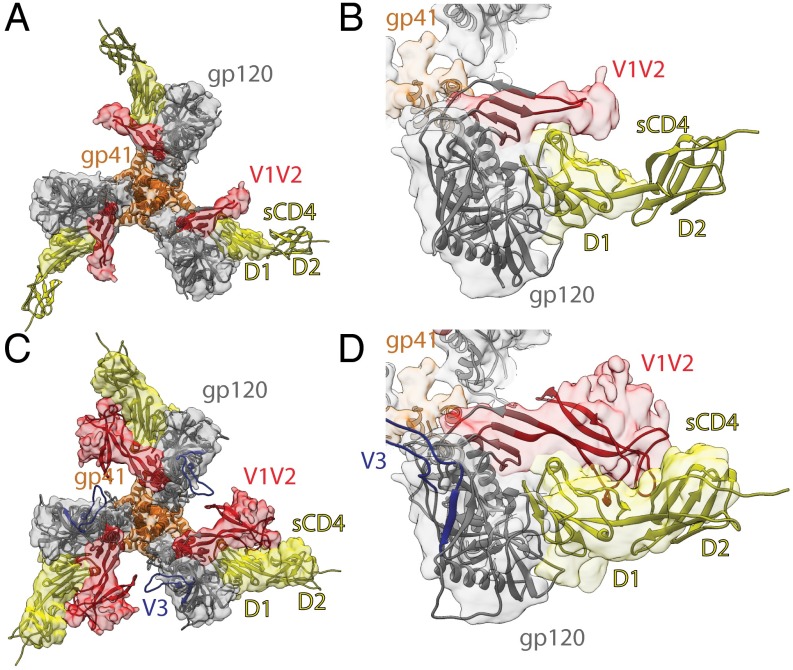
Significance
The HIV-1 envelope (Env) glycoprotein exists in multiple conformations on virion surfaces. Although the closed Env state is well characterized, less is known about open Env conformations stabilized by host receptor (CD4) binding. We solved an 8.9-Å structure of a partially open CD4-bound Env trimer by single particle cryo-EM. In the CD4-bound Env, the gp120 V1V2 loops were displaced by ∼40 Å from their positions at the trimer apex. The displaced V1V2 loops were at the sides of the open trimer in positions adjacent to, and interacting with, the three bound CD4s. These results are relevant to understanding CD4-induced conformational changes leading to coreceptor binding and fusion, and HIV-1 Env conformational dynamics, and describe a target structure relevant to drug design and vaccine efforts.
Keywords: cryo-EM, HIV-1 Env trimer, CD4, HIV-1 coreceptor, conformational change
Abstract
The HIV-1 envelope (Env) glycoprotein, a trimer of gp120–gp41 heterodimers, relies on conformational flexibility to function in fusing the viral and host membranes. Fusion is achieved after gp120 binds to CD4, the HIV-1 receptor, and a coreceptor, capturing an open conformational state in which the fusion machinery on gp41 gains access to the target cell membrane. In the well-characterized closed Env conformation, the gp120 V1V2 loops interact at the apex of the Env trimer. Less is known about the structure of the open CD4-bound state, in which the V1V2 loops must rearrange and separate to allow access to the coreceptor binding site. We identified two anti–HIV-1 antibodies, the coreceptor mimicking antibody 17b and the gp120–gp41 interface-spanning antibody 8ANC195, that can be added as Fabs to a soluble native-like Env trimer to stabilize it in a CD4-bound conformation. Here, we present an 8.9-Å cryo-electron microscopy structure of a BG505 Env–sCD4–17b–8ANC195 complex, which reveals large structural rearrangements in gp120, but small changes in gp41, compared with closed Env structures. The gp120 protomers are rotated and separated in the CD4-bound structure, and the three V1V2 loops are displaced by ∼40 Å from their positions at the trimer apex in closed Env to the sides of the trimer in positions adjacent to, and interacting with, the three bound CD4s. These results are relevant to understanding CD4-induced conformational changes leading to coreceptor binding and fusion, and HIV-1 Env conformational dynamics, and describe a target structure relevant to drug design and vaccine efforts.
The HIV-1 envelope (Env) glycoprotein, a trimer of gp120–gp41 heterodimers, mediates recognition of host receptors and fusion of the viral and target cell membranes (1). Structural flexibility of HIV-1 Env is required for its function in membrane fusion; thus, Env exists in multiple conformational states on the surface of virions (2). Fusion involves several steps: The gp120 portion of Env trimer first binds to the host receptor CD4 to capture a conformational state of Env that exposes the binding site for an HIV-1 coreceptor (CCR5 or CXCR4), which, in turn, leads to gp41-mediated fusion of the viral and host cell membranes. CD4-induced conformational changes within the Env trimer are incompletely understood. The binding of soluble CD4 (sCD4) produces little to no changes in the structures of gp120 cores (gp120 monomers with truncations in the N and C termini and variable V1V2 and V3 loops) (3), but results in rotation of the gp120 protomers within virion-bound Env trimers to create an open conformation distinct from the closed conformation of unliganded virion-bound trimers (4). Single-particle electron microscopy (EM) structures of recombinant native-like soluble Env gp140 trimers (SOSIPs) confirmed that they can adopt the same closed and open architectures as virion-bound Env trimers (5–7), thus the SOSIP substitutions (introduction of a disulfide bond linking gp120 to gp41 and an Ile→Pro mutation in gp41; ref. 6) do not appear to prevent transition to the open state. Despite the plethora of recent crystal and EM structures at atomic and near-atomic resolutions of closed Env trimers, most in complex with broadly neutralizing antibodies (bNAbs) (8–19), only low-resolution structures derived from cryo-electron tomography of HIV-1 virions have been available for sCD4-bound open Env trimers (4–7).
The closed conformation of HIV-1 Env is stabilized by interactions at the trimer apex mediated by the gp120 V1V2 loop (8–19). In the closed state, the V1V2 region shields the binding site for the coreceptor on the V3 loop (11, 16), but V1V2 interactions with V3 cannot be maintained when the gp120 protomers rotate and separate to create the CD4-bound open conformation. The details of V1V2 rearrangements in the open structure of HIV-1 Env trimer have not been addressed: The V1V2 loops were not localized in the Env trimer used in a cryo-EM single particle structure of an open KNH1144 SOSIP bound to the coreceptor-mimicking antibody 17b (7) or in lower-resolution cryoelectron tomography structures of CD4-bound open Env trimers on virions (4, 7). However, computational modeling suggested displacement of V1V2 toward CD4 in CD4-bound Env structures (20, 21), consistent with earlier studies demonstrating involvement of V1V2 in the induction of the epitopes of coreceptor mimic/CD4-induced antibodies such as 17b (22).
A structural description of conformational changes resulting from CD4 binding requires identification of a stable and conformationally homogeneous CD4–Env trimer complex. We previously reported a 16.8-Å negative stain single-particle EM reconstruction of the BG505 SOSIP.664 Env trimer bound to sCD4, 17b, and 8ANC195 (18). The two Fab ligands in this complex bind to distinct epitopes on Env: 8ANC195 binds to a region of the gp120–gp41 interface flanked by N-linked glycans attached to Asn234gp120 and Asn276gp120 (18, 23); and 17b, an HIV-1 coreceptor mimic (24), binds to a CD4-induced (CD4i) epitope (25) that comprises part of the gp120 bridging sheet and overlaps with the coreceptor binding site on gp120 (24). Accordingly, this antibody does not bind or neutralize most HIV-1 strains, the exceptions being sensitive Tier 1 primary isolates in which the coreceptor binding site is exposed in the absence of CD4 (26, 27). Because the coreceptor and 17b binding sites are inaccessible in the closed trimer conformation (11, 16), 17b does not bind to the BG505 SOSIP trimer unless sCD4 is present (6). Because three 17b Fabs cannot be accommodated on a closed Env trimer because of steric clashes (18), a CD4-bound trimer complex might be stabilized by adding 17b and 8ANC195 (to prevent Env closure and rigidify the gp120–gp41 interface, respectively).
Here, we report an 8.9-Å structure of the BG505–sCD4–17b–8ANC195 complex (hereafter referred to as Env–sCD4–17b–8ANC) derived by single-particle cryo-EM. The higher resolution structure reveals an Env structure with a large CD4-induced rotation of the gp120 protomers and an ∼40-Å movement of the V1V2 loop from the apex of the trimer to a position adjacent to sCD4, but relatively minor changes within gp41 from closed trimer structures. Biochemical studies using a BG505 with a truncated V1V2 loop support the model in which sCD4 binding induces V1V2 displacement to expose the coreceptor binding site. These results are relevant to understanding the CD4-induced conformational changes leading to coreceptor binding and fusion that allows HIV-1 entry into CD4+ target cells and HIV-1 Env conformational dynamics, and present a target structure relevant to drug design and vaccine efforts.
Results
Cryo-EM Structure Determination.
BG505 SOSIP.664, a cleaved, soluble native-like HIV-1 Env trimer (6), sCD4 D1D2 (domains 1 and 2 of the CD4 ectodomain), and Fabs from the anti–HIV-1 antibodies 17b and the G52K5 variant of 8ANC195 (23) (hereafter referred to as 8ANC195) were expressed and purified as described (18). The Env–sCD4–17b–8ANC complex was prepared for cryo-EM by first isolating an Env–sCD4–17b ternary complex by size exclusion chromatography (SEC), adding 8ANC195 Fab, using SEC again to isolate the quaternary Env–sCD4–17b–8ANC complex, and then plunge freezing the complex onto EM grids (SI Appendix, Fig. S1A).
Two independent single-particle reconstructions were obtained from 5,175 of 13,268 total particles and 9,606 of 19,355 total particles, respectively, at resolutions of ∼8.9 Å and ∼9.8 Å (calculated by using the 0.143 gold-standard Fourier shell coefficient cutoff criterion) (28) (Figs. 1 and 2 and SI Appendix, Figs. S2 and S3). These relatively high resolutions for a sCD4-bound Env structure were confirmed by feature-based criteria including clear definition of gp41 HR1 α-helices (Fig. 1C) and densities corresponding to BG505 N-linked glycans (Fig. 1D). Regions of the structure that were disordered and/or calculated to be at a lower resolution (29) were areas most distant from the trimer axis of symmetry such as the constant domains (CH and CL) of the Fabs, the D2 domain of sCD4, and density identified as the displaced gp120 V1V2 loop (SI Appendix, Fig. S2C).
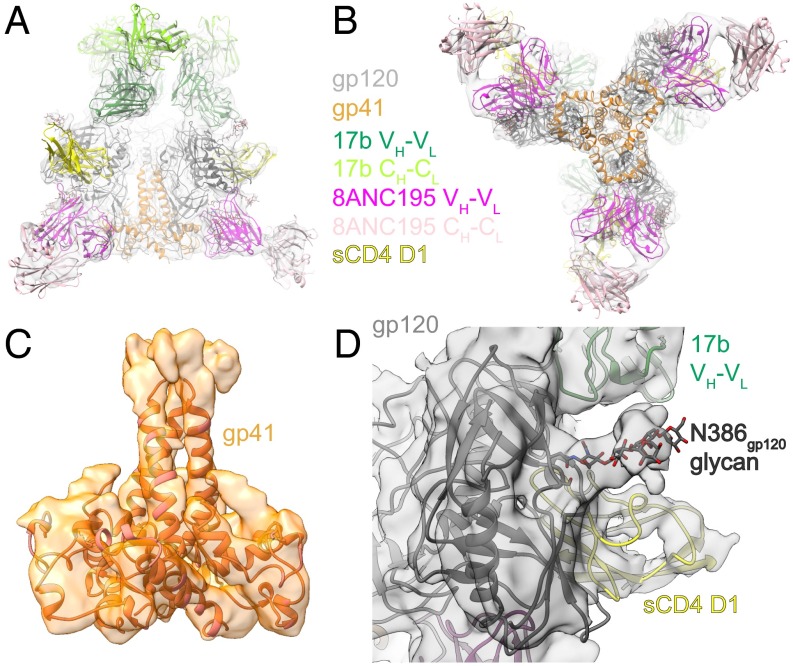
Fig. 1.
An 8.9-Å EM reconstruction of Env–sCD4–17b–8ANC complex. (A and B) Electron density fit by coordinates for gp120 (gray), gp41 (orange), sCD4 D1 (yellow), 17b VH-VL (forest green), 17b CH-CL (chartreuse), 8ANC195 VH-VL (magenta), and 8ANC195 CH-CL (pink). N-linked glycan coordinates are shown as sticks. (A) Side view in which the threefold symmetry axis of the BG505 trimer is vertical. (B) Bottom view looking down the threefold symmetry axis of the BG505 trimer. (C) Side view of density of gp41 portion of the BG505 trimer. (D) Close-up of density near the N-linked glycan attached to BG505 Asn368gp120.
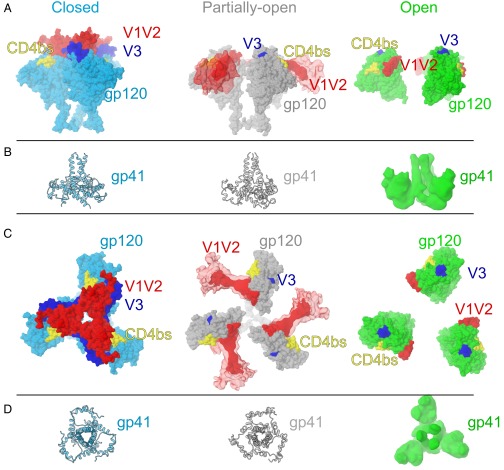
Fig. 2.
CD4-induced structural changes in Env trimers. (A) gp120 surface representations for the closed (PDB ID code 5T3X) (blue), partially open (this study) (gray), and sCD4-bound open Env structures (PDB ID code 3DNO) (green) as seen from the side. V1V2 loops (red) are depicted as surface representations for the closed and open structures and as EM density for the partially open structure. Locations of V3 (blue) and the CD4 binding site (CD4bs) (yellow) are depicted as surface representations. (B) gp41 in ribbon representation (closed and partially open Env structures) or as density (open Env structure) for Env structures. Because gp41 coordinates for an open Env structure were unavailable, we used the density from the single-particle EM structure of an open KNH1144–17b complex (EMDB entry 5462) (7). (C) Top view of gp120 representations shown in A. (D) Top view of gp41 representations shown in A.
Coordinates from crystal structures of individual components of the Env–sCD4–17b–8ANC complex were fit by rigid body docking into cryo-EM density maps. The coordinates of 8ANC195 [Protein Data Bank (PDB) ID code 4P9M] (23), 17b, and sCD4 (PDB 1RZJ) (30) were first docked into their corresponding densities, after which the gp41 coordinates from a BG505 trimer structure (PDB ID code 5CEZ) (9) were fit into density. For fitting gp120 densities, we deleted the V1V2 and V3 coordinates from a closed BG505 trimer structure (PDB ID code 5T3X) (10) and then changed the β2-β3 strand topology to match a CD4-bound gp120 core structure (PDB 1RZJ) (30). These gp120 coordinates were fit individually into protomer densities. After fitting the gp120, 17b, and sCD4 coordinates independently, the complex was compared with the crystal structure of a gp120–sCD4-17b complex (PDB ID code 1RZJ) (30), resulting in root mean square deviations (rmsds) of 1.7 Å for 98 Cα atoms in sCD4 D1 and 1.5 Å for 234 Cα atoms in the 17b VH-VL domain after superimposing the gp120s. The relatively low rmsds for the independently fit sCD4 and 17b VH-VL coordinates demonstrated that the cryoEM reconstruction reproduced known interactions of CD4 and 17b with gp120. The placement of the 8ANC195 VH-VL domains within its epitope at the gp120–gp41 interface was also not greatly shifted from its placement in an 8ANC195-BG505 (closed trimer) crystal structure (PDB ID code 5CJX) (18) (rmsd = 2.3 Å for 238 8ANC195 VH-VL Cα atoms after superimposing the gp120s) (Fig. 3A).
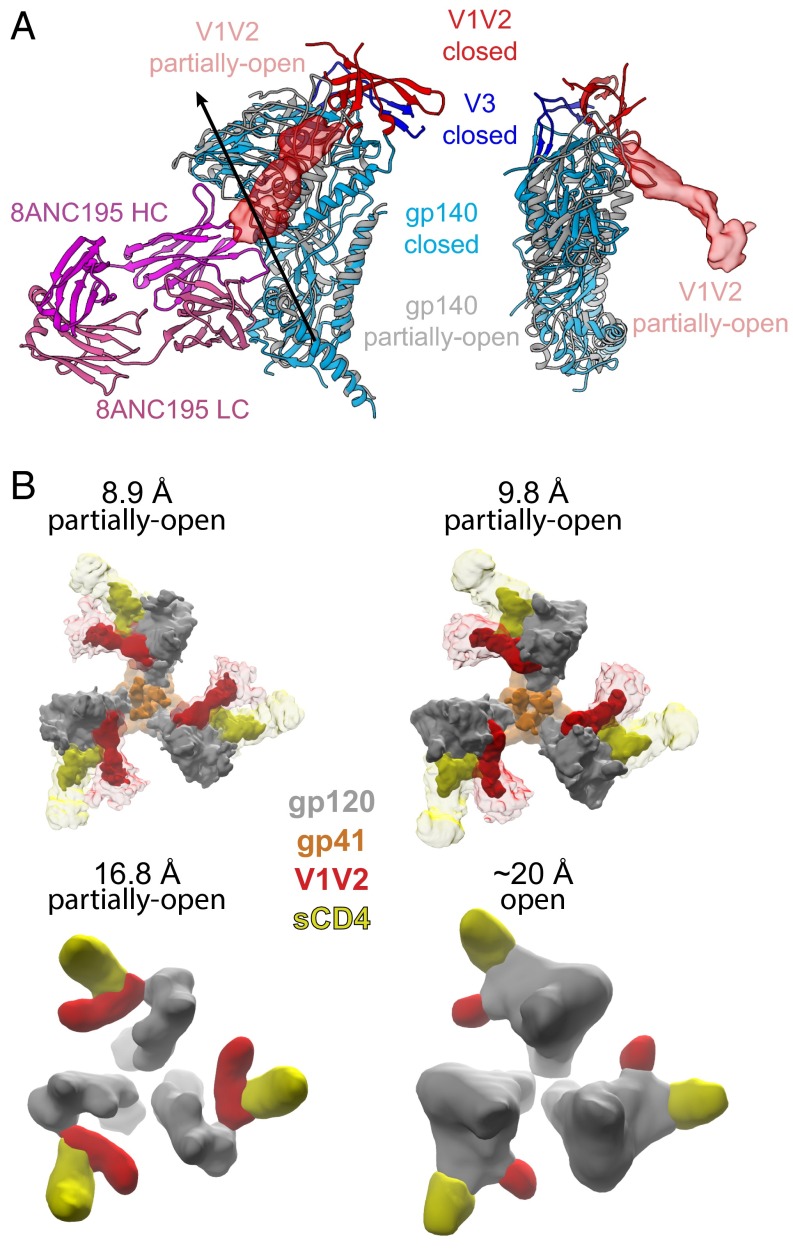
Fig. 3.
Putative V1V2 densities. (A) Superposition of the gp140 (gp120 plus gp41) coordinates from one monomer in a closed BG505 Env trimer structure (PDB ID code 5T3X) (blue) and the gp140 coordinates from the 8.9 Å partially open BG505 Env trimer reported here (gray) as seen from two different orientations. The gp140 monomers were aligned by using the threefold symmetry axis of the gp41 trimer. The arrow in Left marks the axis about which a rotation of 30° and a translation of 2.6 Å relates the gp120 in the closed structure to the gp120 in the partially open structure. The V1V2 loop (red) is depicted as a ribbon for the closed trimer and as EM density for the partially open structure. The V3 loop (dark blue) is depicted as a ribbon for closed trimer and not shown in the partially open structure because it was disordered. The 8ANC195 Fab (magenta heavy chain; light pink light chain) is shown in Left based on its position with respect to the partially open gp140, illustrating that its epitope at the gp120–gp41 interface does not undergo extensive changes. (B) sCD4-proximal densities (red) in four independent CD4-bound Env structures: the 8.9-Å and 9.8-Å cryo-EM reconstructions of the Env–sCD4–17b–8ANC complex (this study), the 16.8-Å negative stain reconstruction of the Env–sCD4–17b–8ANC complex (18), and the ∼20-Å reconstruction of a sCD4-Env trimer structure derived from subtomogram averaging of virion-bound Env spikes (4). High contour level densities in the Env–sCD4–17b–8ANC reconstructions are shown in bright red and bright yellow for V1V2 and sCD4, respectively, with lower contour level densities in lighter colors.
The fitted coordinates and density maps for the 8.9 Å and 9.8 Å reconstructions showed no major differences except for the positions of the Fab CH-CL domains (which are not rigid with respect to the antigen-binding VH-VL domains) (SI Appendix, Fig. S3). Thus, analyses were done by using the 8.9-Å reconstruction, with comparisons to verify features of interest with the independently determined 9.8-Å reconstruction (this work) and the previously described 16.8-Å negative stain reconstruction (18).
Comparison of Env Trimer Conformational States.
The 8.9-Å Env–sCD4–17b–8ANC structure revealed densities for three sCD4, three 17b, and three 8ANC195 Fabs interacting with a threefold symmetric BG505 Env trimer (Fig. 1 A and B). The BG505 Env in this complex adopts a conformation that is more open than the closed conformation in crystal and EM structures of Env trimers (8–19), but less open than the conformation in low-resolution sCD4-bound Env structures (4–7) (Figs. 2 A and B and 3A) and an ∼9-Å cryo-EM reconstruction of the KN1144 SOSIP.681 soluble trimer bound to 17b in the absence of sCD4 (7). The higher resolution and/or improved order of the present reconstruction revealed features that were unresolved in the other open Env structures, including density for several BG505 N-linked glycans (e.g., a well-ordered glycan attached to N386gp120) and density for gp41 and gp120 α-helices (Fig. 1 C and D and SI Appendix, Fig. S4). The localization of gp41 helices allows comparison of the degree of CD4-induced movement of gp120 versus gp41 in open and closed Env structures. Superposition of the gp120s from the Env–sCD4–17b–8ANC structure, a closed trimeric Env (PDB ID code 5T3X) (10), and a sCD4-bound Bal open Env structure (PDB ID code 3DNO) (4) revealed major differences in gp120 orientations (Figs. 2 A and B and 3A). Fig. 2B shows a progression of gp120 displacement from the relatively closely spaced gp120s held together by the V1V2 region in the closed trimeric state (left), to the partially open conformation in the Env–sCD4–17b–8ANC structure with newly identified V1V2 loop displacements (middle), to the fully open conformation in the Bal-17b structure (in which the V1V2 loops were not localized) (right). By contrast to the large differences in gp120s in the closed, partially open, and open Env conformations, the gp41 HR1 α-helices positions were relatively unaffected by CD4-induced Env opening (Fig. 2 A–D).
CD4-Induced V1V2 Loop Displacement.
The BG505 portion of the Env–sCD4–17b–8ANC reconstruction shows no density for the V1V2 and V3 loops in their original positions with respect to gp120, indicating structural rearrangements in addition to the rotation of gp120 monomers that are induced by sCD4 binding. We identified a prominent density near sCD4 as the likely location of the rearranged V1V2 based on several criteria: (i) the density projects toward sCD4 starting from the center of the gp120 β2 and β3 β-strands from which the V1V2 loop emanates, (ii) analogous density is present in independent CD4-bound Env structures (Fig. 3B): the 8.9-Å and 9.8-Å cryo-EM reconstructions of the Env–sCD4–17b–8ANC complex described here, the 16.8 Å negative stain reconstruction of the Env–sCD4–17b–8ANC complex (18), and the ∼20-Å sCD4-bound Env trimer structures derived from subtomogram averaging of virion-bound Env spikes (4, 5), and (iii) the density in the 8.9 Å reconstruction contacts sCD4 D1 (Fig. 3B), consistent with crystal structures of monomeric gp120s showing V1V2 stem interactions with sCD4 (30, 31). We note that analogous density is not present in the open structure of the KNH1144 SOSIP.681 trimer bound to 17b Fab in the absence of sCD4 (7), suggesting that the rearranged V1V2 loop becomes more ordered in open Env structures through interactions with bound sCD4.
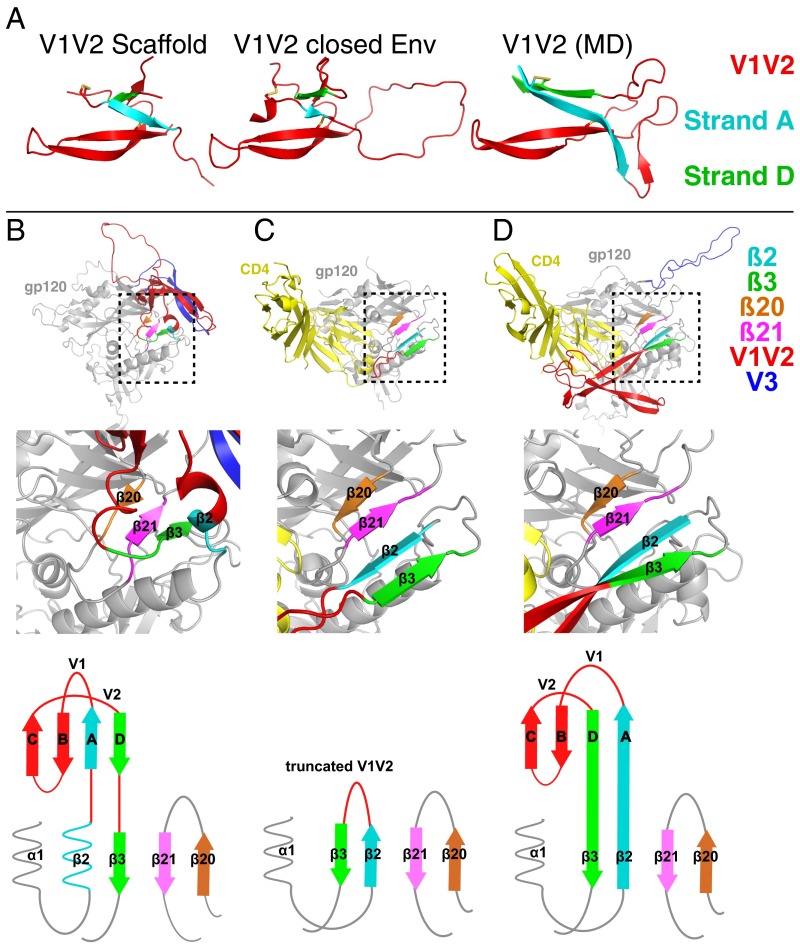
Resolution limitations in the Env–sCD4–17b–8ANC complex structure precluded ab initio building of V1V2 residues into EM density. However, we could use available V1V2 coordinates to interpret the density because evidence suggests that the V1V2 loop is likely to maintain its overall four-stranded Greek key β-sheet folding topology because this fold is preserved in closed Env trimer structures (8–19) and in structures of V1V2-alone scaffolds (32, 33) (Fig. 4A). In addition, EM reconstructions of V1V2-directed bNAbs bound to full-length gp120 monomers showed a variety of binding orientations for V1V2 conformation-specific bNAbs (32), consistent with at least some elements of the disulfide-bonded V1V2 β-sheet structure being maintained despite flexibility between monomeric gp120 and V1V2. In closed Env trimer structures, strand A of the four-stranded Greek key β-sheet structure of the V1V2 loop emanates a region that includes a helical turn (Fig. 4B), but the analogous residues are within a β-strand called β2 in gp120 cores (Fig. 4C). Strand D, the final β-strand of the V1V2 loop, leads into the gp120 β3 β-strand. The environment of the β2-β3 region in the closed Env trimer is rearranged in sCD4-bound gp120 core structures into a four-stranded antiparallel β-sheet comprising strands β20, β21, β2, and β3 (Fig. 4 B and C). A molecular dynamics model of repositioned V1V2 in full-length CD4-bound gp120 assumed this same rearrangement of the β2-β3 region (21) (Fig. 4 A and D).
Fig. 4.
V1V2 loop structures. (A) V1V2 folding topologies in V1V2 scaffold (PDB ID code 5ESV) (32, 33), closed BG505 trimer (PDB ID code 5FYJ) (10, 19), and molecular dynamics model of repositioned V1V2 in full-length CD4-bound gp120 (21). β-strand nomenclature in V1V2 is the same as in ref. 32. Disulfide bonds are shown as yellow sticks. (B–D) gp120s from structures of closed BG505 trimer (PDB ID code 5FYJ) (19) (B), sCD4-bound monomeric gp120 core (truncated V1V2 and V3 loops) (PDB ID code 1RZJ) (30) (C), and the molecular dynamics model of full-length CD4-bound gp120 (21) (D). Top shows structural overviews. Middle shows close-up views of the regions in the boxed areas. Bottom shows topology diagrams of the bridging sheet (adapted from ref. 16). V1V2 is red and V3 is blue. Strands β2 and β3, which precede and follow V1V2 in the gp120 sequence, are cyan and green, respectively. gp120 strands β20 and β21, which form a β-sheet with β2 and β3 in sCD4-bound gp120 structures (31) are orange and magenta, respectively.
We used a lower contour level for interpreting the V1V2 density (light red densities in Figs. 2 and 3) in our EM maps than we used for central portions of the BG505 trimer; the lower level was required to reveal density for less-ordered portions of the complex structure such as sCD4 D2 (SI Appendix, Fig. S2C). At a high contour level, we saw that coordinates for the gp120 and sCD4 D1 from a crystal structure of a V1V2-truncated monomeric gp120 core complexed with sCD4 and 17b (PDB ID code 1RZJ) fit the density well (Fig. 5A). The electron density suggested contacts between V1V2 and sCD4 D1 (Fig. 5B), consistent with monomeric gp120-sCD4 crystal structures (30, 31). At a lower contour level, we found that the coordinates for the molecular dynamics model of full-length gp120 with a rearranged V1V2 loop fit the EM density well (Fig. 5 C and D). We were unable to localize the V3 loop in the EM density of the partially open sCD4-bound Env trimer, but we could rule out the location predicted in the molecular dynamics model or in a crystal structure of a V3 loop-containing monomeric gp120 core (PDB ID code 2QAD) (34) (Fig. 5C and SI Appendix, Fig. S5A), perhaps because the V3 loop position in the crystal structure was influenced by crystal packing (SI Appendix, Fig. S5B).
Fig. 5.
V1V2 fitting to EM density. Densities are gray (gp120), orange (gp41), yellow (sCD4), and red (V1V2). (A) EM density map (high contour level) fit with coordinates of a sCD4-bound monomeric gp120 core (truncated V1V2 and V3 loops) (PDB ID code 1RZJ) (30). (B) Close-up of map and coordinates in A showing putative contacts between V1V2 loop stem and CD4 D1. (C) EM density map (low contour level for V1V2 and sCD4 regions) fit with coordinates of the molecular dynamics model of full-length CD4-bound gp120 (21). The V3 loop does not fit in the density. (D) Close-up of map and coordinates in C.
The Role of V1V2 in 17b Binding.
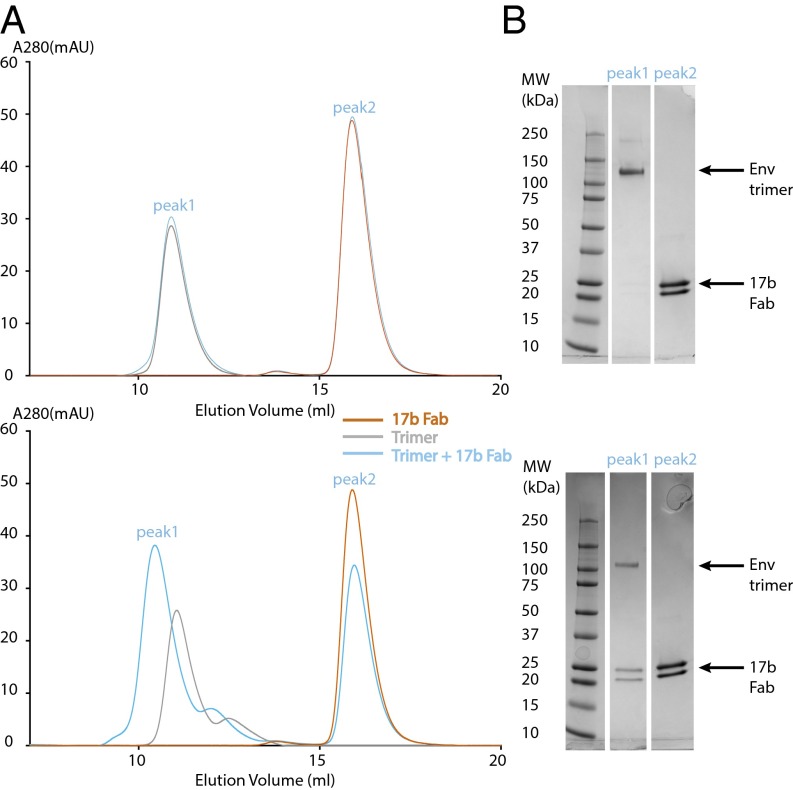
To further investigate the influence of the gp120 V1V2 loop on interactions with sCD4, we constructed a V1V2-truncated version of BG505 SOSIP.664 (BG505-ΔV1V2) analogous to a V1V2-truncated gp120 core (22, 31) with the goal of solving the structure of a V1V2-truncated Env trimer bound to sCD4. Purified BG505-ΔV1V2 appeared trimeric by negative stain EM, and the complex of BG505–ΔV1V2 with sCD4, 17b, and 8ANC195 was stable by SEC (SI Appendix, Fig. S6A). Trimeric BG505-ΔV1V2 and some of the individual ligands could be identified in negative stain EM 2D class averages (SI Appendix, Fig. S6C). However, we could not derive a 3D reconstruction from the class averages to examine structural differences in BG505 resulting from V1V2 truncation, suggesting that the BG505–ΔV1V2 complex with sCD4, 17b, and 8ANC195, which lacked the sCD4–V1V2 interaction, was less structurally homogeneous than the Env-sCD4–17b–8ANC complex.
We used the BG505–ΔV1V2 protein to investigate the effects of the V1V2 loop on 17b binding. Purified BG505 or BG505–ΔV1V2 proteins were incubated with 17b Fab in the absence of sCD4 and subjected to SEC. The unmodified BG505 showed no complex formation with 17b, consistent with previous studies (6, 35), whereas BG505–ΔV1V2 formed an SEC-stable complex with 17b in the absence of sCD4 (Fig. 6). These results are consistent with structural changes in the V1V2 loop upon sCD4 binding allowing binding of 17b in the V1V2 loop-containing Env trimer.
Fig. 6.
Interactions of 17b with BG505 and BG505-ΔV1V2. (A) SEC profile demonstrating that BG505-ΔV1V2, but not BG505, binds 17b Fab. (B) SDS/PAGE analysis of SEC fractions.
Discussion
The HIV-1 Env spike is a conformationally dynamic molecule, both in its native, virion-bound trimeric state, and in soluble native-like SOSIP trimers developed as immunogen candidates that are being used for biochemical and structural studies (36). At least five different conformational states have been identified by EM and/or X-ray crystallography (listed from closed to increasing open conformation categories): (i) closed unliganded and bNAb-bound conformations observed on virions and in SOSIPs (4–19), (ii) unliganded partially open native-like states observed by negative stain EM for SOSIPs other than BG505 SOSIP.664 (37–39), (iii) partially open virion-bound Env trimers complexed with the anti–HIV-1 antibodies b12 or A12 (4), (iv) the partially open sCD4–17b–8ANC195–BG505 structure reported here and in ref. 18, and (v) an open Env conformation induced on virions by sCD4 and 17b binding (4), on BG505 and other SOSIPs by sCD4 and 17b binding (5), or on the KNH1144 SOSIP by binding of either the 17b (7) or Z13e1 (40) antibodies alone. At least some of these conformational states identified through static structural studies are likely to exist on virions, as evidenced by single-molecule fluorescence resonance energy transfer (FRET) studies of Env trimers on HIV-1 virions, which revealed unliganded native Env to be intrinsically dynamic (2). Transitions between low-, intermediate-, and high-FRET states were discovered, with the predominating low-FRET state identified as the closed Env trimer conformation, and the intermediate-FRET state (populated almost exclusively from the high-FRET state) interpreted as a coreceptor-stabilized conformation that was stabilized by simultaneous introduction of sCD4 and 17b (2). Although the high-FRET state could not be precisely identified, the proportions of both the high- and medium-FRET states were increased by sCD4 and 17b addition, suggesting they represent distinct forms of open Env conformational states.
The large conformational differences in the sCD4–17b–8ANC195-bound BG505 trimer with respect to closed Envs, including rotation/separation of the gp120 cores and a >40 Å displacement of the V1V2 loop, suggests that this conformation represents a structural intermediate to coreceptor binding that is closer to completely open sCD4-bound Env structures than to closed structures. The sCD4-induced Env trimer opening observed in this study arose primarily from rigid body rotations of the gp120 monomers as opposed to changes in trimeric gp41 or the gp120–gp41 interface. With respect to trimeric gp41, we observed that the HR1 α-helices in the partially open trimer were slightly closer together than their counterparts in a closed trimer (Fig. 2B), but cannot rule out artifacts from the low resolution of the partially open structure and/or effects of the SOSIP “IP” substitution (I559Pgp41) (6) on sCD4-induced conformation changes. However, the conclusion that the gp120–gp41 interface is relatively unchanged during sCD4-induced trimer opening is supported by the fact that 8ANC195 binds similarly to both closed and sCD4-bound trimers (Fig. 3A). The question remains as to why the BG505 in our structure is only partially open as opposed to completely open. Because the Env–sCD4–17b–8ANC complex was prepared by adding 8ANC195 to a preformed BG505–sCD4–17b complex, we speculate that the BG505–sCD4–17b complex was fully open until incubated with 8ANC195, a bNAb that prefers the closed Env conformation (18); 8ANC195 binding could have induced partial closure of the Env trimer to better bind to its gp120–gp41 epitope, a conformational sequence reminiscent of the high- to intermediate-FRET state conversion described for native Env trimers on virions (2). It is notable that the Env trimer did not completely close, which would have resulted in steric occlusion of the three 17b Fabs at the trimer apex (18) likely leading to 17b Fab dissociation, nor did it even close to the point of creating contacts between 17b Fabs (as evidenced by the cryo-EM map showing no contact between 17b Fabs) (Fig. 1A). Thus, the partially-open Env conformation revealed in the Env–sCD4–17b–8ANC likely represents a conformation accessible to native Env trimers on the pathway toward fusion.
In summary, the cryo-EM structure presented here represents the most detailed glimpse of structural changes occurring during Env-mediated fusion of the HIV-1 and host cell membranes. The presence of 17b, a coreceptor mimicking antibody, suggests that the partially open Env conformation we described is correlated with the coreceptor-bound Env state. Hence, the V1V2 loop movement observed in our complex structure rationalizes why CD4 binding is required for coreceptor binding and subsequent release of the fusion peptide. In addition, the structure provides a potential target for design of antibody- or small molecule-based anti–HIV-1 therapeutics.
Methods
Protein Production and Purification.
His6-tagged Fabs of 17b and the 8ANC195G52K5 variant of 8ANC195 were expressed by transient transfection in HEK293-6E cells (National Research Council of Canada) and purified from cell supernatants by using Ni-NTA chromatography and SEC as described (18). sCD4 D1D2 (domains 1 and 2; residues 1–186 of mature CD4) was produced in baculovirus-infected Hi5 insect cells and was purified by Ni-NTA chromatography and SEC (41). BG505 SOSIP.664, a native-like soluble clade A gp140 trimer (6), was constructed to include “SOS” substitutions (A501Cgp120, T605Cgp41), the IP substitution (I559Pgp41), the N-linked glycan sequence at residue 332gp120 (T332Ngp120), an enhanced gp120–gp41 cleavage site (REKR to RRRRRR), and a stop codon after residue 664gp41 (Env numbering according to HX nomenclature). BG505-ΔV1V2 trimer was constructed by replacing residues 128–194 of the V1V2 loop with a Gly-Ala-Gly linker, as described for previous experiments with a V1V2-truncated gp120 (22). BG505 and BG505-ΔV1V2 proteins were expressed in HEK293-6E cells treated with 5 µM kifunensine (Sigma) by transient transfection of plasmids encoding Env trimer and soluble furin at a ratio of 4:1 as described (18). BG505 SOSIP proteins were isolated from cell supernatants by using a 2G12 immunoaffinity column as described (10). After elution with 3 M MgCl2 followed by immediate buffer exchange into Tris-buffered saline pH 8.0 (TBS), trimers were purified by SEC using a Superdex 200 16/60 column, Mono Q ion exchange chromatography, and a second SEC purification by using a Superose 6 10/300 column (columns from GE Healthcare).
Cryo-EM Data Collection and Processing.
The Env–sCD4–17b–8ANC complex was made by incubating BG505 with excess sCD4 and 17b Fab overnight and then isolated by SEC. After incubating the BG505–sCD4–17b complex with excess 8ANC195 Fab for 2 h at 4 °C, the quaternary complex Env–sCD4–17b–8ANC complex was isolated by SEC. Purified Env–sCD4–17b–8ANC complexes were diluted to 60 μg/mL in TBS and vitrified in liquid ethane by using a Mark IV Vitrobot. Sample grids were prepared by adding 3 µL of complex to glow discharged 400 Mesh Quantifoil R1.2/1.3 copper grids (for the 8.9-Å reconstruction) or to 400 Mesh C-Flat R1.2/1.3 grids (for the 9.8-Å reconstruction). Images were recorded on a Titan Krios electron microscope equipped with Gatan K2 Summit direct detector and an energy filter with a slit width of 20 eV (for the 9.8-Å reconstruction only) by using SerialEM (42). For the 8.9-Å reconstruction, 10-s exposures were divided into 25 subframes, and the dose rate was 3.8 electrons⋅pixel−1⋅subframe−1. For the 9.8-Å reconstruction, 20.25-s exposures were divided into 54 subframes and the dosage rate was 3.7 electrons⋅pixel−1⋅subframe−1. After binning by 2 and motion correction, each image was 4k × 4k and 1.64 Å per pixel (8.9-Å reconstruction) or 4k × 4k and 1.71 Å per pixel (9.8-Å reconstruction).
Both datasets were motion corrected and dose weighted by using Unblur and Summovie (43–45). Motion-corrected micrographs without dose weighting were used for contrast transfer function (CTF) estimations. Motion-corrected micrographs with dose weighting were used for particle picking, and motion-corrected micrographs with dose weighting and restored noise power after filtering were used for all classification and refinement processes.
Particles were picked by using the SWARM method of EMAN2.1 (46), and CTF estimations were done by using CTFFIND4 (47). For the 8.9-Å reconstruction: A total of 808 movies were collected. After motion correction and dose weighting, CTF curves were confidently fit to beyond 6 Å in 360 micrographs; the others were discarded. A total of 13,268 particles were picked. Particles were classified in 2D with Relion (48), resulting in 130 2D classes. Of these, 9 classes from 6,675 particles were selected as “good” classes. For 3D classification, the 16.8 Å Env–sCD4–17b–8ANC negative stain structure [Electron Microscopy Data Bank (EMDB) entry 3086] was used as a reference and the CH-CL domains of the Fabs were masked out; two 3D classes were then produced. After selecting one 3D class as a “good” class, 5,175 particles remained for 3D refinement. After 3D refinement, postprocessing, particle polishing, and gold-standard FSC estimations were done by using Relion (48) following procedures in the tutorial. Density maps were low-pass filtered to 5 Å to remove noise. Local resolution estimations were done by using ResMap (29).
The 9.8-Å structure was produced in the same way with minor differences: 642 movies were collected, only 480 micrographs could be CTF fit to beyond 6 Å, a total of 19,355 particles were picked, 11,915 were retained after 2D classification, and 9,606 particles retained through 3D classification.
After motion correcting, dose weighting, and CTF correcting the two datasets individually, the CTF-corrected (flipped) particles were scaled to the same angstrom-per-pixel value and combined into a single dataset of 32,623 particles; 18,476 were retained after 2D classification, and 17,013 particles were retained through 3D classification. The resolution of a reconstruction calculated from the combined datasets was 9.6 Å (SI Appendix, Fig. S3C), lower than the 8.9-Å resolution calculated for a reconstruction from the first dataset. Because combining the datasets did not improve the resolution beyond 8.9 Å, we kept the datasets separated for structural analyses.
Model Building.
Coordinates from crystal structures were manually fit into cryo-EM density maps as rigid bodies by using University of California, San Francisco Chimera’s Fit in Map function (49), and the complex was further refined by using real-space refinement in PHENIX (50). Coordinates used for fitting or comparisons were gp120 from BG505 SOSIP.664 (closed conformation) (PDB ID code 5T3X), gp41 from BG505 SOSIP.664 (closed conformation) (PDB ID code 5CEZ or 5T3X), sCD4 (PDB ID code 2NXY), 17b Fab (PDB ID code 2NXY), 8ANC195 Fab (PDB ID code 4P9M), 16.8-Å negative stain EM structure of Env–sCD4–17b–8ANC (EMDB entry 3086), BG505-8ANC195 (PDB ID code 5CJX) and X1193.c1 SOSIP.664-PGT122-35O22-VRC01 (PDB ID code 5FYJ) closed conformation complexes, V1V2 scaffold (PDB ID code 5ESV), gp120 with V1V2 model from molecular dynamics (coordinates obtained from Hironori Sato), gp120 core–sCD4–17b complex (PDB ID code 1RZJ), gp120 core with ordered V3 loop (2QAD), open 17b-bound KNH1144 SOSIP.681 (EMDB entry 5462; coordinates for gp120s obtained from Sriram Subramaniam), gp120–sCD4–17b complex (PDB ID code 2NXY), open conformation virion-bound Bal Env-sCD4 (EMD: 5455), and open conformation virion-bound Bal Env-sCD4-17b (EMDB entry 5020 and PDB ID code 3DNO).
The contour levels for EM maps in this study were chosen based on local resolution estimations and fitted coordinates. For rigid body docking, model building, and coordinates visualization, we chose a contour level such that the gp41 HR1 α-helices exactly fit into the density. However, density for the D2 domain of sCD4 could not be visualized at this high contour level. We lowered the contour level such that density for the D2 domain of sCD4 appeared and used this lower contour level to interpret the density corresponding to the rearranged V1V2 loop region. For determining the rotation and translation relating the gp120s in the closed and partially open conformations, the transformation relating gp120 α-helices at positions 60–63, 99–113, 335–349, and 475–483 was calculated by using TM-align (51). The corresponding screw transformation was calculated according to ref. 52 and visualized by using AntibodyDatabase (53).
Negative Stain EM.
The BG505-ΔV1V2–sCD4–17b–8ANC195 quaternary complex was made as described above for the Env–sCD4–17b–8ANC complex. Purified complexes were diluted to 10 μg/mL in TBS immediately before adding 3 µL to a glow discharged ultrathin C film on holey carbon support film, 400 mesh, Cu grids (Ted Pella) and staining with uranyl acetate. Data were collected by using a FEI Tecnai T12 transmission electron microscope operating at 120 keV with a Gatan Ultrascan 2k × 2k CCD camera. Each image was collected by using a 0.5-s exposure at ∼1-µm defocus and 42,000× magnification resulting in 2.5 Å per pixel. For the BG505-ΔV1V2–sCD4–17b–8ANC195 complex, a total of 7,251 particles were picked and CTF corrected by using EMAN2.1 (46). Reference-free 2D classification was performed by using Relion (48).
Supplementary Material
Acknowledgments
We thank Zhiheng Yu, Chuan Hong, and Rick Huang (Janelia Farm) for assistance with cryo-EM data collection and motion correction; Hironori Sato for coordinates of the molecular dynamics model of full-length gp120; Sriram Subramaniam for gp120 coordinates from the KNH1144 SOSIP.681–17b complex; Alasdair McDowall and Songye Chen for training in cryo-EM techniques and data processing; Anthony West for performing alignment calculations; Jost Vielmetter and the Caltech Protein Expression Center for transfections and protein expression; and members of the P.J.B. and G.J.J. laboratories for helpful discussions and critical reading of the manuscript. This research was supported by the National Institutes of Health Grant 2 P50 GM082545-06 (to P.J.B.), National Institute Of Allergy and Infectious Diseases of the National Institutes of Health Grant HIVRAD P01 AI100148 (to P.J.B.), and the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery Grant 1040753 (to P.J.B.). We thank the Gordon and Betty Moore and Beckman Foundations for gifts to Caltech to support electron microscopy. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: Data were deposited into the Protein Data Bank, www.pdb.org (PDB ID code 5THR) and The Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (entry no. EMD-8407).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615939113/-/DCSupplemental.
References
- 1.West AP, Jr, et al. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156(4):633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munro JB, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346(6210):759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci USA. 2012;109(15):5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci USA. 2011;108(28):11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran EE, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8(7):e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20(12):1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garces F, et al. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43(6):1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gristick HB, et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol. 2016;23(10):906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong L, et al. Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 10):2099–2108. doi: 10.1107/S1399004715013917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon YD, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22(7):522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, de Val N, Lyumkis D, Ward AB. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure. 2015;23(10):1943–1951. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351(6277):1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pancera M, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514(7523):455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scharf L, et al. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell. 2015;162(6):1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart-Jones GBE, et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165(4):813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasheed M, Bettadapura R, Bajaj C. Computational Refinement and validation protocol for proteins with large variable regions applied to model HIV Env spike in CD4 and 17b bound state. Structure. 2015;23(6):1138–1149. doi: 10.1016/j.str.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama M, et al. In silico analysis of HIV-1 Env-gp120 reveals structural bases for viral adaptation in growth-restrictive cells. Front Microbiol. 2016;7:110. doi: 10.3389/fmicb.2016.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyatt R, et al. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69(9):5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scharf L, et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Reports. 2014;7(3):785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thali M, et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67(7):3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salzwedel K, Smith ED, Dey B, Berger EA. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: Soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74(1):326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84(3):1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci USA. 2004;101(9):2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorman J, et al. NISC Comparative Sequencing Program Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol. 2016;23(1):81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guttman M, et al. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munro JB, Mothes W. Structure and dynamics of the native HIV-1 Env trimer. J Virol. 2015;89(11):5752–5755. doi: 10.1128/JVI.03187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Taeye SW, et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163(7):1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julien JP, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci USA. 2015;112(38):11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugach P, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol. 2015;89(6):3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris AK, Bartesaghi A, Milne JL, Subramaniam S. HIV-1 envelope glycoprotein trimers display open quaternary conformation when bound to the gp41 membrane-proximal external-region-directed broadly neutralizing antibody Z13e1. J Virol. 2013;87(12):7191–7196. doi: 10.1128/JVI.03284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat Struct Mol Biol. 2010;17(5):608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Brilot AF, et al. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012;177(3):630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campbell MG, et al. Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure. 2012;20(11):1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157(1):281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siciliano B, Khatib O. Springer Handbook of Robotics. Springer; Berlin: 2008. pp. 14–16. [Google Scholar]
- 53.West AP, Jr, et al. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc Natl Acad Sci USA. 2013;110(26):10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.