Significance
Proteasome inhibitors show remarkable anti-multiple myeloma (MM) activity in preclinical and clinical studies. However, resistance develops in the majority of patients, and novel treatments are urgently needed. Histone deacetylase 6 (HDAC6) has been shown to mediate aggresomal protein degradation and could be a potential target for combination treatment to overcome drug resistance. Here we designed and developed an HDAC6-selective small molecule inhibitor, WT161, and used this compound to define mechanisms of anti-MM activity, both alone and in combination with proteasome inhibitors in in vitro and in vivo studies. This study has established the framework for combination treatment of HDAC6 inhibitors with proteasome inhibitors in MM and validates an in vivo quality chemical probe for broad use by the research community.
Keywords: histone deacetylase 6, proteasome inhibitor, multiple myeloma, bortezomib-resistance, WT161
Abstract
Multiple myeloma (MM) has proven clinically susceptible to modulation of pathways of protein homeostasis. Blockade of proteasomal degradation of polyubiquitinated misfolded proteins by the proteasome inhibitor bortezomib (BTZ) achieves responses and prolongs survival in MM, but long-term treatment with BTZ leads to drug-resistant relapse in most patients. In a proof-of-concept study, we previously demonstrated that blocking aggresomal breakdown of polyubiquitinated misfolded proteins with the histone deacetylase 6 (HDAC6) inhibitor tubacin enhances BTZ-induced cytotoxicity in MM cells in vitro. However, these foundational studies were limited by the pharmacologic liabilities of tubacin as a chemical probe with only in vitro utility. Emerging from a focused library synthesis, a potent, selective, and bioavailable HDAC6 inhibitor, WT161, was created to study the mechanism of action of HDAC6 inhibition in MM alone and in combination with BTZ. WT161 in combination with BTZ triggers significant accumulation of polyubiquitinated proteins and cell stress, followed by caspase activation and apoptosis. More importantly, this combination treatment was effective in BTZ-resistant cells and in the presence of bone marrow stromal cells, which have been shown to mediate MM cell drug resistance. The activity of WT161 was confirmed in our human MM cell xenograft mouse model and established the framework for clinical trials of the combination treatment to improve patient outcomes in MM.
Multiple myeloma (MM) is a B-cell malignancy characterized by the proliferation of bone marrow (BM) plasma cells in association with high Ig production (1). Although treatable, MM is considered incurable and has a 5-y overall survival rate of only 45%. The treatment of MM has been transformed in the last decade because of the development of novel therapeutic agents that target MM cells in the BM microenvironment and that can overcome conventional drug resistance (1). Namely, the proteasome inhibitor bortezomib (BTZ), which blocks the degradation of polyubiquitinated misfolded proteins, induces ER stress, and triggers apoptosis of MM cells, has rapidly translated to clinical trials demonstrating remarkable clinical efficacy. The second-generation proteasome inhibitors carfilzomib (CFZ) (2), ixazomib (3), and marizomib (4) also are showing improved pharmacological properties and promising responses. Nonetheless, MM cells develop resistance to BTZ, leading to relapse of disease in most patients (5, 6).
The proteasome serves an important cellular function in clearing abnormal proteins in the cell. Tumor cells, and particularly MM cells that produce high levels of Ig, are more heavily dependent on this clearance mechanism and thus are sensitive to proteasome inhibition (7). The aggresomal protein degradation pathway is an alternative system to proteasomal degradation of ubiquitinated misfolded/unfolded proteins that ultimately induces autophagic clearance of proteins by lysosomal degradation (8). Histone deacetylase 6 (HDAC6) plays a central role in autophagic protein degradation by recruiting ubiquitinated protein cargo for transport to aggresomes. HDAC6 is a member of the class IIb family of HDAC enzymes. HDAC6 possesses two functional deacetylase domains and a zinc finger motif. HDAC6 was initially described as a tubulin deacetylase; however, the literature defines additional substrates, including Hsp90 and p300 (9). HDAC6 modulates cell morphology, adhesion and migration, immune-mediated cell–cell interactions, and tumor cell invasion/metastasis as well as misfolded/unfolded protein degradation and stress response. Inhibition of HDAC6 with nonselective HDAC inhibitors, such as vorinostat (suberoylanilide hydroxamic acid, SAHA) or panobinostat (LBH589), blocks lysosomal protein degradation. To date preclinical and clinical studies have used pan-HDAC inhibitors together with BTZ to inhibit both proteasomal and aggresomal protein degradation and to overcome clinical BTZ resistance (10, 11). However, the side-effect profile of the broad HDAC inhibitors with BTZ, including fatigue, diarrhea, and thrombocytopenia, limits the clinical utility of this combined treatment (10). Thus, targeting HDAC6 selectively could be a potential approach to overcome drug resistance in MM while reducing the overall toxicity seen with the use of less selective HDAC inhibitors.
We previously have shown that proteasomal blockade of protein degradation with BTZ, combined with aggresomal blockade of protein degradation with a prototype HDAC6 inhibitor, tubacin, triggers significant cytotoxicity in MM cells in vitro (4). However, tubacin has limited in vivo bioavailability, and its chemical synthesis is complex, limiting its availability and utility as a chemical probe (12). Therefore the development of a highly potent and selective HDAC6 inhibitor with good bioavailability that can be synthesized in large quantities is highly desired. More importantly, access to a selective HDAC6 inhibitor would enable us to study the combination effects of HDAC6 inhibition with proteasome inhibitors in preclinical in vivo models of MM for translation to clinical trials.
In this study, we minimized the scaffold of tubacin via synthesis of a drug-like library of 400 hydrazones biased for deacetylase inhibition as hydroxamates, which we then tested for HDAC substrate specificity and cellular activity. From these efforts, we identified a potent and selective HDAC6 inhibitor, WT161. The structural basis for its HDAC6 inhibition and selectivity is delineated. Treatment of MM cell lines with WT161 triggers the accumulation of acetylated tubulin and cell death in MM cell lines more potently than tubacin. Additionally, WT161 in combination with BTZ induces synergistic cytotoxicity and overcomes BTZ resistance in vitro. The anti-MM activity of WT161 in combination with BTZ is validated in our murine xenograft model of human MM. The synergistic effect indicates that dual blockade of proteasomal and aggresomal protein degradation could be a potential therapeutic strategy for the treatment of MM and for overcoming proteasome inhibitor resistance in MM.
Results
Discovery of the Potent and Selective HDAC6 Inhibitor WT161.
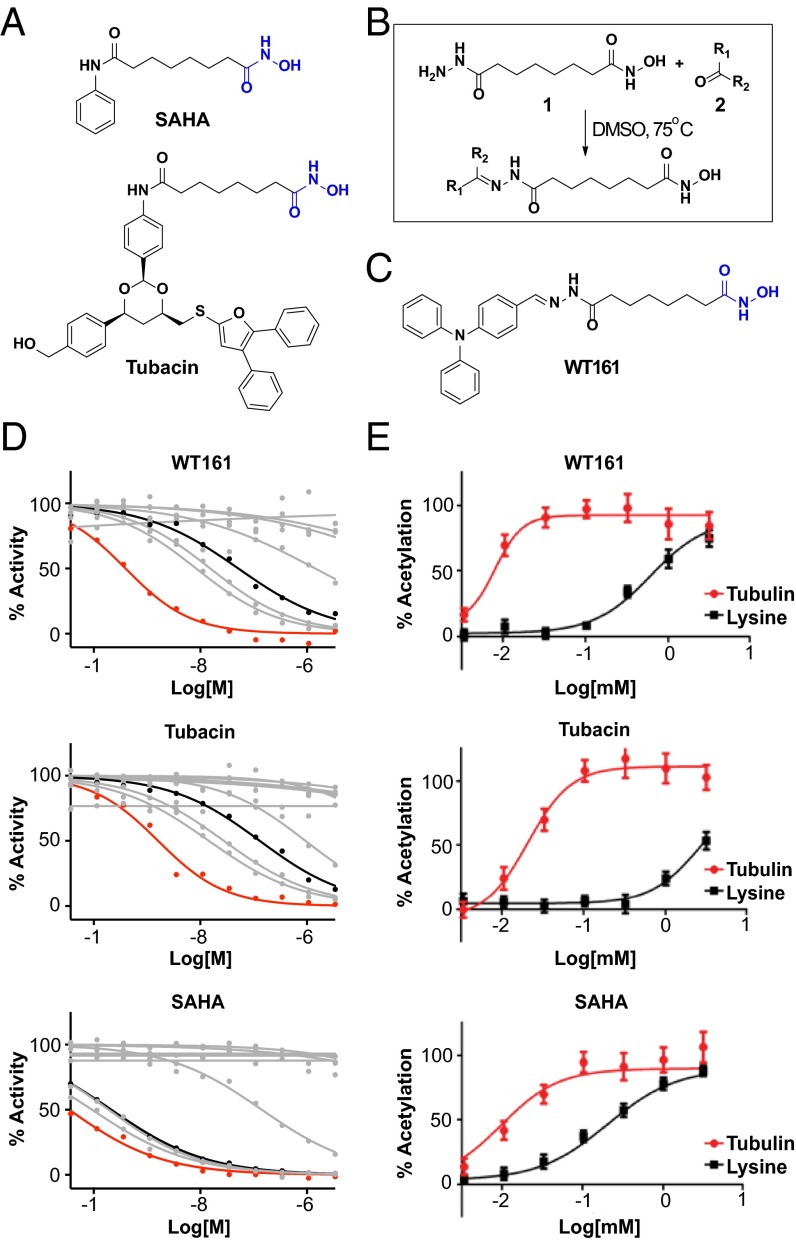
Because the class I, II, and IV HDAC enzymes are zinc-dependent hydrolases, the dominant pharmacophore model of HDAC inhibitors features three key structural elements: a metal-binding domain, a surface recognition element, and a linker bridging these two chemical moieties (13). Improved ligand specificity can be achieved via modulation of the structure of the chelator or via the structure or conformation of the capping feature (14, 15). For example, the 1,3-dioxane–appending group in tubacin is responsible for HDAC6 selectivity compared with the pan-inhibitor SAHA, which has a much smaller aryl surface-binding feature (Fig. 1A) (16).
Fig. 1.
Development of the selective HDAC6 inhibitor WT161. (A) Chemical structures of SAHA and tubacin. The hydroxamic acid is highlighted in blue. (B) The synthesis scheme used to create a hydrazine library that was screened to identify the HDAC6-selective inhibitor WT161. (C) Chemical structure of WT161 (hydroxamic acid is highlighted in blue). (D) Inhibitory activity of WT161, tubacin, and SAHA against HDAC1–9 assessed in a biochemical activity assay. The activity of HDAC6 (red) and HDAC3 (black) are highlighted. The activity curves of the other HDACs are shown in gray. (E) Cells were incubated with increasing concentrations of WT161, tubacin, or SAHA for 4 h before fixation and staining with the antibodies for Ac-α-tubulin and acetylated lysine. Measurements of the cellular effects of WT161, tubacin, and SAHA on tubulin (red) and lysine (black) acetylation were quantified via high-content imaging.
We undertook a chemical strategy to identify more drug-like HDAC6 inhibitors using a highly parallel approach to biased chemical library synthesis (17). We designed a hydroxamic acid building block, compound 1, possessing a reactive hydrazide separated by a six-carbon linear aliphatic linker that mimics the structure of SAHA and tubacin (Fig. 1B). Direct coupling of compound 1 with a library of 400 aldehydes and ketones produced 400 hydrazones in 96-well plate format, which were directly profiled in biochemical and cellular assays without further purification. From the resulting library, we identified compound WT161 as a potent and selective inhibitor of HDAC6 (Fig. 1 C and D and Table S1) (14). Although some potency for HDAC6 was sacrificed as compared to SAHA (SAHA IC50 = 0.03 nM; WT161 IC50 = 0.40 nM), WT161 is still very potent and is more selective against HDAC6 than against the other family members (HDAC3: SAHA IC50 = 0.21 nM; WT161 IC50 = 51.61 nM; tubacin IC50 = 130.90 nM). Biochemically, WT161 is more potent than tubacin and is equivalently selective for HDAC6 (tubacin IC50 = 1.62 nM) and has a dramatically simplified synthesis (three steps, 40% overall yield).
Table S1.
Biochemical inhibitory activity of SAHA, WT161, and tubacin against HDAC1–9
| SAHA | WT161 | Tubacin | ||||
| HDAC isoform | Mean IC50, nM | 95% confidence interval | Mean IC50, nM | 95% confidence interval | Mean IC50, nM | 95% confidence interval |
| HDAC1 | 0.09 | 0.06–0.14 | 8.35 | 5.37–12.97 | 14.5 | 9.99–21.08 |
| HDAC2 | 0.19 | 0.16–0.23 | 15.4 | 12.66–18.70 | 27.8 | 21.95–35.13 |
| HDAC3 | 0.21 | 0.16–0.28 | 51.6 | 36.27–73.44 | 130.9 | 94.37–181.5 |
| HDAC4 | — | — | — | |||
| HDAC5 | — | — | — | |||
| HDAC6 | 0.035 | 0.026–0.046 | 0.40 | 0.28–0.55 | 1.62 | 1.12–2.32 |
| HDAC7 | — | — | — | |||
| HDAC8 | 142.6 | 107.5–189.2 | 1,430 | 881.2–2327 | 943.0 | 644.4–1,382 |
| HDAC9 | — | — | — | |||
—, IC50 >10 μM.
The activity and selectivity of WT161 in cells was confirmed further using a miniaturized assay system we developed to monitor the simultaneous effects on HDAC6 (α-tubulin acetylation) and class I nuclear deacetylases (lysine acetylation), using high-content imaging (15, 18). WT161 selectively inhibits HDAC6 and dramatically increases levels of acetylated α-tubulin (Ac-α-tubulin) with little effect on global lysine acetylation (Fig. S1A). Furthermore, when compared directly with tubacin and SAHA, WT161 was found to increase Ac-α-tubulin in cells more effectively (Fig. 1E). Thus, we identified and validated the small molecule WT161 as a potential chemical probe for HDAC6 inhibition.
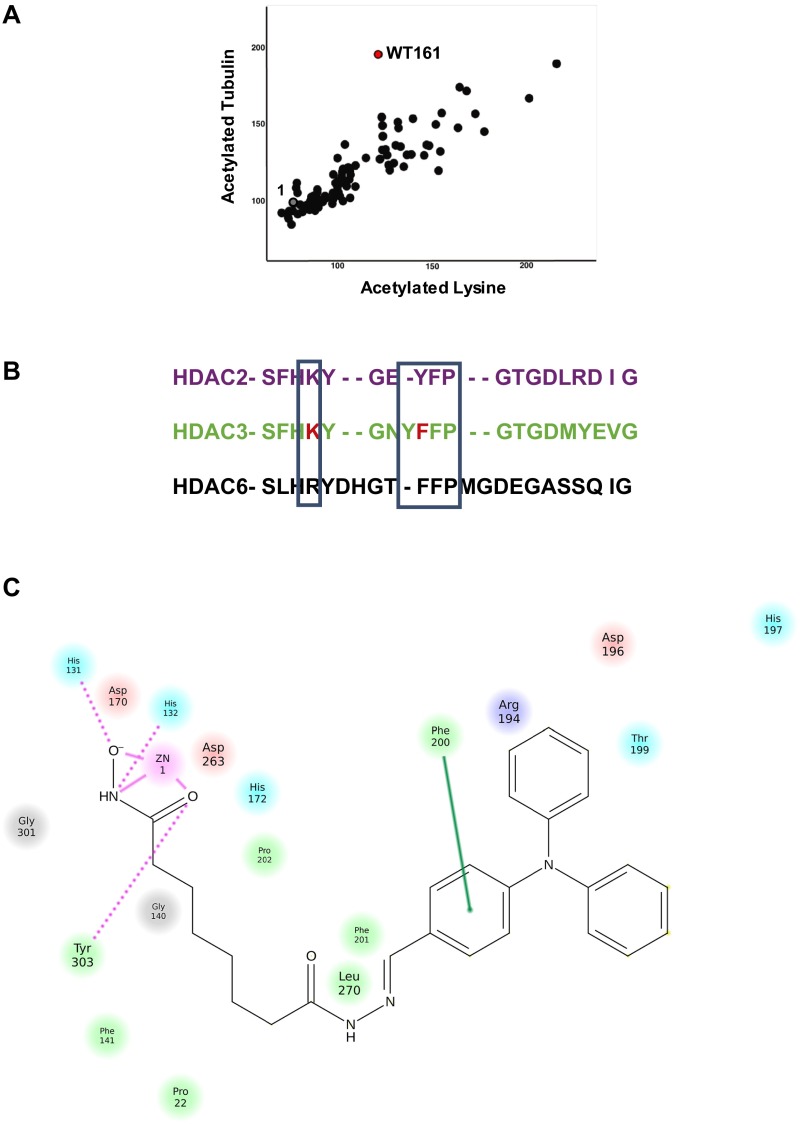
Fig. S1.
Structural basis for selectivity and binding of WT161 to HDAC6. (A) Screening of the hydrazine library in a high-content imaging assay to identify the cellular effects of potential HDAC inhibitors. The activity of WT161 (red) and compound 1 (gray) are shown. (B) Sequence alignment of the key loop region of HDAC2, -3, and -6. (C) Interaction diagram for WT161 bound to HDAC6. Relevant interactions with Arg194 and Phe200 are depicted.
Molecular Recognition of HDAC6 by WT161.
As we have reported previously, the shape of the protein surface around the active site of each HDAC enzyme influences ligand-binding preferences (16). HDAC6 forms a lipophilic pocket (the yellow loop in Fig. 2, Right) containing Phe200 and Phe201 held in place by a cation–π interaction of Phe200 with Arg194. This stable pocket is not conserved at the sequence or structural level in HDAC2 or HDAC3 because the flexibility introduced by the insertion of a tyrosine and the alteration of Arg194 to a lysine reduce the lipophilic contacts (Fig. S1B).
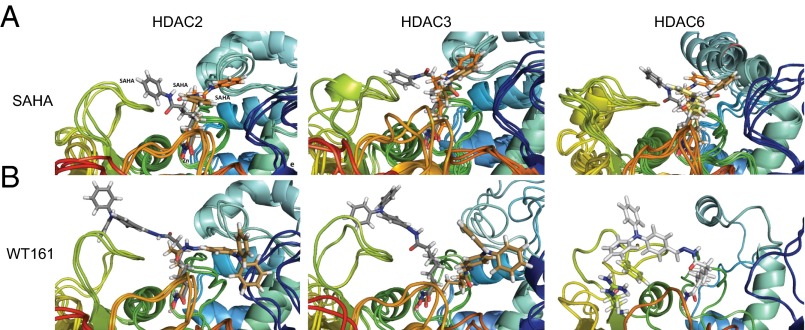
Fig. 2.
Interactions of SAHA and WT161 in the active site pocket of HDAC2, HDAC3, and HDAC6. (A) Snapshots from 30-ns MD simulation are superimposed for SAHA with HDAC2 (Left), HDAC3 (Center), and HDAC6 (Right) with multiple orientations stabilized. (B) Snapshots from 30-ns MD simulation are superimposed for WT161 with HDAC2 (Left) and HDAC3 (Center), with multiple orientations stabilized. Only a single orientation is found for the HDAC6-WT161 complex (Right). The ligand is shown in different colors in the different snapshots.
To study the putative determinants of HDAC inhibitor selectivity, we performed molecular dynamic (MD) simulations of HDAC2, -3, and -6 in the presence of SAHA and WT161. The snapshots of the MD simulations of SAHA in HDAC2, -3, and -6 show that SAHA binds deeply in the active site channel and engages in multiple, nonspecific interactions with all loops surrounding the active site mouth with many possible orientations in all three isoforms (Fig. 2A). This finding indicates that SAHA is not sufficiently lipophilic to engage a specific HDAC isoform in stable interactions, thus explaining its observed lack of selectivity.
In contrast to SAHA and in analogy to tubacin, WT161 exhibits good shape complementarity to HDAC6 via the Y-shaped structure of the lipophilic cap. The cap region fits into the hydrophobic pocket formed by Phe200, Phe201, and Leu270 in HDAC6 and forms a cation–π interaction with the top of the linker and Arg194 (Fig. S1C). Thus, only a single orientation of WT161 bound to HDAC6 is found over the course of the MD simulation (Fig. 2B). These interactions are observed in HDAC6 but not in HDAC2/3, which have a conserved Arg260 that binds more weakly to the lipophilic cap of WT161. Thus, the MD simulation provides the structural basis for the HDAC6 selectivity of WT161.
WT161 Induces Accumulation of Acetylated Tubulin and Cytotoxicity in MM Cells.
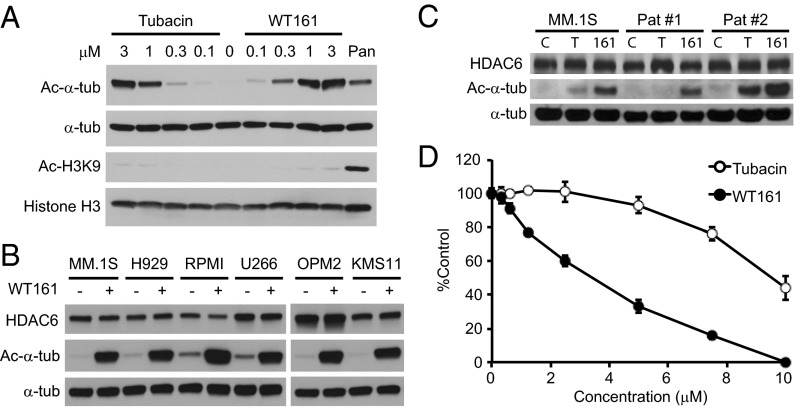
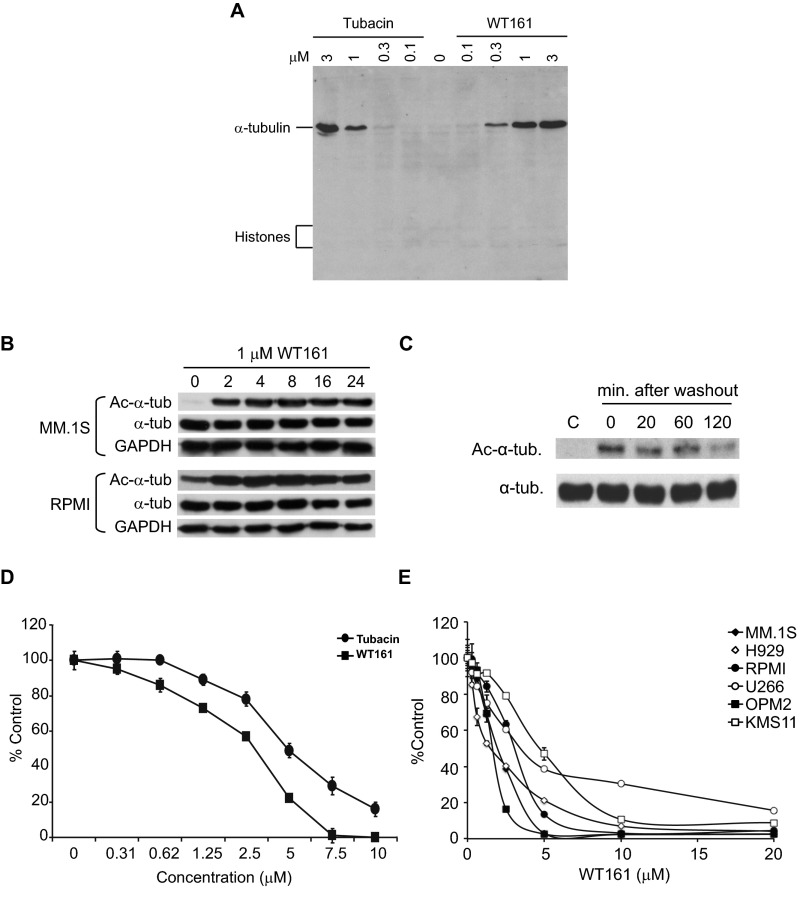
To confirm the activity of WT161 in MM cells, we measured the accumulation of Ac-α-tubulin. Both WT161 and tubacin significantly induce Ac-α-tubulin in a dose-dependent fashion without increasing histone H3K9 acetylation (Fig. 3A and Fig. S2A). WT161 can induce the accumulation of Ac-α-tubulin in MM cell lines in as little as 2 h, and this induction is reversible (Fig. S2 B and C). WT161 similarly induced accumulation of Ac-α-tubulin in a panel of five additional MM cell lines and in patient-derived MM cells that were purified from BM aspirates from two MM patients (Fig. 3 B and C).
Fig. 3.
WT161 is a selective and reversible HDAC6 inhibitor and inhibits MM cell growth. (A) MM.1S cells were cultured with tubacin, WT161, or panobinostat (50 nM) for 6 h. Whole-cell lysates were analyzed by immunoblotting with indicated antibodies. (B) MM cell lines were cultured with or without 1 µM WT161 for 6 h. (C) MM.1S and patient-derived MM cells were cultured with DMSO (control; C), 1 µM tubacin (T), or 1 µM WT161 (161) for 6 h. Whole-cell lysates were subjected to immunoblotting with the indicated antibodies. (D) The relative viability of MM.1S cells cultured with increasing concentrations of tubacin or WT161 for 48 h was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay.
Fig. S2.
WT161 selectively up-regulates Ac-α-tubulin in MM cells. (A) MM.1S cells were cultured with tubacin or WT161 for 6 h. Whole-cell lysates were analyzed by immunoblotting with acetylated lysine antibody. (B) MM.1S and RPMI8226 cells were cultured with WT161 (1 µM) for the indicated times. Whole-cell lysates were analyzed by immunoblotting with indicated antibodies. (C) MM.1S cells were cultured with 1 µM WT161 for 6 h. Cells then were washed with medium and incubated further for the indicated times. Whole-cell lysates were analyzed by Western blotting with the indicated antibodies. (D) RPMI8226 cells were cultured with increasing concentrations of WT161 or tubacin for 48 h. Cell growth inhibition was assessed by MTT assay. Data represent the mean ± SD from triplicate cultures. (E) MM cell lines were cultured with the indicated concentrations of WT161 for 48 h. Cell growth was assessed by MTT assay. Error bars indicate the SD; n = 3.
We previously have shown that HDAC6 inhibition by either tubacin or siRNA triggers growth inhibition in MM cells (4). WT161 inhibited cell growth more potently than tubacin (Fig. 3D and Fig. S2D). The IC50 values of WT161 and tubacin in MM1.S cells were calculated as 3.6 µM and 9.7 µM, respectively. WT161 induced significant toxicity in all MM cell lines tested, with IC50s between 1.5 and 4.7 µM (Fig. S2E). Overall, these data demonstrate that WT161 is a more potent inhibitor of HDAC6 than tubacin.
WT161 Enhances Proteasome Inhibitor-Induced Cytotoxicity in MM cells.
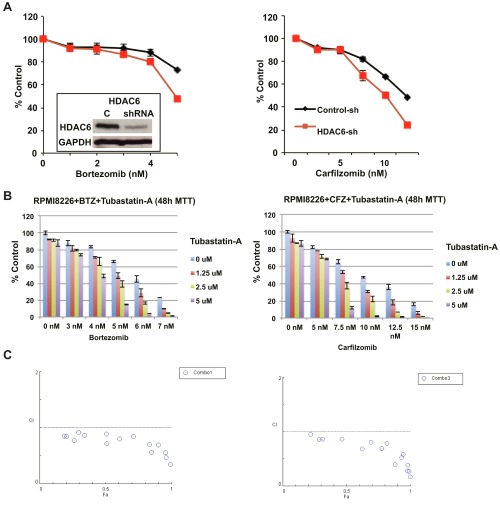
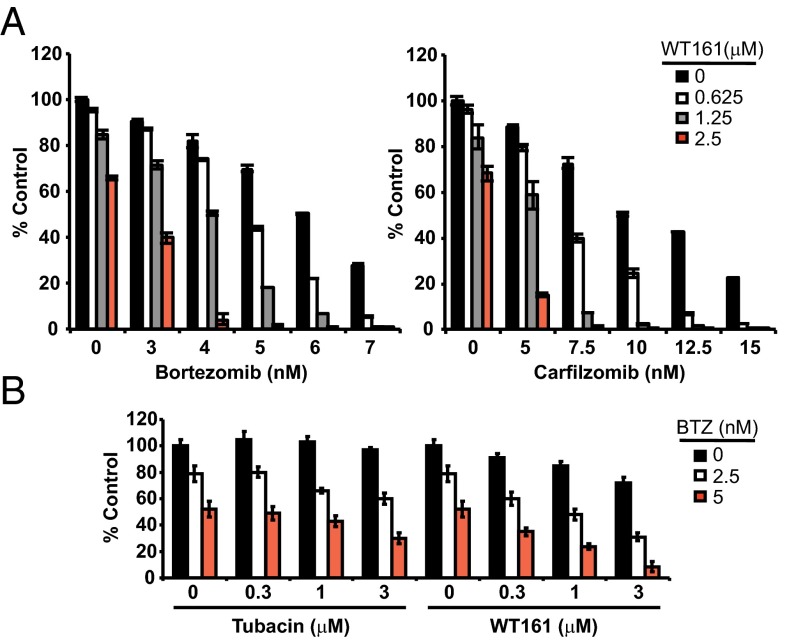
To investigate the cytotoxic effect of WT161 combined with the first- and second-generation proteasome inhibitors BTZ and CFZ, we first examined the effect of selective HDAC6 knockdown using lentiviral HDAC6 shRNA. HDAC6 knockdown alone inhibited cell growth, and the inhibition was significantly enhanced by both BTZ and CFZ treatments (Fig. S3A). If HDAC6 knockdown can enhance BTZ and CFZ cytotoxicity, we would expect WT161 to exhibit similar effects. Indeed, both BTZ and CFZ triggered growth inhibition that was further enhanced with increasing concentrations of WT161 (Fig. 4A). To demonstrate the synergy between dual proteasomal and aggresomal inhibition further, we tested BTZ and CFZ in combination with an additional HDAC6-selective inhibitor, tubastatin A (Fig. S3B). Although tubastatin A was slightly less potent than WT161, it displays synergism with both BTZ and CFZ, as calculated by isobologram analysis with a combination index (CI) less than 1 (Fig. S3C).
Fig. S3.
Both genetic and chemical inhibition of HDAC6 enhances proteasome inhibitor-induced cytotoxicity. (A) RPMI8226 cells were infected with lentiviral control or HDAC6 shRNAs. (Inset) Whole-cell lysates were subjected to immunoblotting to confirm HDAC6 knockdown. Cells were cultured for 48 h with the indicated concentrations of BTZ (Left) or CFZ (Right). Relative cell growth was assessed by MTT assay; n = 4. (B) RPMI8226 cells were cultured with BTZ (Left) or CFZ (Right) in the presence of Tubastatin A (1.25–5 µM) for 48 h. Cell growth inhibition was assessed by MTT assay. Data represent the mean ± SD from triplicate cultures. (C) The CI was calculated using CompuSyn software.
Fig. 4.
HDAC6 inhibition enhances BTZ-induced cytotoxicity in MM cells. (A) RPMI8226 cells were cultured for 48 h with BTZ (Left) or CFZ (Right) in the absence or presence of WT161; n = 3. (B) MM.1S cells were cultured for 24 h with tubacin or WT161 in the absence or presence of BTZ. All data represent mean ± SD.
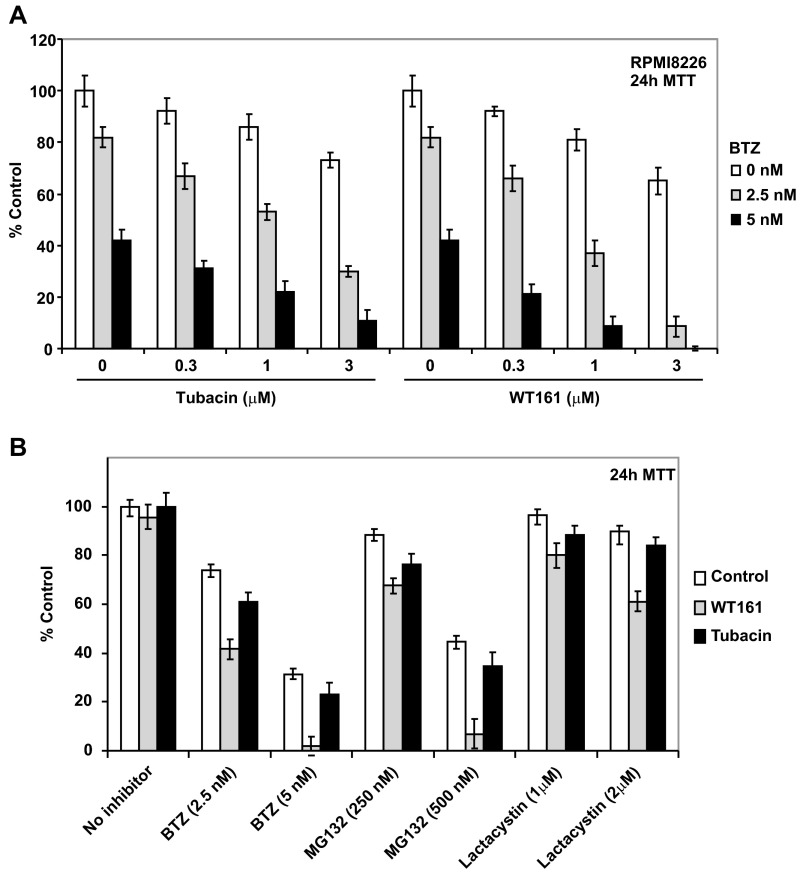
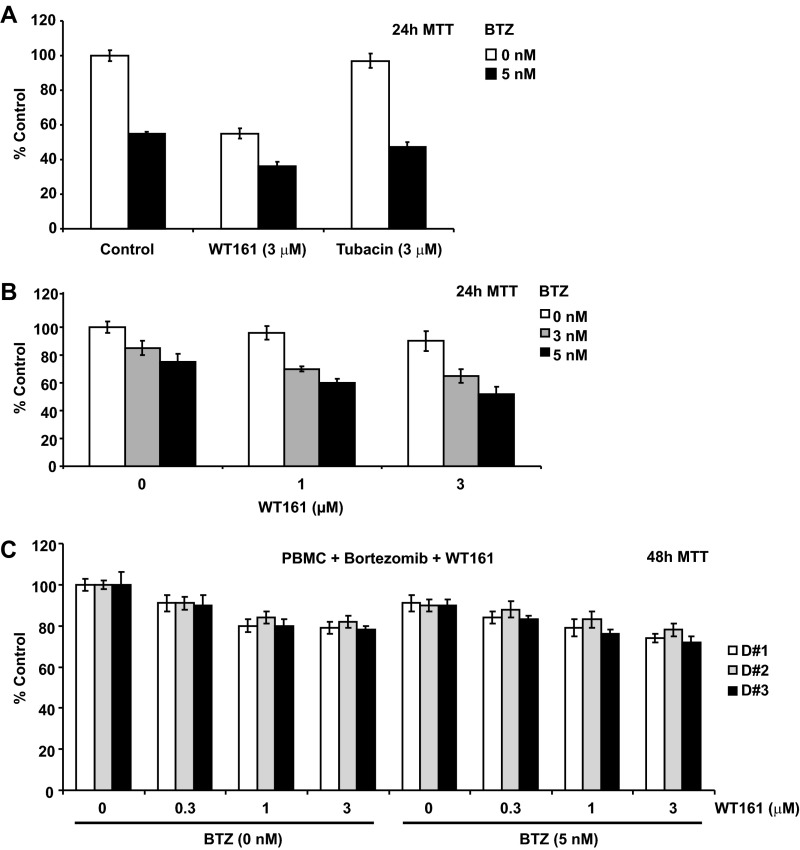
Accordingly, as shown in Fig. 4B and Fig. S4A, WT161 enhanced BTZ-induced cytotoxicity more strongly than tubacin. Moreover, WT161 enhanced the cytotoxicity induced by other classes of proteasome inhibitors (MG132 and lactacystin) more potently than tubacin (Fig. S4B). Last, WT161 enhanced BTZ-induced cytotoxicity in patient MM cells (Fig. S5 A and B). Importantly, this combination treatment did not induce toxicity in peripheral blood mononuclear cells (PBMCs) derived from healthy volunteers, thus suggesting a favorable therapeutic index (Fig. S5C).
Fig. S4.
The combination of BTZ with WT161 is more cytotoxic than with tubacin. (A) RPMI8226 cells were cultured with increasing concentrations of tubacin (Left) or WT161 (Right) in the absence or presence of 2.5 nM or 5 nM BTZ for 24 h. Cell growth inhibition was assessed by MTT assay. Data represent the mean ± SD, n = 3. (B) MM.1S cells were cultured with the indicated concentrations of the proteasome inhibitors BTZ, MG132, or lactacystin in the absence or presence of 1 µM tubacin or 1 µM WT161 for 24 h. Cell growth inhibition was assessed by MTT assay. Data represent mean ± SD; n = 3.
Fig. S5.
WT161 with BTZ induces significant cytotoxicity in patient tumor cells but not in normal PBMCs. (A and B) Primary tumor cells from MM patients were treated with BTZ at the indicated concentrations in the absence or presence of WT161 or tubacin for 24 h. A indicates the comparison of WT161 vs. Tubacin (fixed concentration) in the presence of BTZ (one dose), whereas B shows the combination of WT161 with BTZ at different concentrations. Moreover, these MM tumor cells are from different patients. (C) PBMCs from three healthy volunteers (D#1, D#2, and D#3) were cultured with 5 nM BTZ in the absence or presence of increasing concentrations of WT161 for 48 h. Cell growth inhibition was assessed by MTT assay. Data represent mean ± SD from triplicate experiments.
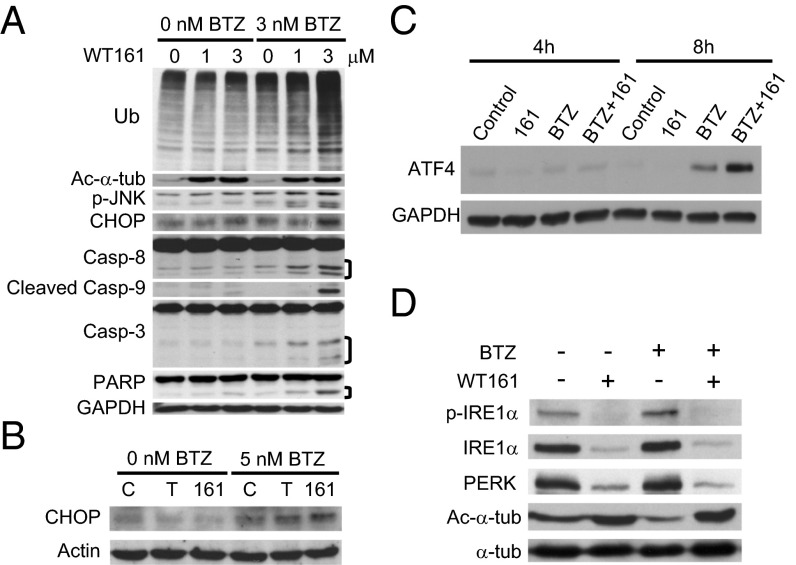
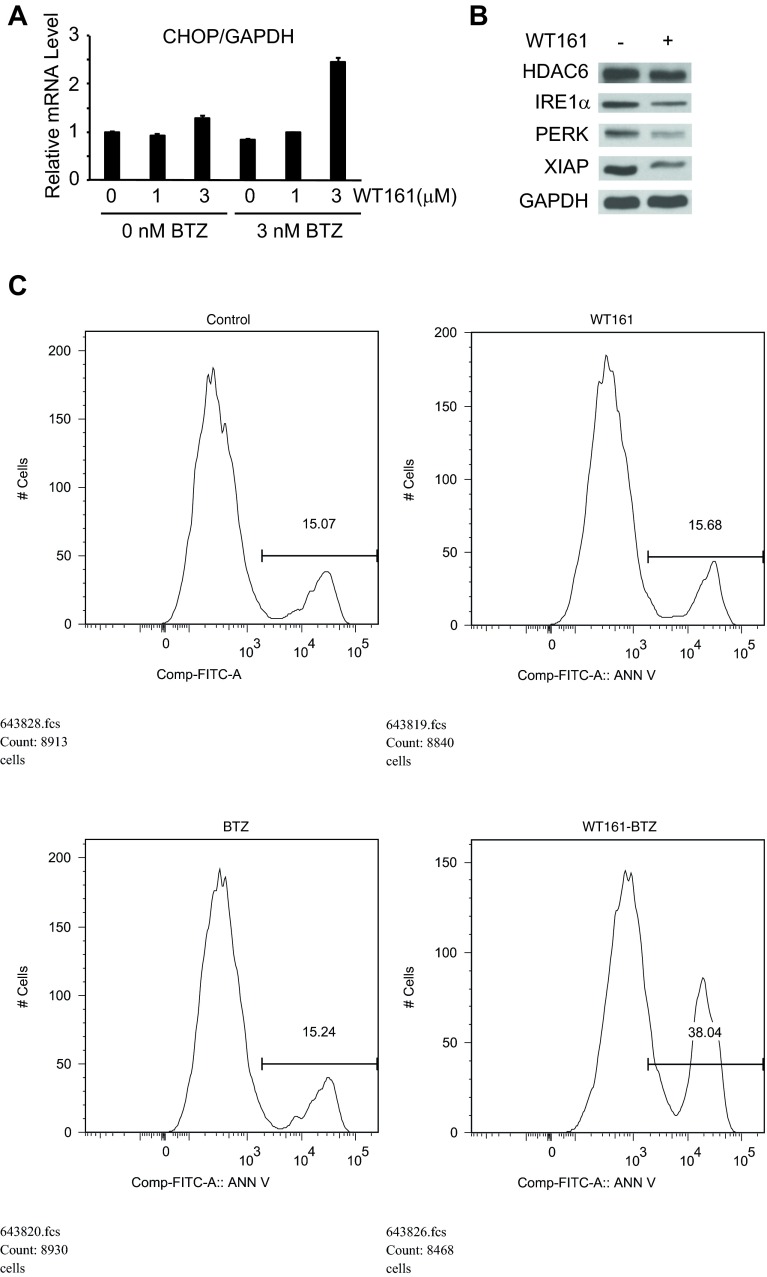
We next examined the molecular mechanisms whereby WT161 and BTZ induce synergistic cytotoxicity in MM cells. Consistent with our previous studies (4), treatment of MM.1S cells with BTZ and WT161 triggered increased accumulation of polyubiquitinated proteins and activation of the stress-activated protein kinase JNK relative to treatment with either drug alone (Fig. 5A). Accumulation of polyubiquitinated proteins induces endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) to restore normal ER function. As evident in MM1.S cells and in cells from MM patients treated ex vivo, treatment with both BTZ and WT161 triggered increased expression of CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP), a proapoptotic protein induced by the UPR (Fig. 5 A and B and Fig. S6A). Activating transcription factor 4 (ATF4) directly activates CHOP, and it, too, was markedly up-regulated after treatment of BTZ with WT161 (Fig. 5C) (19). If the ER stress is prolonged, apoptotic cell death ensues (19). We observed down-regulation of ER stress sensor proteins inositol-requiring enzyme 1α (IRE1α) and protein kinase R (PRKR)-like ER kinase (PERK) with WT161, both alone and with BTZ, in patient MM cells (Fig. 5D), suggesting that WT161 treatment impairs the UPR to augment further ER stress-induced death signaling. Finally, the antiapoptotic protein X-linked inhibitor of apoptosis (XIAP) was down-regulated with WT161 treatment (Fig. S6B), and combination treatment induced apoptosis as determined by the cleavage of caspases 3, 8, and 9 and poly(ADP-ribose) polymerase (PARP) and as confirmed by annexin V positivity (Fig. 5A and Fig. S6C) (20). Together, these results demonstrate that inhibition of proteasomal and aggresomal protein degradation with BTZ and WT161, respectively, effectively activates cell-stress signaling, resulting in apoptotic cell death in MM cell lines and in patient-derived cells.
Fig. 5.
WT161 enhances BTZ-induced ER stress and apoptotic cell death. (A) MM.1S cells were cultured with increasing concentrations of WT161 for 16 h in the presence or absence of BTZ. (B) Patient-derived MM cells were treated with DMSO (control; C), 2 µM tubacin (T), or 2 µM WT161 (161) in the absence or presence of BTZ for 24 h. (C) MM.1S cells were cultured with vehicle control, WT161 (3 μM), BTZ (3 nM), or BTZ+WT161 for 4 h and 8 h. Whole-cell lysates were subjected to immunoblotting with the indicated antibodies. (D) Patient-derived MM cells were treated with 5 nM BTZ ± 2 µM WT161 for 24 h. Whole-cell lysates were subjected to SDS/PAGE and immunoblotting with the indicated antibodies.
Fig. S6.
WT161 with BTZ induces apoptosis. (A) MM.1S cells were cultured with increasing concentrations of WT161 for 16 h in the presence or absence of BTZ and were analyzed by real-time quantitative PCR to assess CHOP mRNA levels. (B) Patient-derived MM cells were treated with 0 or 1 µM WT161 for 24 h. (C) MM.1S cells were treated with WT161 (3 μM) with or without BTZ (3 nM) for 16 h and were stained for annexin V.
Overcoming BM Microenvironment and BTZ-Induced Resistance.
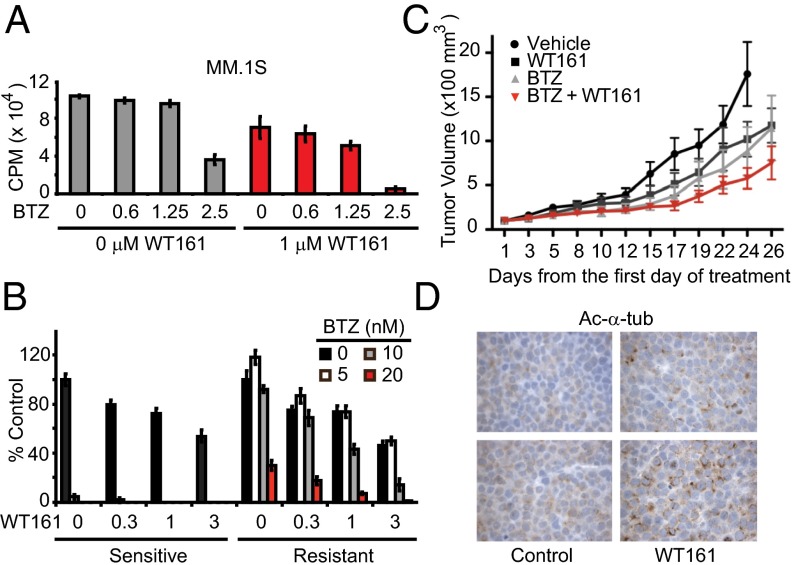
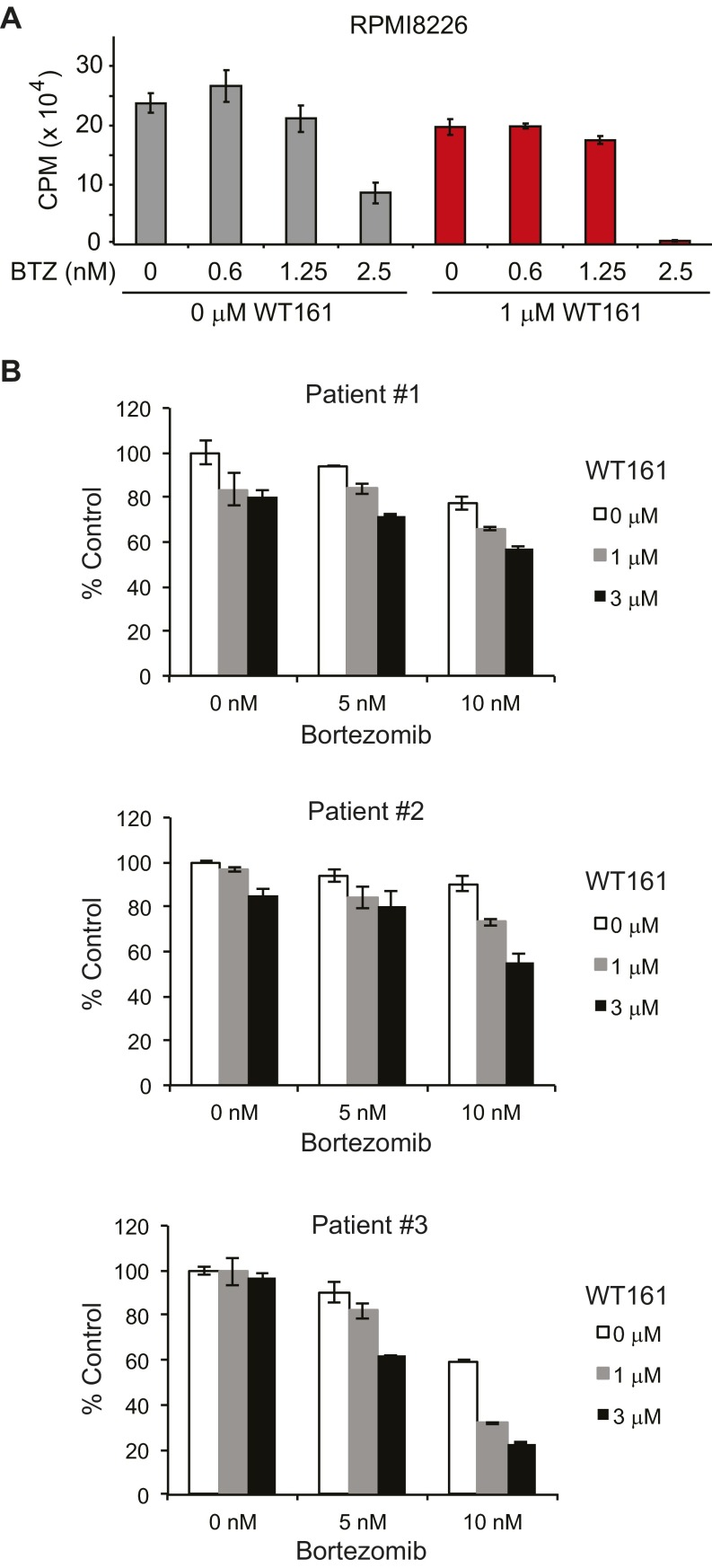
The BM microenvironment promotes MM cell proliferation and confers resistance to apoptosis (1). For example, dexamethasone-induced apoptosis in MM cells is completely abrogated by coculture with BM stromal cells (BMSCs). In MM–BMSC cocultures, BTZ alone inhibited cell growth, and the inhibition was further enhanced with WT161 (Fig. 6A and Fig. S7A).
Fig. 6.
Anti-MM activity of WT161 overcomes resistance and decreases tumor burden in vivo. (A) MM.1S cells were cocultured with BMSCs ± WT161 with increasing concentrations of BTZ (measured in nanomolars) for 24 h. Cell proliferation was assessed by [3H]-thymidine uptake; n = 3. CPM, counts per minute. (B) ANBL-6 (BTZ-sensitive) and ANBL-6-V5R (BTZ-resistant) cells were cultured with increasing concentrations of WT161 for 24 h in the absence or presence of increasing concentrations of BTZ. Cell growth was assessed by MTT assay; n = 3. (C) SCID mice were injected s.c. with 5 × 106 MM.1S cells and were treated with vehicle, WT161 (50 mg/kg, i.p.), BTZ (0.5 mg/kg, i.v.), or BTZ + WT161 for 26 d. Tumor volume (100 × mm3) was measured and calculated versus time (days); n = 9 mice per group. All data represent mean ± SD. (D) Immunostaining of Ac-α-tubulin in xenograft tissue from two control and two WT161-treated mice. Representative results are shown.
Fig. S7.
WT161 with BTZ is effective in overcoming resistance. (A) RPMI8226 cells were cocultured with BMSCs ± WT161 with increasing concentrations of BTZ for 24 h. Cell proliferation was assessed by [3H]-thymidine uptake; n = 3. (B) Myeloma tumor cells from patients resistant to BTZ treatment were treated with WT161 in the presence or absence of BTZ. Cytotoxicity was assessed by MTT assay from triplicate cultures. Data represent the mean ± SD.
Although BTZ has achieved responses and markedly improved outcomes in MM, drug resistance develops leading to relapse of disease in most patients. We therefore examined whether the combination of BTZ with WT161 can overcome acquired BTZ resistance. The BTZ-sensitive ANBL-6 cell line and the subline ANBL-6-V5R, which exhibits decreased response to BTZ, were cultured with BTZ, in the presence or absence of WT161. ANBL-6-V5R cells showed resistance to BTZ treatment compared with parental ANBL-6 BTZ-sensitive cells; importantly, this resistance was completely abrogated with increasing concentrations of WT161 (Fig. 6B). Additionally, WT161 was able to enhance cytotoxicity in patient-derived cells from three individuals who had developed resistance to BTZ treatment (Fig. S7B). Therefore, WT161 supplementation to BTZ treatment may abrogate resistance conferred by the microenvironment and BTZ-acquired resistance.
In Vivo Anti-MM Activity of WT161 with BTZ.
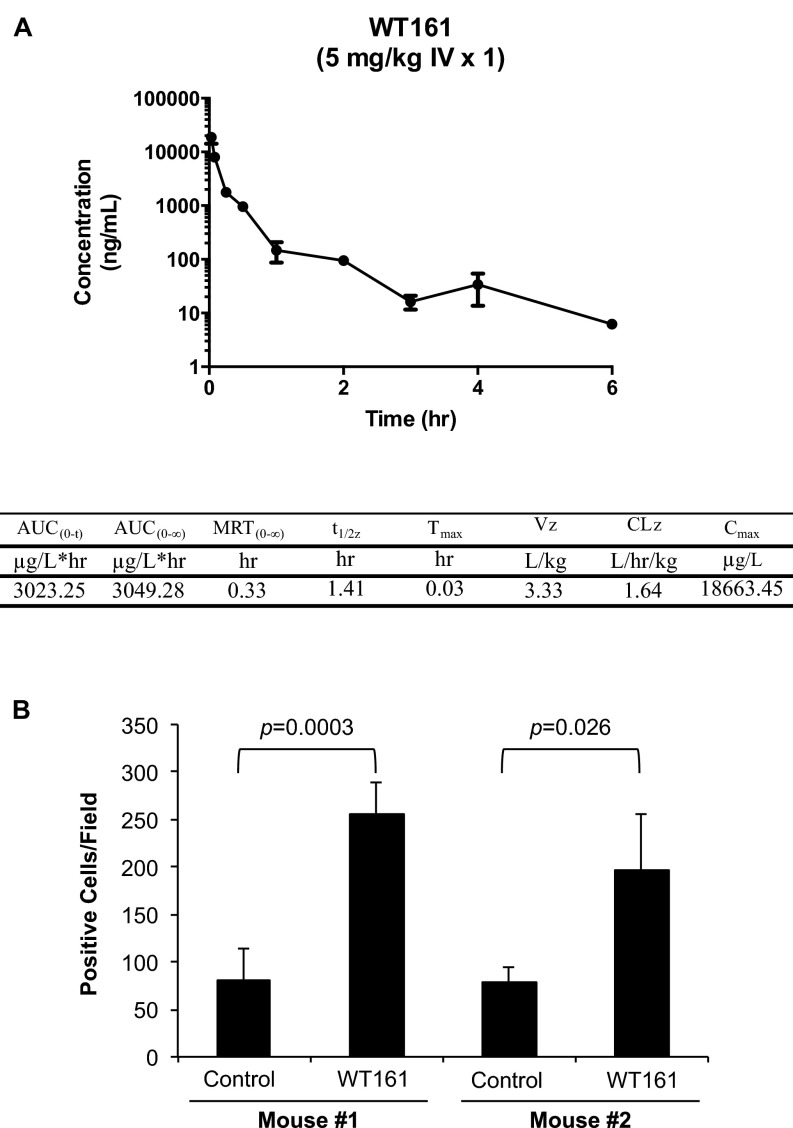
Finally, the relatively simple synthesis of WT161 provided sufficient material to examine the effect of WT161 with BTZ treatment in vivo. First, we evaluated the pharmacokinetic (PK) properties of WT161 (Fig. S8A). With reasonable half-life in mice (1.4 h) and drug exposure [maximum concentration (Cmax) = 18 mg/L], WT161 was formulated for i.p. administration. The i.p. dose, 50–100 mg/kg, was empirically determined based on the in vitro activity of WT161 in MM cell lines (IC50 = 1.5–4.7 µM) and drug exposure information obtained from the PK study (21). We observed toxicity with WT161 at 100 mg⋅kg−1⋅d−1 i.p., but at 50 mg⋅kg−1⋅d−1 i.p. WT161 was well tolerated as a single agent and in combination with BTZ. Next, we tested WT161 in combination with BTZ in a murine xenograft model of human MM (MM.1S). Tumor-bearing mice were randomized into four treatment groups: control, BTZ, WT161, or BTZ + WT161. Although there was no significant difference in tumor growth between the control and BTZ-treated (P = 0.07) or WT161-treated (P = 0.095) cohorts, BTZ combined with WT161 demonstrated a significant antitumor effect (P = 0.0078) (Fig. 6C). Moreover, there was no significant difference between BTZ or WT161 treatment and the combinatorial treatment (P = 0.327 and P = 0.079, respectively). The on-target activity of WT161 in vivo was confirmed by assessing Ac-α-tubulin levels in resected tumor samples (Fig. 6D and Fig. S8B). In conclusion, these data demonstrate that simultaneous inhibition of the proteasome and aggresome by BTZ and WT161, respectively, triggers significant anti-MM activities both in vitro and in vivo and represents a viable therapeutic option for the treatment of MM, especially proteasome inhibitor-resistant MM.
Fig. S8.
WT161 exhibited adequate PK exposure and efficacy in mice. (A) Male CD-1 mice (n = 3) were injected i.v. with WT161 at 5 mg/kg. Mean plasma concentration–time profiles were used to calculate drug exposure [area under the curve (AUC) = 3,049 ng⋅L−1⋅h−1], t1/2 = approximately 1.4 h, and Cmax = 18,663 ng/L. CLz, clearance; MRT, mean residence time extrapolated to infinity; t1/2z, terminal elimination half-life; Tmax, time to maximum concentration; Vz, volume of distribution at terminal phase. (B) Quantitative analysis of Ac-α-tubulin–positive cells in MM1.S xenograft tumors from two control and two WT161-treated mice. Data represent the mean ± SD.
Discussion
For target validation of HDAC6 in MM and for broader use by the biological community, we endeavored to create a potent, selective, and bioavailable HDAC6 inhibitor. WT161 was identified via biochemical and cellular screening from a hydrazone library containing ∼400 molecules. Biochemically, WT161 is most selective against HDAC6, and in cells WT161 effectively demonstrated the predicted cellular effects of HDAC6 inhibition, namely the accumulation of acetylation on α-tubulin but not histones. Our proof-of-concept work demonstrated that HDAC6 inhibition by either siRNA knockdown or tubacin was cytotoxic to MM cells, so we tested WT161 for similar effects. Notably, we determined that WT161 can induce acetylation of α-tubulin and cell death efficiently in MM cell lines and in patient samples earlier and at lower doses than tubacin (4).
The structural origin of the HDAC6 selectivity of WT161 was traced to the differences in the shape of the protein surface adjacent to the binding site. The HDAC6 protein contains a large lipophilic pocket adjacent to the active site that is unique to this protein. The large pendent unit of tubacin fills this lipophilic pocket but precludes it from binding to alternate HDAC family proteins. Based on this design principle, we developed WT161 to have a zinc-binding motif and a hydroxymate head to bind the enzyme’s active site connected by a linker to a large triphenyl amine motif designed to bind with the lipophilic pocket of HDAC6. The simplified triphenyl amine structure has successfully achieved better shape complementarity than tubacin and avoids the complicated synthesis of this pendent unit. WT161, which has no stereocenters, can be synthesized in large quantities for further in-depth in vitro and in vivo experimentation.
Resistance mechanisms are the largest hurdle to treating and effectively curing MM and can persist initially or emerge in the course of treatment. Thus, to examine the synergistic effects of proteasomal and aggresomal inhibition in MM, we tested WT161 with the proteasome inhibitors BTZ and CFZ. Excitingly, WT161 was able to enhance both BTZ and CFZ cytotoxic effects in MM cell lines and patient samples, with no effect on PBMCs. With this combinatorial treatment, we observed the accumulation of ubiquitinated proteins, ER stress and induction of the UPR, activation of stress signaling (JNK activation), and cleavage of caspases followed by apoptotic cell death. Interestingly, WT161 as a single agent does not induce ER stress, the UPR, or ER stress-mediated apoptosis. We did observe down-regulation of the antiapoptotic protein XIAP and the ER stress-sensor proteins PERK and IRE1α with WT161 after 24-h treatment. This down-regulation is also observed after 24 h of exposure of MM cells to CFZ (22). At first glance, this effect seems counterintuitive, but activation of PERK and IRE1α not only promotes cellular adaptation to/survival of ER stress but also actively inhibits the ER stress-induced apoptotic program. Thus, we believe down-regulation is required to impair the UPR and to augment ER stress-induced apoptosis. Many candidate proteins are involved in orchestrating the switch from the protective UPR signaling to proapoptotic signaling. Some of these proteins, such as P58IPK, GADD34, and TRB3, are involved in shutting down the PERK-mediated pathway (19). In the combination treatment, BTZ induces ER stress and the UPR, and WT161 down-regulates antiapoptotic proteins and the proteins required for adaptation and survival of ER stress. These distinct activities combine to tip the balance toward apoptosis more strongly than when either agent is used alone.
Additionally, components of the BM microenvironment are well known to play critical roles in MM cell survival and environment-mediated drug resistance (1). Using a cellular assay to mimic MM in its microenvironment, we observed that the WT161–BTZ combination treatment could overcome the resistance conferred by BMSCs. Finally, the combination of BTZ and WT161 was effective against BTZ-resistant MM cell lines and samples derived from patients who had developed BTZ resistance. The molecular mechanisms conferring BTZ resistance in MM have not yet been fully elucidated, but our results suggest that HDAC6 inhibitors in combination with BTZ could overcome clinical BTZ resistance in MM.
The easy and cost-effective synthesis of WT161 enabled us to study HDAC6 inhibition in combination with proteasome inhibition in a preclinical mouse model of human MM. We observed a significant inhibitory effect on tumor growth with the WT161–BTZ combinatorial treatment. Our studies provide the framework for clinical evaluation of combined targeted therapy to achieve dual blockade of proteasomal and aggresomal protein degradation and to improve patient outcomes in MM. Importantly, WT161 has been further refined to develop ACY-1215 (ricolinostat), a hydroxamic acid HDAC6 inhibitor with similar anti-MM activities, which has moved rapidly from preclinical to promising phase I/II clinical trials in relapsed/refractory MM (23, 24). Unlike preclinical studies, single-agent activity of ricolinostat in clinical trials is possibly restrained by its limited PK properties. Specifically, the Cmax of ricolinostat in serum at a dose of 160 mg reached 1.1 μM, which may not be sufficient to induce direct anti-MM activities. A single dose of WT161 at 5 mg/kg i.v. reaches a Cmax in serum of ∼40 μM.
HDAC6/aggresome inhibition may be more broadly applicable to cancers other than MM. Malignant cells generally have higher protein synthesis rates than their normal counterparts, making them more prone to protein aggregation and perhaps more sensitive to proteasome inhibitor-induced apoptosis. Inhibition of proteasome activity has been demonstrated to induce proapoptotic ER stress in pancreatic carcinoma (25), head and neck cancer (26), and nonsmall cell lung carcinoma (27).
Mechanistic and translational biology has moved in vivo, and so must the field of chemical biology. The availability of tubacin has provided significant insights into HDAC6 biology. Since the development of tubacin, additional HDAC6 inhibitors have been described, such as tubastatin A and, more recently, N-hydroxy-4-{[N-(2-hydroxyethyl)-2-phenylacetamido]methyl} benzamide (HPB) and an aminopyrrolidinone HDAC6 inhibitor (28–30). When these molecules are validated in vivo, we anticipate an informative and structurally diverse set of chemical probes of HDAC6 to drive further mechanistic exploration and definitive clinical translation in cancer and nonmalignant diseases.
Materials and Methods
All experiments with patient samples were performed according to a protocol approved by the Institutional Review Board of Dana-Farber Cancer Institute. All animal studies were conducted according to protocols approved by the Animal Ethics Committee of the Dana-Farber Cancer Institute. Mice used in this study were handled in compliance with the NIH Guide for the Care and Use of Laboratory Animals (31).
Reagents and General Synthetic Procedure.
Tubacin was synthesized in the J.E.B. laboratory (32). BTZ, CFZ, tubastatin A, and panobinostat were purchased from Selleck Chemicals. Antibodies used in this study were purchased directly from the vendors listed in SI Materials and Methods. Detailed synthetic methods can be found in SI Materials and Methods and as previously reported by Tang et al. (33). All reactions were performed and monitored by LCMS. The intermediates and final product were fully characterized with proton and carbon-13 NMR (1H NMR and 13C NMR) spectra and high-resolution mass spectra (HRMS). Compounds were biochemically profiled against HDAC1–9 as previously reported (14).
Cell Lines.
MM.1S, NCI-H929, RPMI8226, and U266 cells were obtained from American Type Culture Collection (ATCC). The KMS11 cell line was obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank. OPM-2 cells were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (German Collection of Microorganisms and Cell Cultures). ANBL-6 and ANBL-6-VR5 cell lines were kindly provided by Robert Orlowski, MD Anderson Cancer Center, Houston, TX.
Statistical Analysis.
The statistical significance of differences observed in drug-treated versus control cultures was determined using the Wilcoxon signed-ranks test.
SI Materials and Methods
Instrumentation.
Proton and carbon-13 NMR (1H NMR and 13C NMR) spectra were recorded with a Varian inverse probe 600 INOVA spectrometer at the Harvard Medical School East Quad NMR Facility. Chemical shifts are reported in parts per million on the δ scale and are referenced from the residual protium in the NMR solvent (DMSO-d6: δ 2.50) for 1H NMR and the carbon resonances of the solvent (DMSO-d6: δ 40.0) for 13C NMR. Data are reported as follows: chemical shift [multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad), coupling constant(s) in Hertz, integration]. High-resolution mass spectra (HRMS) were recorded on a Bruker APEX 4.7 Tesla Fourier transform mass spectrometer using electrospray ion source (ESI) at the Instrumentation Facility of the Department of Chemistry, Massachusetts Institute of Technology. The intermediates and final product were purified with a CombiFlash RF system (Teledyne Isco). Organic solutions were concentrated on Büchi R-205 rotary evaporators.
Synthetic Procedure.
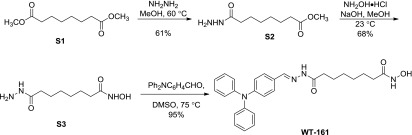
Scheme S1.
Synthesis of the WT161.
Methyl 8-Hydrazinyl-8-Oxooctanoate, Compound S2.
Hydrazine (0.78 mL, 25.0 mmol, 1 equivalent) was added to a solution of dimethyl suberate, compound S1 (10.1 g, 50.0 mmol, 2.0 equivalent), in methanol (25 mL, 2.0 M) at 23 °C under nitrogen. The mixture then was heated to 60 °C. After 16 h, the reaction mixture was cooled to 23 °C, and solvents were removed under reduced pressure. The residue was purified by flash column chromatography (40 g silica gel, 15% methanol-methylene chloride) to afford methyl 8-hydrazinyl-8-oxooctanoate, compound S2 (3.06 g, 61%) as a white solid.
1H NMR (600 MHz, DMSO-d6, 25 °C) δ 8.87 (br, 1H), 4.11 (br, 2H), 3.56 (s, 3H), 2.26 (t, J = 7.2 Hz, 2H), 1.97 (t, J = 7.2 Hz, 2H), 1.50–1.42 (m, 4H), 1.25–1.18 (m, 4H).
13C NMR (150 MHz, DMSO-d6, 25 °C) δ 174.0, 172.2, 51.8, 33.8, 33.7, 28.7, 28.6, 25.5, 24.8.
HRMS (ESI) calculated for C9H18N2O3Na [M+Na]+: 225.1210; found: 225.1201 m/z.
TLC (15% methanol-methylene chloride), retention factor: 0.32 (iodine vapor).
8-Hydrazinyl-N-Hydroxy-8-Oxooctanamide, Compound S3.
Hydroxylamine hydrochloride (1.98 g, 28.00 mmol, 2.00 equivalent) was added to NaOH solution in methanol (2.28 g, 56.00 mmol, 4.00 equivalent in 14.00 mL methanol, 4.0 M). The mixture was stirred at 23 °C for 5 min and then was added to a solution of compound S2 in methanol (2.88 g, 14.00 mmol, 1 equivalent in 14.00 mL methanol, 1.00 M). The reaction mixture was stirred at 23 °C. After 16 h, hydroxylamine hydrochloride salt (1.98 g, 28.00 mmol, 2.00 equivalent) was added to neutralize the excess base. The precipitate was filtered, and the resulted filtrate was concentrated under reduced pressure. The residue was recrystallized from methanol to afford 8-hydrazinyl-N-hydroxy-8-oxooctanamide, compound S3 (1.58 g, 55%) as a white solid.
1H NMR (400 MHz, DMSO-d6, 25 °C) δ 10.30 (br, 1H), 8.88 (s, 1H), 8.62 (br, 1H), 4.21 (br, 2H), 1.96 (t, J = 8.0 MHz, 2H), 1.90 (t, J = 8.0 Hz, 2H), 1.51–1.36 (m, 4H), 1.27–1.12 (m, 4H).
13C NMR (100 MHz, DMSO-d6, 25 °C) δ 172.0, 169.5, 33.8, 32.7, 28.8, 28.7, 25.6, 25.5.
HRMS (ESI) calculated for C8H17N3O3Na [M+Na]+: 226.1162, found 226.1158 m/z.
(E)-8-(2-(4-(Diphenylamino)Benzylidene)Hydrazinyl)-N-Hydroxy-8-Oxooctanamide, WT161.
4-(Diphenylamino)-benzaldehyde (134.0 mg, 0.5 mmol, 1.0 equivalent) was added to the solution of compound S3 (101.0 mg, 0.5 mmol, 1 equivalent) in DMSO (5.0 mL, 0.1 M), at 23 °C. The mixture was heated to 75 °C for 24 h. The crude product was purified by reverse-phase preparatory HPLC (20 mL/min, 5–95% CH3CN-H2O with 0.1% formic acid, 25-min gradient) to provide WT161 (217 mg, 95%) as a yellow solid.
1H NMR (400 MHz, DMSO-d6, 25 °C) δ 11.16 (s, 1H), 11.05 (s, 1H), 10.34–10.25 (m, 2H), 8.61 (br, 2H), 8.05 (s, 1H), 7.86 (s, 1H), 7.50 (t, J = 8.0 Hz, 4H), 7.36–7.27 (m, 8H), 7.13–7.00 (m, 12H), 6.92 (d, J = 8.0 Hz, 4H), 2.54 (t, J = 8.0 MHz, 2H), 2.50–2.45 (m, 8H), 2.14 (d, J = 8.0 Hz, 2H), 1.95–1.85 (m, 4H), 1.58–1.40 (m, 8H), 1.31–1.17 (m, 8H).
13C NMR (100 MHz, DMSO-d6, 25 °C) δ 174.5, 169.5, 149.2, 148.9, 147.1, 147.0, 145.9, 142.5, 130.2, 130.1, 128.6, 128.3, 128.1, 125.3, 125.1, 124.4, 124.2, 122.3, 121.9, 32.7, 32.2, 28.9, 28.8, 25.5, 25.4, 24.6.
HRMS(ESI) calculated for C27H31N4O3 [M+H]+: 459.2391; found: 459.2393 m/z.
High-Content Imaging Assay.
This assay was adapted from previously described cytoblot assays (15, 18). A431 cells were plated at 2,500 cells per well in 50 μL media in 384-well clear-bottomed plates (Corning 3712;) and were incubated overnight at 37 °C. Compounds were added using a Janus Workstation and were incubated at 37 °C for 4 h. Cells then were fixed in 3.7% formaldehyde in PBS at RT for 10 min. After two rinses with blocking solution (1% BSA in PBS), 1% SDS in PBS was added for 2 min. After one washing, cells were incubated with blocking solution for 1 h at RT. Cells then were incubated either for 1 h at RT or overnight at 4 °C in 10 μL of primary antibody for acetylated tubulin (Sigma T7451) and acetylated lysine (Cell Signaling 9441L) at a 1:500 dilution in blocking solution. After rinsing with blocking solution, cells were incubated for 1 h at RT in 10 μL of secondary fluorescent antibody (Invitrogen A-21202, A-21244) and nuclear staining (Invitrogen H3570) solution at 1:1,000 dilution in blocking solution. Cells were washed twice; then 50 μL of PBS was added to each well. Images were acquired on a high-content screening microscope (ImageXpress Micro; Molecular Devices), and image analysis (MetaXpress3.0; Molecular Devices) was performed to obtain an average acetylated tubulin and acetylated lysine signal per cell. Dose–response data (normalized to SAHA and DMSO) were generated (GraphPad Prism) by normalization of maximum and minimum fluorescence.
Biochemical HDAC Assay.
The inhibition of HDAC proteins (HDAC1–9) was determined using a homogenous, kinetic assay, as reported previously (14).
Computational Studies.
MD simulations were carried out using the PMEMD version included in the AMBER10 suite of programs (34) after careful relaxation of the system using minimization and equilibration protocols in analogy to our previous work (16). The initial coordinates for the protein atoms were taken from homology models of HDAC3 and HDAC6 and the X-ray structure of HDAC2 [Protein Data Bank (PDB) ID code 3MAX]. The homology models were generated using Prime, with the X-ray structures of HDAC2 as a template for HDAC3, and of HDAC7 (PDB ID code 3C10) for HDAC6. The ligand was placed in the active site pocket by superposition of the binding moiety with available structures of SAHA bound to the HDAC8 isoform. According to previous computational studies (35), the hydroxamic ligand was modeled in a negatively charged state. The ionizable residues were set to their normal ionization states at pH 7, except for the His residue that is hydrogen bonded to the negatively charged hydroxamate ligand, which has been modeled as positively charged. The protein atoms and all the water molecules of the crystal structure were surrounded by a periodic box of TIP3P water molecules that extended 10 Å from the protein. Na+ counterions were placed by LEaP to neutralize the system. A bonded approach was used to model the Zn-containing active site.
The ff03 version of the all-atom AMBER force field was used to model the protein, and the general AMBER force field (GAFF) was used for the organic ligand. Atom-centered partial charges were derived by using the AMBER antechamber program (restrained electrostatic potential methodology), after geometry optimization at the B3LYP/6–31G* level. In the MD simulation protocol, the time step was chosen to be 2 fs, and the SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. A nonbonded cutoff of 10.0 Å was used, and the nonbonded pair list was updated every 25 time steps. Langevin dynamics was used to control the temperature (300 K) using a collision frequency of 1.0/ps, with isotropic position scaling to maintain the pressure (1 atm). Periodic boundary conditions were applied to simulate a continuous system. To include the contributions of long-range interactions, the Particle-Mesh-Ewald (PME) method was used with a grid spacing of ∼1 Å combined with a fourth-order B-spline interpolation to compute the potential and forces between grid points. The trajectories were analyzed using the PTRAJ module of AMBER.
Patient MM Cells and BM Stromal Cells.
Patient MM cells were purified from BM aspirates by negative selection using an antibody mixture according to the manufacturer's protocol (RosetteSep Separation System; STEMCELL Technologies). Mononuclear cells separated by Ficoll-Hypaque density sedimentation from BM aspirates were also used to establish long-term BM stromal cells. All experiments with patient samples were performed according to a protocol approved by the Institutional Review Board of Dana-Farber Cancer Institute.
All MM cell lines were cultured in RPMI 1640 medium containing 10% FBS (Sigma Chemical Co.), 2 μM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO). ANBL and ANBL-VR5 cells were maintained with the addition of IL-6 (1 ng/mL) and IL6 + BTZ (5 nM), respectively. Cell growth was assessed by measuring MTT dye absorbance or the uptake of [3H]-thymidine, as previously described (36).
Immunoblotting.
Cells cultured with WT161, BTZ, and CFZ were harvested, washed, and lysed using lysis buffer: 50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, 5 mM EDTA, 5 mM EGTA, 5 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and complete protease inhibitor mixture (Roche Diagnostics). Whole-cell lysates were subjected to SDS/PAGE, transferred to nitrocellulose membrane (Bio-Rad Laboratories), and immunoblotted with specific antibodies. Antibodies against acetylated α-tubulin (Sigma), p-IRE1α (Affinity Bioreagents), and actin (Santa Cruz Biotechnology) were purchased. Antibodies against HDAC6, acetylated H3K9, histone H3, acetylated lysine, α-tubulin, GAPDH, ubiquitin, p-JNK, CHOP, caspase 8, caspase 3, PARP, XIAP, PERK, IRE1α, and GAPDH were purchased from Cell Signaling Technology.
Statistical Analysis.
Biochemical inhibition of HDAC enzymes by small-molecule inhibitors was measured as described above. Data were analyzed by logistic regression with determination of IC50 and 95% confidence intervals (GraphPad Prism). IC50 values were derived from nonlinear curve fit of the dose–response data using the least-squares (ordinary) fit, variable slope model. The bottom was constrained to 0, the top was constrained to 100, and the means of replicate data are depicted. Statistically significance of differences observed in drug-treated versus control cultures were determined using the Wilcoxon signed-ranks test. The minimal level of significance was P < 0.05. The interaction between WT161 and BTZ was analyzed by isobologram analysis using the CalcuSyn software program (Biosoft) to determine whether the combination was additive or synergistic; a CI <1.0 indicates a synergistic effect, as previously described (36). For in vivo experiments, tumor volumes were compared using one-way ANOVA and Bonferroni post hoc tests.
Flow Cytometric Analysis.
For annexin V-PI staining, MM.1S cells cultured for 16 h with BTZ (3 nM) and/or tubacin/WT161 (3 µM) were washed with PBS and processed according to the manufacturer’s protocol (FITC Annexin V Apoptosis Detection Kit I; BD Pharmingen).
BMSC Cocultures.
To evaluate the effect of combined WT161 and BTZ treatment on the growth of MM cells in the context of the BM microenvironment, MM.1S and RPMI8226 cells were cultured in BMSC-coated 96-well plates in the presence or absence of WT161 and/or BTZ. Cell proliferation was measured by [3H]-thymidine (Perkin-Elmer) uptake. All experiments were performed in quadruplicate.
HDAC6 Knockdown by Lentiviral shRNA.
Scrambled and HDAC6 pLKO shRNA vectors were kindly provided by the Broad Institute RNAi Consortium. Recombinant lentivirus was produced by transient transfection of 293T cells following a standard protocol. After 48 h, MM cells were incubated for 6 h with culture supernatants from 293T cells containing crude virus and then were washed with medium. A second transfection was repeated on the next day. Forty-eight hours later MM cells were counted and examined for cell proliferation and by immunoblotting analysis.
Real-Time Quantitative PCR.
Real-time quantitative PCR was performed on an ABI Prism 7300 Sequence Detection System (Applied Biosystems). Primer sequences were as follows: human CHOP, 5′-AGAACCAGGAAACGGAAACAGA-3′ and 5′-TCTCCTTCATGCGCTGCTTT-3′; and GAPDH, 5′- CTCCTCTGACTTCAACAGCGACAC-3′ and 5′-TGCTGTAGCCAAATTCGTTGTCAT-3′. Thermal cycling conditions were 10 min at 95 °C, 40 cycles at 95 °C for 15 s, followed by 1 min at 60 ′C.
PK Studies.
Male CD1 mice (n = 3) were injected i.v. with one dose of WT161 at 5 mg/kg. The concentration of WT161 in plasma was determined by LC-MS/MS via serial bleeds at different time points. Blood plasma concentration versus time was plotted and used to determine the half-life of the compound using noncompartmental analysis.
Xenograft Murine Model.
Beige nude X-linked immune-deficient (XID) (BNX) mice (5–6 wk old) were obtained from the Frederick Cancer Research and Development Center. All animal studies were conducted according to protocols approved by the Animal Ethics Committee of the Dana-Farber Cancer Institute. The mice were inoculated s.c. in the right flank with 5 × 106 MM.1S cells in 100 µL RPMI 1640 and 100 µL Matrigel basement membrane matrix (Becton Dickinson). When tumors were measurable (∼100 mm3), mice were assigned into cohorts receiving vehicle (control), BTZ (0.5 mg/kg, i.v.), WT161 (50 mg/kg, i.p.), or BTZ + WT161. WT161 was administered for five consecutive days each week, and BTZ was given on a twice-weekly schedule. Caliper measurements of the longest perpendicular tumor diameters were performed on alternate days to estimate the tumor volume, using the following formula representing the 3D volume of an ellipse: 4π/3 × (width/2)2 × (length/2). Animals were killed when tumors reached 2 cm3 or earlier if the animal appeared moribund to prevent unnecessary morbidity. Tumor growth was evaluated using caliper measurements from the first day of treatment until the day the animal was killed, which was day 15 for controls, day 32 for animals treated daily, and day 30 for animals treated weekly. Mice used in this study were handled in compliance with the NIH Guide for the Care and Use of Laboratory Animals (31).
Immunohistochemistry.
Paraffin-embedded tumor sections were incubated with Ac-α-tubulin (Sigma Aldrich). The primary antibodies were visualized with the corresponding biotinylated antibody coupled to streptavidin–peroxidase complex (Vector Laboratories). All antibodies, conditions, and reactivities were tested in positive control slides. Histologic micrographs were taken using a Leica DM200 microscope (aperture HC PLANs 10×/22, objective lenses: N PLAN 100×/1.25 oil) and a SPOT Insight QE Model camera with SPOT Advanced acquisition software (Diagnostic Instruments).
To quantitate immunohistochemical staining, at higher magnification cells from four fields of resected tumor (control and WT161-treated) with strong or moderate brown staining were counted as positive Ac-α-tubulin cells. All assessments were performed by three independent investigators.
Acknowledgments
This study was supported by NIH Grants SPORE CA10070, P01 78378, and R01 CA50947 (to K.C.A.), R01 CA178264 (to T.H. and K.C.A.), R01 GM38627 (to S.L.S.), K08 CA128972 (to J.E.B.), T32 HL07623-18 (to J.E.B.), and R01 CA152314 (to J.E.B. and O.W.); a Burroughs-Wellcome Foundation Career Award for Medical Scientists (to J.E.B.); the Doris Duke Charitable Foundation (J.E.B.); and National Science Foundation Grant TG-CHE090124 (to O.W.). K.C.A. is an American Cancer Society Clinical Research Professor.
Footnotes
Conflict of interest statement: T.H. is a consultant for Acetylon Pharmaceuticals. R.M. has financial interests in SHAPE Pharmaceuticals and Acetylon Pharmaceuticals and is the inventor on intellectual property licensed to these two entities. K.C.A. is an advisor for Celgene, Millennium Pharmaceuticals, and Gilead Sciences and is a Scientific Founder of OncoPep, Acetylon Pharmaceuticals, and C4 Therapeutics. J.E.B. is a Scientific Founder of SHAPE Pharmaceuticals, Acetylon Pharmaceuticals, Tensha Therapeutics, and C4 Therapeutics and is the inventor on intellectual property licensed to these entities. As of January 1, 2016, J.E.B. is an employee of the Novartis Institutes of Biomedical Research.
This article is a PNAS Direct Submission. R.P. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608067113/-/DCSupplemental.
References
- 1.Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7(8):585–598. doi: 10.1038/nrc2189. [DOI] [PubMed] [Google Scholar]
- 2.Badros AZ, et al. Carfilzomib in multiple myeloma patients with renal impairment: Pharmacokinetics and safety. Leukemia. 2013;27(8):1707–1714. doi: 10.1038/leu.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Gomez A, et al. Preclinical activity of the oral proteasome inhibitor MLN9708 in Myeloma bone disease. Clin Cancer Res. 2014;20(6):1542–1554. doi: 10.1158/1078-0432.CCR-13-1657. [DOI] [PubMed] [Google Scholar]
- 4.Hideshima T, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci USA. 2005;102(24):8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stessman HA, et al. Profiling bortezomib resistance identifies secondary therapies in a mouse myeloma model. Mol Cancer Ther. 2013;12(6):1140–1150. doi: 10.1158/1535-7163.MCT-12-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichter DI, et al. Sequence analysis of β-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood. 2012;120(23):4513–4516. doi: 10.1182/blood-2012-05-426924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hideshima T, Richardson PG, Anderson KC. Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol Cancer Ther. 2011;10(11):2034–2042. doi: 10.1158/1535-7163.MCT-11-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17(3):351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18(5):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Dimopoulos M, et al. Vorinostat or placebo in combination with bortezomib in patients with multiple myeloma (VANTAGE 088): A multicentre, randomised, double-blind study. Lancet Oncol. 2013;14(11):1129–1140. doi: 10.1016/S1470-2045(13)70398-X. [DOI] [PubMed] [Google Scholar]
- 11.San-Miguel JF, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: A multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15(11):1195–1206. doi: 10.1016/S1470-2045(14)70440-1. [DOI] [PubMed] [Google Scholar]
- 12.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6(3):159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 13.Sternson SM, Wong JC, Grozinger CM, Schreiber SL. Synthesis of 7200 small molecules based on a substructural analysis of the histone deacetylase inhibitors trichostatin and trapoxin. Org Lett. 2001;3(26):4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 14.Bradner JE, et al. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci USA. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estiu G, et al. Structural origin of selectivity in class II-selective histone deacetylase inhibitors. J Med Chem. 2008;51(10):2898–2906. doi: 10.1021/jm7015254. [DOI] [PubMed] [Google Scholar]
- 17.Vegas AJ, et al. Fluorous-based small-molecule microarrays for the discovery of histone deacetylase inhibitors. Angew Chem Int Ed Engl. 2007;46(42):7960–7964. doi: 10.1002/anie.200703198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haggarty SJ, Koeller KM, Wong JC, Butcher RA, Schreiber SL. Multidimensional chemical genetic analysis of diversity-oriented synthesis-derived deacetylase inhibitors using cell-based assays. Chem Biol. 2003;10(5):383–396. doi: 10.1016/s1074-5521(03)00095-4. [DOI] [PubMed] [Google Scholar]
- 19.Gorman AM, Healy SJ, Jäger R, Samali A. Stress management at the ER: Fegulators of ER stress-induced apoptosis. Pharmacol Ther. 2012;134(3):306–316. doi: 10.1016/j.pharmthera.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Desplanques G, et al. Impact of XIAP protein levels on the survival of myeloma cells. Haematologica. 2009;94(1):87–93. doi: 10.3324/haematol.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korting HC, Schäfer-Korting M. The Benefit/Risk Ratio: A Handbook for the Rational Use of Potentially Hazardous Drugs. CRC; Boca Raton: 1999. [Google Scholar]
- 22.Kikuchi S, et al. Class IIa HDAC inhibition enhances ER stress-mediated cell death in multiple myeloma. Leukemia. 2015;29(9):1918–1927. doi: 10.1038/leu.2015.83. [DOI] [PubMed] [Google Scholar]
- 23.Hideshima T, et al. Induction of differential apoptotic pathways in multiple myeloma cells by class-selective histone deacetylase inhibitors. Leukemia. 2014;28(2):457–460. doi: 10.1038/leu.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santo L, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119(11):2579–2589. doi: 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nawrocki ST, et al. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 2005;65(24):11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- 26.Fribley A, Wang CY. Proteasome inhibitor induces apoptosis through induction of endoplasmic reticulum stress. Cancer Biol Ther. 2006;5(7):745–748. doi: 10.4161/cbt.5.7.2971. [DOI] [PubMed] [Google Scholar]
- 27.Morgillo F, et al. Antitumor activity of bortezomib in human cancer cells with acquired resistance to anti-epidermal growth factor receptor tyrosine kinase inhibitors. Lung Cancer. 2011;71(3):283–290. doi: 10.1016/j.lungcan.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Butler KV, et al. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132(31):10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JH, et al. Creation of a histone deacetylase 6 inhibitor and its biological effects [corrected] Proc Natl Acad Sci USA. 2015;112(39):12005–12010. doi: 10.1073/pnas.1515882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin X, et al. Design and synthesis of orally bioavailable aminopyrrolidinone histone deacetylase 6 inhibitors. J Med Chem. 2015;58(6):2809–2820. doi: 10.1021/jm502011f. [DOI] [PubMed] [Google Scholar]
- 31. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 32.Wong JC, Hong R, Schreiber SL. Structural biasing elements for in-cell histone deacetylase paralog selectivity. J Am Chem Soc. 2003;125(19):5586–5587. doi: 10.1021/ja0341440. [DOI] [PubMed] [Google Scholar]
- 33.Tang W, Luo T, Greenberg EF, Bradner JE, Schreiber SL. Discovery of histone deacetylase 8 selective inhibitors. Bioorg Med Chem Lett. 2011;21(9):2601–2605. doi: 10.1016/j.bmcl.2011.01.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Case DA, et al. 2008. AMBER 10. (University of California, San Francisco, San Francisco.)
- 35.Wang D, Helquist P, Wiest O. Zinc binding in HDAC inhibitors: A DFT study. J Org Chem. 2007;72(14):5446–5449. doi: 10.1021/jo070739s. [DOI] [PubMed] [Google Scholar]
- 36.Hideshima T, et al. Biologic sequelae of IkappaB kinase (IKK) inhibition in multiple myeloma: Therapeutic implications. Blood. 2009;113(21):5228–5236. doi: 10.1182/blood-2008-06-161505. [DOI] [PMC free article] [PubMed] [Google Scholar]