent modulation of unfolding rates and global stabilities.
Results
Seven Ancestral Sequences of RNase H.
We evaluated the folding and unfolding kinetics of seven ancestral proteins of the RNase H family along the lineages of mesophilic ecRNH and thermophilic ttRNH. Anc1 is the last common ancestor between ecRNH and ttRNH. AncA, B, C, and D are ancestors along the mesophilic lineage, and Anc2 and 3 are ancestors along the thermophilic lineage (Fig. 1). The thermostabilities of these ancestors have pronounced divergent trends, with Tm increasing along the thermophilic lineage and decreasing along the mesophilic lineage (16). Because the folding landscapes of both extant proteins have been extensively characterized in the cysteine-free background (21, 23), we engineered cysteine-free variants of the ancestral proteins (Dataset S1). (Each cysteine-free variant is denoted by an asterisk after its name.) The absence of cysteines does not affect RNase H activity (24), and circular dichroism (CD) spectra of the ancestors show that the mutations do not perturb the overall fold (Fig. 2A).
Fig. 1.
Phylogenetic tree of the reconstructed RNase H family. Branches are labeled with their branch lengths (average number of substitutions per site). Circles represent the reconstructed RNase H ancestors. Anc1 is the last common ancestor between ecRNH and ttRNH. Two ancestors—Anc2 and Anc3—exist along the thermophilic lineage leading to ttRNH. Four ancestors—AncA, AncB, AncC, and AncD—exist along the mesophilic lineage leading to ecRNH. Modified from ref. 16.
Fig. 2.
Ancestral proteins are well folded and display divergent trends in global stability. (A) CD spectra of ancestral and extant proteins. MRE, mean residue ellipticity. (B) Urea denaturation melts of ancestral proteins fitted to a two-state linear extrapolation model and plotted as fraction folded vs. [urea]. (C) ΔGunf obtained from a two-state fit of the urea denaturation melts plotted over evolutionary distance. Red indicates thermophilic lineage; blue, mesophilic lineage; black, Anc1*; gray triangles, alternate sequences of Anc1*. For AncB*, ΔGunf values calculated from the global fit from kinetic data are plotted, because AncB* does not exhibit two-state behavior at equilibrium (Fig. S1).
Global Stability Diverges in RNase H History.
For comparison with our kinetic studies, the stability of each ancestral protein was determined by equilibrium urea denaturation monitored by CD at 222 nm at 25 °C. All proteins showed a cooperative transition and were fitted by a two-state linear extrapolation model (Fig. 2B) (25). A two-state fit of AncB* revealed a lower m-value (i.e., the dependence of ΔG on [urea]) than was expected for RNase H, and further kinetic analysis (see below) revealed that this protein is three-state at equilibrium, with a notable population of the canonical RNase H folding intermediate (Fig. S1) (26). Therefore, the global stability of AncB* was determined from a three-state fit of the kinetic data (see below) (Fig. S1). In sum, these results show the same trends in stability (ΔGunf) obtained previously via thermal and guanidinium chloride denaturation; Anc1* has stability between that of ecRNH* and ttRNH*, and ΔGunf gradually increases along the thermophilic lineage to ttRNH* (Fig. 2C) (16). In the mesophilic lineage, there is an initial drop in ΔGunf, which is then maintained throughout the lineage to ecRNH*.
Fig. S1.
Global fit of ancestral RNase H kinetic data to a three-state model. The burst-phase amplitude (data, gray triangle; fit, green line), final amplitude (data, black square; fit, blue line), and observed rates (data, black circle; fit, red line) of each ancestral protein were globally fit to a three-state on-pathway model to extract the kinetic and thermodynamic parameters of the folding landscape. MRE (mean residue ellipticity) at 222 nm.
Kinetic Studies: The Three-State Folding Model Is Preserved.
To characterize the folding mechanism of the ancestral RNases H, we monitored the folding and unfolding kinetics as a function of [urea] by CD, using both stopped-flow and manual mixing techniques. At low [urea], all ancestors showed a large signal change (burst phase) within the dead time of the stopped flow (∼15 ms), followed by a slower observable phase that fit well by a single exponential (Fig. S2). This burst-phase amplitude is indicative of a folding intermediate, consistent with the three-state folding model of ecRNH* and ttRNH* (21, 23). All unfolding data were accounted for by a single observable exponential phase. The resulting chevron plots, ln(kobs) vs. [urea], show the classic rollover at low [urea] owing to the presence of a stable folding intermediate (Figs. 3A and 4) (27). The denaturant dependence of the burst-phase amplitude and the final amplitude yielded a cooperative transition for the folding intermediate and native state, respectively (Fig. 3B). We globally fit the burst-phase amplitude, final amplitude, and observed rates of each ancestor to a three-state on-pathway model to extract kinetic and thermodynamic parameters of the folding landscape (Fig. 3 A and B, Table 1, and Fig. S1) (27). Remarkably, all of the ancestors are fit well by this model, indicating that the overall folding trajectory of RNase H is robust over billions of years of evolution.
Fig. S2.
Folding kinetics of Anc1*. Sample kinetic trace of Anc1* folding in 3.3 M [urea]. A large burst phase forms within the dead time of the stop-flow instrument. Black dots indicate the observed CD signal; red line, fit of the observed kinetic phase to a single exponential; red arrow, expected CD signal for unfolded Anc1*.
Fig. 3.
Anc1* folds through a three-state pathway with a folding intermediate. (A) Chevron plot of Anc1*. The red line indicates a global fit to a three-state model. Previously published chevron plots of ecRNH* (blue) and ttRNH* (orange) are also shown (21, 23). (B) Urea dependence of the burst-phase amplitude (data, gray triangle; fit, green line) and final amplitude (data, black circles; fit, blue line) of Anc1*, globally fit to a three-state model. MRE (mean residue ellipticity) at 222 nm.
Fig. 4.
Chevron plot of ancestral RNases H. Chevron plot of ancestral proteins along the mesophilic (A) and thermophilic (B) lineages. The published chevrons for ecRNH* (A) and ttRNH* (B) are shown as well (21, 23).
Table 1.
Kinetic and thermodynamic parameters of ancestral RNases H
| Parameter | ttRNH*† | Anc3* | Anc2* | Anc1* | AncA* | AncB* | AncC* | AncD* | ecRNH*‡ |
| Equilibrium data | |||||||||
| ΔGunf, kcal mol−1 | 12.8 | 12.5 ± 0.3 | 10.5 ± 0.4 | 10.2 ± 0.2 | 9.4 ± 0.2 | 8.6 ± 0.1 | 9.3 ± 0.4 | 9.3 ± 0.3 | 9.7 ± 0.4 |
| munf, kcal mol−1 M−1 | 2.0 | 1.85 ± 0.04 | 1.95 ± 0.04 | 1.91 ± 0.03 | 2.22 ± 0.10 | 1.76 ± 0.01 | 2.21 ± 0.13 | 2.20 ± 0.18 | 2.10 ± 0.09 |
| Cm, M | 6.4 | 6.71 ± 0.01 | 5.38 ± 0.01 | 5.34 ± 0.06 | 4.17 ± 0.09 | 4.89 ± 0.02 | 4.12 ± 0.04 | 4.23 ± 0.04 | 4.62 ± 0.04 |
| Kinetic data | |||||||||
| kIN, s−1 | 0.6 ± 0.1 | 4.1 ± 1.1 | 14.1 ± 5.8 | 10.4 ± 2.3 | 9.7 ± 3.5 | 0.77 ± 0.11 | 2.8 ± 0.9 | 0.13 ± 0.05 | 0.74 ± 0.02 |
| mIN, kcal mol−1 M−1 | 0.36 ± 0.04 | 0.26 ± 0.06 | 0.31 ± 0.10 | 0.21 ± 0.06 | 0.50 ± 0.15 | 0.43 ± 0.03 | 0.47 ± 0.10 | 0.45 ± 0.12 | 0.45 ± 0.01 |
| kNI, s−1 | 4 ± 6 × 10−6 | 2.1 ± 1.1 × 10−4 | 1.1 ± 0.5 × 10−3 | 1.6 ± 0.5 × 10−3 | 2.4 ± 0.7 × 10−4 | 1.1 ± 0.4 × 10−4 | 1.7 ± 0.6 × 10−4 | 7.3 ± 2.5 × 10−6 | 1.7 ± 0.1 × 10−5 |
| mNI, kcal mol−1 M−1 | −0.4 ± 0.1 | −0.24 ± 0.08 | −0.23 ± 0.04 | −0.25 ± 0.03 | −0.35 ± 0.05 | −0.40 ± 0.03 | −0.39 ± 0.04 | −0.41 ± 0.03 | −0.42 ± 0.03 |
| ΔGUI, kcal mol−1 | 6.2 ± 0.9 | 5.9 ± 0.7 | 4.6 ± 0.8 | 5.2 ± 0.4 | 2.4 ± 0.3 | 3.8 ± 0.8 | 3.9 ± 0.9 | 3.6 ± 0.7 | 3.5 ± 0.1 |
| mUI, kcal mol−1 M−1 | 1.2 ± 0.2 | 1.37 ± 0.09 | 1.41 ± 0.12 | 1.40 ± 0.08 | 1.37 ± 0.20 | 0.96 ± 0.10 | 1.34 ± 0.17 | 1.39 ± 0.13 | 1.24 ± 0.08 |
| ΔGunf, kcal mol−1 | 13 ± 1 | 11.7 ± 0.9 | 10.2 ± 1.0 | 10.4 ± 0.6 | 8.7 ± 0.6 | 9.1 ± 0.9 | 9.7 ± 1.0 | 9.4 ± 0.9 | 9.9 ± 0.1 |
| munf, kcal mol−1 M−1 | 2.0 ± 0.2 | 1.87 ± 0.13 | 1.95 ± 0.16 | 1.86 ± 0.10 | 2.21 ± 0.25 | 1.79 ± 0.11 | 2.23 ± 0.20 | 2.25 ± 0.18 | 2.11 ± 0.09 |
Folding Is Robust to Reconstruction Uncertainty.
A reconstructed ancestor is an inferred maximum likelihood (ML) sequence based on a phylogenetic relationship of extant proteins and thus cannot be verified to be the true ancestral sequence at the phylogenetic node. As such, it is important to address whether uncertainty in ASR affects our measured biophysical parameters. Numerous ASR studies have addressed the effect of uncertainty by a variety of approaches, and in general, the measured parameters have proven robust (15–19, 28–31). For the RNase H family, we generated alternative sequences of the last common ancestor, Anc1*, which had the greatest uncertainty of all of the reconstructed ancestors (16). Sampling of the alternate sequences from the posterior probability contained between 3 and 11 amino acid changes from the ML sequence. We previously found that thermostability was robust, because the alternative Anc1 proteins had similar Tm values as the ML Anc1 protein (16). To assess how uncertainty affects folding, we obtained urea melts and chevrons of these alternative Anc1* proteins (Fig. S3). The kinetics and thermodynamic properties were very similar to those obtained for the ML Anc1* (Fig. 2C, Fig. S3, and Table S1). Thus, the properties of the RNase H folding landscape appear to be robust to the reconstruction.
Fig. S3.
Alternate sequences of Anc1* have similar thermodynamic and kinetic properties as the ML Anc1*. (A) CD spectra of alternate sequences of Anc1*. MRE, mean residue ellipticity. (B) Urea denaturation melts of five alternate sequences of Anc1* fitted to a two-state equilibrium model and plotted as fraction folded vs. [urea]. (C) Chevron plot (ln kobs vs. [urea]) of five alternate sequences of Anc1*.
Table S1.
Kinetic and thermodynamic parameters of alternate sequences of Anc1
| Parameter | Alt0* | Alt3* | Alt5* | Alt6* | Alt9* |
| Equilibrium data | |||||
| ΔGunf, kcal mol−1 | 9.8 ± 0.1 | 10.4 ± 0.3 | 10.3 ± 0.3 | 9.8 ± 0.4 | 10.0 ± 0.5 |
| munf, kcal mol−1 M−1 | 1.76 ± 0.03 | 1.72 ± 0.03 | 1.61 ± 0.05 | 1.73 ± 0.10 | 1.83 ± 0.10 |
| Cm, M | 5.59 ± 0.06 | 6.07 ± 0.05 | 6.42 ± 0.04 | 5.63 ± 0.06 | 5.44 ± 0.08 |
| Kinetic data | |||||
| kIN, s−1 | 6.6 ± 2.6 | 7.9 ± 1.2 | 5.9 ± 1.0 | 5.5 ± 0.9 | 4.2 ± 0.8 |
| mIN, kcal mol−1 M−1) | 0.14 ± 0.10 | 0.14 ± 0.10 | 0.10 ± 0.04 | 0.11 ± 0.15 | 0.09 ± 0.04 |
| kNI, s−1 | 4.3 ± 2.9 × 10−4 | 5.8 ± 5.0 × 10−4 | 6.4 ± 1.5 × 10−4 | 5.5 ± 4.2 × 10−4 | 4.8 ± 4.1 × 10−4 |
| mNI, kcal mol−1 M−1 | −0.29 ± 0.06 | −0.24 ± 0.07 | −0.27 ± 0.06 | −0.22 ± 0.06 | −0.29 ± 0.07 |
| ΔGUI, kcal mol−1 | 4.8 ± 0.6 | 4.7 ± 0.4 | 5.4 ± 0.3 | 4.8 ± 0.5 | 5.1 ± 0.4 |
| mUI, kcal mol−1 M−1 | 1.44 ± 0.13 | 1.33 ± 0.08 | 1.45 ± 0.10 | 1.50 ± 0.09 | 1.52 ± 0.14 |
Errors are in SD.
The Rate-Limiting Barrier Increases Over Evolutionary Time.
There are clear evolutionary trends in folding and unfolding kinetics. The rate constant to cross the rate-limiting barrier decreases over evolutionary time in both the folding and unfolding directions. Notably, this occurs along both the mesophilic and thermophilic lineages (Fig. 5 B and C). Anc1*, the last common ancestor, folds and unfolds significantly faster than the extant proteins. In the mesophilic lineage, there is a 14-fold decrease in the folding rate constant (kIN) and a 96-fold decrease in the unfolding rate constant (kNI) over the rate-limiting barrier. Similarly, in the thermophilic lineage, there is a 17-fold decrease in the folding rate constant (kIN) and a 400-fold decrease in the unfolding rate constant (kNI). The decrease in unfolding rate is particularly interesting, because it means that the extant proteins, ecRNH* and ttRNH*, are significantly more kinetically stable than their common ancestor. This universal decrease in the folding and unfolding rate constants along the two lineages contrasts with the divergent trend observed for global stability.
Fig. 5.
Evolutionary trends in kinetic and thermodynamic parameters of the RNase H folding landscape. (A) Three-state energy landscape of RNase H. U, unfolded state; I, folding intermediate; N, native state; kIN, folding rate constant over the rate-limiting barrier; kNI, unfolding rate constant over the rate-limiting barrier. Red circles indicate thermophilic lineage; blue circles, mesophilic lineage; black circle, Anc1*; gray triangles, alternate sequences of Anc1*. (B) Folding rate constant over evolutionary time. (C) Unfolding rate constant over evolutionary time. (D) Correlation of changes in the stability of the intermediate to changes in global stability. (E) Correlation of changes in the rate-limiting barrier to changes in global stability.
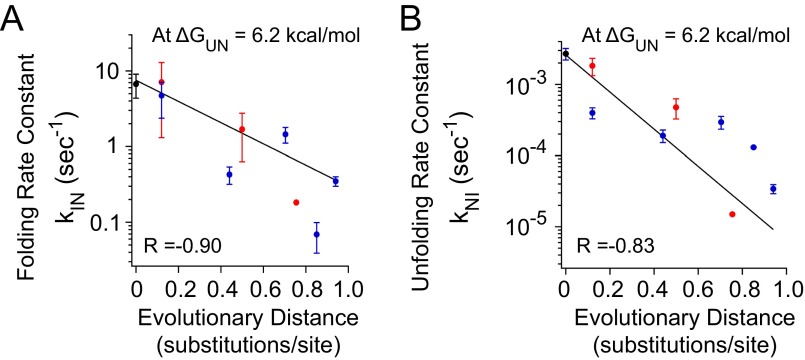
The observed trends in folding and unfolding kinetics are derived from experiments carried out at 25 °C; however, the environmental growth temperatures likely vary across the ancestors. Unfortunately, extrapolation of folding and unfolding rates to different temperatures is nontrivial, because the temperature dependence of the folding and unfolding rates is unknown. Nonetheless, we have reason to believe that RNases H have similar global stabilities at their functioning temperature (16, 32). At 37 °C, ecRNH has a stability of ΔGunf = 6.8 kcal/mol, and at 68 °C, ttRNH has a stability of ΔGunf = 5.6 kcal/mol. Therefore, we chose the average of the two, ΔGunf = 6.2 kcal/mol, and calculated folding and unfolding rate constants over the rate-limiting barrier for each ancestor under conditions in which their stabilities are the same (Fig. S4). We found that the trends in folding and unfolding are present regardless of whether the analysis is done at the same temperature (25 °C) or at the same stability (6.2 kcal/mol).
Fig. S4.
Trends in folding and unfolding rates at an iso-stability point, ΔGunf = 6.2 kcal/mol. (A) Folding rate constant, kIN, when ΔGunf = 6.2 kcal/mol over evolutionary time. (B) Unfolding rate constant, kNI, when ΔGunf = 6.2 kcal/mol over evolutionary time.
Discussion
In this study, we used ASR to investigate how a protein’s folding pathway evolved. Because protein folding is crucial to proper cellular function (33), we reasoned that there are selective pressures that shape how proteins fold, and that such pressures may result in evolutionary trends in folding mechanisms or rates. Therefore, we characterized the folding properties of ancestral proteins of the RNase H family. We found that the overall RNase H folding trajectory is remarkably robust to sequence changes and has been maintained throughout billions of years of evolution. Strikingly, both lineages have evolved to fold and unfold more slowly, resulting in more kinetically stable extant proteins.
Evolution of Kinetic Stability in the RNase H Family.
Modern RNases H are more kinetically stable than their ancestors, as evidenced by a >100-fold decrease in the unfolding rate relative to their common ancestor. For both RNase H lineages, the evolution of kinetic stability was achieved by decreasing the unfolding rate while either increasing (thermophilic lineage) or decreasing (mesophilic lineage) thermodynamic stability. This pattern may be the result of selection to increase kinetic stability over time. Kinetic stability would prevent frequent access to the unfolded state, which is particularly critical in the presence of competing kinetic processes, such as aggregation and proteolysis, and has been proposed to be a biophysical parameter under selection (34, 35). The biological function and lifetime of proteins in vivo has been linked to their degree of kinetic stability (36, 37) and particularly high unfolding barriers have been observed for proteins that reside in harsh conditions (38–40). The observed trend also might arise from selection for other traits linked to kinetic stability.
In addition, we observed a smaller decrease in the folding rate in both the thermophilic and mesophilic lineages. Whereas the extant homologs have an ∼10-fold slower folding rate than their common ancestor, they still fold efficiently and reversibly, reaching their native folded state within seconds. It is possible that changes in the folding rate may well be a result of neutral drift, and that there may be no selective pressure to optimize folding rates beyond a biologically necessary range (8, 41). Modern proteins can be computationally redesigned to fold faster, demonstrating that their folding rates have not been maximized over evolution (42). Nevertheless, the observed directional trend in the folding rate suggests, rather than a random scatter, some selective pressure that either directly or indirectly affects folding rate. Alternatively, it could be that random mutations generally decrease the folding rate, similar to how mutations in general destabilize proteins (43). Studies on the evolution of folding rates have been limited (4), and additional work across a large number of protein families will be needed to establish generalizable trends.
A Folding Intermediate Uncouples Kinetics and Thermodynamics.
All of the RNase H ancestors fold via a three-state pathway with an intermediate that forms within the dead time of our stop-flow instrument. Given that such intermediates are marginally stable and readily perturbed by mutations (44, 45), this conservation is significant. In fact, a single amino acid change (I53D) is known to destabilize the intermediate of ecRNH* such that it is no longer populated and the protein folds, albeit very slowly, through a two-state mechanism (45). The presence and the energetics of such folding intermediates can have a large impact on folding rates (10, 45, 46), and, moreover, these partially folded states can be gateways for misfolding and aggregation (47). Whether folding intermediates are productive or counterproductive for folding is a matter of debate, and it has been suggested that proteins evolve to fold cooperatively in a two-state manner (48).
Why, then, does the RNase H folding intermediate persist over evolution? Our results show that the intermediate allows for sequence changes to tune the height of the rate-limiting barrier independently from global thermodynamic stability. For a simple protein folding through a two-state pathway, changes in thermodynamic stability are directly reflected in the kinetics: ΔGunf = −RTln (ku/kf). For the three-state pathway observed for RNase H, the outcome is more complex, because the stability of the partially folded intermediate can change independently of the rate-limiting barrier. Thus, global stability will be the sum of the stability of the intermediate, ΔGUI, and the log ratio of the folding and unfolding rates over the rate-limiting barrier, ΔGunf = ΔGUI + −RTln (kNI/kIN) (Fig. 5A). Given the observed decrease in both rate constants, kIN and kNI, along the two RNase H lineages, the thermodynamic stability of the folding intermediate is primarily responsible for the divergent trend in global stability. As such, changes in global stability strongly correlate with changes in stability of the intermediate (Fig. 5D), whereas there is only a weak correlation with stability changes contributed by the rate-limiting step (Fig. 5E). Thus, the physical chemistry of the three-state energy landscape allows independent modulation of the height of the unfolding barrier and global stability to yield parallel trends in kinetics across lineages where thermodynamic stability diverges. The presence of a folding intermediate, despite decreasing cooperativity and introducing roughness in the landscape, may be evolutionarily beneficial, allowing for an independent response of the landscape to different selective pressures on kinetics and thermodynamics.
ASR as a Tool to Decipher the Role of Between Sequence and Topology on Folding.
All of the RNase H proteins in our study retain the canonical RNase H fold (16) and fold through a three-state pathway. This is consistent with suggestions that topology, rather than sequence, is likely the determinant of the folding trajectory for this protein (49) and possibly proteins in general (50). In addition, the robust folding of the RNase H ancestors is consistent with evidence indicating that ASR generates proteins of evolutionary significance, unlike consensus proteins, which, despite being likened to ancestral proteins, have been found to fold poorly in some instances (18, 51).
Previous work on extant RNases H has established that the “core” region of the protein (helices A–D and strands 4 and 5) folds early and is structured in the intermediate, whereas the periphery (helix E and strands 1–3) remains unfolded until the rate-limiting step (23, 52–54) (Fig. S5). Although our kinetic studies reported in this paper do not reveal structural information about the folding intermediate, this region is highly conserved in sequence, so we anticipate that the same core comprises the folding intermediate for all of the ancestral proteins (Fig. S5). Even so, it is likely that there are at least small changes in the structure of the folding intermediate, as indicated by the small variation in the kinetic m-values across the ancestors (Table 1). Techniques such as pulsed-labeling hydrogen exchange will be important to identify any differences in the acquisition of structure during folding (54).
Fig. S5.
Sequence identity of RNase H proteins for core region or periphery region. (A) Crystal structure of E. coli RNase H (Protein Data Bank ID code 2RN2). In blue is the “core” region of the protein, which includes HelixA-D and strands 4 and 5 and corresponds to residues 43–121. In gray is the periphery region of the protein, which includes HelixE and strands 1, 2, and 3 and corresponds to residues 1–42 and 122–155. (B) Pairwise percent sequence identity of RNase H homologs and ancestors for the core (Lower Left) and the periphery (Upper Right) regions. The core and periphery regions for each RNase H sequence were determined from a multiple sequence alignment using the E. coli RNase H residues as cutoffs.
In addition, the observed “decoupling” of kinetics and thermodynamics implies that there must be sequence changes that affect the transition state without affecting other features of the landscape, and vice versa. Changes in the unfolding m-value indicate that not just the height, but also the position of the transition state may shift over evolution. Identifying sequences that differentially alter particular features of the folding landscape will help us understand how folding is encoded in the sequence and provide tools to engineer specific features of the folding landscape.
In summary, by coupling ASR with biophysical characterization, we have determined the evolutionary history of a protein’s folding trajectory. The folding intermediate plays a critical role in the RNase H family by decoupling changes in thermodynamic and kinetic stabilities, and presents a possible adaptive advantage for partially folded states. There are robust trends in the unfolding rates and folding rates, which may reflect constraints on the energy landscape, and it will be intriguing in the future to investigate how they contribute to organismal fitness. Our findings lay a foundation for the identification of evolutionary pressures that drive the emergence of specific biophysical properties.
Materials and Methods
Expression and Purification of RNase H Ancestors.
The cysteine-free variants of RNase H ancestors were generated by site-directed mutagenesis and were sequence-verified by Sanger sequencing. The proteins were expressed and purified as described previously (23). The purity and mass of the purified proteins were confirmed by SDS/PAGE and mass spectrometry.
CD Equilibrium Experiments.
CD spectra and urea melts were measured with an Aviv 410 CD spectrometer. Signals from 200 nm to 300 nm were obtained for buffer only and for buffer with 0.4 mg/mL protein using a 1-mm path length cuvette at 25 °C. The buffer conditions were 20 mM sodium acetate and 50 mM potassium chloride, pH 5.5 (RNase H buffer). For urea melts, samples containing 40 μg/mL protein and varying [urea] in RNase H buffer were equilibrated, and the signal at 222 nm was measured in a 1-cm path length cuvette at 25 °C with stirring. Melts were obtained in triplicate. The data were fitted to a two-state model with a linear free-energy extrapolation (25).
CD Kinetic Experiments.
Refolding was initiated either by stopped-flow or manual mixing. For stopped-flow mixing, unfolded protein (8 mg/mL) in high urea was diluted 1:11 into final refolding conditions (RNase H buffer, varying [urea]), and the CD signal at 222 nm was monitored with an Aviv 202 stopped-flow CD spectrometer using a 1-mm path length cuvette with a dead time of ∼15 ms at 25 °C. Refolding reactions were repeated eight times and averaged. For manual mixing, 1.5 mg/mL unfolded protein in high [urea] was diluted 1:30 into final refolding conditions, and the CD signal at 222 nm was monitored in an Aviv 410 CD spectrometer using a 1-cm path length cuvette. Dead times ranged from about 8–12 seconds. Unfolding kinetics were similarly measured by manual mixing, but starting from folded protein in low [urea] with rapid dilution to final unfolding conditions. All kinetic data were adequately fit by single exponential. Rates, burst-phase amplitudes, and final amplitudes were globally fit to a three-state model with a rapidly forming on-pathway intermediate (21, 23, 27).
Supplementary Material
Acknowledgments
We thank Charlotte Nixon and the members of the S.M. laboratory for helpful comments and discussion. This work was funded by National Institutes of Health Grant GM050945 (to S.M.) and a National Science Foundation Graduate Research Fellowship (to S.A.L.). M.J.H. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611781113/-/DCSupplemental.
References
- 1.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338(6110):1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 2.Mirny LA, Shakhnovich EI. Universally conserved positions in protein folds: Reading evolutionary signals about stability, folding kinetics and function. J Mol Biol. 1999;291(1):177–196. doi: 10.1006/jmbi.1999.2911. [DOI] [PubMed] [Google Scholar]
- 3.Smock RG, Yadid I, Dym O, Clarke J, Tawfik DS. De novo evolutionary emergence of a symmetrical protein is shaped by folding constraints. Cell. 2016;164(3):476–486. doi: 10.1016/j.cell.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debès C, Wang M, Caetano-Anollés G, Gräter F. Evolutionary optimization of protein folding. PLOS Comput Biol. 2013;9(1):e1002861. doi: 10.1371/journal.pcbi.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallik S, Kundu S. Co-evolutionary constraints of globular proteins correlate with their folding rates. FEBS Lett. 2015;589(17):2179–2185. doi: 10.1016/j.febslet.2015.06.032. [DOI] [PubMed] [Google Scholar]
- 6.Hobbs JK, Prentice EJ, Groussin M, Arcus VL. Reconstructed ancestral enzymes impose a fitness cost upon modern bacteria despite exhibiting favourable biochemical properties. J Mol Evol. 2015;81(3-4):110–120. doi: 10.1007/s00239-015-9697-5. [DOI] [PubMed] [Google Scholar]
- 7.Louis JM, Roche J. Evolution under drug pressure remodels the folding free-energy landscape of mature HIV-1 protease. J Mol Biol. 2016;428(13):2780–2792. doi: 10.1016/j.jmb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durão P, et al. Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat Chem Biol. 2015;11(2):148–155. doi: 10.1038/nchembio.1715. [DOI] [PubMed] [Google Scholar]
- 9.Nickson AA, Wensley BG, Clarke J. Take home lessons from studies of related proteins. Curr Opin Struct Biol. 2013;23(1):66–74. doi: 10.1016/j.sbi.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson N, Capaldi AP, James R, Kleanthous C, Radford SE. Rapid folding with and without populated intermediates in the homologous four-helix proteins Im7 and Im9. J Mol Biol. 1999;286(5):1597–1608. doi: 10.1006/jmbi.1998.2548. [DOI] [PubMed] [Google Scholar]
- 11.Kwa LG, et al. The folding of a family of three-helix bundle proteins: Spectrin R15 has a robust folding nucleus, unlike its homologous neighbours. J Mol Biol. 2014;426(7):1600–1610. doi: 10.1016/j.jmb.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr Opin Struct Biol. 2010;20(3):360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms MJ, Thornton JW. Evolutionary biochemistry: Revealing the historical and physical causes of protein properties. Nat Rev Genet. 2013;14(8):559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler LC, Lim SA, Marqusee S, Harms MJ. The thermostability and specificity of ancient proteins. Curr Opin Struct Biol. 2016;38:37–43. doi: 10.1016/j.sbi.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451(7179):704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- 16.Hart KM, et al. Thermodynamic system drift in protein evolution. PLoS Biol. 2014;12(11):e1001994. doi: 10.1371/journal.pbio.1001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akanuma S, et al. Experimental evidence for the thermophilicity of ancestral life. Proc Natl Acad Sci USA. 2013;110(27):11067–11072. doi: 10.1073/pnas.1308215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs JK, et al. On the origin and evolution of thermophily: Reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol Biol Evol. 2012;29(2):825–835. doi: 10.1093/molbev/msr253. [DOI] [PubMed] [Google Scholar]
- 19.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian β-lactamases. J Am Chem Soc. 2013;135(8):2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Jimenez R, et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 2011;18(5):592–596. doi: 10.1038/nsmb.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollien J, Marqusee S. Comparison of the folding processes of T. thermophilus and E. coli ribonucleases H. J Mol Biol. 2002;316(2):327–340. doi: 10.1006/jmbi.2001.5346. [DOI] [PubMed] [Google Scholar]
- 22.Raschke TM, Marqusee S. The kinetic folding intermediate of ribonuclease H resembles the acid molten globule and partially unfolded molecules detected under native conditions. Nat Struct Biol. 1997;4(4):298–304. doi: 10.1038/nsb0497-298. [DOI] [PubMed] [Google Scholar]
- 23.Raschke TM, Kho J, Marqusee S. Confirmation of the hierarchical folding of RNase H: A protein engineering study. Nat Struct Biol. 1999;6(9):825–831. doi: 10.1038/12277. [DOI] [PubMed] [Google Scholar]
- 24.Goedken ER, Keck JL, Berger JM, Marqusee S. Divalent metal cofactor binding in the kinetic folding trajectory of Escherichia coli ribonuclease HI. Protein Sci. 2000;9(10):1914–1921. doi: 10.1110/ps.9.10.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Street TO, Courtemanche N, Barrick D. Protein folding and stability using denaturants. Methods Cell Biol. 2008;84(07):295–325. doi: 10.1016/S0091-679X(07)84011-8. [DOI] [PubMed] [Google Scholar]
- 26.Spudich G, Marqusee S. A change in the apparent m value reveals a populated intermediate under equilibrium conditions in Escherichia coli ribonuclease HI. Biochemistry. 2000;39(38):11677–11683. doi: 10.1021/bi000466u. [DOI] [PubMed] [Google Scholar]
- 27.Baldwin RL. On-pathway versus off-pathway folding intermediates. Fold Des. 1996;1(1):R1–R8. doi: 10.1016/S1359-0278(96)00003-X. [DOI] [PubMed] [Google Scholar]
- 28.Groussin M, et al. Toward more accurate ancestral protein genotype-phenotype reconstructions with the use of species tree-aware gene trees. Mol Biol Evol. 2015;32(1):13–22. doi: 10.1093/molbev/msu305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akanuma S, Yokobori S, Nakajima Y, Bessho M, Yamagishi A. Robustness of predictions of extremely thermally stable proteins in ancient organisms. Evolution. 2015;69(11):2954–2962. doi: 10.1111/evo.12779. [DOI] [PubMed] [Google Scholar]
- 30.Hanson-Smith V, Kolaczkowski B, Thornton JW. Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol Biol Evol. 2010;27(9):1988–1999. doi: 10.1093/molbev/msq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bar-Rogovsky H, et al. Assessing the prediction fidelity of ancestral reconstruction by a library approach. Protein Eng Des Sel. 2015;28(11):507–518. doi: 10.1093/protein/gzv038. [DOI] [PubMed] [Google Scholar]
- 32.Hollien J, Marqusee S. A thermodynamic comparison of mesophilic and thermophilic ribonucleases H. Biochemistry. 1999;38(12):3831–3836. doi: 10.1021/bi982684h. [DOI] [PubMed] [Google Scholar]
- 33.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Ruiz JM. Protein kinetic stability. Biophys Chem. 2010;148(1-3):1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Godoy-Ruiz R, et al. Natural selection for kinetic stability is a likely origin of correlations between mutational effects on protein energetics and frequencies of amino acid occurrences in sequence alignments. J Mol Biol. 2006;362(5):966–978. doi: 10.1016/j.jmb.2006.07.065. [DOI] [PubMed] [Google Scholar]
- 36.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99(3):1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaswal SS, Sohl JL, Davis JH, Agard DA. Energetic landscape of alpha-lytic protease optimizes longevity through kinetic stability. Nature. 2002;415(6869):343–346. doi: 10.1038/415343a. [DOI] [PubMed] [Google Scholar]
- 38.Wittung-Stafshede P. Slow unfolding explains high stability of thermostable ferredoxins: Common mechanism governing thermostability? Biochim Biophys Acta. 2004;1700(1):1–4. doi: 10.1016/j.bbapap.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Kelch BA, Agard DA. Mesophile versus thermophile: Insights into the structural mechanisms of kinetic stability. J Mol Biol. 2007;370(4):784–795. doi: 10.1016/j.jmb.2007.04.078. [DOI] [PubMed] [Google Scholar]
- 40.Kelch BA, et al. Structural and mechanistic exploration of acid resistance: Kinetic stability facilitates evolution of extremophilic behavior. J Mol Biol. 2007;368(3):870–883. doi: 10.1016/j.jmb.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 41.Zeldovich KB, Shakhnovich EI. Understanding protein evolution: From protein physics to Darwinian selection. Annu Rev Phys Chem. 2008;59:105–127. doi: 10.1146/annurev.physchem.58.032806.104449. [DOI] [PubMed] [Google Scholar]
- 42.Scalley-Kim M, Baker D. Characterization of the folding energy landscapes of computer-generated proteins suggests high folding free energy barriers and cooperativity may be consequences of natural selection. J Mol Biol. 2004;338(3):573–583. doi: 10.1016/j.jmb.2004.02.055. [DOI] [PubMed] [Google Scholar]
- 43.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369(5):1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 44.Connell KB, Horner GA, Marqusee S. A single mutation at residue 25 populates the folding intermediate of E. coli RNase H and reveals a highly dynamic partially folded ensemble. J Mol Biol. 2009;391(2):461–470. doi: 10.1016/j.jmb.2009.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spudich GM, Miller EJ, Marqusee S. Destabilization of the Escherichia coli RNase H kinetic intermediate: Switching between a two-state and three-state folding mechanism. J Mol Biol. 2004;335(2):609–618. doi: 10.1016/j.jmb.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 46.Khorasanizadeh S, Peters ID, Roder H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nat Struct Biol. 1996;3(2):193–205. doi: 10.1038/nsb0296-193. [DOI] [PubMed] [Google Scholar]
- 47.Jahn TR, Radford SE. Folding versus aggregation: Polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008;469(1):100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fersht AR. Optimization of rates of protein folding: The nucleation-condensation mechanism and its implications. Proc Natl Acad Sci USA. 1995;92(24):10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connell KB, Miller EJ, Marqusee S. The folding trajectory of RNase H is dominated by its topology and not local stability: A protein engineering study of variants that fold via two-state and three-state mechanisms. J Mol Biol. 2009;391(2):450–460. doi: 10.1016/j.jmb.2009.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker D. A surprising simplicity to protein folding. Nature. 2000;405(6782):39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 51.Risso VA, Gavira JA, Gaucher EA, Sanchez-Ruiz JM. Phenotypic comparisons of consensus variants versus laboratory resurrections of Precambrian proteins. Proteins. 2014;82(6):887–896. doi: 10.1002/prot.24575. [DOI] [PubMed] [Google Scholar]
- 52.Rosen LE, Connell KB, Marqusee S. Evidence for close side-chain packing in an early protein folding intermediate previously assumed to be a molten globule. Proc Natl Acad Sci USA. 2014;111(41):14746–14751. doi: 10.1073/pnas.1410630111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosen LE, Marqusee S. Autonomously folding protein fragments reveal differences in the energy landscapes of homologous RNases H. PLoS One. 2015;10(3):e0119640. doi: 10.1371/journal.pone.0119640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu W, et al. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2013;110(19):7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.