Significance
The hemochorial placenta is a dynamic structure endowed with responsibilities controlling the extraction of maternal resources, ensuring fetal development and preserving maternal health. A healthy placenta exhibits plasticity and can adapt to environmental challenges. Such adaptations can be executed through instructive actions on trophoblast stem cells, influencing their abilities to expand and differentiate into specialized cells that accommodate the challenge. Hypoxia, when appropriately timed, promotes invasive trophoblast-directed uterine spiral artery remodeling. Hypoxia activates hypoxia inducible factor-dependent expression of lysine demethylase 3A, modifying the histone landscape on key target genes, including matrix metallopeptidase 12, which acts to facilitate trophoblast invasion and uterine vascular remodeling. Plasticity and adaptations at the maternal–fetal interface safeguard placental development and the healthy progression of pregnancy.
Keywords: placenta, hypoxia, trophoblast invasion, epigenetics, plasticity
Abstract
The hemochorial placenta develops from the coordinated multilineage differentiation of trophoblast stem (TS) cells. An invasive trophoblast cell lineage remodels uterine spiral arteries, facilitating nutrient flow, failure of which is associated with pathological conditions such as preeclampsia, intrauterine growth restriction, and preterm birth. Hypoxia plays an instructive role in influencing trophoblast cell differentiation and regulating placental organization. Key downstream hypoxia-activated events were delineated using rat TS cells and tested in vivo, using trophoblast-specific lentiviral gene delivery and genome editing. DNA microarray analyses performed on rat TS cells exposed to ambient or low oxygen and pregnant rats exposed to ambient or hypoxic conditions showed up-regulation of genes characteristic of an invasive/vascular remodeling/inflammatory phenotype. Among the shared up-regulated genes was matrix metallopeptidase 12 (MMP12). To explore the functional importance of MMP12 in trophoblast cell-directed spiral artery remodeling, we generated an Mmp12 mutant rat model using transcription activator-like nucleases-mediated genome editing. Homozygous mutant placentation sites showed decreased hypoxia-dependent endovascular trophoblast invasion and impaired trophoblast-directed spiral artery remodeling. A link was established between hypoxia/HIF and MMP12; however, evidence did not support Mmp12 as a direct target of HIF action. Lysine demethylase 3A (KDM3A) was identified as mediator of hypoxia/HIF regulation of Mmp12. Knockdown of KDM3A in rat TS cells inhibited the expression of a subset of the hypoxia–hypoxia inducible factor (HIF)-dependent transcripts, including Mmp12, altered H3K9 methylation status, and decreased hypoxia-induced trophoblast cell invasion in vitro and in vivo. The hypoxia-HIF-KDM3A-MMP12 regulatory circuit is conserved and facilitates placental adaptations to environmental challenges.
Vascular remodeling is an important pregnancy-associated adaptation in hemochorial placentation and is orchestrated, in part, from the contributions of invasive trophoblast cells (also termed extravillous trophoblast) (1–3). These cells invade into the uterus and restructure spiral arteries turning them into flaccid low resistance vessels facilitating the flow of maternal resources to the placenta and then to the fetus. Failure of trophoblast cell invasion and vascular remodeling is associated with pathological conditions such as preeclampsia, intrauterine growth restriction, and preterm birth (3–5). Invasive trophoblast cells arise from trophoblast stem (TS)/progenitor cell populations and can be classified based on their entry into the uterine parenchyma (5, 6). Endovascular invasive trophoblast cells enter uterine spiral arteries, facilitate removal of the endothelium, acquire a pseudovascular phenotype, and restructure the spiral artery, whereas interstitial invasive trophoblast cells migrate into a specialized uterine stroma, termed decidua, and infiltrate areas surrounding the spiral arteries (2, 4, 7, 8). These seminal events in hemochorial placentation are conserved in the rat and human (9–11).
Placentation is a malleable process responsive to a range of stimuli present in the maternal environment (6, 12). As in other tissues, low oxygen is a potent driver of vascular development at the maternal-fetal interface. Hypoxia exposure can redirect placental organization and promote development of the invasive trophoblast cell lineage and uterine spiral artery remodeling, representing adaptive responses conserved in the rat, monkey, and human (13–16). Cellular responses to oxygen deficits are mediated by hypoxia inducible factor (HIF), a transcription factor consisting of a heterodimer composed of oxygen sensitive HIF1A or HIF2A and a constitutively active HIF1B (also known as aryl hydrocarbon receptor nuclear translocator, ARNT) (17, 18). Mouse mutagenesis experiments have demonstrated the importance of key components of the HIF signaling pathway to the regulation of placentation (19–24). HIF acts on targets directly promoting transcription and cellular adaptations to low oxygen and indirectly via modification of the epigenetic landscape (18). Lysine demethylase 3A (KDM3A; also known as JMJD1A) is a hypoxia/HIF responsive histone 3 lysine 9 (H3K9) demethylase implicated in epigenetic regulation of cell differentiation and tissue homeostasis (25, 26). Structural changes associated with tissue plasticity are engineered via activation of matrix metalloproteinases (MMPs) (27, 28). MMPs are responsive to environmental signals and promote the movement of cells through tissue matrices, facilitate structural changes in blood vessel integrity, and contribute to morphogenesis of the placenta (18, 27, 29–31).
Mechanisms underlying the orchestration of placental adaptations and plasticity are not well understood. In this study, we reveal a central and conserved role for HIF-KDM3A-MMP12 signaling in regulating adaptations at the placentation site.
Results
Hypoxia-Dependent Transcript Responses in TS Cells and at the Placentation Site.
Hypoxia signaling regulates the development of the invasive trophoblast cell lineage and uterine spiral artery remodeling (13–16). As a first step toward identifying potential adaptive mechanisms downstream of hypoxia exposure, we profiled transcriptomes of rat TS cells (32) exposed to low oxygen and placentation sites from rats exposed to hypoxic conditions using DNA microarray technology (Gene Expression Omnibus repository: GSE80339 and GSE80340; www.ncbi.nlm.nih.gov/geo/).
TS cells maintained in the stem state were exposed to 0.5% oxygen for 24 h, harvested, and profiled. Under these conditions, TS cells are induced to move through extracellular matrices (15). Cell number was not significantly affected by the oxygen tensions investigated (Fig. S1A). Low oxygen conditions resulted in robust effects on gene expression: 838 transcripts were down-regulated (≤1.5-fold) and 786 transcripts were up-regulated (≥1.5-fold) in response to low oxygen. Many down-regulated transcripts were associated with the stem state (Cdh1, Cdx2, Bmp4, Id2, Satb1, Rbm3, and Lin28), whereas up-regulated transcripts were characteristic of proteins contributing to cell migration (Cul7 and Loxl2), extracellular matrix remodeling (Mmp12, Mmp9, Plau, Dpp3, and Plod2), and inflammatory (Il33 and Cd200) phenotypes and adaptive responses to hypoxia (Egln1, Bhlhe40, Ppp1r3c, Vegfa, Ankrd37, and Kdm3a; Fig. 1 A–F and Dataset S1). The involvement of HIF signaling in the transcriptomic responses to hypoxia was evaluated in TS cells expressing HIF1B short hairpin RNAs (shRNAs) or control shRNAs. Down-regulated transcripts showed a mix of HIF dependence, whereas all of the up-regulated transcripts examined were dependent on HIF signaling (Fig. 1F). Thus, low oxygen interfered with maintenance of the TS cell stem state and promoted differentiation consistent with an HIF-driven invasive trophoblast cell phenotype.
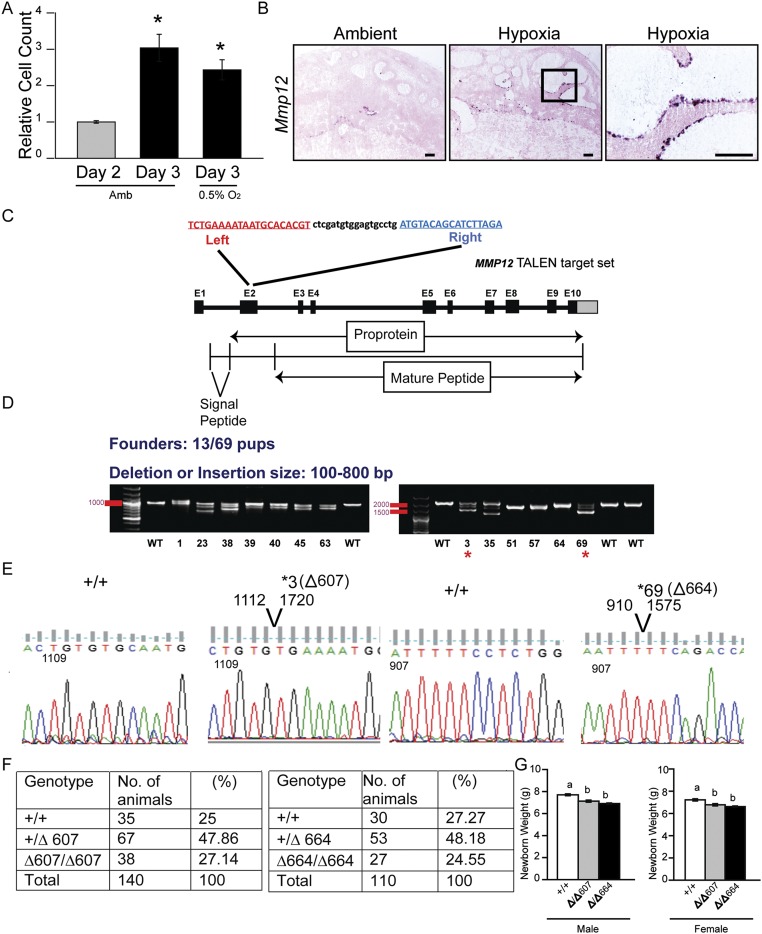
Fig. S1.
Effects of low oxygen culture conditions on TS cell numbers and TALEN targeting of exon 2 within the rat Mmp12 locus. (A) Low oxygen effects on TS cell numbers. Note that 24-h exposure to ambient (Amb) or low oxygen (0.5% O2) conditions showed similar growth responses (n = 4/group, Mann–Whitney test, *P < 0.05). (B) In situ hybridization analysis of Mmp12 transcripts in gd 13.5 placentation sites from pregnant rats exposed to ambient or hypoxia conditions. (Scale bar, 250 μm.) (C) Schematic representation of the rat Mmp12 gene and the TALEN target site within exon 2 (NC_005107.4). Diagrammatic organization of the MMP12 protein. (D) PCR-based identification of Mmp12 mutant founders (13 founders identified from 69 offspring). Founder numbers 3 and 69 were used for expansion and characterization. (E) DNA sequence analysis showing two founder strains possessing deletions of 607 bp (Δ607) or 664 bp (Δ664). (F) Mendelian ratios generated from breeding +/Δ607 males x +/Δ607 females (Left) and +/Δ664 males x +/Δ664 females (Right). (G) Assessment of postpartum day 1 neonatal weights of WT (+/+), Δ/Δ 607, and Δ/Δ664 pups, sexed at birth (WT: males, n = 39, females, n = 33; Δ/Δ 607: males, n = 44, females n = 23; Δ/Δ664: males, n = 44, females, n = 43 females). Different letters above bars signify differences among means (ANOVA with Dunnett’s test, P < 0.05).
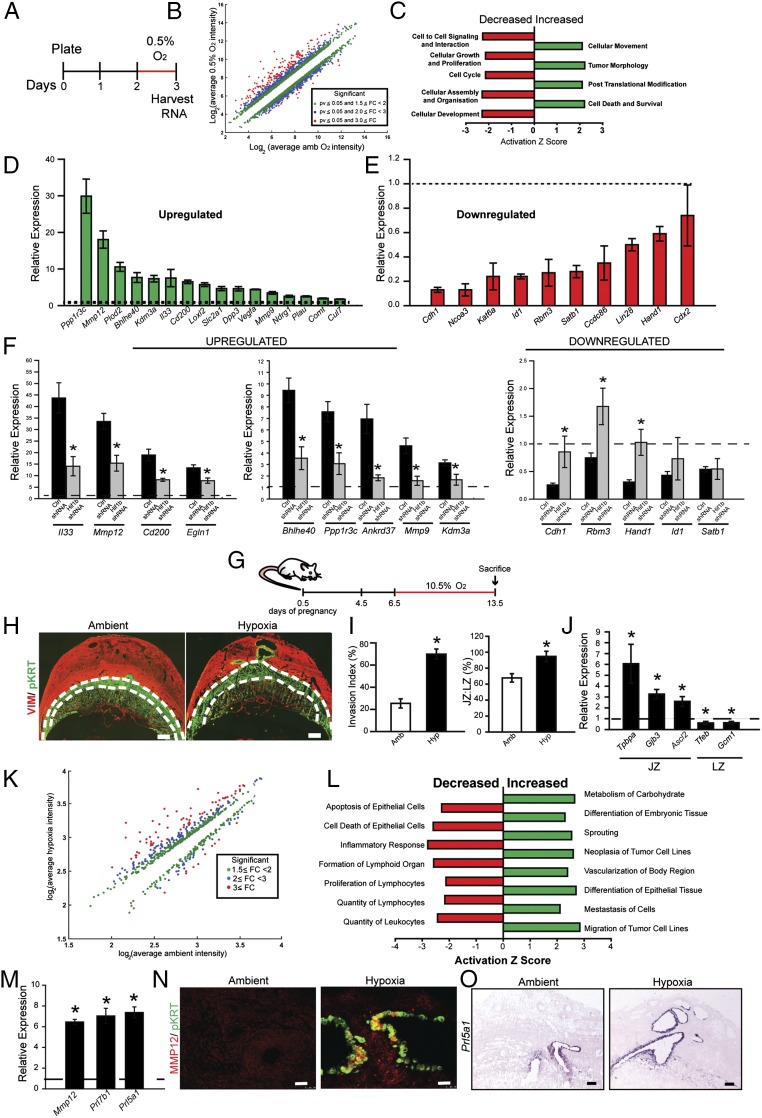
Fig. 1.
Hypoxia-dependent responses of TS cells and the placentation site. (A) Schematic representation of TS cell exposure to low oxygen tension (0.5% O2 for 24 h) before harvesting RNA for DNA microarray analysis. (B) Scatter plot presentation of in vitro hypoxia-responsive transcripts. (C) Presentation of pathway analysis of transcripts differentially expressed following TS cell exposure to 0.5% O2. (D and E) Validation of select differentially expressed transcripts by qRT-PCR. All hypoxia responses are significantly different from ambient control (n = 5/group; P < 0.05). (F) Examination of the dependence of hypoxia-dependent transcript changes on HIF signaling. TS cells were exposed to 0.5% O2 in the presence of control (Ctrl) or Hif1b shRNAs. RNA was harvested and transcript levels assessed by qRT-PCR (n = 4/group; ANOVA with Student–Newman–Keuls test, *P < 0.05). Dashed lines represent the ambient control values. (G) Schematic representation of in vivo maternal exposure to hypoxia (10.5% O2). (H) Representative cross sections of gd 13.5 placentation sites immunostained for vimentin and cytokeratin from pregnant rats exposed to ambient (Amb) or hypoxia (Hyp, 10.5% O2). The junctional zone (devoid of vimentin staining) is demarcated by the dashed white lines. (Scale bar, 1 mm.) (I) Quantification of invasion of trophoblast cells into the uterine mesometrial compartment and ratio of junctional and labyrinth zones (ambient, n = 10; hypoxia, n = 12; *P < 0.05). (J) Relative expression of transcripts associated with the junctional zone (JZ) and labyrinth zone (LZ) (n = 8/group, *P < 0.05). Dashed lines represent the ambient control values. (K) Scatter plot presentation of maternal hypoxia-responsive transcripts in the metrial gland. (L) Presentation of pathway analysis of differentially expressed transcripts in the metrial glands following hypoxia exposure. (M) Validation of selected differentially expressed transcripts by qRT-PCR (n = 10/group, *P < 0.05). Dashed lines represent the ambient control values. (N) Immunohistochemical analysis of MMP12 and pan cytokeratin (pKRT) staining in tissue sections from pregnant rats exposed to ambient or hypoxia conditions. (Scale bar, 50 μm.) (O) In situ hybridization analysis of Prl5a1 transcripts in placentation sites from pregnant rats exposed to ambient or hypoxia conditions. (Scale bar, 250 μm.) Data presented in D–F, I, J, and M were analyzed with Mann–Whitney test.
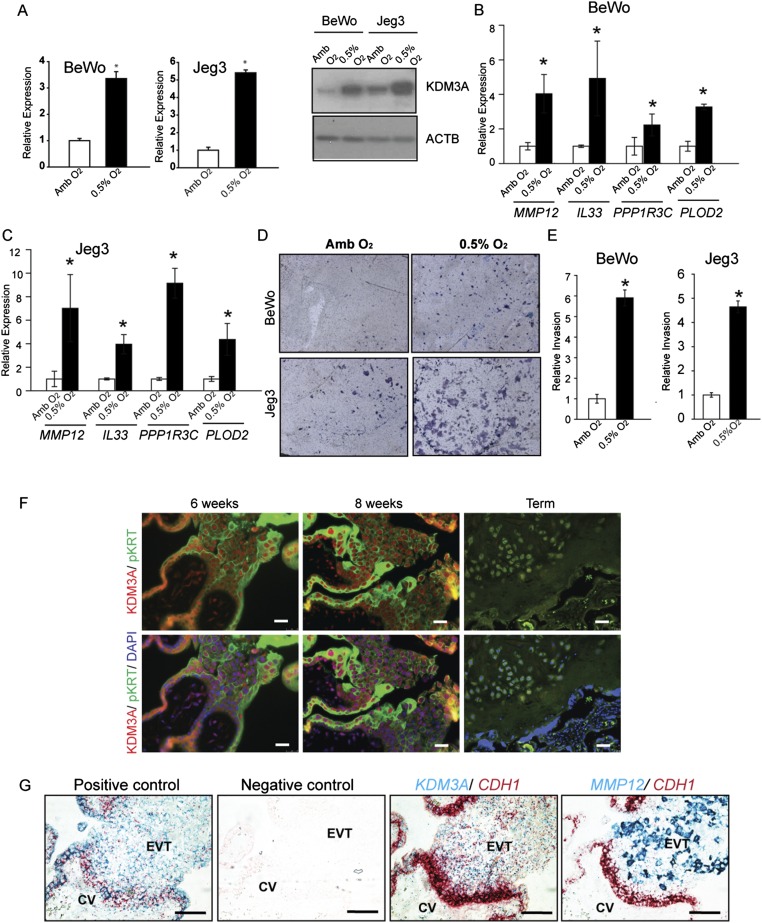
Because low oxygen promoted TS cell differentiation toward the invasive trophoblast lineage, we sought to identify an in vivo correlate of differentiated invasive trophoblast cells. Hypoxia-exposed gestation day (gd) 13.5 metrial gland tissue contains a prominent population of differentiated invasive endovascular trophoblast cells (14). Rats were exposed to ambient (21% oxygen) or hypoxic environments (10.5% oxygen) from gd 6.5 to 13.5. Animals were euthanized at gd 13.5, placentation sites were prepared for assessment of intrauterine trophoblast invasion and spiral artery remodeling or alternatively dissected, and transcript expression was investigated (14, 15). Pregnancy-associated uterine spiral artery remodeling is defined by trophoblast cell intravasation of spiral arteries, their replacement of endothelial cells lining the vessel, and subsequent restructuring the underlying extracellular matrix and dissolution of the tunica media (2, 15). Hypoxia stimulated intrauterine endovascular trophoblast invasion, the preferential allocation of trophoblast cells within the placenta to the junctional zone, and some alterations in the expression of transcripts associated with the junctional zone (Tpbpa, Gjb3, and Ascl2) and labyrinth zone (Tfeb and Gcm1; Fig. 1 G–J). Transcriptional responses to hypoxia were examined in more detail from the metrial gland (Fig. 1 K–M). The metrial gland is a heterogeneous tissue, composed of an assortment or stromal, endothelial, and immune cell populations, and represents the site of intrauterine endovascular trophoblast cell invasion of the spiral arteries (15, 33). A subset of hypoxia-responsive transcripts identified by DNA microarray analysis of the metrial gland exhibited overlap with transcripts up-regulated by low oxygen tension in TS cells, including members of the prolactin family (Prl5a1 and Prl7b1) and MMP12 (Fig. 1M, Dataset S1, and Table S1). Prl5a1 and Mmp12 expression was restricted to endovascular trophoblast (Fig. 1 N and O and Fig. S1B). MMP12 is a metalloelastase and has been implicated in human trophoblast-directed uterine spiral artery remodeling (30, 34). We used the overlap of in vitro and in vivo profiles and the conservation in rat and human placentation to guide our analysis, which led to a focus on MMP12. In the rat placentation site, MMP12 expression was activated by hypoxia and restricted to invasive endovascular trophoblast cells (Fig. 1N and Fig. S1B). Thus, MMP12 was viewed as a candidate effector of hypoxia-activated uterine spiral artery remodeling.
Table S1.
Effects of maternal hypoxia on metrial gland gene expression
| Protein | Gene symbol | Function | Microarray fold change (Hyp/Amb) | qRT-PCR ± SEM (Hyp/Amb) |
| TSH releasing hormone | Trh | Hormone | 31.04 | 9.97 ± 1.162* |
| Matrix metallopeptidase 10 | Mmp10 | ECM remodeler | 8.75246 | 10.31 ± 1.415* |
| HtrA serine peptidase 1 | Htra1 | Protease | 4.78358 | 7.08 ± 0.883* |
| Placenta-specific 1 | Plac1 | Unknown | 4.45683 | 4.43 ± 0.459* |
| Secretory leukocyte peptidase inhibitor | Slpi | Inhibitor of proteases | 4.35631 | 3.92 ± 0.634* |
| Bone morphogenetic protein 6 | Bmp6 | Growth factor | 3.64587 | 9.92 ± 1.007* |
| Cathepsin Q | Ctsq | Protease | 3.60704 | 3.50 ± 0.371* |
| Prolactin family 2, subfamily a, member 1 | Prl2a1 | Hormone | 3.60029 | 5.62 ± 0.898* |
| Integrin, β 6 | Itgb6 | Cell adhesion receptor/signaling | 3.50875 | 2.54 ± 0.259* |
| Prolactin family 4, subfamily a, member 1 | Prl4a1 | Hormone | 3.3691 | 3.47 ± 0.457* |
| Coagulation factor III (thromboplastin, tissue factor) | F3 | Cell surface glycoprotein/coagulation cascade | 0.47 | 0.66 ± 0.0891* |
| Epithelial cell adhesion molecule | Epcam | Cell adhesion molecule | 0.29 | 0.4 ± 0.0809* |
| WAP four-disulfide core domain 10 | Wfdc10 | Protease inhibitor | 0.28 | 0.4 ± 0.0965* |
| Insulin-like growth factor binding protein 1 | Igfbp1 | Growth factor binding protein | 0.26 | 0.33 ± 0.0478* |
Mann–Whitney rank sum test (P < 0.05).
MMP12 and Hypoxia-Activated Uterine Spiral Artery Remodeling by Trophoblast Cells.
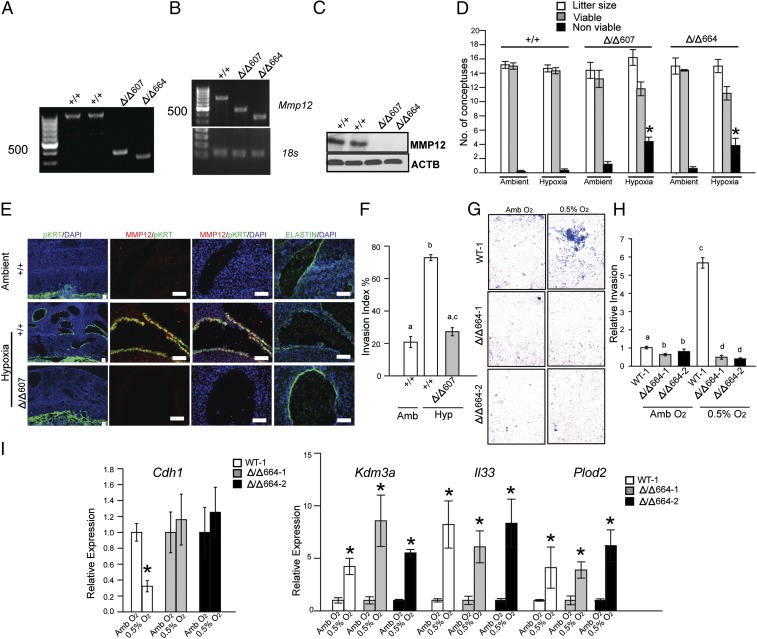
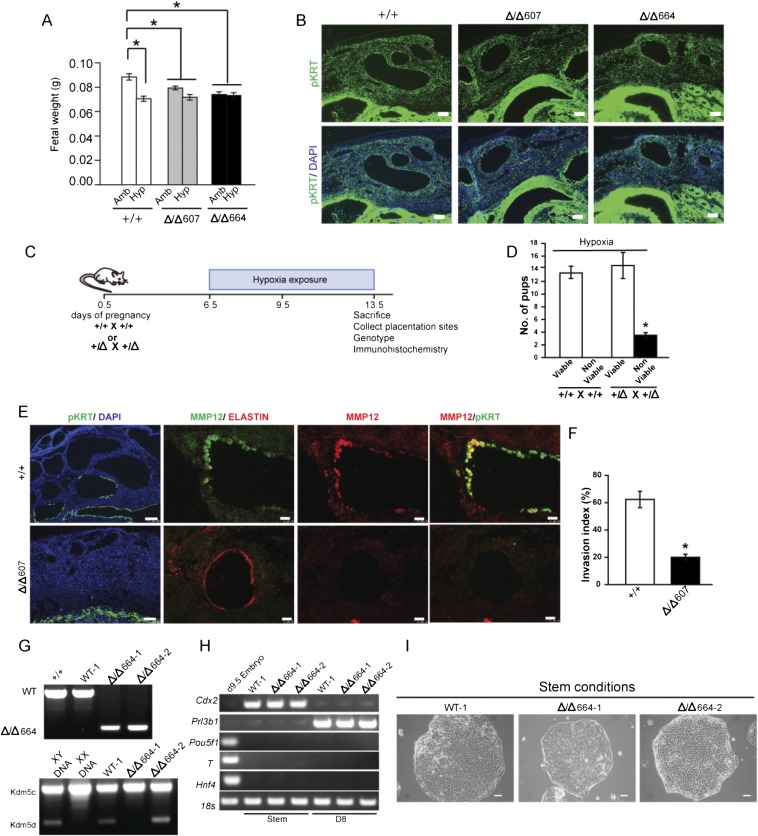
To test the involvement of MMP12 in uterine spiral artery remodeling, Mmp12 mutant rats were generated using transcription activator-like nucleases (TALEN)-mediated genome editing (Fig. S1 C–G). Thirteen founder animals were identified with deletions ranging from 100 to 800 bp. Two deletions encompassing exon 2 (607-bp deletion, referred to as Δ/Δ607; 664-bp deletion, referred to as Δ/Δ664) were further characterized. Each mutation caused nonsense nucleotide frameshifts and premature stop codons, resulting in the interference of MMP12 protein expression, as tested in the spleen and the placentation site (Fig. 2 A–C). Heterozygous × heterozygous breeding generated expected Mendelian ratios of progeny genotypes (Fig. S1F). Mmp12-null × Mmp12-null pregnancies yielded modest but significant decreases in litter size and postpartum day 1 neonatal body weights (Fig. S1G and Fig. S2A). MMP12 deficiency did not adversely affect placentation or intrauterine trophoblast invasion at gd 18.5 (Fig. S2B). However, an MMP12 deficit did affect the capacity of the placenta to adapt to hypoxia. WT × WT and Mmp12-null × Mmp12-null rat mating combinations were challenged with hypoxia (10.5% oxygen) from gd 6.5 to 13.5. Pregnancy outcomes, fetal weights, and intrauterine trophoblast invasion were assessed. Mmp12-null pregnancies showed impaired adaptations to hypoxia and significant increases in fetal death (Fig. 2D). Disruption of MMP12 expression interfered with hypoxia-activated trophoblast invasion and uterine spiral artery remodeling, including impairment of uterine spiral artery-associated elastin degradation (Fig. 2E). Similar results were obtained from WT and Mmp12-null placentation sites generated from an Mmp12 heterozygous × Mmp12 heterozygous breeding scheme (Fig. S2 C–F). Shapiro and colleagues noted deficits in Mmp12-null mouse macrophage invasive behavior (35), which prompted an evaluation of the direct role of MMP12 on trophoblast cell invasive properties. TS cell lines were established from WT and Mmp12-null blastocysts (Fig. S2 G–I). Both TS cell populations exhibited FGF4-driven proliferation and mitogen removal-dependent differentiation; however, responses to low oxygen tension differed. Unlike WT TS cells, Mmp12-null TS cells showed attenuated low oxygen-activated invasive properties and a failure of Cdh1 down-regulation when exposed to low oxygen (Fig. 2 G–I). Other low oxygen-activated transcriptional behaviors examined did not differ between WT and Mmp12-null TS cells (Fig. 2I). Collectively, the results indicate that MMP12 is an effector of hypoxia-activated endovascular trophoblast invasion and uterine spiral artery remodeling.
Fig. 2.
MMP12 and hypoxia-activated trophoblast-directed uterine spiral artery remodeling. (A) Genotyping of WT (+/+) and Mmp12 homozygous mutant rat strains generated by genome editing. (B) RT-PCR analysis for Mmp12 and 18s RNAs from spleens of WT (+/+) and Mmp12 mutant (Δ/Δ607 and Δ/Δ664) rats. (C) Western blotting for MMP12 and ACTB from spleens of WT (+/+) and Mmp12 mutant (Δ/Δ607 and Δ/Δ664) rats. (D) Effects of ambient and hypoxia (10.5% O2) conditions on litter size and the numbers of viable and nonviable conceptuses (+/+, n = 7; Δ/Δ 607, n = 5; Δ/Δ 664 n = 5; *P < 0.05). (E) Immunohistochemical analyses (pan cytokeratin, pKRT; MMP12; elastin) of the mesometrial placentation sites from WT (+/+) and Mmp12 mutant (Δ/Δ607) rats exposed to ambient or hypoxia conditions. (F) Quantification of trophoblast cell invasion into the uterine mesometrial compartment. Different letters above bars signify differences among means (n = 5/group, *P < 0.05). (G) Effects of low oxygen (0.5% O2) on invasive behavior of WT (WT-1) and two Mmp12-null (Δ/Δ664–1 and Δ/Δ664–2) TS cell populations. Images are representative filters. (H) Quantification of invasion through Matrigel. Different letters above bars signify differences among means (n = 5/group, P < 0.05). (I) qRT-PCR analysis of hypoxia responsive transcripts in WT (WT-1; white bars) and two Mmp12-null (Δ/Δ664–1; gray bars and Δ/Δ664–2; black bars) TS cell populations. Comparisons were between ambient (Amb) and 0.5% O2 conditions for each genotype (n = 3/group, Mann–Whitney test, *P < 0.05). Data presented in D, F, and H were analyzed with ANOVA and Holm–Sidak (D) or Newman–Keuls tests (F and H).
Fig. S2.
Role of MMP12 in placental and fetal adaptations to hypoxia. (A) Fetal weights at gd 13.5 from WT and Mmp12 mutant (Δ/Δ607 and Δ/Δ664) rat pregnancies exposed to ambient (Amb) or hypoxia (Hyp) conditions (Ambient, WT: n = 36; Hypoxia, WT: n = 44; Ambient, Δ/Δ607: n = 49; Hypoxia, Δ/Δ607: n = 41; Ambient, Δ/Δ664: n = 35; Hypoxia, Δ/Δ664: n = 41; *P < 0.05). (B) Immunohistochemical analyses (pKRT) of gd 18.5 WT (+/+), Δ/Δ607, and Δ/Δ664 placentation sites. (C) Schematic representation of in vivo maternal exposure to hypoxia (10.5% O2) and analyses. WT and Mmp12 mutant conceptuses were generated by +/Δ607 male × +/Δ607 female breeding. (D) Effects of hypoxia (10.5% O2) on the numbers of viable and nonviable conceptuses from WT × WT and heterozygous × heterozygous breeding. Heterozygous × heterozygous pregnancies exposed to hypoxia exhibited a higher number of nonviable conceptuses than did WT × WT (+/+) pregnancies (n = 5/group; *P < 0.05). (E) Immunohistochemical analyses (pan cytokeratin, pKRT; MMP12; elastin) of placentation sites from WT (+/+) and Mmp12 mutant (Δ/Δ607) conceptuses exposed to hypoxic conditions. (F) Quantification of trophoblast cell invasion into the uterine mesometrial compartment (n = 5/group, Mann–Whitney test, *P < 0.05). (G) Genotyping of WT (WT-1) and Mmp12-null (Δ/Δ664–1 and Δ/Δ664–2) TS cells for WT and Δ/Δ664 alleles (Top) and sex chromosome determination of WT and Mmp12-null TS cells (Bottom). WT-1 and Δ/Δ664–2 TS cells are X,Y and Δ/Δ664–1 TS cells are X,X. (H) Conventional RT-PCR analysis of embryonic and trophoblast-specific markers in stem and differentiated WT-1, Δ/Δ664–1, and Δ/Δ664–2 TS cells. (I) Representative images of stem state colonies for WT-1, Δ/Δ664–1, and Δ/Δ664–2 TS cells. Data presented in A and D were analyzed with ANOVA and Student–Newman–Keuls test.
KDM3A and Hypoxia-HIF Signaling in Trophoblast Cells.
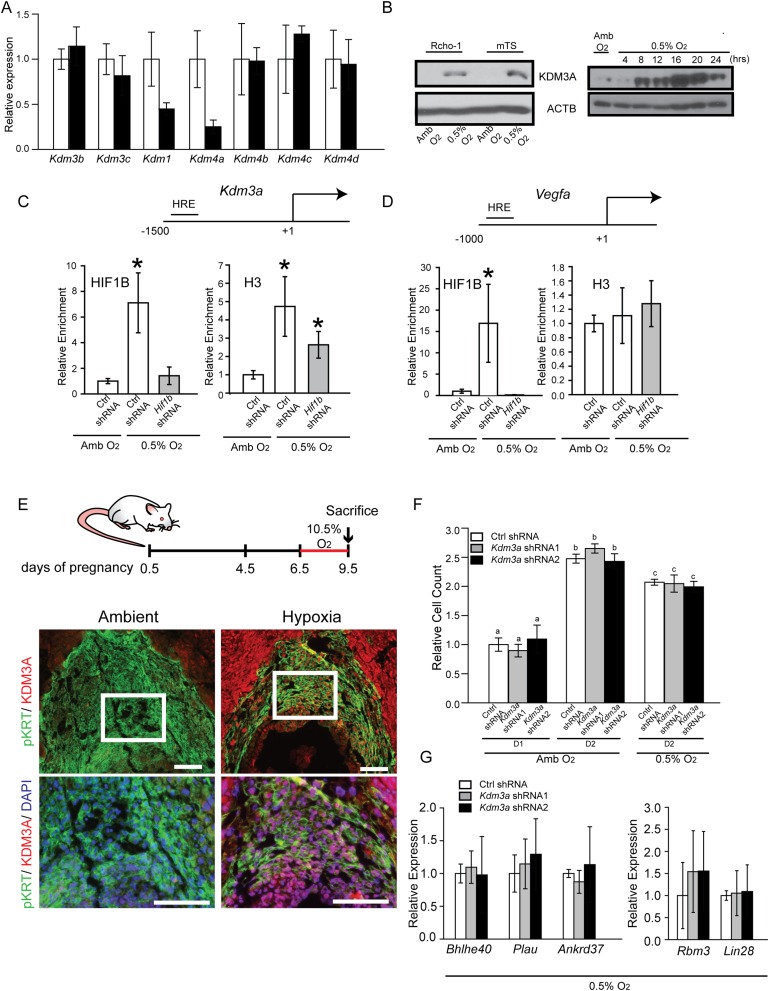
The above experimentation implicated a link between hypoxia, HIF, and the regulation of MMP12; however, evidence did not support Mmp12 as a direct target of HIF action. Conserved HIF binding motifs were not present within regulatory DNA associated with the Mmp12 gene and HIF ChIP sequencing datasets did not support a direct interaction of HIF with the Mmp12 locus (36–39). Consequently, potential intermediaries were explored. Perusal of the DNA microarray profile generated from TS cells exposed to ambient or low oxygen tension yielded a HIF-dependent candidate mediator, KDM3A (Fig. 1 D and F). This responsiveness to low oxygen was not shared by two closely related jumonji domain containing family members, Kdm3b and Kdm3c, or other histone H3K9 demethylases (Fig. S3A). KDM3A is a direct target of HIF action in an assortment of cell types (40–43), including TS cells (Fig. 3 A–E and Fig. S3 B–D), and possesses the capacity to promote gene activation through demethylation of histone H3K9, e.g., removal of a repressive histone mark (25, 26). Accordingly, the involvement of KDM3A in hypoxia-activated trophoblast responses was investigated.
Fig. S3.
KDM3A, hypoxia, and placental expression. (A) qRT-PCR analysis of known histone H3K9 demethylases in rat TS cells maintained in ambient conditions (white bars) or following 24-h exposure to 0.5% O2 (black bars). Statistical analysis: n = 3/group, not significant. (B) KDM3A expression in Rcho-1, mouse TS (mTS), and rat TS cells. (Left) Western blot analysis of KDM3A expression after Rcho-1 TS cells and mouse TS cells exposed to Amb or 0.5% O2 for 24 h. (Right) Western blot analysis for KDM3A in rat TS cell exposed to 0.5% O2 for various time intervals. ACTB was used as a loading control for Western blots. (C) ChIP analysis for HIF1B and histone H3 at a conserved hypoxia response element (HRE) within the Kdm3a promoter. Rat TS cells were infected with lentiviral vectors containing control (Ctrl) or Hif1b shRNAs and exposed to ambient (Amb) or 0.5% O2 culture conditions (n = 4, *P < 0.05). (D) ChIP analysis for HIF1B and histone H3 at a conserved HRE within the Vegfa promoter. Rat TS cells were infected with lentiviral vectors containing Ctrl or Hif1b shRNAs and exposed to Amb or 0.5% O2 culture conditions (n = 4, *P < 0.05). (E) In vivo trophoblast KDM3A expression in response to maternal hypoxia. Rats were exposed to ambient or hypoxic (10.5% O2) conditions from gd 6.5 to 9.5 and euthanized, and placentation sites were dissected. Immunohistochemistry analysis (pan cytokeratin, pKRT; KDM3A) was performed on gd 9.5 placentation site tissue sections. White boxed areas shown in the upper images are presented in the lower panel. (Scale bars, 100 µm.) (F) Effects of Amb or 0.5% O2 culture conditions on cell numbers for control shRNA (Ctrl shRNA)-, Kdm3a shRNA1-, and Kdm3a shRNA2-expressing TS cells (n = 4/group, *P < 0.05). (G) Effects of Kdm3a knockdown on hypoxia responsive transcripts in TS cells (n = 4, not significant). Data presented in B was analyzed with Mann–Whitney test and C, D, F, and G were analyzed with ANOVA and Student–Newman–Keuls test.
Fig. 3.
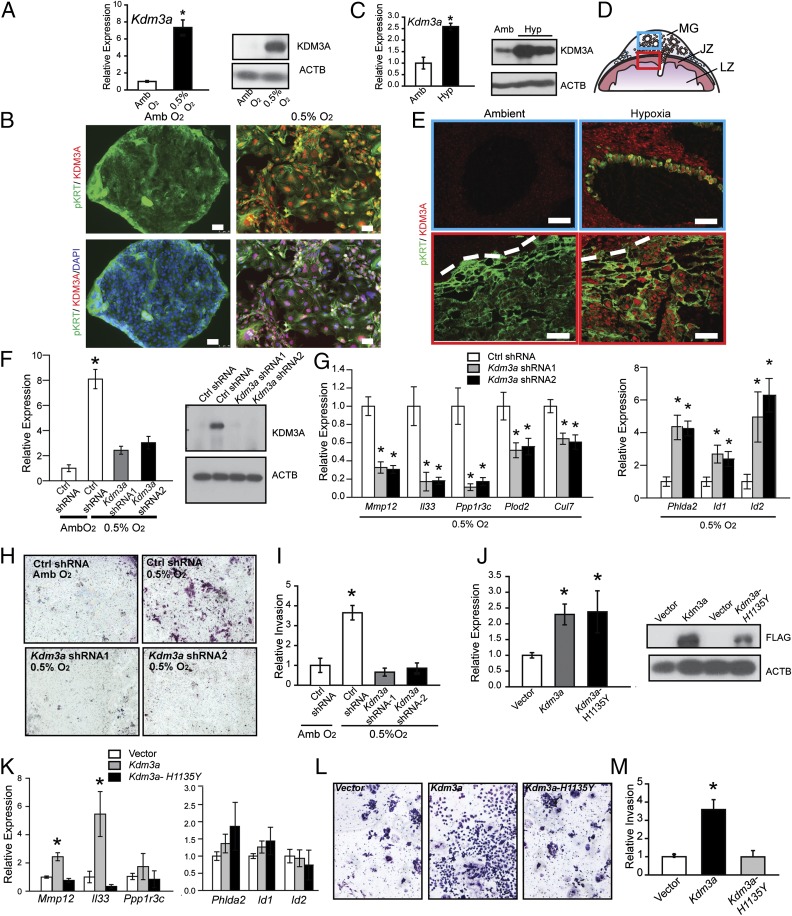
KDM3A and hypoxia signaling in trophoblast cells. (A) Kdm3a transcript (qRT-PCR; Left) and protein (Western blot; Right) responses in TS cells cultured under ambient (Amb) or low oxygen (0.5% O2). Statistical analysis: n = 5/group, Mann–Whitney test, *P < 0.05. (B) Immunocytochemical staining for pan-cytokeratin (pKRT) and KDM3A in TS cells cultured under Amb or 0.5% O2. (Scale bar, 50 µm.) (C) In vivo placental Kdm3a transcript (qRT-PCR; Left) and protein (Western blot; Right) responses to maternal hypoxia (10.5% O2 from gd 6.5 to 13.5). (D) Schematic representation of a midgestation placentation site, consisting of the metrial gland (MG), junctional zone (JZ), and labyrinth zone (LZ). The blue box corresponds to the metrial gland region (Upper) of E and the red box corresponds to the junctional zone (Lower) of E. (E) Immunohistochemistry analysis (pan cytokeratin, pKRT; KDM3A) of gd 13.5 placentation site sections from pregnant rats exposed to ambient and hypoxic conditions. The dashed line in the bottom panels represents the border between the decidua and chorioallantoic placenta. (Scale bar, 100 µm.) (F) qRT-PCR and Western blot validation of Kdm3a shRNAs. TS cells expressing control (Ctrl) or Kdm3a shRNAs were examined in Amb or 0.5% O2 culture conditions (n = 4, *P < 0.05). (G) Effects of Kdm3a knockdown on hypoxia responsive transcripts in TS cells (n = 4, *P < 0.05). (H) Effects of Kdm3a knockdown on 0.5% O2 activated invasive behavior of TS cells. Images are representative filters. (I) Quantification of invasion through Matrigel (n = 3, Ctrl shRNA + 0.5% O2 vs. all other treatments, *P < 0.05). (J) Ectopic transcript (qRT-PCR; Left) and protein (Western blot, Right) expression of control (vector) and WT (Kdm3a) and mutant (Kdm3a-H1135Y) Kdm3a constructs stably transfected into TS cells. (K) Effects of ectopic expression of control (vector), WT (Kdm3a), and mutant Kdm3a (Kdm3a-H1135Y) constructs stably transfected into TS cells on hypoxia responsive transcripts (n = 4, Kdm3a vs. vector or Kdm3a-H1135Y, *P < 0.05). (L) Effects of ectopic expression of control (vector), WT (Kdm3a), and mutant Kdm3a (Kdm3a-H1135Y) constructs stably transfected into TS cells on invasive behavior. Images are representative filters. (M) Quantification of invasion through Matrigel (n = 4, Kdm3a vs. vector or Kdm3a-H1135Y, *P < 0.05). Data presented in F, G, I–K, and M were analyzed with ANOVA and Student–Keuls test. ACTB was used as a loading control for the Western blots shown in A, C, F, and J.
Initially, in vivo responsiveness of KDM3A expression in placentation sites of pregnant rats exposed to normoxic or hypoxic conditions from gd 6.5 to 9.5 or from gd 6.5 to 13.5 was evaluated. At gd 9.5, KDM3A protein was up-regulated by hypoxia at the site of trophoblast progenitors (ectoplacental cone; Fig. S3E). KDM3A mRNA and protein were up-regulated at gd 13.5 in endovascular invasive trophoblast and within the junctional zone of hypoxia exposed placentation sites (Fig. 3 C–E).
Next, loss-of-function and gain-of-function experiments were performed in TS cells (Fig. 3 F–M). Knockdown of KDM3A using specific shRNAs did not affect proliferation of TS cells in ambient or low oxygen tensions (Fig. S3F) but did inhibit low oxygen-activated TS cell differentiation-dependent movement through extracellular matrices and the activation of several low oxygen-responsive transcripts (Mmp12, Il33, Ppp1r3c, etc.; Fig. 3 G–I). Not all low oxygen/HIF dependent transcripts were responsive to KDM3A manipulation (Fig. S3G). In contrast to the knockdown experiments, ectopic expression of KDM3A activated some low oxygen-responsive transcripts (Mmp12 and Il33) and stimulated movement of TS cells through extracellular matrices, both independent of exposure to low oxygen (Fig. 3 J–M). These gain-of-function actions were compromised in TS cells expressing a mutant KDM3A lacking demethylase activity (Fig. 3 J–M).
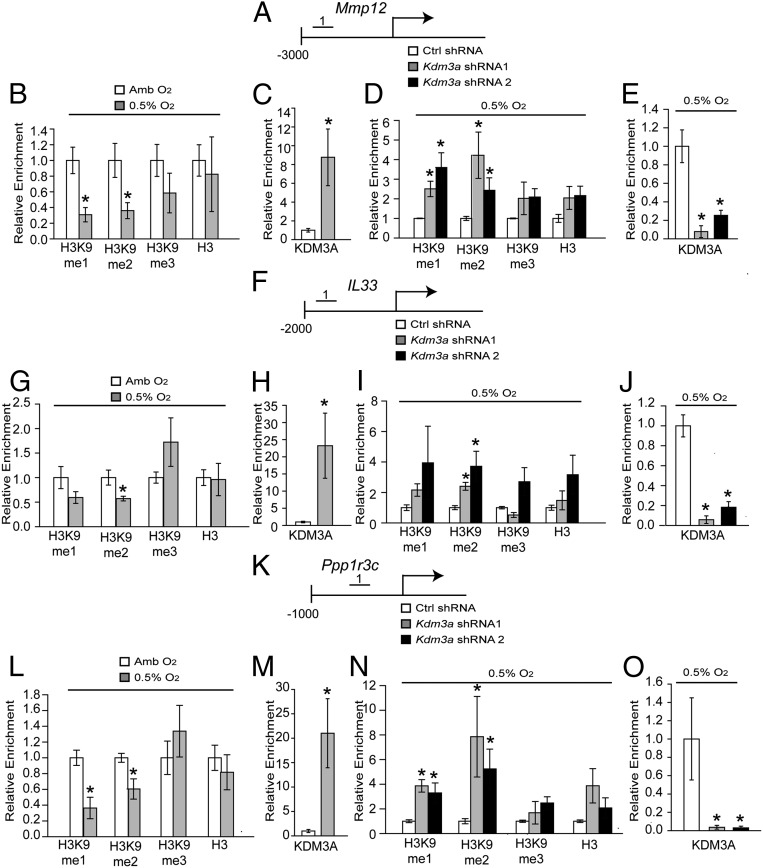
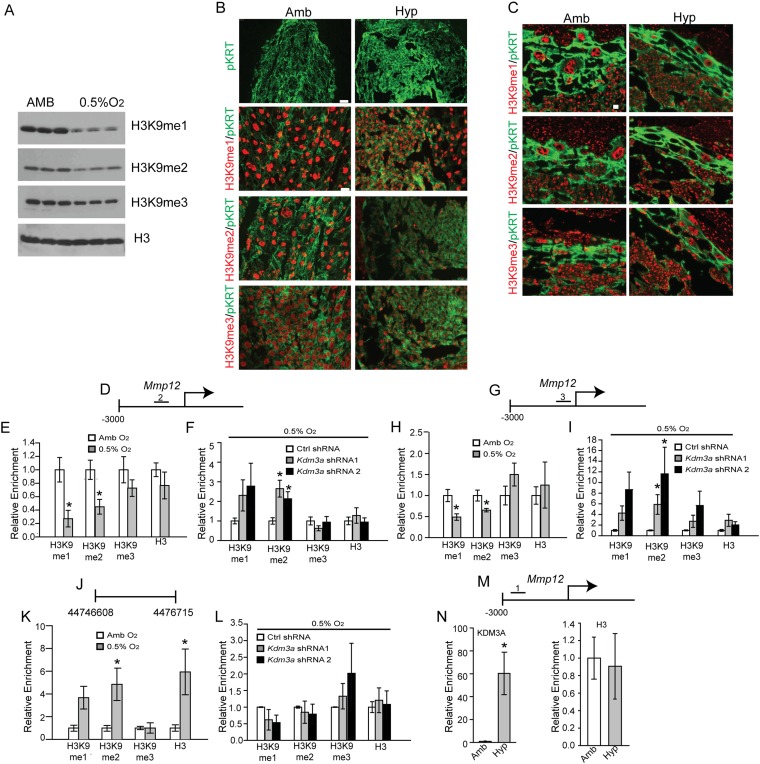
Because KDM3A acts as a histone H3K9 demethylase, we explored the H3K9 methylation landscape, including global- and gene-specific (Mmp12, Il33, and Ppp1r3c) H3K9 methylation in TS cells exposed to ambient or low oxygen conditions (Fig. 4). TS cells exposed to low oxygen and placentation sites exposed to maternal hypoxia exhibited generalized decreases in histone H3K9 monomethylation (H3K9-me1) and H3K9 dimethylation (H3K9-me2; Fig. S4 A–C). More specifically, low oxygen exposure led to significant decreases in histone H3K9 monomethylation (H3K9-me1) and H3K9 dimethylation (H3K9-me2) associated with regulatory regions of select hypoxia-HIF-KDM3A responsive genes but not control regions of the genome (Fig. 4 B, G, and L and Fig. S4 E, H, and K). Additionally, these shifts in H3K9 methylation status were not observed in KDM3A knockdown TS cells exposed to low oxygen (Fig. 4 D, I, and N and Fig. S4 F and I). Furthermore, KDM3A accumulated at these same putative regulatory regions associated with the Mmp12, Il33, and Ppp1r3c genes in low oxygen exposed TS cells (Fig. 4 C, E, H, J, M, and O) and at the Mmp12 locus in hypoxia exposed gd 13.5 junctional zone tissue (Fig. S4N). Conserved hypoxia response element (HRE) motifs could not be identified within the Mmp12, Il33, and Ppp1r3c regulatory regions binding KDM3A.
Fig. 4.
Histone H3K9 methylation landscape associated with KDM3A targets in hypoxia-exposed TS cells. (A) Schematic layout of the rat Mmp12 gene and the location of one of the regions [No. 1: −2681 to −2568 bp upstream transcription start site (TSS)] surveyed by ChIP analysis in B–E. (B) ChIP analyses for histone H3K9 methylation (monomethylation, me1; dimethylation, me2; trimethylation, me3) and histone 3 (H3) at the Mmp12 locus in ambient (Amb) and low oxygen (0.5% O2) exposed TS cells. (C) ChIP analysis for KDM3A at the Mmp12 locus in Amb and 0.5% O2 exposed TS cells. (D) ChIP analyses for histone H3K9 methylation and H3 at the Mmp12 locus in 0.5% O2 exposed TS cells treated with control (Ctrl) or Kdm3a shRNAs. (E) ChIP analysis for KDM3A at the Mmp12 locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. (F) Schematic layout of the rat Il33 gene and the location of one of the regions (No. 1: −1372 to −1272 bp upstream of TSS) surveyed by ChIP analysis in G–J. (G) ChIP analysis for histone H3K9 methylation and H3 at the Il33 locus in Amb and 0.5% O2 exposed TS cells. (H) ChIP analysis for KDM3A at the Il33 locus in Amb and 0.5% O2 exposed TS cells. (I) ChIP analyses for histone H3K9 methylation and H3 at the Il33 locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. (J) ChIP analysis for KDM3A at the Il33 locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. (K) Schematic layout of the rat Ppp1r3c gene and the location of one of the regions (No. 1: −417 to −316 bp upstream of TSS) surveyed by ChIP analysis in L–O. (L) ChIP analyses for histone H3K9 methylation and H3 at the Ppp1r3c locus in Amb and 0.5% O2 exposed TS cells. (M) ChIP analysis for KDM3A at the Ppp1r3c locus in Amb and 0.5% O2 exposed TS cells. (N) ChIP analyses for histone H3K9 methylation and H3 at the Ppp1r3c locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. (O) ChIP analysis for KDM3A at the Ppp1r3c locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. Statistical analyses: n = 4; control vs. hypoxia exposed TS cell experiments: Mann–Whitney test, *P < 0.05; shRNA experiments: ANOVA with Dunnett’s test vs. the control, *P < 0.05).
Fig. S4.
Regulatory landscape associated with KDM3A target genes. (A) Western blot analysis for histone H3K9 methylation (monomethylation, me1; dimethylation, me2; trimethylation, me3) in lysates from rat TS cells exposed to ambient (Amb) or low oxygen (0.5% O2). (B) Immunolocalization of H3K9me1, H3K9me2, and H3K9me3 on gd 9.5 placentation sites from rats exposed to Amb or hypoxic (10.5% O2) conditions from gd 6.5 to 9.5. (C) Immunolocalization of H3K9me1, H3K9me2, and H3K9me3 on gd 13.5 placentation sites from rats exposed to Amb or 10.5% O2 from gd 6.5 to 13.5. (D) Schematic layout of the rat Mmp12 gene and location No. 2 (−920 to −765 bp upstream of TSS) within the 5′ flanking region surveyed by ChIP analysis in E and F. (E) ChIP analyses for histone H3K9 methylation and histone 3 (H3) at the Mmp12 locus in Amb and 0.5% O2 exposed TS cells. (F) ChIP analyses for histone H3K9 methylation and H3 at the Mmp12 locus in 0.5% O2 exposed TS cells treated with control (Ctrl) or Kdm3a shRNAs. (G) Schematic layout of the rat Mmp12 gene and location No. 3 (−414 to −276 bp upstream of TSS) within the 5′ flanking region surveyed by ChIP analysis in H and I. (H) ChIP analyses for histone H3K9 methylation and H3 at the Mmp12 locus in Amb and 0.5% O2 exposed TS cells. (I) ChIP analyses for histone H3K9 methylation and H3 at the Mmp12 locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. (J) Schematic layout of the gene desert location (Rnor_6.0, 44,746,608–44,746,715) surveyed by ChIP analysis in K and L. (K) ChIP analyses for histone H3K9 methylation and H3 at a gene desert location in Amb and 0.5% O2 exposed TS cells. (L) ChIP analyses for histone H3K9 methylation and H3 at the gene desert locus in 0.5% O2 exposed TS cells treated with Ctrl or Kdm3a shRNAs. (M) Schematic layout of the rat Mmp12 gene and location No. 1 (−2,681 to −2,568 bp upstream of the TSS) within the 5′ flanking region surveyed by ChIP analysis in N. (N) In vivo ChIP analyses for KDM3A and H3 in junctional zone tissue from gd 13.5 pregnant rats exposed to Amb or hypoxic (10.5% O2; Hyp) conditions. Statistical analyses: n = 4; control vs. hypoxia exposed TS cell experiments: Mann–Whitney test, *P < 0.05; shRNA experiments: ANOVA with Dunnett’s test vs. the control, *P < 0.05.
Thus far, the results supported a role for KDM3A as a mediator of hypoxia-HIF actions on trophoblast cells.
Conservation of Hypoxia-Dependent Responses in Human Trophoblast Cells.
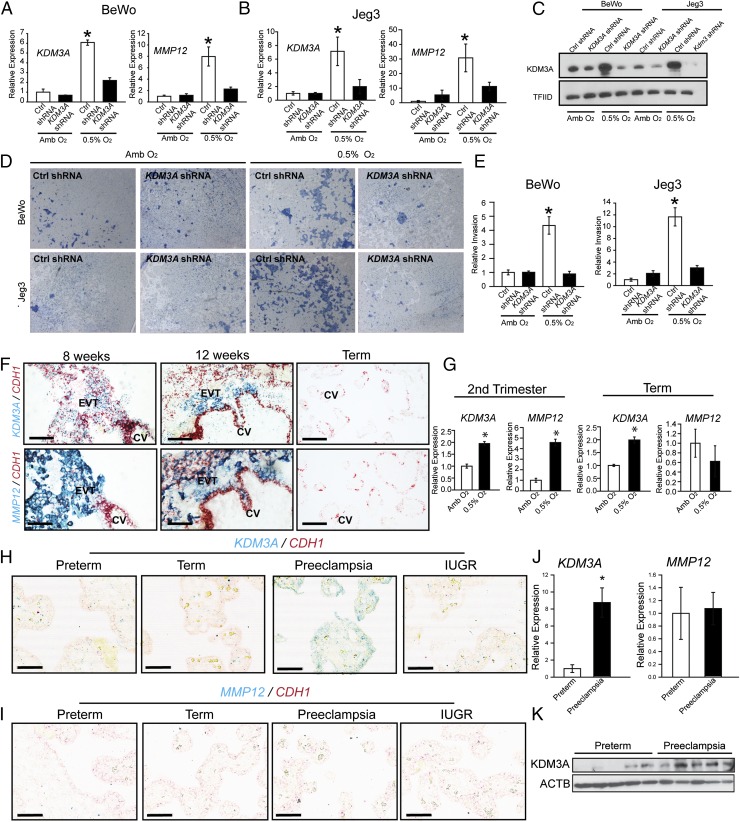
Next, we determined whether components of the hypoxia signaling pathway are conserved in human trophoblast cells (Fig. 5). First, we investigated responses of transformed trophoblast cell populations to low oxygen (Fig. 5 A–C). Each cell population responded to low oxygen with increases in KDM3A, MMP12, IL33, PPP1R3C, and PLOD2 expression (Fig. 5 A–C and Fig. S5 A–C). Low oxygen conditions also stimulated the movement of trophoblast cells through extracellular matrices (Fig. 5 D and E and Fig. S5 D and E). These low oxygen-activated responses were disrupted in KDM3A knockdown human trophoblast cells. Low oxygen stimulated KDM3A expression in primary second trimester and term trophoblast cells, whereas low oxygen stimulated MMP12 expression in primary second trimester trophoblasts but not primary term trophoblasts (Fig. 5G). The distribution of KDM3A transcripts and protein was also examined in the human placenta. KDM3A was localized to subpopulations of trophoblast cells within extravillous columns of first trimester human placentas, which represent the source of invasive trophoblast cells (Fig. 5F and Fig. S5 F and G). MMP12 transcripts were also localized to the extravillous columns (Fig. 5F and Fig. S5G). Thus, a nexus between KDM3A and MMP12 was established in the developing human placenta.
Fig. 5.
Conservation of hypoxia-dependent responses in human trophoblast cells. (A) Effects of KDM3A knockdown on low oxygen (0.5% O2) activated KDM3A and MMP12 transcript levels in BeWo human trophoblast cells (qRT-PCR; n = 3, *P < 0.05). (B) Effects of KDM3A knockdown on 0.5% O2 activated KDM3A and MMP12 transcript levels in Jeg3 human trophoblast cells (qRT-PCR; n = 3, *P < 0.05). (C) Effects of KDM3A knockdown on 0.5% O2 activated KDM3A protein in BeWo and Jeg3 human trophoblast cells. (D) Effects of KDM3A knockdown on 0.5% O2 induced invasive behavior of BeWo and Jeg3 human trophoblast cells. Images are representative Insets. (E) Quantification of invasion through Matrigel (n = 3, *P < 0.05). (F) In situ hybridization localization of CDH1 (red), KDM3A (blue, Top), and MMP12 (blue, Bottom) transcripts in first trimester (8 and 12 wk) and term human placental tissues. (Scale bar, 100 μm.) CV, chorionic villus; EVT, extravillous trophoblast. (G) Second trimester and term primary human trophoblast cell responses to 0.5% O2. KDM3A and MMP12 transcripts were measured by qRT-PCR (n = 3/group, Student t test, *P < 0.001). (H) In situ hybridization localization of CDH1 (red) and KDM3A (blue), transcripts in replicate representative placenta sections from preterm, term, preeclampsia, and intrauterine growth restriction (IUGR) pregnancies (low magnification: scale bar, 300 µm; high magnification: scale bar, 60 µm). (I) In situ hybridization localization of CDH1 (red) and MMP12 (blue), transcripts in replicate representative placenta sections from preterm, term, preeclampsia, and IUGR pregnancies. (Scale bar, 300 µm.) Data presented in A, B, and E were analyzed with ANOVA and Student–Newman–Keuls test. (J) qRT-PCR for Kdm3a and Mmp12 transcripts in placentas from preterm control and preeclamptic pregnancies (n = 6/group, Student t test, *P < 0.002). (K) Western blotting for KDM3A protein in placentas from preterm control and preeclamptic pregnancies (n = 5/group).
Fig. S5.
Hypoxia responses of human trophoblast cells and protein and transcript localization in human placental tissues. (A) Effects of low oxygen (0.5% O2) on KDM3A transcript (qRT-PCR) and protein (Western blot) expression in BeWo and Jeg3 human trophoblast cells. Amb, ambient. (B and C) Effects of 0.5% O2 on MMP12, IL33, PPP1R3C, and PLOD2 expression in BeWo (B) and Jeg3 (C) human trophoblast cells. Statistical analysis for A–C: n = 3/group; Mann–Whitney test, *P < 0.0.5. (D) Effects of 0.5% O2 on invasive behavior of BeWo and Jeg3 human trophoblast cells. Images are representative Insets. (E) Quantification of invasion through Matrigel (Student t test, *P < 0.05). (F) First trimester (6 and 8 wk) and term human placental tissues were processed for immunohistochemistry using KDM3A and pan cytokeratin (pKRT) antibodies. (Scale bar, 25 μm.) (G) In situ hybridization analysis of first trimester human placental tissue, including positive [POLR2A (red) and PPIB (blue)] and negative (DapB) controls, and colocalization of KDM3A (blue, third image) and MMP12 (blue, fourth image) with CDH1 (red). (Scale bar, 100 μm.)
KDM3A and MMP12 Expression in Diseased Human Placental Tissues.
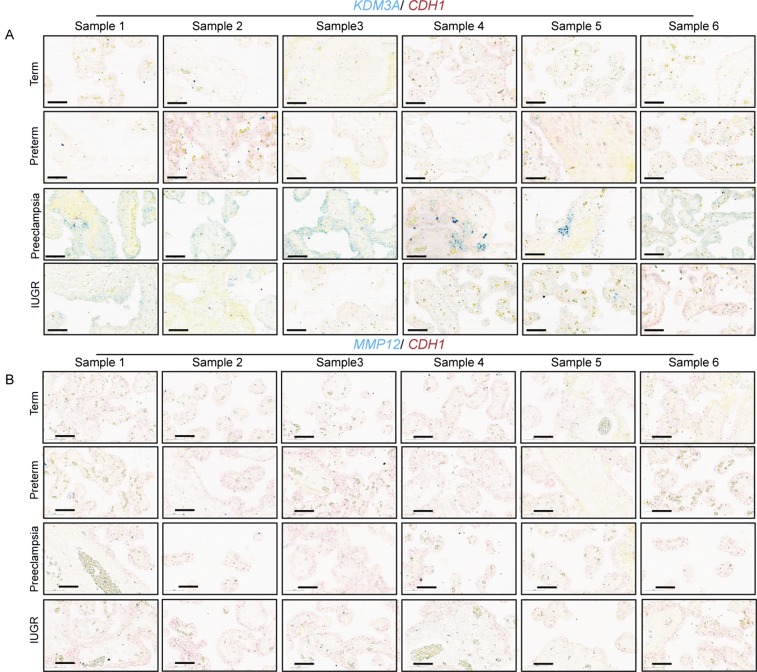
Preeclampsia is a multifaceted disease that can be associated with placental hypoxia and a failure of trophoblast-directed uterine spiral artery remodeling (3–5, 44–46). Analyses of placental tissue from preterm and term controls, intrauterine growth restriction, and preeclamptic pregnancies indicated that KDM3A expression was up-regulated in preeclampsia (Fig. 5H and Fig. S6A). This observation was further supported by quantitative RT-PCR (qRT-PCR) and Western blot analyses (Fig. 5 J and K). A finding consistent with the known hypoxia status of preeclamptic placental tissue (44–46) and the hypoxia-dependence of KDM3A expression (40–43). However, an up-regulation of MMP12 did not accompany the up-regulation of KDM3A in placental tissues from preeclamptic pregnancies (Fig. 5 H–K and Fig. S6), in contrast to the coordinated expression of KDM3A and MMP12 expression in first trimester placental tissue (Fig. 5F and Fig. S5G). A limitation of this experiment is that the analysis was performed on placental specimens obtained at delivery, whereas the hypoxia-HIF-KDM3A-MMP12 regulatory circuit we described is associated with an early pregnancy event. Nevertheless, the idea that MMP12 is dysregulated in preeclampsia is not unique to the present research; it is also supported by transcriptome profiling of chorionic villus samples (10–12 wk of gestation) from pregnancies destined to develop preeclampsia (47).
Fig. S6.
KDM3A and MMP12 expression in diseased human placental tissues. (A) In situ hybridization localization of CDH1 (red) and KDM3A (blue), transcripts in replicate placenta sections from preterm, term, preeclampsia, and IUGR pregnancies (n = 6 for each). (Scale bar, 60 μm.) (B) In situ hybridization localization of CDH1 (red) and MMP12 (blue), transcripts in replicate placenta sections from preterm, term, preeclampsia, and IUGR pregnancies (n = 6 for each). (Scale bar, 60 μm.) The integrity and effectiveness of CDH1 and MMP12 probes were examined in first trimester placental tissue as a control.
KDM3A and Hypoxia-Activated Trophoblast-Directed Uterine Spiral Artery Remodeling.
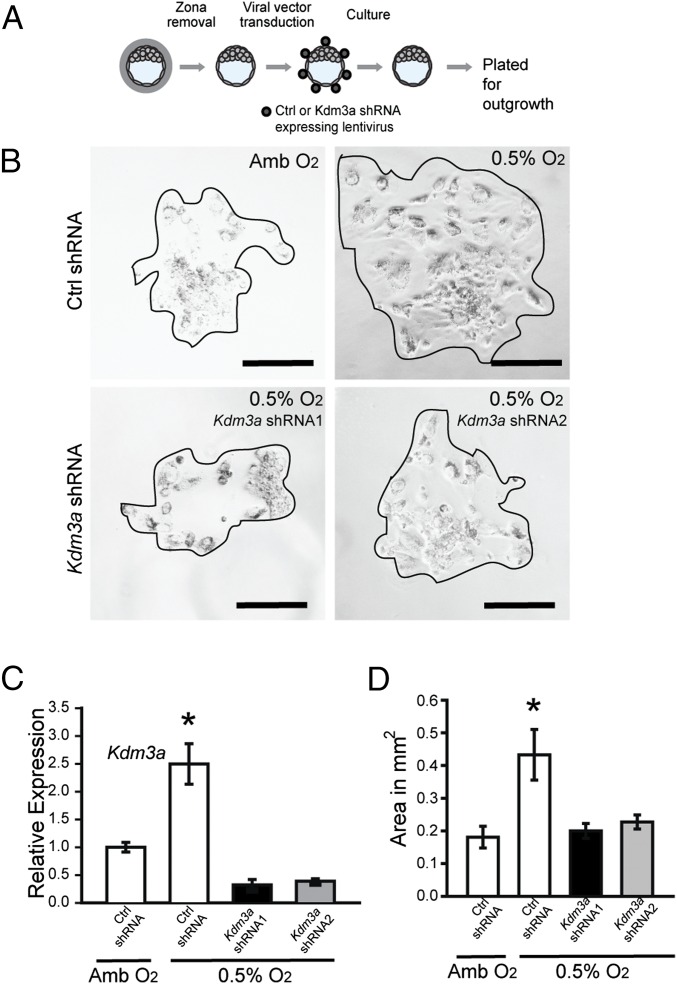
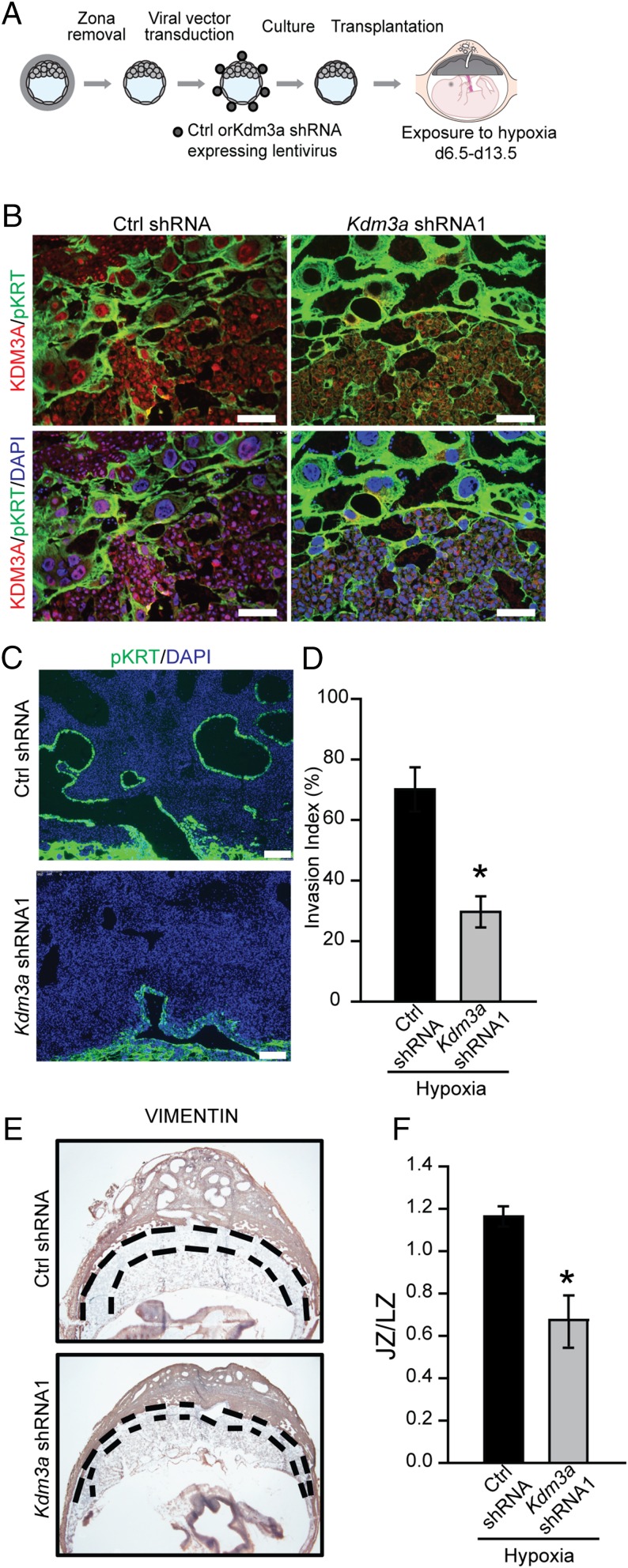
The provocative findings supporting a conserved role for KDM3A in trophoblast cell responses to low oxygen prompted an in vivo assessment of its involvement in hypoxia-activated placental adaptations. A loss-of-function strategy using lentiviral trophoblast-specific delivery of KDM3A shRNAs was used. Blastocysts were infected with KDM3A shRNAs or control shRNAs and examined ex vivo (Fig. 6A) or following transfer to pseudopregnant hosts (Fig. 7A). KDM3A shRNAs effectively down-regulated low oxygen-activated Kdm3a expression and in vitro blastocyst outgrowth (Fig. 6 B–D). Blastocyst outgrowth is a function of cell number, cell spreading, and cell movement, which were not distinguished in the assay. In vivo analysis demonstrated that KDM3A shRNAs down-regulated hypoxia-activated KDM3A expression (Fig. 7B) and interfered with hypoxia-activated trophoblast invasion and trophoblast-directed uterine spiral artery remodeling (Fig. 7C). Evidence also supported a role for KDM3A in hypoxia-induced expansion of the junctional zone (Fig. 7D). Our findings indicate that hypoxia acts through HIF and KDM3A to promote adaptations in placental development.
Fig. 6.
Effects of oxygen tension and KDM3A expression on blastocyst outgrowth. (A) Schematic showing experimental plan for lentiviral transduction of blastocysts and outgrowth assay. Blastocysts were transduced with control (Ctrl) or Kdm3a shRNA and cultured for 72 h to allow hatching from the zona pellucida. The attached blastocysts were exposed to ambient (Amb) or low oxygen (0.5% O2) for 24 h and analyzed. (B) Representative images of blastocyst outgrowths from Ctrl shRNA and exposed to Amb, Ctrl shRNA and exposed to 0.5% O2, and Kdm3a shRNAs and exposed to 0.5% O2. (C) Measurement of Kdm3a transcripts in control and knockdown cultures was measured by qRT-PCR. Asterisks indicate significant differences among groups (n = 6/group; *P < 0.05). (D) The bar graph shows quantification of outgrowth area in square millimeters. The area of the outgrowth was measured using Image J software (Ctrl shRNA + Amb, n = 6; Ctrl shRNA + 0.5% O2, Kdm3a shRNA1 + 0.5% O2, n = 10; Kdm3a shRNA2 + 0.5% O2, n = 10; *P < 0.05). Data presented in C and D were analyzed with ANOVA and Student–Newman–Keuls test.
Fig. 7.
KDM3A and hypoxia-activated trophoblast-directed uterine spiral artery remodeling. (A) Schematic showing experimental plan for lentiviral transduction of blastocysts and in vivo transfer to pseudopregnant recipient animals. (B) Representative images of immunolocalization of KDM3A and pan cytokeratin (pKRT) on gd 13.5 placentation sites expressing control (Ctrl) shRNA or Kdm3a shRNA exposed to 10.5% O2 tension from gd 6.5 to 13.5. (C) Immunolocalization of pKRT on gd 13.5 placentation sites expressing Ctrl shRNA or Kdm3a shRNA exposed to 10.5% O2 tension from gd 6.5 to 13.5. (D) Quantification of the depth of cytokeratin-positive cell penetration into the uterine mesometrial vasculature (n = 6/group; *P < 0.05). (E) Localization of vimentin in placentation sites following Ctrl shRNA or Kdm3a shRNA transduction. (Scale bar, 1 mm.) Dashed black lines demarcate the location of the junctional zone (JZ) relative to the underlying labyrinth zone (LZ). (F) Ratio of cross-sectional areas of JZ vs. LZ from Ctrl shRNA and Kdm3a shRNA transduced placentation sites (n = 5/group; *P < 0.05). Data presented in D and F were analyzed with Student t test.
Discussion
During development, the hemochorial placenta is constructed in response to stimuli present in the maternal environment and optimized to facilitate nutrient delivery to the fetus. Plasticity in placental organization can be achieved via differential regulation of TS cell proliferation and differentiation. Among the myriad of potential signals emanating from the maternal environment, oxygen delivery plays a significant role. Eukaryotic cells have evolved mechanisms for adapting to low oxygen exposure (18). TS cells use hypoxia-signaling pathways to control placentation (15, 21–23, 48). The proximal response of TS cells to hypoxia is their differentiation into invasive trophoblast cells capable of targeting uterine spiral arteries and expanding nutrient flow to the placenta and fetus (14, 15). In the present report, hypoxia is shown to activate a HIF-KDM3A-MMP12 signaling cascade that promotes trophoblast invasion and trophoblast-directed uterine spiral artery remodeling.
TS cell adaptations to hypoxia are hierarchically regulated. Low oxygen exposure elicits a broad spectrum of changes in the TS cell transcriptome highlighted by activation of genes associated with cell movement, invasion, and vascular remodeling. A subset of the hypoxia-induced changes in gene expression is dependent on HIF signaling, especially those genes activated by hypoxia, and a more restricted cohort of the HIF-dependent genes is linked to the actions of KDM3A. Thus, a path can be constructed from hypoxia to HIF activation to increases in KDM3A expression to alterations in the histone methylation status of genes promoting development of the invasive trophoblast lineage.
Hypoxia, HIF, and KDM3A have been implicated in regulating epithelial mesenchymal transition and cell invasion in disease states, including cancers (18, 25, 26, 42, 43). HIF binds to HREs at the Kdm3a locus and stimulates Kdm3a transcription (42, 43). KDM3A targets histone H3K9me1 and H3K9me2 for demethylation and thus extends and amplifies HIF signaling (42, 43). Early events in the development of the trophoblast lineage have also been associated with the methylation status of histone H3K9 (49). The histone H3K9me2 mark is associated with gene repression, and thus its removal is linked to gene activation (50). Loss of histone H3K9me2 modifications at several loci corresponding to hypoxia-HIF-KDM3A targets correlate with increases in target gene expression. Effectively, KDM3A regulates key downstream events in the development of the invasive trophoblast lineage and especially those necessary for trophoblast-directed uterine spiral artery remodeling, representing critical adaptations at the placentation site.
KDM3A is specifically targeted to its sites of action within the genome. HIF1A can recruit KDM3A to some of its target genes, including the Glut3 locus, resulting in H3K9me2 demethylation and transcriptional up-regulation (43). Under hypoxic conditions in TS cells, KDM3A may be delivered by HIF transcription factors to its target genes in some instances, whereas arrival at other loci may be independent of HIF (Mmp12 and Il33). This conclusion is based on the observed differential outcomes of loss-of-function and gain-of-function KDM3A manipulations in TS cells and the absence of conserved HIF binding motifs in regulatory regions binding KDM3A. In addition to HIF, there is evidence for other types of transcription factors (e.g., nuclear receptors and OCT1) guiding KDM3A to its genomic targets in various nontrophoblast cells (51–53). Mechanisms controlling KDM3A arrival at its target loci within the TS cell genome are yet to be elucidated.
Among the hypoxia-HIF-KDM3A targets in trophoblast cells, we focused our attention on MMP12 as a downstream effector. MMP12 was shown to be essential for placental adaptations to hypoxia, where it promoted endovascular invasion of trophoblast cells. These adaptive responses and actions of MMP12 are conserved (30, 34). Trophoblast-derived MMP12 contributes to uterine spiral artery remodeling in the human (30, 34). MMP12 acts on trophoblast cell invasive behavior in a cell autonomous mode. Mmp12-null TS cells exhibited disruptions in invasion, an observation reminiscent of defective invasive properties associated with MMP12 deficient macrophages (35), and consistent with in vivo findings presented in this report. Even though Mmp12-null TS cells did not invade through an extracellular matrix in response to low oxygen, some other cellular responses were consistent with the abilities of the mutant TS cells to respond to low oxygen. However, unlike WT TS cells, a down-regulation of Cdh1 in response to low oxygen was not observed. CDH1 encodes a protein pivotal to epithelial cell–cell adhesion and in general opposes invasive behaviors accompany cell invasion (54, 55), Previously, Aplin and colleagues provided compelling evidence that elastin-derived peptides promote human trophoblast cell migration and invasion (56). Thus, an MMP12 product of elastin degradation or MMP12-dependent actions on other cellular/extracellular substrate(s) could represent the active factor driving trophoblast migration and endovascular invasion critical to hypoxia-activated placental adaptations in the rat. Although MMP12 is vital to hypoxia activated trophoblast invasion, it is dispensable for interstitial and endovascular trophoblast invasion during the last week of gestation in the rat. These latter events may be influenced by the actions of other MMPs possessing substrate specificities that overlap with MMP12.
In conclusion, our observations connect hypoxia-dependent establishment of the invasive trophoblast lineage to HIF, KDM3A, and MMP12. HIF signaling is a key regulator of KDM3A expression. KDM3A acts on the epigenetic landscape of target loci, including the Mmp12 gene. MMP12 is a critical downstream effector of hypoxia-HIF-KDM3A signaling, controlling trophoblast invasion and trophoblast-directed uterine spiral artery remodeling. The hypoxia-HIF-KDM3A-MMP12 regulatory pathway is conserved and facilitates placental adaptations to environmental challenges. Interruption of the hypoxia-HIF-KDM3A-MMP12 signaling cascade may be at the origin of some placental diseases.
Methods
The University of Kansas Medical Center Animal Care and Use Committee approved all protocols performed. Animals and tissue collection, rat TS and human trophoblast cell cultures, DNA microarray, RT-PCR, immunohistochemistry, in situ hybridization, morphometric measurements, Western blot analysis, generation of the Mmp12 mutant rat model, shRNA constructs and production of lentivirus, ectopic expression of KDM3A, Matrigel invasion assay, chromatin immunoprecipitation analysis, ex vivo lentiviral trophoblast shRNA delivery and analyses, and statistical analyses are provided in SI Methods and primer and shRNA sequences are provided in Tables S2–S4.
Table S2.
Primer sequences
| Symbol | Gene ID/accession no. | Forward primer | Reverse primer | Amplicon size (bp) |
| Conventional RT-PCR analyses | ||||
| Mmp12 (genotyping) | 117033 | TACCTCCCCCGATAAAGTGC | GGGTGTGCATGTGTGTTTGT | 976 |
| Kdm5c (genotyping) | 317432 | TTTGTACGACTAGGCCCCAC | CCGCTGCCAAATTCTTTGG | 692 |
| Kdm5d (genotyping) | 100312983 | TTGGTGAGATGGCTGATTCC | CCGCTGCCAAATTCTTTGG | 250 |
| T | NM_001106209 | GCTCATCGGAACAACTCTCC | CTCCGAGGCTAGACCAGTTG | 91 |
| Pou5f1 | NM_013633 | GAGGTGGAACCTGGAGAACA | GCCGGTTACAGAACCACACT | 122 |
| Hnf4 | NM_022180 | GTGTCGTTACTGCAGGCTCA | GCTGTCCTCGTAGCTTGACC | 109 |
| Cdx2 | NM_023963 | CCCTAGGAAGCCAAGTGAAA | CTGCGGTTCTGAAACCAAAT | 187 |
| Prl3b1 | NM_012535 | CAAGTCCACCAGGCAAAATC | CATGCACCGATACAGGACAC | 311 |
| Mmp12 | NM_053963 | TTGATGAACTGGACTCTG | GAAGGTATCTGTAGGTAGG | 757 |
| qRT-PCR | ||||
| Ppp1r3c | NM_001012072 | TACCTGAACGACCATCTA | AAAGTGTGGCAGAAAGTT | |
| Mmp12 | NM_053963 | GCTGGTTCGGTTGTTAGG | GTAGTTACACCCTGAGCATAC | |
| Plod2 | NM_175869 | AGGAAGATCATTGCCCCTCT | ATCTCTGATCGCAGCGTCTT | |
| Bhlhe40 | NM_053328 | TGAAGCGACACCTCAAAG | GGTCTCAAGGGGTATGTTC | |
| Il33 | NM_001014166 | CAACGACCAATCTGTTAG | CGGAGTAGCACCTTATCT | |
| Kdm3a | NM_175764.2 | CTGAGATTCCTGAGCAAGTTATTC | AGCCGAAGACTGTTTACATCC | |
| Cd200 | NM_031518 | GTCTACTTGAATCTGATGTTA | TGTAAATACTGAGGACCC | |
| Loxl2 | NM_001106047 | GCAGAGAAGACTTACAACC | AGATGTGTGCTTCAGTTC | |
| Slc2a1 | NM_138827 | GCTCAGTGTCATCTTCATC | ACCCTCTTCTTTCATCTCC | |
| Dpp3 | NM_053748 | AGAGTTACATTGGCTTCA | CTTTGCACTCATGTCTTT | |
| Vegfa | NM_031836 | TTAGTGGTTTCAATGGTCTG | GGTCCTGGCAAAGAGAAG | |
| Mmp9 | NM_031055 | TACTGCTGGTCCTTCTGA | CCGTCCTTGAAGAAATGC | |
| Ndrg1 | NM_001011991 | CCCACTAAACTCATTCCT | TCAGCATTCAGAACTCTAA | |
| Plau | NM_013085 | TTGGTTCAGACTGTGAGAT | GCACTGTTCGTGAGAAAT | |
| Comt | NM_012531 | CTGACTTCCTGGCGTATG | TTCTCCAAGCCGTCTACA | |
| Cul7 | XM_002727163 | CAAGTCATCAGACCTTCC | TTCACATCCGTTTCCATC | |
| Cdh1 | NM_031334 | TTCTGAAGACTCCCGATT | ATGGACGAGAAAACTGGT | |
| Ncoa3 | XM_215947 | AGTGTATCGTTTCTCATT | TACATCCATTCTGTTCTC | |
| Kat6a | NM_001100570 | AATCTTGTATGCTATGGTTA | TTATCCTGATGCTGAGTA | |
| Id1 | NM_012797 | CTGAACGGCGAGATCAGTG | GGAGTCCATCTGGTTCCTCA | |
| Rbm3 | NM_053696 | ATATGGAAGTGGAAGATA | TTGTCATAATTGTCTCTG | |
| Satb1 | NM_001012129.2 | GCCACTGGTATTTCCTAC | TCCTTCTGACTTCCTGTT | |
| Ccdc86 | NM_001006974.2 | CAACGAGAGCAGTCATCCA | TGGGATCACCTCCTCCTTAG | |
| Lin28 | NM_001109269 | AAACAAGTGTCAAACCAAGATTA | GCCCTGTGGAAATAACCT | |
| Hand1 | NM_021592 | AGCAAGCGGAAAAGGGAGT | CGGCCTCTTCTCACTGATTT | |
| Cdx2 | NM_023963 | CAGGAGGAAAGCTGAGTTGG | TGCTGCTGTTGCAACTTCTT | |
| Egln1 | XM_008772679.1 | GACCTGTCACCTAACTGAG | AATAGCATGAAGAGGTTTACAAA | |
| Ankrd37 | NM_001108400 | TGAGACAGAAGCGGAGTT | TGCCCAACAAGACATCATC | |
| Tpbpa | NM_172073 | GCAAGAGCAGAAGGGTAAAGAAGG | TTTCTATGTCGAGCTCCTCCTCCT | |
| Gjb3 | NM_019240 | TGTGAACCAGTACTCCACCGCATT | GCTGCCTGGTGTTACAGTCAAAGT | |
| Ascl2 | NM_031503 | GATGGGGCAGATGTTTGACT | GCAAGAAACGCAGGTAGGTC | |
| Tfeb | NM_001025707 | ACTCAGTTTCTCCTTATGC | AGACAGGTCCATGAAGTA | |
| Gcm1 | NM_017186 | AGAGGCAAGAAGAGCCATGA | TCCCTGACTTGGGATTTCAC | |
| Prl7b1 | NM_153738 | AACAATGCCTCTGGCCACTGC | AGGCCATTGATGTGCTGAGACAGT | |
| Prl5a1 | NM_138527 | TCCACACCAGACATTCCAGA | TTTCCAGGAAGCCAACATTC | |
| Phlda2 | NM_001100521.1 | AGAGACCCACGGGAGTAG | AGCAGGTAACCAATATAAATACG | |
| Id2 | NM_013060 | GGACATCAGCATCCTGTCCT | AAAAAGGAAAAAGTCCCCAAA | |
| Trh | NM_013046.3 | GGTGCTGCCTTAGACTCCTG | TTCTCCCAAGTCTCCCCTCT | |
| Mmp10 | NM_133514.1 | AATTGTTCCTGCATGTGCTGTGGC | TTCTGTGCTCAGGTGATGCTCTGT | |
| Htra1 | NM_031721.1 | GCGGGCAAAACAAATGTAAT | GGCAGGGACAGATGTTGACT | |
| Plac1 | NM_001024894.1 | ACTATGCTTCCAAGGGCTCA | CGCCCATGTTACTGCTAGGT | |
| Slpi | NM_053372.1 | GAATCCTGTTCCCATTCGTG | TTCCCACACATACCCTCACA | |
| Bmp6 | NM_013107.1 | TTTGTGGAGGTGCCTTCTGT | GTATGGCGTGGGAATGCTAT | |
| Ctsq | NM_139262.1 | TCATCCTCTGCTTGGGAGTT | TCCACTCTTGCCATTGAACA | |
| Prl2a1 | NM_053791.2 | GCTTCCAAAACCCAGCAGTA | AAGGATGGCAGGTTGTTCAC | |
| Itgb6 | NM_001004263.1 | TTGCTCCTCAAAGCTTGGTT | CCGAGAGGTCCATGAGGTAA | |
| Prl4a1 | NM_017036.2 | GCTCCTGGATGCCATAAAGA | TATTGGGCGATTGCAAGAAT | |
| F3 | NM_013057.2 | ACAAATGCACTGGAACCACA | TCCCCATGAGTTCCAAAGAG | |
| Epcam | NM_138541.1 | ATTTGCTCCAAACTGGCATC | TCGTCACACTCGGGATCATA | |
| Wfdc10 | NM_001109461.1 | GTCCAGCGAAAACGGAGATA | GGCGTCTCCCACACACTATT | |
| Igfbp1 | NM_013144.1 | GCGGTAGTGCCTAGAACGAG | TGGGATTCGATGAGGAAGTC | |
| Lsd1 | NM_001130098.1 | ATTATTATAGGCTCAGGTGTT | CAGAAGTGTGACATCCAT | |
| Kdm4a | NM_001107966.1 | AGCCTTCCCTTCCCAGATTA | CGAGGCGTCCTTCTACAGAC | |
| Kdm4b | NM_001044236.2 | ACAGCTGGGGTAGCTGGTCT | CTTGCTCCTGGCCTTAGTTG | |
| Kdm4c | NM_001106663.3 | TGACACCAAGCTCTCAGGAA | AACGTGACTCCTTGCCTGTT | |
| Kdm4d | NM_001079712.1 | TAGCCCCACCATATCCTCTG | GGACTGGGACTGAAGCTCTG | |
| Kdm3b | XM_008772060.1 | AAAGGAAAGAAGAAGAGA | CATTACTGTCTACTTCAC | |
| Kdm3c | NM_001191719.1 | GTTACTTAGTCACATTCCT | TTCCTCTCCAATTCTTCT | |
| MMP12 (human) | NM_002426.5 | TGGTTTTTGCCCGTGGAGCTCAT | GAATGGCCAATCTCGTGAACAGCA | |
| IL33 (human) | NM_001199640.1 | GGCTGAGAATTACCATACAAGG | AGTGTTTTTCAGATGGGATGA | |
| PLOD2 (human) | NM_182943.2 | GCGTTCTCTTCGTCCTCATC | CATGAAGCTCCAGCCTTTTC | |
| KDM3A (human) | NM_001146688.1 | ACTCTTCAAGTCAACTGT | ATGTCACAAGGTTATCGT | |
| PPP1R3C (human) | NM_005398.5 | CATGTTGGGCAAGGAAAAGT | CCCACTCCTCtGACTTGAGC | |
| 18S rRNA | M11188 | GCAATTATTCCCCATGAACG | GGCCTCACTAAACCATCCAA |
Table S4.
ChIP primer sequences
| Target gene | Forward primer | Reverse primer |
| Mmp12 region 1 | TCCTGGGCCTCTGACTCTTA | AGTCAGCTGCCTGGAACATT |
| Mmp12 region 2 | AAGGATTCGTGTATTCTAGCA | ACGCCATTAGCAGAGTTT |
| Mmp12 region 3 | TGCTGAGTCATTTTGCAAGC | ATCCCACCACAAGCAGAGTC |
| Il33 region 1 | TGACTTCCCAGACGGGTAAC | CCATTATTCACCAGGCAAGC |
| Ppp1r3c region 1 | TTGGATACACAACGCAAGGA | GTGGGTCTGTGCAGAGTTGA |
| Kdm3a HRE | AGGATCATGAGCGCCTATTG | TCTAACTGCGTACGCCTGAA |
| Vegfa HRE | GCCAGACTCCACAGTGCATA | TTCCCAGGGGAGAAGAATTT |
| Gene desert | GGGGGATAATGATTGCAAAA | GCGTGGACAGAGATCTAGGC |
SI Methods
Animals and Tissue Collection.
Holtzman Sprague–Dawley rats were acquired from Envigo. Animals were housed in an environmentally controlled facility with lights on from 0600 to 2000 hours and allowed free access to food and water. Virgin female rats 8–10 wk of age were cohabited with adult males (>3 mo of age) of the same strain. Mating was assessed by inspection of vaginal lavages. The presence of sperm in the vaginal lavage was considered gd 0.5. Rat embryos were collected by flushing uteri with M2 medium (Millipore) at gd 4.5.
Pregnant rats were placed in a hypoxic [10.5% (vol/vol) oxygen] gas-regulated chamber (BioSpherix, Lacona, NY) from gd 6.5–13.5. Pregnant rats exposed to ambient conditions (normoxia) served as controls. Rat placental tissues were collected on gd 13.5. Placentation site dissections were performed as previously described (57). Tissues for histological analysis were frozen in dry-ice cooled heptane and stored at −80 °C. Tissue samples for protein or RNA extraction were frozen in liquid nitrogen and stored at −80 °C until processed. The University of Kansas Medical Center (KUMC) Animal Care and Use Committee approved protocols for the care and use of animals.
Paraffin-embedded human placental tissue and tissue microarrays containing placental tissue samples from preterm, term, preeclampsia, and IUGR pregnancies were obtained from the Research Centre for Women’s and Children’s Health Biobank (Mount Sinai Hospital, Toronto, CA). Clinical details of the tissue microarray are provided in Dataset S1. Tissue collections were performed with consent and were approved by the University of Toronto and the KUMC human research ethics review committees.
Rat TS and Human Trophoblast Cell Cultures.
Blastocyst-derived rat TS cells (32), mouse TS cells (58), Rcho-1 TS cells (59), and human trophoblast cells were used to evaluate the effects of low oxygen on trophoblast cell behavior. Rat TS cells were cultured in basal culture medium [RPMI 1640 (Cellgro), 20% (vol/vol) FBS (Sigma Aldrich), 100 µM 2-mercaptoethanol (Sigma-Aldrich), 1 mM sodium pyruvate (Cellgro), 50 µM penicillin, and 50 U/mL streptomycin (Cellgro)] supplemented with 70% rat embryonic fibroblast conditioned medium, FGF4 (25 ng/mL; Sigma-Aldrich), and heparin (1 µg/mL; Sigma-Aldrich). New rat TS cells were established from WT and Mmp12-null blastocysts, characterized, and maintained based on procedures previously described (32). Sex chromosome composition of rat TS cells was performed on genomic DNA using PCR for Kdm5c (X chromosome) and Kdm5d (Y chromosome) genes (60). Primers can be found in Table S2. Mouse TS cells were cultured under the same conditions with the substitution embryonic fibroblast conditioned medium from mouse instead of rat. Rcho-1 TS cells were maintained in basal culture medium without conditioned medium, FGF4, and heparin.
Human trophoblast cell lines (BeWo and JEG3) were obtained from the American Type Culture Collection and cultured in DMEM culture medium supplemented with 10% (vol/vol) FBS, 50 µM penicillin, and 50 U/mL streptomycin (Cellgro). Primary cultures of human trophoblast (20–22 wk of gestation) were obtained from ScienCell and cultured according to the vendor’s recommendations. Term trophoblast cells were processed from placental villous tissue obtained with consent from the University of Kansas Hospital under a protocol approved by the KUMC Institutional Review Board. Trophoblast cells were processed according to the method of Kliman et al. (61).
Cells were exposed to ambient or low oxygen tensions (0.5%) and 5% CO2 at 37 °C using a NAPCO Series 8000WJ incubator (Thermo Scientific) for 24 h and then harvested for cell and molecular analyses. Pre-equilibration of culture medium to the low oxygen tension was not performed before the initiation of the experiments.
DNA Microarray.
Affymetrix 230 2.0 DNA microarray chips (Affymetrix) were probed with cDNAs generated from rat TS cells grown under oxygen replete conditions versus low (0.5%) oxygen conditions or alternatively from dissected gd 13.5 metrial gland tissue harvested from pregnant rats exposed to normoxic versus hypoxic [10.5% (vol/vol) oxygen] conditions between gd 6.5 and 13.5. Each treatment group was repeated in triplicate. RNA samples were hybridized to the Affymetrix 230 2.0 DNA microarray chip using the GeneChip Hybridization Oven 640 (Affymetrix). Washing and staining of hybridized chips were conducted using the GeneChip Fluidics Station 450 (Affymetrix). Chips were scanned using the Affymetrix GeneChip Scanner 3000 (Affymetrix) with autoloader by the KUMC Biotechnology Support Facility. Hybridization signals were normalized with internal controls using the Mas5 algorithm in Expression Console (Affymetrix) and fold change computed. Significant differences were determined by paired two-tailed Student t tests. Microarray data were processed for functional analysis using Ingenuity Pathway Analysis. Probe sets included in the analysis were restricted to those changing at least 1.5-fold between group comparisons with signal strengths of ≥500 for the maximal value.
RT-PCR.
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen). cDNAs were synthesized from total RNA (1 µg) for each sample using SuperScript 2 reverse transcriptase (Invitrogen), diluted five times with water, and subjected to qRT-PCR to estimate mRNA levels. Primers were designed using Beacon primer designer (Applied Biosystems). Conventional RT-PCR and qRT-PCR primer sequences are presented in Table S2. Conventional RT-PCR was performed for 30–35 cycles (94°C for 30 s; 55–60 °C for 30 s; 72 °C for 30 s). Real-time PCR amplification of cDNAs for qRT-PCR was carried out in a reaction mixture (20 µL) containing SYBR GREEN PCR Master Mix (Applied Biosystems,) and primers (250 nM each). Amplification and fluorescence detection were carried out using the ABI 7500 real-time PCR system (Applied Biosystems). Cycling conditions included an initial hold step (95 °C for 10 min) and 40 cycles of a two-step PCR (92 °C for 15 s and then 60 °C for 1 min), followed by a dissociation step (95 °C for 15 s, 60 °C for 15 s, and then 95 °C for 15 s). The comparative cycle threshold method was used for relative quantification of the amount of mRNA for each sample normalized to 18S RNA, and 18S RNA was stable among the conditions and tissues tested.
Immunohistochemistry.
Immunocytochemical analyses were performed on 10-µm frozen tissue sections using indirect immunofluorescence detection using goat anti-mouse IgG tagged with Alexa 488 (Invitrogen; cat. no. A11029; 1:1,000 dilution), goat anti-mouse IgG tagged with Alexa 568 (Invitrogen; cat. no. A11031; 1:400 dilution), or goat anti-rabbit IgG tagged with cyanine 3 (Cy3; Jackson ImmunoResearch Laboratories; cat. no. 111-165-003; 1:250 dilution). Nuclei were visualized with DAPI (Molecular Probes). Primary antibodies to human KDM3A (Novus; cat. no. NBP1-49601), mouse KDM3A (62), MMP12 (AbCam; cat. no. ab66157), pan cytokeratin (Sigma-Aldrich; cat. no. F3418), and vimentin (Sigma-Aldrich; cat. no. V6630) were used in the analyses. Negative controls were performed with normal rabbit serum or isotype-specific control mouse IgG and did not exhibit positive reactivity in the tissue sections. Processed tissue sections were inspected and images recorded with a Leica MZFLIII stereomicroscope equipped with a charge-coupled device (CCD) camera (Leica).
In Situ Hybridization.
Detection of Prl5a1 in situ hybridization was performed as previously described (33). Ten-micrometer cryosections were prepared and stored at −80 °C until used. A plasmid containing a cDNA for PRL family 5, subfamily a, member 1 (Prl5a1) was used as a template to synthesize sense and antisense digoxigenin-labeled riboprobes according to the manufacturer's instructions (Roche Molecular Biochemicals). Tissue sections were air dried and fixed in ice cold 4% (wt/vol) paraformaldehyde in PBS. Prehybridization, hybridization, and detection of alkaline phosphatase-conjugated antidigoxigenin were performed. Detection of KDM3A, CDH1, and MMP12 transcripts was performed on paraffin-embedded human placenta tissue sections using the RNAScope 2-plex chromogenic assay (Advanced Cell Diagnostics), according to the manufacturer’s instructions. Probes were prepared to detect KDM3A (NM_018433.5; target region: 450–1,443), CDH1 (NM_004360.3; target region: 263–1,255), MMP12 (NM_002426.4; target region: 295–1,598), POLR2A (positive control, NM_000937.4; target region: 2,514–3,433), PPIB (positive control, NM_000942.4; target region: 139–989); and DapB (negative control, EF191515; target region: 414–862).
Morphometric Measurements.
Morphological measurements of the depth of invasion and sizes of placental compartments were performed with NIH Image J software as previously described (14, 15). The depth of intrauterine trophoblast cell invasion was used to determine an invasion index (distance of endovascular cytokeratin positive cell location relative to the trophoblast giant cell layer of the chorioallantoic placenta/total distance from the trophoblast giant cell layer to the outer mesometrial surface of the uterus) and used in part as a surrogate for assessing trophoblast-directed uterine spiral artery remodeling. The thickness of the junctional zone was estimated from cross-sectional area measurements of vimentin immunostained placentation sites. Measurements were expressed as the ratio of junctional zone to labyrinth zone cross-sectional areas. Depth of intrauterine trophoblast cell invasion and chorioallantoic zone measurements were all made from a histological plane at the center of each placentation site perpendicular to the flat fetal surface of the placenta. Sample sizes for the analyses were at least five placentation sites from at least five different animals per treatment group.
Western Blot Analysis.
KDM3A, ACTB, histone H3, histone H3K9 methylation modifications (monomethylated: H3K9me1, dimethylated: H3K9me2, trimethylated: H3K9me3), and MMP12 were monitored by Western blot analysis. Rat TS cell lysates were prepared in RIPA buffer (10 mM Tris⋅HCl, pH 7.2, 1% Triton X-100 or 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 5 mM EDTA, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/mL aprotinin). Protein concentrations were determined by the DC protein assay (Bio-Rad). Proteins were separated by SDS/PAGE and transferred onto nitrocellulose membranes. Immunoreactive proteins were detected with rabbit antibodies to human KDM3A (Novus; cat. no. NBP1-49601), mouse KDM3A (62), ACTB (Sigma-Aldrich; cat. no. A1978), TFIID (Santa Cruz Biotechnology; cat. no. sc-56794), MMP12 (AbCam; cat. no. ab66157), and FLAG (Sigma-Aldrich; cat. no. F3165). Antibodies to histone H3 and histone H3K9 methylated isoforms were obtained from Abcam (histone H3, cat. no. ab1791; H3K9me1, cat. no. ab9045; H3K9me2, cat. no. ab1220; H3K9me3, cat. no. ab8898). Immunoreactive proteins were visualized by enhanced chemiluminescence according to the manufacturer’s instructions (Amersham Biosciences).
Generation of the Mmp12 Mutant Rat Model.
Targeted mutations were generated using TALEN genome editing. TALEN constructs specific for the rat Mmp12 gene were designed, assembled, and validated by the Genome Engineering Center, Washington University. Selected TALENs were targeted to exon 2 of the Mmp12 gene (target sequence: TCTGAAAATAATGCACACGTctcgatgtggagtgcctgATGTACAGCATCTTAGA; corresponding to nucleotides 281–300 and 319–335, NM_053963.2; Fig. S1). Genome editing constructs were microinjected into the cytoplasm of single-cell rat embryos. Injected embryos were transferred to oviducts of day 0.5 pseudopregnant rats. Offspring were screened for mutations at specific target sites within the Mmp12 gene. For initial screening, genomic DNA was purified from tail-tip biopsies using the E.Z.N.A. tissue DNA kit (Omega Bio-Tek). Potential mutations within 5,000 bp at each target locus were screened by PCR and precise boundaries of deletions determined by DNA sequencing (Genewiz). Founders were backcrossed with WT rats to demonstrate germ-line transmission. Two mutant rat strains possessing a 607- or 664-bp deletion including all of exon 2 of the Mmp12 gene were characterized (Fig. S1). Genotyping was performed using PCR on genomic DNA purified from tail-tip biopsies using the REDExtract-N-Amp tissue PCR kit (Sigma-Aldrich) with specific sets of primers shown in Table S2. The mutant Mmp12 rat model is available at the Rat Resource & Research Center (University of Missouri).
shRNA Constructs and Production of Lentivirus.
shRNA constructs cloned into the pLKO.1 vector were obtained from Open Biosystems. Several shRNAs were tested for each gene evaluated. shRNA sequences used in the analyses are provided in Table S3. Lentiviral packaging vectors were purchased from Addgene and included pMDLg/pRRE (plasmid 12251), pRSV-Rev (plasmid 12253), and pMD2.G (plasmid 12259). Lentiviral particles were produced as previously reported (63). In brief, 293FT cells (Invitrogen) were transiently transfected using Lipofectamine 2000 (Invitrogen) with the following plasmids: shRNA containing transducing vector, packaging system plasmids (pMDLg/pRRE and pRSV-Rev), and a VSVG envelope plasmid (pMD2.G). After transfection, cells were maintained in 50% DMEM with high glucose (Cellgro), 45% Opti-MEM I (Invitrogen), and 5% (vol/vol) FBS. Culture supernatants containing lentiviral particles were harvested every 24 h for 2–3 d. Supernatants were centrifuged to remove cell debris, filter sterilized, concentrated by ultracentrifugation, and stored at −80 °C until used.
Table S3.
shRNA sequences
| Target | shRNA sequence |
| Control shRNA | CCTAAGGTTAAGTCGCCCTCGCTCTAGCGAGGGCGACTTAACCTTAGG |
| Hif1b shRNA | GAGAAGTCAGAAGGTCTCTTTTCTCGAGTAAAGAGACCTTCTGACTTCTC |
| Kdm3a shRNA-1 | CATGATCAGAGCTGGTATTTACTCGAGTAAATACCAGCTGATCATG |
| Kdm3a shRNA-2 | CTTCGAAGTTTCATTGGATTTCTCGAGAAATCCAATGAAACTTCGAAG |
| KDM3A shRNA (human) | CTTTGATTGTGAAGCATTTACTCGAGTAAATGCTTCACAATCAAAGCT |
Ectopic Expression of KDM3A.
A WT FLAG-tagged mouse Kdm3a pcDNA3 expression vector (64) was generously provided by Christopher Mack, University of North Carolina, Chapel Hill, NC. An enzymatically dead version of KDM3A was generated from full-length mouse Kdm3a (Open Biosystems-GE Dharmacon) by introducing a point mutation within the catalytic domain (H1135Y) using PCR-based site directed mutagenesis and subcloned into pcDNA3. Empty and FLAG-tagged pcDNA3 vectors expressing WT or mutant Kdm3a were transfected into rat TS cells using Lipofectamine 2000 (LifeTechnologies) transfection reagent according to manufacturer’s instructions. Cells were selected with Geneticin (LifeTechnologies) for 5 d before the initiation of the experiments.
Matrigel Invasion Assay.
Invasive properties of rat TS cells were assessed as previously described (15). Rat TS cells were placed in Matrigel invasion chambers (BD Biosciences) at a density of 2 × 104 cells per chamber. Cells were allowed to invade for 24 h. Membranes were collected and stained with Diff-Quick (Dade Behring). Invading cells were visualized by light microscopy and counted.
ChIP.
ChIP analysis was performed according to a previously published procedure (42). Briefly, TS cells were grown in 150-mm dishes for 2 d in ambient conditions and then exposed to 0.5% oxygen for 24 h. Cells were then fixed with 1% formaldehyde, and purified nuclear lysates were sonicated using a Diagenode Bioruptor (Diagenode) to prepare DNA fragments at a size of ∼500 bp. Lysates were immunoprecipitated with 3 µg of appropriate antibodies obtained from AbCam (ARNT, cat. no. ab2; histone H3, cat. no. ab1791; H3K9me1, cat. no. ab9045; H3K9me2, cat. no. ab1220; H3K9me3, cat. no. ab8898; KDM3A, cat. no. NB100-77282) or with rabbit or mouse IgG (BD Biosciences). Immunoprecipitated chromatin fragments were eluted from agarose beads. DNA–protein interactions were reverse cross-linked and purified using a QiaQuick PCR purification kit (Qiagen). Purified DNA fragments were assessed by qPCR using primers designed to putative regulatory regions of target or control genes (Table S4) and SYBRgreen PCR master mix (Applied Biosystems). Putative regulatory regions were examined throughout 5 kb upstream and 1 kb downstream of the transcription start site for the target genes and at putative CpG islands. The presence of conserved hypoxia response elements (HREs) was determined using the MultiTF program (https://multitf.dcode.org/). Relative occupancy/enrichment was normalized to input samples by use of the ΔΔCT method.
Ex Vivo Lentiviral Trophoblast shRNA Delivery and Analyses.
Rat embryos were transduced with lentiviral particles as previously described (63) containing control or Kdm3a shRNAs. Briefly, blastocysts collected on gd 4.5 were treated with Pronase (Sigma-Aldrich; 10 mg/mL for 10 min) to remove zonae pellucidae and incubated with concentrated lentiviral particles (750 ng of p24/mL) for 4.5 h. Transduced blastocysts were cultured ex vivo for blastocyst outgrowth analysis or transferred to uteri of day 3.5 pseudopregnant rats for subsequent in vivo evaluation of gene knockdown and placentation site phenotypes.
The blastocyst outgrowth analysis was performed as reported before (33). Blastocysts were obtained from gd 4.5 rats by flushing them from uteri with 0.1 mL DMEM (Life Technology). Embryos were cultured in basal culture medium at 37 °C in a 5% CO2 incubator for 72 h to allow hatching from the zona pellucida. The attached blastocysts were exposed to ambient or 0.5% oxygen for 24 h in a NAPCO Series 8000WJ incubator (ThermoScientific) and then fixed in 4% (wt/vol) paraformaldehyde solution. Outgrowth surface areas were visualized using light microscopy and measured using NIH Image J software.
In vivo analysis was based on assessing adaptations to hypoxia. On gd 6.5, rats were placed in a hypoxic [10.5% (vol/vol) oxygen] gas-regulated chamber until gd 13.5. Rat placental tissues were collected on gd 13.5 for histologic, morphometric, and biochemical analyses.
Statistical Analyses.
Data analyses were performed using SigmaPlot 11.2 statistical software package (Systat Software). Specific details of the analyses are presented in the figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Khursheed Iqbal for providing valuable feedback and suggestions during the course of the research, Dr. Christopher Mack (University of North Carolina) for providing a Kdm3a expression vector, Dr. Sumedha S. Gunewardena for assistance with the bioinformatic analysis, Mr. Clark Bloomer for assistance with the DNA microarray analysis, and Stacy McClure for administrative assistance. This work was supported by NIH Grants HD020676 and HD079363. G.X.R. was supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been dposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE80339 and GSE80340).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612626113/-/DCSupplemental.
References
- 1.Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23(1):3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 2.Harris LK. Review: Trophoblast-vascular cell interactions in early pregnancy: How to remodel a vessel. Placenta. 2010;31(Suppl):S93–S98. doi: 10.1016/j.placenta.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Maltepe E, Fisher SJ. Placenta: The forgotten organ. Annu Rev Cell Dev Biol. 2015;31:523–552. doi: 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- 4.Kaufmann P, Black S, Huppertz B. Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol Reprod. 2003;69(1):1–7. doi: 10.1095/biolreprod.102.014977. [DOI] [PubMed] [Google Scholar]
- 5.Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta. 2006;27(9-10):939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Soares MJ, Chakraborty D, Kubota K, Renaud SJ, Rumi MA. Adaptive mechanisms controlling uterine spiral artery remodeling during the establishment of pregnancy. Int J Dev Biol. 2014;58(2-4):247–259. doi: 10.1387/ijdb.140083ms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damsky CH, Fisher SJ. Trophoblast pseudo-vasculogenesis: Faking it with endothelial adhesion receptors. Curr Opin Cell Biol. 1998;10(5):660–666. doi: 10.1016/s0955-0674(98)80043-4. [DOI] [PubMed] [Google Scholar]
- 8.Rai A, Cross JC. Development of the hemochorial maternal vascular spaces in the placenta through endothelial and vasculogenic mimicry. Dev Biol. 2014;387(2):131–141. doi: 10.1016/j.ydbio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Pijnenborg R, Robertson WB, Brosens I, Dixon G. Review article: Trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta. 1981;2(1):71–91. doi: 10.1016/s0143-4004(81)80042-2. [DOI] [PubMed] [Google Scholar]
- 10.Pijnenborg R, Vercruysse L. Animal models of deep trophoblast invasion. In: Pijnenborg R, Brosens I, Romero R, editors. Placental Bed Disorders: Basic Science and Its Translation to Obstetrics. Cambridge Univ Press; Cambridge, UK: 2010. pp. 127–139. [Google Scholar]
- 11.Soares MJ, Chakraborty D, Karim Rumi MA, Konno T, Renaud SJ. Rat placentation: An experimental model for investigating the hemochorial maternal-fetal interface. Placenta. 2012;33(4):233–243. doi: 10.1016/j.placenta.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myatt L. Placental adaptive responses and fetal programming. J Physiol. 2006;572(Pt 1):25–30. doi: 10.1113/jphysiol.2006.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadyrov M, Schmitz C, Black S, Kaufmann P, Huppertz B. Pre-eclampsia and maternal anaemia display reduced apoptosis and opposite invasive phenotypes of extravillous trophoblast. Placenta. 2003;24(5):540–548. doi: 10.1053/plac.2002.0946. [DOI] [PubMed] [Google Scholar]
- 14.Rosario GX, Konno T, Soares MJ. Maternal hypoxia activates endovascular trophoblast cell invasion. Dev Biol. 2008;314(2):362–375. doi: 10.1016/j.ydbio.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakraborty D, Rumi MA, Konno T, Soares MJ. Natural killer cells direct hemochorial placentation by regulating hypoxia-inducible factor dependent trophoblast lineage decisions. Proc Natl Acad Sci USA. 2011;108(39):16295–16300. doi: 10.1073/pnas.1109478108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. Increased depth of trophoblast invasion after chronic constriction of the lower aorta in rhesus monkeys. Am J Obstet Gynecol. 1993;169(1):224–229. doi: 10.1016/0002-9378(93)90172-f. [DOI] [PubMed] [Google Scholar]
- 17.Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Dev Cell. 2009;17(6):755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Semenza GL. Oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med. 2010;2(3):336–361. doi: 10.1002/wsbm.69. [DOI] [PubMed] [Google Scholar]
- 19.Gnarra JR, et al. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc Natl Acad Sci USA. 1997;94(17):9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev Biol. 1997;191(2):297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- 21.Adelman DM, Gertsenstein M, Nagy A, Simon MC, Maltepe E. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 2000;14(24):3191–3203. doi: 10.1101/gad.853700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowden Dahl KD, et al. Hypoxia-inducible factors 1alpha and 2alpha regulate trophoblast differentiation. Mol Cell Biol. 2005;25(23):10479–10491. doi: 10.1128/MCB.25.23.10479-10491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maltepe E, et al. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development. 2005;132(15):3393–3403. doi: 10.1242/dev.01923. [DOI] [PubMed] [Google Scholar]
- 24.Takeda K, et al. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26(22):8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melvin A, Rocha S. Chromatin as an oxygen sensor and active player in the hypoxia response. Cell Signal. 2012;24(1):35–43. doi: 10.1016/j.cellsig.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock RL, Dunne K, Walport LJ, Flashman E, Kawamura A. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015;7(5):791–811. doi: 10.2217/epi.15.24. [DOI] [PubMed] [Google Scholar]
- 27.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8(3):221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solberg H, Rinkenberger J, Danø K, Werb Z, Lund LR. A functional overlap of plasminogen and MMPs regulates vascularization during placental development. Development. 2003;130(18):4439–4450. doi: 10.1242/dev.00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris LK, et al. Trophoblast- and vascular smooth muscle cell-derived MMP-12 mediates elastolysis during uterine spiral artery remodeling. Am J Pathol. 2010;177(4):2103–2115. doi: 10.2353/ajpath.2010.100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plaks V, et al. Matrix metalloproteinase-9 deficiency phenocopies features of preeclampsia and intrauterine growth restriction. Proc Natl Acad Sci USA. 2013;110(27):11109–11114. doi: 10.1073/pnas.1309561110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asanoma K, et al. FGF4-dependent stem cells derived from rat blastocysts differentiate along the trophoblast lineage. Dev Biol. 2011;351(1):110–119. doi: 10.1016/j.ydbio.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ain R, Canham LN, Soares MJ. Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: Novel endocrine phenotype and regulation. Dev Biol. 2003;260(1):176–190. doi: 10.1016/s0012-1606(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 34.Harris LK, et al. BeWo cells stimulate smooth muscle cell apoptosis and elastin breakdown in a model of spiral artery transformation. Hum Reprod. 2007;22(11):2834–2841. doi: 10.1093/humrep/dem303. [DOI] [PubMed] [Google Scholar]
- 35.Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci USA. 1996;93(9):3942–3946. doi: 10.1073/pnas.93.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benita Y, et al. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37(14):4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mole DR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284(25):16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia X, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA. 2009;106(11):4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schödel J, et al. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283(52):36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard PJ, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416(3):387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 42.Krieg AJ, et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30(1):344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mimura I, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;32(15):3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caniggia I, Grisaru-Gravnosky S, Kuliszewsky M, Post M, Lye SJ. Inhibition of TGF-beta 3 restores the invasive capability of extravillous trophoblasts in preeclamptic pregnancies. J Clin Invest. 1999;103(12):1641–1650. doi: 10.1172/JCI6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajakumar A, Brandon HM, Daftary A, Ness R, Conrad KP. Evidence for the functional activity of hypoxia-inducible transcription factors overexpressed in preeclamptic placentae. Placenta. 2004;25(10):763–769. doi: 10.1016/j.placenta.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Tal R. The role of hypoxia and hypoxia-inducible factor-1alpha in preeclampsia pathogenesis. Biol Reprod. 2012;87(6):134. doi: 10.1095/biolreprod.112.102723. [DOI] [PubMed] [Google Scholar]
- 47.Founds SA, et al. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30(1):15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou S, Xie Y, Puscheck EE, Rappolee DA. Oxygen levels that optimize TSC culture are identified by maximizing growth rates and minimizing stress. Placenta. 2011;32(6):475–481. doi: 10.1016/j.placenta.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proc Natl Acad Sci USA. 2010;107(24):10783–10790. doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 52.Shakya A, Kang J, Chumley J, Williams MA, Tantin D. Oct1 is a switchable, bipotential stabilizer of repressed and inducible transcriptional states. J Biol Chem. 2011;286(1):450–459. doi: 10.1074/jbc.M110.174045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abe Y, et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat Commun. 2015;6:7052. doi: 10.1038/ncomms8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harris LK, Jones CJ, Aplin JD. Adhesion molecules in human trophoblast: A review. II. extravillous trophoblast. Placenta. 2009;30(4):299–304. doi: 10.1016/j.placenta.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Schneider MR, Kolligs FT. E-cadherin’s role in development, tissue homeostasis and disease: Insights from mouse models: Tissue-specific inactivation of the adhesion protein E-cadherin in mice reveals its functions in health and disease. BioEssays. 2015;37(3):294–304. doi: 10.1002/bies.201400141. [DOI] [PubMed] [Google Scholar]
- 56.Desforges M, Harris LK, Aplin JD. Elastin-derived peptides stimulate trophoblast migration and invasion: A positive feedback loop to enhance spiral artery remodelling. Mol Hum Reprod. 2015;21(1):95–104. doi: 10.1093/molehr/gau089. [DOI] [PubMed] [Google Scholar]
- 57.Ain R, Konno T, Canham LN, Soares MJ. Phenotypic analysis of the rat placenta. Methods Mol Med. 2006;121:295–313. doi: 10.1385/1-59259-983-4:293. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 59.Faria TN, Soares MJ. Trophoblast cell differentiation: Establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology. 1991;129(6):2895–2906. doi: 10.1210/endo-129-6-2895. [DOI] [PubMed] [Google Scholar]
- 60.Clapcote SJ, Roder JC. Simplex PCR assay for sex determination in mice. Biotechniques. 2005;38(5):702–706, 704, 706. doi: 10.2144/05385BM05. [DOI] [PubMed] [Google Scholar]
- 61.Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF., 3rd Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 1986;118(4):1567–1582. doi: 10.1210/endo-118-4-1567. [DOI] [PubMed] [Google Scholar]
- 62.Kuroki S, et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341(6150):1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- 63.Lee DS, Rumi MA, Konno T, Soares MJ. In vivo genetic manipulation of the rat trophoblast cell lineage using lentiviral vector delivery. Genesis. 2009;47(7):433–439. doi: 10.1002/dvg.20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101(12):e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.